Abstract
We report the zygotic encystment of geographically dispersed isolates in the dinoflagellate species complex Alexandrium tamarense, in particular, successful mating of toxic Group I and nontoxic Group III isolates. However, hypnozygotes produced in Group I/III co-cultures complete no more than three divisions after germinating. Previous reports have suggested a mate recognition mechanism whereby hypnozygotes produced in co-cultures could arise from either homotypic (inbred) or heterotypic (outbred) gamete pairs. To determine the extent to which each occurs, a nested PCR assay was developed to determine parentage of individual hypnozygotes. The vast majority of hypnozygotes from pairwise Group I/III co-cultures were outbred, so that inviability was a result of hybridization, not inbreeding. These findings support the assertion that complete speciation underlies the phylogenetic structure of the Alexandrium tamarense species complex. Additionally, the ribosomal DNA (rDNA) copy numbers of both hybrid and single ribotype hypnozygotes were reduced substantially from those of haploid motile cells. The destruction of rDNA loci may be crucial for the successful mating of genetically distant conjugants and appears integral to the process of encystment.
The inviability of Group I/III hybrids is important for public health because the presence of hybrid cysts may indicate ongoing displacement of a nontoxic population by a toxic one (or vice versa). Hybrid inviability also suggests a bloom control strategy whereby persistent, toxic Group I blooms could be mitigated by introduction of nontoxic Group III cells. The potential for hybridization in nature was investigated by applying the nested PCR assay to hypnozygotes from Belfast Lough, Northern Ireland, a region where Group I and III populations co-occur. Two hybrid cysts were identified in 14 successful assays, demonstrating that Group I and III populations do interbreed in that region. However, an analysis of mating data collected over an 18-year period indicated a leaky pre-mating barrier between ribosomal species (including Groups I and III). Whether the observed selectivity inhibits hybridization in nature is dependent on its mechanism. If the point of selectivity is the induction of gametogenesis, dissimilar ribotypes could interbreed freely, promoting displacement in cases where hybridization is lethal. If instead, selectivity occurs during the adhesion of gamete pairs, it could enable stable coexistence of A. tamarense species. In either case, hybrid inviability may impose a significant obstacle to range expansion. The nested PCR assay developed here is a valuable tool for investigation of interspecies hybridization and its consequences for the global biogeography of these important organisms.
Keywords: Dinoflagellates, Hybridization, Biogeography, Harmful algal blooms, Genotypes, UK, Northern Ireland, Belfast Lough
1. Introduction
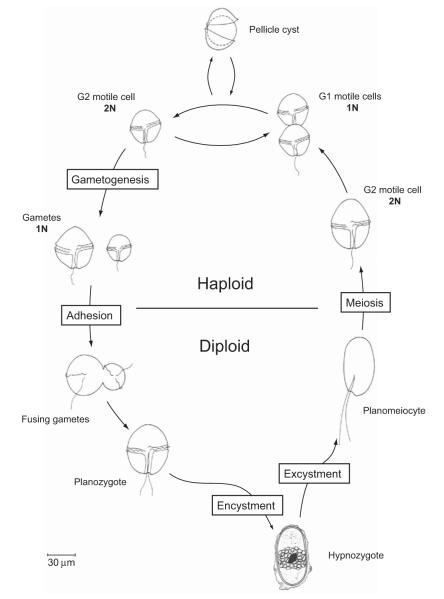
Mating is an integral part of the life cycle of many dinoflagellates and is associated in some species with the formation of hypnozygotic cysts. These cysts are highly resistant to environmental stresses and are a likely vector for the global expansion and persistence of many harmful dinoflagellate blooms (Anderson, 1989; Hallegraeff, 1993). The great majority of dinoflagellates proliferate as motile, haploid cells. Under certain conditions, some species cease mitotic division and undergo gametogenesis, transforming to non-dividing gametes that are competent to conjugate and fuse with one another. Importantly, the process of gametogenesis does not involve meiotic division because the progenitor motile cells are already haploid. Upon fusion, gamete pairs form planozygotes – swimming diploids that may persist for days or weeks before either returning to haploid, mitotic cell growth or metamorphosing to the hypnozygote form (Fig. 1; Figueroa and Bravo, 2005; Figueroa et al., 2006; Pfiester and Anderson, 1987).
Fig. 1.
Life cycle diagram of the A. tamarense species complex. Though the hypnozygote stage may be bypassed by other encysting dinoflagellates, such an alternate pathway of zygote maturation has not been described for A. tamarense.
Dinoflagellate species are categorized as either homothallic or heterothallic depending on whether clonal cultures can be induced to form zygotes. Heterothallic classification supposes that all pairs of gametes are heterotypic – that is, they are formed by fusion of non-sibling cells – whereas homothallic species may form hetero- or homotypic pairs. Species that have a hypnozygote stage are said to be homothallic if encystment can be induced in clonal culture, or heterothallic if encystment requires mixing of two compatible clones. However, conjugation in clonal cultures of putatively heterothallic species, including the subject of this study, the A. tamarense species-complex, is sometimes observed with low-grade or no subsequent production of cysts (Blackburn et al., 2001; Destombe and Cembella, 1990). Moreover, conjugation and encystment are dissociated in numerous other dinoflagellates so that encystment may not be the sole pathway for zygote maturation (Figueroa and Bravo, 2005; Figueroa et al., 2006; Uchida, 2001). Still, it is the hypnozygote stage of A. tamarense that confers resistance to prolonged stresses, and hypnozygotes may be the only abundant cell type within A. tamarense populations for long periods (Anderson and Wall, 1978). While non-encysting zygotes may benefit from genetic recombination, failure to encyst is likely catastrophic in regions where sexual cycles occur in phase with extended periods of poor growth conditions (e.g. high latitude winter). Though zygosis and encystment may be separable in other species, these processes are strongly intertwined among the A. tamarense species.
Alexandrium tamarense species are similar to many ciliates in that their mating system consists of more than two self-incompatible mating types (Destombe and Cembella, 1990). Mating types in any eukaryotic microbe must affect one of two control points in the sexual cycle: the transition to sexuality (gametogenesis); or, the physical coupling (adhesion) of compatible gametes (Fig. 1). A prime example of the latter type of control is the unicellular chlorophyte Chlamydomonas reinhardtii, a species whose gametes are either plus or minus type. These types each express specific and complimentary flagellar agglutinins that ensure heterotypic pairing (Adair et al., 1983; Musgrave et al., 1981). While conjugation by A. tamarense clones indicates expression of similar recognition factors, these factors may not differentiate distinct cell genders. In species where mating types act by reciprocally spurring gametogenesis, homotypic conjugation (i.e. inbreeding) may be possible. Well-elucidated examples include the hypotrich ciliate genus Euplotes (Dini and Nyberg, 1993) and the human fungal pathogen Cryptococcus neoformans (Lin et al., 2005). In both C. neoformans and Euplotes, mature gametes are capable of initiating fusion with either similar or dissimilar gamete types. In the ciliate E. crassus, gametogenesis may also be induced non-reciprocally by co-culture with clones of its congener species E. minuta. In these interspecies co-cultures, only E. crassus clones are sexually induced so that all conjugants are homotypic pairs of E. crassus cells (Dini et al., 1990). If interspecies mating within the A. tamarense species complex was similar to that observed in Euplotes, the heterothallic classification of A. tamarense species might not preclude them from homotypic encystment. Further, stimulation of inbreeding could be most profound when compatible clones are genetically distinct and stimulation of gametogenesis is non-reciprocal.
In this study, the extent and implications of homotypic and heterotypic encystment among genetically distinct groups of the A. tamarense species complex were assessed both in culture and in nature. Members of the A. tamarense complex are globally distributed and may produce saxitoxin and congener compounds, which, in turn, cause paralytic shellfish poisoning (PSP) in human and animal shellfish consumers. Most toxic clones used here are Group I and most nontoxic clones are Group III, where group designations are derived from phylogenetic relationships among the clones’ large subunit ribosomal DNA sequences (LSU rDNA). These two groups occur in close geographic proximity along the northern coasts of Ireland and Great Britain (Lilly et al., 2007). The group designations (I and III) refer to two of five distinct clades that are largely isolated in their global distribution. Each of the groups is consistent with respect to the toxicity of its members: all tested Group I and IV clones produce PSP toxins, and all Group II, III and V clones do not. Sequence divergence is less than 2% within the clades but up to 11% between them, comparable to that between the closely related species A. tropicale and A. affine. Groups I and III, the focal groups in this study, have approximately 6% divergence and are among the most closely related of the clades. Morphotype designations A. catenella, A. fundyense and A. tamarense are commonly used to differentiate isolates within the A. tamarense complex, but we do not regard these as valid species because they are inconsistent with the complex’s ribosomal phylogeny. All three morphotypes occur but do not cluster within Groups I and IV. Only the A. tamarense morphotype is known within Groups II, III and V (Lilly et al., 2007; Scholin et al., 1995).
Mating intercompatibility among the A. tamarense ribosomal clades was assessed through an analysis of encystment and germination data accumulated over 18 years. Germination studies demonstrated complete post-zygotic lethality among hypnozygotes resulting from pairwise co-cultures of toxic Group I and nontoxic Group III isolates. Most of these hypnozygotes failed to germinate, and those that did germinate completed no more than three cell divisions. Because Group I/III progeny were inviable, a method was needed to identify hypnozygotes as Group I, III or, if present, Group I/III hybrids. Therefore, we developed a quantitative nested PCR assay for genotyping single hypnozygotes, and used the assay to confirm outbreeding in pairwise Group I/III co-cultures. The assay was also used to analyze hypnozygotes in sediment samples from Belfast Lough, a region in Northern Ireland where both Group I and III blooms are known to occur.
2. Methods
2.1. Clonal cultures
One hundred nineteen clonal isolates were evaluated for hypnozygote formation in clonal culture and in pairwise co-culture with other clones. Clones were isolated from throughout the world, represented all major clades (62 Group I, 7 Group II, 39 Group III, 7 Group IV, and 1 Group V isolate), and included A. tamarense, A. fundyense, and A. catenella morphotypes. Additionally, two A. affine clones and one A. tropicale clone were evaluated (see Supplementary Table S1 and Figs. S1 and S2). In many cases, cultures are no longer maintained and LSU rDNA sequence is not available for direct determination of a given clone’s group affiliation. In these cases, group membership was assigned using LSU rDNA sequence data from clones that were isolated from the same field expeditions, or inferences were made based on available toxicity data and the known biogeographical distributions of the A. tamarense clades. Most clones were isolated between 1980 and 1995. The oldest clone, PGT183, was isolated in 1957. The youngest, a set of 15 Group I and 8 Group III clones were isolated from slurry germinations of Belfast Lough sediments in 2006.
2.2. Encystment evaluation and hypnozygote storage
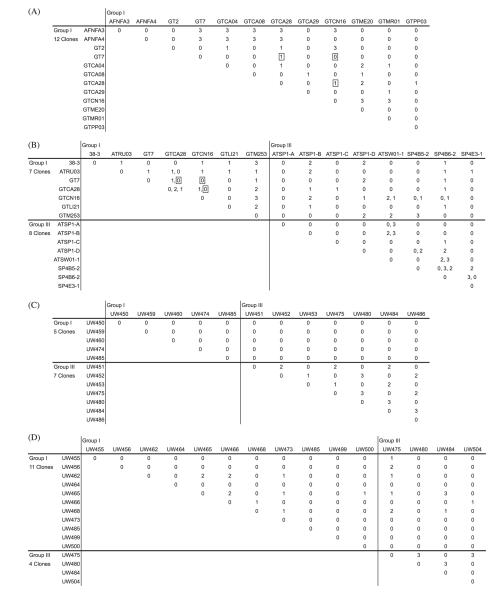
A screen for encystment in pairwise co-cultures was initiated in 1989. The last experiments compiled here were completed in 2007. All 119 isolates were tested for clonal encystment in N-limited culture, but all possible pairwise combinations were not. Four subsets of 12–15 clones were tested in all possible pairwise combinations (Fig. 2A-D). Two of these subsets were derived from a set of Belfast Lough isolates, and the others from American and European (‘cosmopolitan’) collections of isolates. One Belfast and one cosmopolitan subset were biased by selecting isolates that would lead to a higher proportion of successful encystment, while the other two subsets (one Belfast and one cosmopolitan) were unbiased. Experiments for both Belfast subsets were completed within one year of the isolation of clones (2007); the experiments for the unbiased and biased cosmopolitan subsets were completed in 1990 and 2005, respectively.
Fig. 2.
Matrices of cyst yields from all pairwise co-cultures of clone subsets: (A) unbiased cosmopolitan (1990 results only), (B) biased cosmopolitan (all yields observed, 1990–2006), (C) unbiased Belfast Lough, and (D) biased Belfast Lough. GTM253 (biased cosmopolitan subset, B) is clonal isolate GTM253-17. Yields from each pairwise co-culture were scored 0–3 on the basis of presence and abundance of hypnozygotes after >1 month incubation in N-limited medium (see text). Three co-cultures (combinations of GT7, GTCA28, and GTCN16) from the unbiased cosmopolitan subset were repeated in 2005 and are boxed in both A (1990) and B (2005 result only).
Cultures were grown in 25 mL volumes of modified f/2 (replete) or N-limited (encystment) medium (Guillard and Ryther, 1962) in 50 mL borosilicate culture tubes at 20 °C, on a 14:10-h L:D cycle with 150–200 μmol m−2 s−1 photon flux density (Anderson et al., 1984). Medium modifications included: (1) elimination of NaSiO3, (2) the reduction of CuSO4 to 10−8 M, and (3) addition of H2SeO3 to the trace metal mixture (10−8 M final concentration). In the N-limited medium, NH4 was substituted for NaNO3 as the nitrogen source at a final concentration of 25 μM. Additionally, 16 Belfast co-cultures were repeated in phosphate-limited medium that was prepared as described by Anderson and Lindquist (1985). Inoculum cultures maintained in replete medium were grown to mid/late-exponential phase and transferred into N-limited or phosphate-limited medium at an approximate cell density of 100–500 cells mL−1 for each clone. After inoculation, the cultures were incubated for 30–40 days to maximize the opportunity for complete hypnozygote formation. The cell/cyst deposits at the bottom of the culture tubes were sampled by Pasteur pipette and loaded into a Palmer-Maloney chamber. The contents of the chamber were scanned under 100×magnification to determine if hypnozygotes were present. Cyst yields from N-limited and phosphate-limited cultures were scored semi-quantitatively as 0, 1, 2 or 3. The ratings indicate that 0, 1–50, 50–100, or greater than 100 cysts were found per 25 mL N-limited culture. Hereafter, we refer to co-cultures scored 0 as ‘negatives’ and those scored 1, 2, or 3 as ‘positives’. Some positives with higher yields (scored 2 and 3) were repeated to produce cysts for germination and genotyping experiments. Samples for germination and genotyping experiments were harvested into cryovials and stored in anoxic sediment as described by Anderson et al. (2003). We have previously found that exposure to anoxic sediment substantially prolongs germinability of stored culture samples compared to storage within the original culture tubes and incubation chambers.
2.3. Hypnozygote viability
For germination studies, single vials of hypnozygotes were removed from storage 1 to 31 months after collection. Vials were sorted on ice and under red-light to reduce premature germination (Binder and Anderson, 1986). Hypnozygote samples were disaggregated by brief sonication and 1–237 cysts isolated by micropipette to separate wells of 96-well tissue culture plates containing 130–200 μL replete medium. In some cases, more than one cyst was placed in a well because the sonication step failed to eliminate clumping entirely. Plates were incubated at 4, 15 or 20 °C on a 14:10-h L:D cycle with approximately 150 μmol m−2 s 1 photon flux density. In all, hypnozygotes from 4 separate Group I only co-cultures, 7 Group III only co-cultures, and 10 Group I/III co-cultures were examined. Of these, germinations of 7 co-cultures (including 4 Group I/III co-cultures) were attempted at multiple maturation periods (1–31 months) and temperatures (4, 15 or 20 °C). Isolated hypnozygotes were monitored for excystment weekly or biweekly for at least 1 month. Because hypnozygotes from Group I/III co-cultures never yielded viable cultures, the total number of cells arising from these cysts was carefully observed under a stereomicroscope so that all cells including those trapped in the air/water interface could be counted.
2.4. Real-time quantitative PCR assay
Two real-time quantitative PCR (qPCR) assays were developed for a genotyping assay and also for estimation of the LSU rDNA copy number within Group I and III cysts. Consensus alignments of Group I and III LSU rDNA D1–D2 hypervariable regions were constructed and primers were designed to anneal to two regions of dissimilarity between the respective sequences (Fig. 3; Group I forward primer: 5′-GTG TTG CAC TTG CTT GAC AAG AGC-3′; Group I reverse primer: 5′-CAT CCC CAA GCA CAG GAA CAC AC-3′; 200 nucleotide amplicon; Group III forward primer: 5′-GGT GAG ATT GTA GTG CTT GCT TGA CAA TAG-3′; Group III reverse primer: 5′-AAG GAA GGA AGC AAC CTC AAA CAC ATG-3′; 220 nucleotide amplicon). Specific amplification of targets was verified by gel electrophoresis of PCRs with the respective primer pairs and genomic DNA templates.
Fig. 3.
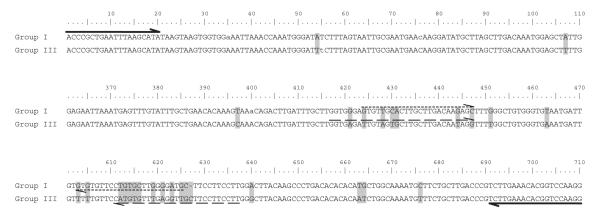
Sequence alignment of Group I and III D1–D2 rDNA consensus sequences (segments 111–360 and 471–600 omitted). Conserved differences are highlighted in gray. Overline and underline arrows indicate forward/reverse positions of D1R/D2C (solid), Group I (short dashed), and Group III (long dashed) primer pairs. Consensus sequences were constructed from 18 Group I and 6 Group III clones. Polymorphisms in the respective consensus sequences are denoted by lower case letters. Divergence between the consensus sequences is 6%.
All qPCR assays were performed in 25 μL reactions by detection of SYBR Green dye binding of product (Stratagene FullVelocity real-time qPCR chemistry, 400 nM forward and reverse primers, 5 μl template, and 1 μl 1:103 dilution BioRad 1 mM fluorescein standard in DMSO). A fast two-step thermocycler program essentially as described in the FullVelocity manual was used (40 cycles; 10 s 95 °C, 30 s 63 °C; BioRad iCycler IQ real-time PCR detection system). Specific amplification of qPCR product was confirmed by melt curve analysis on each assay plate. Fluorescence thresholds were generally set according to default preferences of BioRad iCycler analytical software but were sometimes modified to exclude reaction failures (as marked by grossly aberrant slopes in log RFU/cycle plots).
D1R–D2C primer PCR products (Scholin et al., 1994) of Group I and III templates were used as standards. Primers and other reactants from D1R–D2C amplifications were removed by product concentration on Qiagen QIAquick PCR Purification microcentrifuge columns. D1R–D2C products were then eluted and quantified using a NanoDrop ND-1000 spectrophotometer. Serial dilutions of these standards were made across 6 orders of magnitude, and all diluted standards were amplified in triplicate on each assay plate. Only technical replicates within 0.5 cycles of standard means were used for standard curve estimates. Standard curves were calculated from linear regression of log amplicons and threshold cycle values (Ct). Amplification efficiencies were calculated as the mean fraction of template molecules copied in each successive round of amplification.
2.5. Single hypnozygote genotyping by nested PCR
The parentage of individual hypnozygotes produced in Group I/III co-cultures was assayed by nested PCR. In brief, individual hypnozygotes were isolated directly to a D1R–D2C PCR reactant mixture. Hypnozygotes were lysed during isolation and denaturing steps of PCR amplification. In the last step of the method, the D1R–D2C amplicons were interrogated using the Group I and III qPCR assays to estimate the numbers of Group I and III ‘primary’ amplicons (i.e. the approximately 700 bp fragments amplified during D1R–D2C PCR). This approach increased sensitivity compared to a single amplification, multiplex qPCR assay and also enabled qPCR replicates from single cysts.
In developing the nested PCR assay, micropipette isolation of hypnozygotes was found to yield inconsistent results with low detectable numbers of primary PCR amplicons. The performance of the assay improved dramatically when cysts were isolated using a laser pressure catapult system. Sample preparation for laser catapulting was as follows: first, (1) Director laser microdissection slides (Expression Pathology, Gaithersburg, MD) were conditioned by an overnight wash in 1 N HCl and then treated with 0.01% poly-l-lysine; (2) for hypnozygotes from sediment samples, sediment slurries were disaggregated by sonication and sieving, then enriched for hypnozygotes by density centrifugation over a modified Nalco-sucrose step gradient (Schwinghamer et al., 1991); (3) hypnozygote suspensions, either from sediment samples or stored pellets, were diluted in 5–6 volumes deionized water, then sonicated using a Microsonix Sonicator 3000 (output level 2.5, 1 min) to disaggregate clumps; (4) suspensions were then dried to the Director slides in a 55 °C oven; lastly, (5) dried samples were rinsed successively in coplin jars of deionized water, 70% ethanol, and 100% ethanol for >5 min each wash. All slides were stored in 100% ethanol until hypnozygote isolation using a Zeiss PALM inverted microscope system.
Hypnozygote cysts retained normal morphology when wet with ethanol but could also be identified by a characteristic ‘raisin-like’ appearance when dried. Individual cysts were laser pressure catapulted to PCR tube caps that had been pre-loaded with ~25 μL D1R–D2C (primary PCR) reactant mixture (600 nM D1R primer, 600 nM D2C primer, 1X NEB ThermoPol buffer, 2.5 U NEB Taq polymerase, 200 μM dNTPs). The caps were positioned <1 mm above the slide surface, so that the great majority of cysts were captured effectively using the AutoLPC function of the Zeiss PALM system. After launching a cyst, caps were re-positioned >2 cm above the slide then replaced into their PCR tube and stored on ice until thermocycling. The primary PCR itself was limited to 20 cycles to prevent product skew associated with plateau phase of amplification (TM init=96 °C, 5 min; TM cycle=94 °C, 40 s; TA=50 °C, 1 min; TE=72 °C, 1.5 min). Primers, reactants and other debris were removed from the primary PCR products using Qiagen QIAquick microcentrifuge columns and the products were eluted in 50 μL Qiagen EB buffer (10 mM Tris–HCl, pH 8.5). The purified products were then suitable for use as template in the Group I and III qPCR assays.
Genotyping was performed on hypnozygotes from 10 co-culture samples (Group I only co-cultures: 38–3+GTM253-17, GTLI21+GTM253-17; Group III only co-cultures: ATSW01-1+SP4B6-2, SP4B5-2+SP4B6-2; and mixed Group I/III co-cultures: 38-3+ATSP1-B, 38-3+SP4B6-2, ATSW01-1+GTM253-17, ATSP1-D+GTM253-17, GTM253-17+SP4B5-2, and a polyclonal co-culture of 4 Group I and 5 Group III isolates). Cell debris was also isolated from each of the Group I/III co-culture samples in order to assess contamination by adsorption of free Group I and III templates to cysts. Lastly, hypnozygotes from 2 sediment samples collected from within Belfast Lough, Northern Ireland were also examined after analysis of isolates from slurry germinations revealed that both Group I and III hypnozygotes were present in the embayment.
2.6. Hypnozygote rDNA copy number estimation
We estimated the genomic copy number of LSU rDNA loci within Group I, III and hybrid hypnozygotes. Experiments completed by our labs have established that the copy number in motile cells is quite high and can vary substantially: Group III LSU rDNA copy numbers range from approximately 7×104–1.5×105 and Group I rDNA copy numbers from approximately 5×105–1×106 (experiments not described further in the present work, see also Galluzzi et al., 2009). The Zeiss PALM system was used to collect samples of 484–1518 hypnozygotes from which DNA was extracted by a modification of the Invitrogen ChargeSwitch gDNA Micro Tissue kit.
Slides of co-culture or sediment material were prepared as described for single hypnozygote genotyping by nested PCR. After the final ethanol wash, slides were allowed to dry while set on the inverted microscope of the Zeiss PALM system. Batches of 100–300 cysts were rapidly mapped, then serially catapulted to a 500 μL PCR tube cap with ~70 μL Invitrogen proteinase K/Lysis Buffer L15. The number of cysts captured could then be assessed by raising the focal plane of the microscope to the lysis buffer surface. The lysis buffer (containing captured cysts) was transferred to PCR tubes by brief centrifugation and the process repeated 4–6 times for a single co-culture or sediment sample. Capture efficiency was only evaluated for 1–2 batches (PCR tube caps) each sample because the efficiency was consistently high (>85%) and because counting of cysts could be time consuming, causing evaporative loss of lysis buffer. Following capture, samples were incubated overnight in a 55 °C temperature block and vortexed in a suspension of 500 μM zirconium beads 3×1 min. PCR tubes were then punctured and lysate collected by centrifugation at 2000g for 10 min. RNAse A treatment, wash and purification steps were followed according to the manufacturer’s instructions. Extracted DNA was eluted from the ChargeSwitch magnetic beads in 150 μL Elution Buffer E5 (Tris-EDTA, pH 8.5) after brief warming in the 55 °C temp block.
In all, 8 samples were isolated for copy number estimation. Seven co-culture samples were examined (Group I only: 38-3+GTM253-17, GTLI21+GTM253-17; Group III only: SP4B5-2+SP4B6-2; Group I/III: 38-3+ATSP1-B, GTM253-17+ATSP1-D, GTM253-17+ATSW01-1, and GTM253-17+SP4B5-2). A Group I sample was also taken from naturally produced hypnozygote cysts collected in the fall of 2006 from Casco Bay, ME.
2.7. Statistical analysis of genotyping data
The significance of differences in the amplicon ratio of hypnozygotes from pairwise same-group and out-group co-cultures of Group I and III isolates was tested using a simple randomization procedure (Solow, 1990). Optimal threshold values of the Group I:III amplicon ratio for separating the three groups of hypnozygotes were found by minimizing the cross-validated misclassification rate. The significance of this rate was assessed by randomizing group designations (i.e. Group I only, Group III only, or Group I/III co-culture) and repeating the classification procedure (including threshold optimization) a total of 10,000 times. The same approach was used in separate assessments of the significance of Group I/hybrid, Group III/hybrid, and Group I/III differences.
The randomization procedure demonstrated that significant differences exist in the amplicon ratio between hypnozygotes from pairwise same-group and out-group co-cultures (P<0.0001, see Section 3.4), so that hypnozygotes are predominantly formed from heterotypic gamete pairs. The misclassification rate was substantially higher for cysts with low total numbers of amplicons, likely reflecting poor lysis efficiency. The number of amplicons detected is also related to the amplification efficiency during the primary PCR which was estimated to be 80% by qPCR of amplified and unamplified Group I and III genomic DNA standards (data not shown, 1.820≅130,000-fold total amplification). It is assumed that the primary PCR efficiency was similar in the cyst genotyping experiments. Using nominal estimates of the Group I and III rDNA copy numbers and the estimated primary PCR efficiency, the expected amplicon sums were estimated at thresholds of 0.01, 0.1%, 1%, 10% and 100% lysis efficiency. The sums were evaluated for all mixtures of Group I and III cyst copy numbers under the constraint that the sum be the product of the cyst lysis efficiency and one whole cyst equivalent. In no case did a single cyst genotyping experiment yield more than the number of amplicons expected from complete lysis of a Group I cyst (i.e. the maximum amplicon sum expected based on results from hypnozygote copy number estimation; 100% lysis efficiency ×45,000 Group I rDNA copies ×1.820 primary PCR amplification=5.7×109 amplicons in the primary product). Only 1 of PCR 75 controls from laser catapult isolation of debris exceeded the number of copies expected from 0.01% lysis of a hypnozygote (data not shown). Accordingly, individual cyst genotyping experiments were considered successful if the sum of amplicons exceeded 0.01% efficiency (though ratios derived from larger numbers of amplicons are reported with greater confidence).
To refine and extend the analysis, the results of cyst genotyping experiments with less than 0.01% efficiency were omitted and probability density functions (pdfs) of the log amplicon ratio:
were estimated by kernel density estimation. Kernel density estimation is a standard nonparametric approach to fitting a probability density function to data (Silverman, 1986). To apply kernel density estimation, it is necessary to select a bandwidth that controls the smoothness of the estimated density. Here, a common bandwidth was selected to minimize the cross-validated misclassification rate, where each cyst was classified to the group for which the estimated probability density of its log amplicon ratio was greatest. The classification thresholds for this rule correspond to the values of x at which the estimated probability density functions intersect.
The estimated pdfs can be used to assess the group membership probabilities for a cyst from an unknown group. Let P(x|g) be the estimated probability density function for group g and assume equal prior group membership probabilities. By Bayes’s Theorem, the posterior group membership probabilities are given by
for g=Group I, Group III, and hybrid. The classification rule described above assigns a cyst to the group for which this probability is greatest. The posterior group membership probabilities for the cysts of known parentage are shown in Fig. 5C. These probabilities were also calculated for cysts of unknown parentage that were collected from the polyclonal Group I/III co-culture and the two Belfast Lough sediment samples.
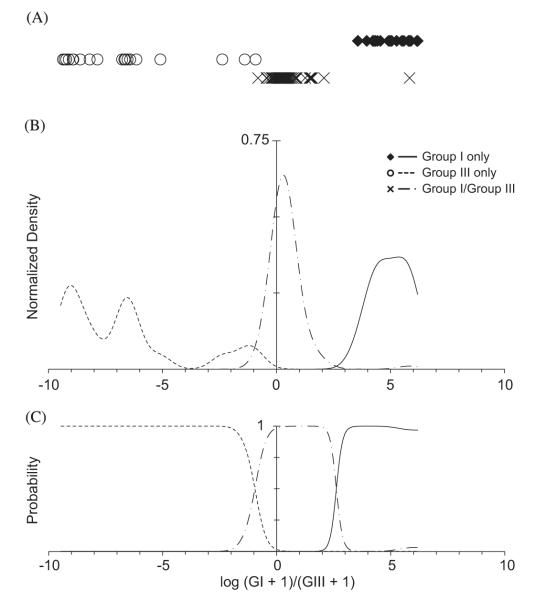
Fig. 5.
Probability density estimation of log ratios from successful ‘known’ genotyping experiments. (A) Linear scatter of successful ‘known’ experiments: Group I only co-culture – black, solid diamonds; Group III only co-culture – open circles; Group I/III co-culture–black ‘×’ symbols. (B) Gaussian kernel estimation of probability densities. The intersection of the estimated Group I (solid line) and the hybrid (dash-dot line) densities is the Group I/hybrid classification threshold; the intersection of the estimated hybrid density and the estimated Group III density (dotted line) is the hybrid/Group III classification threshold (see Fig. 4). (C) Estimated probabilities of Group I, hybrid and Group III cyst parentage by log ratio given naïve prior probability of hypnozygote parentage. Probabilities are calculated by Bayes’s theorem.
3. Results
3.1. Mating
Encystment results from 896 co-cultures were compiled (12.8% of the possible pairwise combinations from the 119 isolates; see Supplementary Table S1 and Fig. S2). Of these, 224 co-cultures produced hypnozygotes (25% positives). Only five isolates produced hypnozygotes when grown clonally: two Group I isolates and three Group III isolates (Supplementary Fig. S2). Co-cultures of isolates from the same ribosomal sequence group (‘same-group co-cultures’) were more likely to be positives than out-group co-cultures (37.2% of same-group pairs versus 17.8% of out-group pairs). No crosses between A. catenella and A. fundyense morphotype isolates were positive but Group IV A. catenella did produce hypnozygotes in co-culture with Group I, III and V isolates of A. tamarense morphotype. Of the 21 possible out-group permutations among A. tamarense Groups I–V, A. affine and A. tropicale, 13 were tested and 9 produced hypnozygotes. A significant caveat to these results is that our effort is dominated by co-cultures of Group I and III isolates; 41.0% of the co-culture experiments consist of 1 Group I and 1 Group III isolate, 23.8% are Group I only, and 12.7% are Group III only (see Table 1; also, Supplementary Fig. S2). Among the 12 other out-group co-cultures, 7 have been attempted in fewer than 15 isolate combinations, including only 9 out-group combinations that did not include 1 Group I or III isolate.
Table 1.
Pairwise co-culture effort and number of positives (hypnozygote production).
| Groups | Isolate pairs tested | Positives |
|---|---|---|
| I, I | 214 | 68 (31.8%) |
| III, III | 114 | 55 (48.2%) |
| IV, IV | 5 | 1 |
| I, II | 35 | 1 (2.9%) |
| I, III | 367 | 86 (23.4%) |
| I, IV | 22 | 2 |
| I, V | 28 | 2 |
| I, A. affine | 28 | 1 |
| I, A. tropicale | 13 | 2 |
| II, IV | 14 | 0 |
| III, IV | 50 | 5 (10.0%) |
| III, V | 1 | 0 |
| III, A. affine | 1 | 0 |
| IV, V | 2 | 1 |
| IV, A. affine | 1 | 0 |
| IV, A. tropicale | 1 | 0 |
Three of seven possible same-group combinations (I, III and IV) and 13 of 21 possible out-group combinations were tested. Positive ratios are reported in parentheses for group combinations that were tested using more than five unique pairs of isolates.
Because the co-culture dataset is not exhaustive and experiments were pursued non-randomly, a randomization test was used to compare the ratios of positives to total combinations from same-group and out-group experiments (hereafter termed ‘positive ratios’). Group assignments of the 119 isolates were permuted 1000 times to construct probability density estimates of differences between same-group and out-group positive ratios. In approximately half of these trials, the positive ratio of out-group crosses exceeded that of same-group co-cultures and in no case was the difference greater than 12% (substantially less than the 19.4% difference found in our dataset). In a similar randomization analysis restricted to results from Group I same-group, Group III same-group, and Group I/III co-cultures, the difference between same-group and out-group ratios also never exceeded the same-group/out-group differences in the true dataset (31.8% and 48.2% for Group I same-group and Group III same-group co-cultures, respectively and 23.4% for Group I/III co-cultures; Table 1).
Encystment yields among positives varied substantially, including among replicates of some co-cultures, but were generally low. In all, 93 combinations (41.5%) were scored ‘1’, 53 (23.7%) were ‘2’, 41 (18.3%) were ‘3’, where ratings represent 0, 1–50, 50–100, or greater than 100 cysts per 25 mL culture. The remaining positives (37 total) had variable yields (Supplementary Fig. S2). The highest yields were observed in Group III same-group combinations followed by Group I same-group combinations (mean ratings of 0.47 and 0.39, respectively; N-limited co-cultures only and excluding variable yield positives). The highest average yield among out-group co-cultures was Group I/III (0.33). Yields in phosphate-limited trials were sometimes lower than yields from N-limited trials but no differences were observed in encystment compatibility (Table 2).
Table 2.
Comparison of positive ratios and average overall and positive yield ratings from four clone subsets that were tested in all pairwise combinations.
| Cosmopolitan | Belfast Lough | |||
|---|---|---|---|---|
| Unbiased | Biased | Unbiased | Biased | |
| Positive ratio | 0.250 | 0.429 | 0.159 | 0.181 |
| Average yield rating | 0.694 | 0.616 | 0.394 | 0.305 |
| Average positive yield rating | 2.78 | 1.44 | 2.17 | 1.68 |
Positive ratio is the proportion of co-cultures that were found to produce hypnozygotes. Average yield rating is the mean of yield ratings (0, 1, 2 or 3) from all subset co-cultures. Average positive yield rating is the mean of all subset cocultures rated 1, 2 or 3. See text for further description of biased and unbiased subsets.
Among the four clone subsets, the two unbiased subsets had the highest yield ratings among positive combinations (2.78 and 2.17, Fig. 2A and C), indicating more robust mating interactions versus biased subsets that had higher positive ratios. Yields in two of three co-cultures were reduced in the 2005 cosmopolitan subset from yields in the 1990 cosmopolitan subset (GT7/GTCA28 and GTCA28/GTCN16 co-cultures; Fig. 2A and B). Also noteworthy, was the appearance of clonal encystment (auto-encystment) activity by GTCA28 after the completion of the 1990 experiments. The suggestion that yields decrease in time from the cosmopolitan subsets is contradicted by the Belfast subset data (Fig. 2 C and D; experiments completed within 1 year of clone isolations). While the yields from positive Group III combinations were higher than same-group yields in the complete dataset (2.23 vs. 1.85), yields from Belfast Group I combinations were quite low (1.38; or less than the average yield from all positive out-group combinations, 1.57).
Assignment of mating types within the four subsets was attempted by two different methods. In the first, we assumed a dioecious mating system in which the mating type of each isolate is fixed. If the assumptions of this model hold, all isolates should be assigned one of two mutually incompatible mating types, + or −. Of the four subsets, only one, the unbiased Belfast subset, can be divided in this manner. However, in this case, the only positives were Group III combinations. Similarly, all Group III isolates from the biased Belfast subset (excluding Group I/III co-culture results) can be assigned mating types without ambiguity, but this is unsurprising given that three of the four Group III isolates are shared with the unbiased subset (Fig. 2 C and D). More noteworthy from the Belfast results is that all but one combination of + and − Group III clones were positive for encystment. Neither of the cosmopolitan subsets can be sorted into only + and − mating types, either as a complete set or when results from Group I and III isolates are considered separately (Supplementary Fig. S3A–D).
The failure of the dioecious/fixed mating-type model contradicts one or both of its assumptions: that the number of mating types is limited to two, and that mating type is fixed within clonal cultures of each isolate. We addressed the former criterion by also considering only negative co-culture results. Clones of the respective subsets were divided into a minimal number of groups whose members are mutually incompatible. As occurs when only considering positives, some clones are poorly constrained and may be segregated to more than one group. Using this approach, the unbiased cosmopolitan and biased Belfast subsets can be solved using a three mating-type system, whereas the biased cosmopolitan subset requires four. When the biased cosmopolitan subset is broken into matrices of only Group I or III same-group co-cultures, the Group I clones still require four mating types but the Group III clones may be segregated into three (see Supplementary Fig. S3 and S4).
3.2. Germination
Six Group I isolates and 13 Group III isolates were co-cultured in 21 different pairwise combinations and in three polyclonal combinations and the resulting cysts used for germination trials (Table 3; Supplementary Table S1). Cysts were formed, stored, and incubated numerous times over several years. The longest storage time was 31 months for cysts from Group I/III co-cultures, 17 months for Group I co-cultures, and 10 months for Group III co-cultures. However, hypnozygotes from all three group combinations remained ungerminated for more than 3 years when stored in cryovials as described.
Table 3.
Germination results of hypnozygotes collected from co-culture of Group I only, Group III only and Group I/III isolates.
| Germination trial conditions |
||||||
|---|---|---|---|---|---|---|
| Group(s) | Co-culture | Total trials |
Overall excystment success |
Temperature (°C) |
Age at isolation (Mos.) |
Excystment success |
| I | 38-3, GTM253-17 | 1 | 2.7% | 15 | 3 | 3/110 |
|
| ||||||
| GTCN16, GTMR01 | 14 | 10.0% (22/219) | 4 | 7/15 | ||
| 6 | 1/33 | |||||
| 4 | 7 | 2/16, 1/15, 3/11, 0/8 |
||||
| 8 | 0/18* | |||||
| 9 | 3/20 | |||||
| 17 | 4/19 | |||||
|
| ||||||
| 15 | 4 | 0/21* | ||||
| 7 | 0/1, 0/21* | |||||
| 8 | 1/4 | |||||
|
| ||||||
| GTLI21, GTM253-17 | 1 | 0.0% | 15 | 3 | 0/116* | |
|
| ||||||
| 38-3, GTCN16, GTLI21, GTM253-17 | 1 | 3.0% | 15 | 3 | 5/162* | |
|
| ||||||
| III | ATSP1-B, ATSW01-1 | 1 | 2.0% | 15 | 3 | 2/100 |
|
| ||||||
| ATSW01-1, SP3B8-3 | 1 | 76.2% | 15 | 1 | 16/21 | |
|
| ||||||
| ATSW01-1, SP4B6-2 | 1 | 46.3% | 15 | 3 | 76/162 | |
|
| ||||||
| ATSW01-1, SP4E6-2 | 3 | 14.3% (4/28) | 4 | 9 | 1/5 | |
| 10 | 3/16 | |||||
|
| ||||||
| 15 | 7 | 0/7* | ||||
|
| ||||||
| SP3B8-3, SP4E3-1 | 1 | 3.8% | 15 | 4 | 1/26 | |
|
| ||||||
| SP4E5-4, SP4E6-2 | 3 | 37.5% (3/8) | 4 | 9 | 2/4 | |
|
| ||||||
| 15 | 4 | 1/3 | ||||
| 7 | 0/1* | |||||
|
| ||||||
| ATSP1-B, ATSP1-D, ATSW01-1, SP4B5-2, SP4B6-2 | 1 | 42.2% | 15 | 3 | 100/237 | |
|
| ||||||
| I, III | 2TOW1, PGT183 | 1 | 0.0% | 20 | 5 | 0/7* |
|
| ||||||
| 38-3, SP4B6-2 | 1 | 61.4% | 15 | 3 | 51/83* | |
|
| ||||||
| 38-3, GT5-6 | 1 | 0.0% | 20 | 5 | 0/19* | |
|
| ||||||
| 3TOW1, PGT183 | 1 | 0.0% | 20 | 5 | 0/9* | |
|
| ||||||
| GTCN16, SP4C7-3 | 1 | 100.0% | 4 | 6 | 1/1* | |
|
| ||||||
| GTCN16. SP5B2-3 | 6 | 10.9% (11/101) | 4 | 5 | 1/11* | |
| 6 | 0/13*, 2/18*, 5/23* |
|||||
| 9 | 2/11* | |||||
|
| ||||||
| 15 | 4 | 1/25* | ||||
|
| ||||||
| GTM253-17, ATSW01-1 | 1 | 5.1% | 15 | 3 | 5/98* | |
|
| ||||||
| GTM253-17, SP4B5-2 | 1 | 0.0% | 15 | 3 | 0/76* | |
|
| ||||||
| GTMR01, GT5-6 | 23 | 60.3% (117/194) | 4 | 6 | 1/8*, 0/10* | |
| 7 | 5/10*, 16/17*, 1/2* |
|||||
| 8 | 3/3* | |||||
| 10 | 8/9* | |||||
| 12 | 6/18*, 1/5* | |||||
| 14 | 2/2* | |||||
| 16 | 0/1* | |||||
| 18 | 9/11* | |||||
| 20 | 1/2* | |||||
| 22 | 2/5* | |||||
| 23 | 1/4* | |||||
| 30 | 1/1* | |||||
| 31 | 3/3* | |||||
| UR | 7/13* | |||||
|
| ||||||
| 15 | 6 | 0/1* | ||||
|
| ||||||
| 20 | 8 | 19/32* | ||||
| 9 | 14/19* | |||||
| 10 | 14/15* | |||||
| 15 | 3/3* | |||||
|
| ||||||
| GTMR01, SP4E5-4 | 7 | 23.1% (12/52) | 4 | 3 | 0/5* | |
| 5 | 2/4* | |||||
| 6 | 1/19*, 1/2* | |||||
| 9 | 7/14* | |||||
|
| ||||||
| 15 | 5 | 0/2* | ||||
| 7 | 1/6* | |||||
|
| ||||||
| GTMRO1, SP5B3-1 | 3 | 0.0% (0/14) | 4 | 5 | 0/3* | |
| 6 | 0/5* | |||||
|
| ||||||
| 15 | 4 | 0/6* | ||||
|
| ||||||
| MR16G6, GT5-6 | 1 | 6.7% | 20 | 5 | 1/15* | |
|
| ||||||
| 38-3, GTCN16, GTLI21, GTM253-17, ATSP1-B, ATSP1-D, ATSW01-1, SP4B5-2, SP4B6-2 |
1 | 58.4% | 15 | 3 | 115/197 | |
Excystment success is the proportion of hypnozygotes germinated (as demonstrated by the presence of germling cells or an archeopyle/empty hypnozygote test). Co-cultures that did not yield any viable hypnozygotes are shaded. Trials in which germlings did not complete more than 3 divisions are marked by an asterisk.
Overall, 590 hypnozygotes resulting from Group I same-group combinations, 582 from Group III same-group combinations, and 866 hypnozygotes from Group I/III combinations (including 197 hypnozygotes from a polyclonal co-culture) were monitored for germination (Table 3). Among co-cultures from which 20 or more cysts were isolated, excystment success (defined as the proportion of hypnozygotes that gave rise to a germling cell) ranged from 0% to 76.2%. However, some excystment was observed in all ribosomal group combinations and incubation temperatures examined. Approximately half of the germling cells from hypnozygotes produced in Group I same-group and Group III same-group co-cultures were viable (i.e., completed >3 divisions after excystment), even though excystment success was low (5.1% and 34.5% overall average for Group I same-group and Group III same-group combinations, respectively; Table 5). A precise count of viable germlings from same-group hypnozygotes was impossible because the cysts were sometimes isolated to tissue culture wells in clumps. When more than two hypnozygotes in a single well excysted, the well was frequently overgrown with motile cells so that it was not possible to determine the success of the respective germlings. Germination trials of hypnozygotes from pairwise Group I/III out-group co-cultures did not present similar difficulty because the germlings never completed more than three divisions. Overall excystment success of these hypnozygotes was 29.5%. For comparison, a much greater proportion of hypnozygotes from the polyclonal Group I/III co-culture excysted (58.4%), and slightly less than a third of these divided more than three times.
Table 5.
Summary of findings.
| Gulf of Maine | Group I | Group III | Group I/III hybrids | Abnormal rDNA/cyst | |
|---|---|---|---|---|---|
| Encystment | Yes | Yes | Yes | Yes | Yes |
| rDNA/cyst | 45,000 | 45,000 | 30,000 | 30,000-40,000 | 120,000 and 8500 |
| Excystment rate (%) | 90* | 5.1 | 34.5 | 29.5 | 0 (0/192) |
| Return to haploid mitosis | Yes | Yes | Yes | No | No |
All cysts produced in culture had substantially lower excystment rates than those recovered from natural sediments (Anderson and Keafer, 1987; Matrai et al., 2005). Both hybrid cysts and those with abnormal rDNA abundance failed to resume mitotic reproduction when stimulated to germinate, but only those with abnormal rDNA abundance failed to excyst.
(=excystment rate commonly observed in naturally occurring Group I hypnozygotes during peak phase of their circannual germination rhythm).
3.3. Hypnozygote rDNA copy number estimation
DNA extractions from bulk samples of hypnozygotes were analyzed using Group I amplicon and Group III amplicon qPCR assays (8 samples, ranging from 484 to 1518 cysts; Table 4). The mean efficiency of the respective assays was 75.4% and 84.3%. Cross reactivities between the Group I primers and Group III templates, and the Group III primers and Group I templates were both negligible, though the former was higher (0.9% versus 0.009% of the true number of opposite standard templates ‘detected’).
Table 4.
Copy number estimates of Groups I and III-type LSU rDNA loci in Group I, Group III and hybrid hypnozygotes.
| Groups/Clones | N | GI copy number | GIII copy number |
|---|---|---|---|
| Group I only | (45,000) | - | |
| 38-3, GTM253-17 | 744 | 44,588 | N/T |
| GTLI21, GTM253-17* | 1518 | 119,207 | N/T |
| Gulf of Maine Sediment | 608 | 44,454 | N/T |
| Group III only | - | (30,000) | |
| SP4B5-2, SP4B6-2** | 484 | N/T | 30,136 |
| Group I, Group III | |||
| 38-3, ATSP1-B** | 948 | 28,513 | 2,718 |
| GTM253-17, ATSP1-D** | 664 | 35,059 | 4,462 |
| GTM253-17, ATSW01-1 | 642 | 32,169 | 6,533 |
| GTM253-17, SP4B5-2* | 769 | 7,692 | 760 |
N is the estimated number of hypnozygotes collected by laser pressure catapult. Approximations of Group I and III copy numbers used for Fig. 4 boundaries are italicized (see text). Hypnozygotes that did not excyst are marked by an asterisk.
Hypnozygotes for which no germination trials were attempted are marked by two asterisks.
The rDNA copy number estimates for Group I hypnozygotes were 44,454, 44,588, and 119,207 (Table 4). The latter estimate was from hypnozygotes formed during co-culture of clones GTLI21 and GTM253-17, and is considered aberrant because no cysts from this co-culture germinated (Table 5). The nominal estimated copy number was 45,000 in samples from another Group I co-culture (38-3/GTM253-17) and from Group I hypnozygotes collected from a Gulf of Maine sediment sample. Only one sample of Group III hypnozygotes was collected (SP4B5-2/SP4B6-2) but no germination trials were completed. The estimated Group III amplicon copy number was nominally 30,000 (Table 4).
Four hypnozygote samples from Group I/III co-cultures were analyzed for copy number and two were used in germination trials. The Group I rDNA copy number estimates were 5–10 fold greater than those for Group III in all four samples. This difference is consistent with that found during the genotyping experiments (described in Section 3.4). In three of the four samples, the sum of the Groups I and III copy estimates was intermediate between the nominal copy numbers of Group I and III hypnozygotes. In the one case where rDNA copy number was less than 30,000 (GTM253-17/SP4B5-2, estimated copy sum=8453), hypnozygotes did not excyst (n=76). In the one other Group I/III sample tested for both germination and copy number (GTM253-17/ATSW01-1), 38,703 ribosomal copies were detected per cyst and 5 of 98 hypnozygotes excysted (Tables 3–5).
3.4. Single cyst genotyping
The nested PCR assay effectively differentiated hypnozygotes formed by Group I and III parents. In an initial analysis, only hypnozygotes from pairwise co-cultures were considered because such cysts would have known parentage if A. tamarense clones formed only heterotypic pairs (total of 282 experiments: 42 from Group I co-culture, 24 from Group III co-culture, and 216 from Group I/III co-culture). Ratios between the number of Group I and III amplicons were used because both types were frequently detected (likely due to limited cross-reactivity between the respective templates and primers). If A. tamarense isolates were to behave as true heterothallic cells (no homotypic conjugation), the ratio of Group I to III amplicons detected should be heavily skewed toward Group I for Group I hypnozygotes, heavily skewed toward Group III for Group III hypnozygotes, and intermediate between Groups I and III for hybrids.
Heterotypic encystment was tested by a randomization analysis of the genotyping results from pairwise co-cultures of Group I and III clones (described in Section 2.7). The misclassification rate in the true dataset was 11.7%, but was never lower than 22.0% in several 10,000 randomization trials so that the low error rate in the true dataset is highly significant (P<0.0001). Similarly, when the pairwise data were themselves considered in pairs (e.g. Group I and hybrid data only, not Group III), the misclassification rates were significantly lower than expected under a null hypothesis of no difference between groups (P<0.0001 in each case). Thus, genotyping data from pairwise cultures strongly support a mating mechanism whereby homotypic conjugant pairs are prevented from encysting. Following from this finding, hypnozygotes produced in pairwise co-cultures are considered to be of known parentage (Group I, Group III or hybrid). Accordingly, hypnozygotes from pairwise co-culture are hereafter referred to as ‘knowns’, and those isolated from polyclonal co-cultures and sediment samples are referred to as ‘unknowns’.
In estimation of the Group I, Group III and hybrid probability density functions of x, only assays that exceeded the 0.01% lysis contour were used because misclassifications were rare beyond this threshold (Fig. 4A and B). Overall, 31.5% of assays exceeded the 0.01% threshold (137/435), but most were of knowns (113 versus 24). Intersections between the pdfs were x= −0.96 (Group III/hybrid boundary) and x=2.61 (hybrid/Group I boundary;Fig. 5B), so that the range classified as hybrids is approximately 1:9 to 407:1, Group I to Group III amplicons. This range reflects incomplete cyst lysis during isolation and consequent sub-sampling of individual cysts’ rDNAs. The hybrid probability density is greatest where Group I:Group III is approximately 2:1, a skew that is comparable to that observed in copy number estimates from unamplified hypnozygote genomic DNA (approximately 8:1; Table 4). The bias toward Group III in the genotyping data suggests a difference in D1R–D2C amplification efficiency between Group I and III templates that is similar to the variance observed between the efficiencies in the two qPCR assays. However, as the misclassification rate was quite low under the thresholds defined by the pdf intersections (1.8%, 2/113), such a bias during D1R–D2C amplification did not substantially affect the classification skill of the assay.
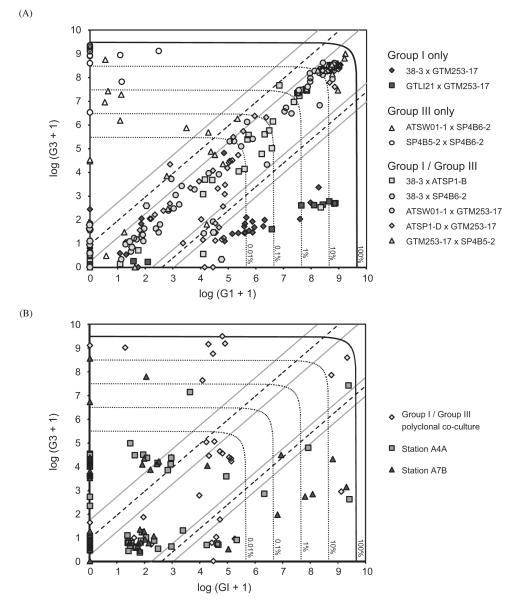
Fig. 4.
Single hypnozygote genotyping by nested PCR. (A) Results of hypnozygotes from pairwise co-culture of Group I and III isolates (‘knowns’). Lysis efficiency contours (0.01%, 0.1%, 1%, 10% and 100%) are expected numbers of amplicons after primary D1R–D2C PCR. Group I/hybrid and hybrid/Group III thresholds are indicated by dashed diagonal lines; gray diagonal lines indicate regions about the thresholds where classification probability is less than 95% (see text, Fig. 5). (B) Results of hypnozygotes from a polyclonal Group I/III co-culture and two sediment samples taken within Belfast Lough (survey stations A4A and A7B). Lysis efficiency contours and classification boundaries are drawn as in (A).
Of the 24 successful assays of unknowns, 4 hypnozygotes were classified hybrid, 9 Group I, and 11 Group III. Only 2 of 10 polyclonal unknowns were classified hybrid, but this was expected because multiple compatible clones of both types were inoculated in these co-cultures. Seven of the eight other successful polyclonal assays were Group III, a result that is consistent with higher growth rates and stationary densities observed in the Group III isolates. Only one unknown was classified with probability less than 95%, a hybrid that was isolated from Belfast Lough sediment core A7B (x=2.46, estimated probability 76.5%). A second hybrid was isolated from a Belfast sediment core A4A and was classified with probability greater than 99%. Lysis efficiency of the A4A hybrid exceeded 10% (estimated >3000 template molecules), further supporting its classification. Group I and III single-type cysts were also detected in the Belfast sediment samples, though more Group I were found than Group III (8 versus 4).
4. Discussion
4.1. Speciation within the A. tamarense complex
An authoritative delineation of species within the A. tamarense complex has been lacking since the discovery that the group’s morphotypes are uncorrelated with its ribosomal phylogeny (Scholin and Anderson, 1994). Subsequent investigations using various molecular and morphological characters have repeatedly confirmed the divergence of the five A. tamarense clades, the clades’ global geographic separation, and also their close kinship to sister species, A. tropicale, A. tamiyavanichi, and A. affine (John et al., 2003; Leaw et al., 2005; Lilly et al., 2007; MacKenzie et al., 2004). In the present study, limited mating compatibility was found across most of these groups, a finding that is similar to reports of broad mating compatibility among globally dispersed isolates of another PSP dinoflagellate, Gymnodinium catenatum (Blackburn et al., 2001; Oshima et al., 1993). However, unlike G. catenatum, hybrids of two closely related A. tamarense clades failed to yield any viable progeny. The latter supports the assertion that complete speciation underlies the phylogenetic topology of the A. tamarense complex. Such has been proposed previously by Lilly et al. (2007) on the basis of ribosomal divergence between the complex’s clades and its uncontested sister species. However, the mating and germination data presented here are only conclusive with respect to two of the five clades (Groups I and III). It remains to be shown whether the progeny of other mating-compatible clades are viable.
Even so, if species are defined as populations that can recombine genetically in nature, the A. tamarense clades are largely speciated solely on the basis of their present-day biogeography. Expansive intermingling of clades is limited to Groups I and III along the northern coasts of Ireland and Great Britain and to Groups I and IV (both toxic) in the northwest Pacific (Lilly et al., 2007). Our demonstration of hybrid cysts in Belfast Lough, Northern Ireland, confirms that mating is possible between cells of different clades within a natural setting. However, because these hybrids do not yield viable progeny, gene flow between Group I and III populations is not possible. There are also small, isolated populations that commingle with the larger, interconnected populations of other ribosomal species. These isolated populations most likely arose as a result of global shipping (e.g. a population of Group IV, an Asian clade, in Thau Lagoon, France; Lilly et al., 2002). Such human-assisted dispersal provides a mechanism to renew interaction between clades, but is unlikely to have affected the evolution of the A. tamarense complex because large-scale shipping did not begin until the last century.
The present-day biogeography of the A. tamarense complex and its genetic structure both indicate a sustained period of allopatry (geographic isolation) beginning 23–45 MYA (John et al., 2003). With establishment of allopatry, genetic divergence of the clades occurred according to selection and drift in each of the their respective ranges. Such divergence is correlated with the development of incompatibilities that limit sexual interaction or that causes inviability upon inter-group mating. Groups I and III represent the latter case because encystment by Group I/III conjugants does not yield viable progeny. The timing of lethality is highly significant because contact between populations of Groups I and III may cause local elimination of the less abundant population, particularly in the absence of pre-mating barriers between these species. Elimination would occur because the probability that an individual Group I or III gamete will form a hybrid is proportional to the probability that it encounters gametes of the other clade. The more abundant population will have greater mating success than the less abundant one, so that, in subsequent mating cycles, numerical differences tend to be compounded and cells of the less abundant species become increasingly likely to form fatal pairs. It follows that pre-mating barriers are necessary for stable co-existence because they limit (or abolish) gametic contact between species (Paterson, 1993).
For sympatric dinoflagellates, pre-mating barriers may have a variety of mechanisms including: (1) different receptivities to biological signals, (2) variation in gamete aggregation patterns (e.g. depths of thin layer formations), and (3) variation in the timing of sexual induction. The reduced positive encystment ratios in out-group combinations of Alexandrium (as compared to same-group combinations; Supplementary Fig. S2) suggest that the biological signals for mating are only partially shared. This limited reactivity between clades may be caused by non-transitive relationships among mating factors. As an example, a ‘male’ signal from one group could interact positively with a ‘female’ signal from another. However, this positive interaction does not ensure similar reactivity between a ‘female’ signal of the first group and a ‘male’ signal of the second. It is also possible that the lower positive ratio among out-group co-cultures is due to incompatibilities that occur after mating but before encystment (i.e. during the planozygote stage; Fig. 1). Such a post-conjugation mechanism is considered less likely because many out-group combinations do encyst successfully. It is therefore assumed that non-transitive relationships between pre-mating factors are more likely than between post-conjugal ones. If this assumption were violated (i.e. the lower positive ratio were due to zygotic rather than mating incompatibilities), spatial or temporal pre-mating barriers that limit contact between A. tamarense clades could still exist in areas where their ranges overlap. However, such ecological barriers have not been shown. Further, the presence of hybrids in Belfast Lough indicates that such barriers, if they do exist, are incomplete (at least between Groups I and III).
The sum of these observations is that while broad mating intercompatibility has been retained within the A. tamarense complex, it does not ensure gene flow between commingling populations of divergent clades. This is because post-zygotic incompatibility like that shown in Group I/III hybrids may prevent the proliferation of genetic intermediates. Further, in the absence of pre-mating barriers, post-zygotic lethality can provide a mechanism for elimination of less abundant clades (sensu species) through fatal hybridization. These findings create an incongruity within our study in that the two focal clades have been distinguished as separate species but their hybrids are used to establish normal, intraspecies mating behavior. The approach has been borne out by its results as virtually no inbred cysts were detected in Group I/III pairwise co-cultures. Obligate hybridization demonstrates an inability of clonal siblings to conjugate with one another and encyst.
4.2. Post-zygotic lethality of Group I/III hybrids
Initially, the assessment of post-zygotic lethality was complicated because of the uncertainty in cyst parentage and the low germinability of hypnozygotes produced in culture. Although germination success was improved by storage of hypnozygotes in anoxic sediment, excystment rates remained low (26.5% across all trials, Table 3). Three possible causes of low excystment were considered: (1) that hypnozygotes were not adequately stimulated to germinate; (2) that hypnozygotes were commonly subject to inbreeding depression; or (3) that maturation of hypnozygotes was impaired under the conditions used in this study. The first cause was rejected because hypnozygotes from natural sediments may be stimulated to excyst at rates >90% under similar conditions (Anderson and Keafer, 1987). Likewise, our finding that cysts are predominantly outbred in pairwise Group I/III co-cultures negated the possibility of inbreeding depression. With respect to impaired maturation, the biotic and abiotic conditions in culture were unavoidably different from those experienced in nature. Therefore we conclude that the chosen conditions, though they prolong germinability, also cause germination rates to be low. We have no ready solution for improving the germinability of cultured cysts other than to follow Figueroa et al. (2005), who demonstrated improved germinability after isolation to un-enriched medium. A similar protocol might improve excystment success for Group I hypnozygotes but it remains unclear why natural Group I cysts will germinate efficiently in enriched medium.
In any case, low overall germinability of cultured cysts does not diminish the observation of inviability among hybrids. A much greater proportion of hybrid hypnozygotes than Group I hypnozygotes excysted in the 669 hybrid trials and, most importantly, no germlings from 152 hybrid excystments completed more than one mitotic division. The lethality of Group I/III hybrids must result from genetic recombination because it does not occur until after the completion of zygotic stages. The viability of the zygotic stages is surprising given that the estimated rDNA copy number of Group I motile cells is 5-fold greater than that for Group III motile cells (0.5–1×106 and 1–2×105 copies/cell, respectively). If these copy numbers are fixed in the respective haploid genomes, as many as 2×106 copies per Group I zygote and 4×105 copies per Group III zygote should be recovered upon cyst lysis. Instead, the apparent rDNA copy number of the hypnozygotes is as much as 90-fold lower than expected and nearly equivalent between Group I and III cysts. Erdner et al. (2010) have estimated similarly low copy numbers for Group I hypnozygotes, corroborating our results. The reductions in rDNA copy number by both Group I and III zygotes to nearly equivalent values may be crucial for the successful maturation observed in hybrids.
In the two cases where hypnozygote copy numbers deviated from the majority of other measurements (GTLI21/GTM253-17 and GTM253-17/SP4B5-2), hypnozygotes uniquely failed to excyst. In the former case, the apparent copy number was substantially greater than that found in other Group I cysts so that excystment failure was associated with incomplete rDNA copy reduction. Conversely, the estimated copy number from GTM253-17/SP4B5-2 hybrid hypnozygotes was 4- to 5-fold lower than that of hybrids that did excyst successfully. In this case, an over-reduction of rDNA loci was associated with excystment failure. Combined, the copy estimates from viable and inviable hypnozygotes suggest that the low rDNA copy number of hypnozygotes results from destruction of loci during the process of encystment rather than recalcitrance of DNA during extraction. Such origin regulation of rDNA loci is prominent in the sexual cycle of ciliates (Tower, 2004). However, the scale of regulation we report for A. tamarense is much less (compare to several thousand-fold in Tetrahymena thermophila).
The destruction of rDNA loci (and presumably other genes) during encystment may enable the progression of hybrids through the sexual cycle by masking disequilibria between distinct motile cell genomes. In the absence of origin regulation, differences in homolog structure or regulation might be catastrophic to any of the steps involved in the diploid phase of the sexual cycle. Tolerance of over-abundant loci in the early diploid stages may provide resilience to disequilibria upon hybrid conjugation. In turn, reductions to near equivalent copy numbers during subsequent zygote stages could be critical for successful completion of meiosis. Hybrid lethality is likely caused by genetic disequilibrium because its onset occurs after the completion of meiosis (and within the first germling mitotic cell cycle). Cell death is unavoidable once motile cells begin to re-establish higher gene copy numbers.
4.3. Mating system models and interactions between ribosomal species
A curious aspect of sexual processes in nature is the ubiquity of dioecious (two mating-type) systems. Such binary cell recognition is easily evolved from models of primitive homothallism, and is also a sufficient mechanism to reduce inbreeding depression (Hoekstra, 1987). Gametic dimorphism (or anisogamy) is also ubiquitous among eukaryotes and has been reported in some A. tamarense cultures (Anderson and Lindquist, 1985; Turpin et al., 1978). If sexual conjugation is limited to unlike gametes, such dimorphism confirms a system that both prevents homotypic pairing and that also consists of only two mating types. That anisogamous gamete pairs form within A. tamarense cultures suggests that these species are dioecious, but has been contradicted by attempts to classify clones as one of two mating types (Supplementary Figs. S3 and S4; Destombe and Cembella, 1990). A critical assumption underlying these assessments was that mating types are fixed within clones. The occurrence of auto-encystment by some clonal cultures partly undermines this assumption since it must have occurred via a mating-type switch. However, auto-encystment was limited to 4 of 119 isolates and cyst yields in each of these cases were low (Supplementary Fig. S2). Together, the rarity of auto-encystment and its low yields suggest that mating-type switches by individual cells are uncommon, even within stocks of auto-encysting clones. Further, if mating-type switching were frequent, a much greater proportion of all pairwise combinations would have produced cysts than was observed (Table 1).
Given the evidence of a multiple mating-type system, it was also surprising that the positive encystment ratios remained low even among same-group combinations taken from restricted geographical areas (e.g. consider the unbiased Belfast Lough subset where less than 40% of same-group combinations were positive). Most multiple mating-type organisms have mating reactivity rates that approach 100% with increasing numbers of mating types (Bull and Pease, 1989). From the attempts to classify clone genders in the four isolate subsets, a minimum of four mating types must exist so that the positive ratio of same-group combinations should approach 75%. Instead, the maximum positive ratio observed for any clade was 48.4% (Group III; Table 1), much less than would be expected for a freely interbreeding species having four mating types, but very near the 50% expected under a dioecious model.
The lower positive encystment ratios among out-group combinations in A. tamarense is similar to broad but diminished inter-reactivity that has been described between populations of numerous ciliate genera (Dini and Nyberg, 1993). For this reason and because ciliates are closely allied to dinoflagellates within the Alveolate protist lineage, previous authors have speculated that the multiple mating-type systems of heterothallic dinoflagellates are like those of ciliates (Blackburn et al., 2001; Destombe and Cembella, 1990). A comparison to the ciliate genus Euplotes is therefore instructive because its mating system has been described in considerable molecular detail, and because its species are also known to have more than two mating types. As mentioned previously, the mating-type system of Euplotes stimulates transition to the sexual cycle, but in contrast to A. tamarense, does not prevent homotypic conjugation. Individual Euplotes cells express their mating type through one of numerous pheromones that inhibit gametic differentiation when bound in homologous pairs (as would occur in clonal culture). When unlike mating types interact in polyclonal cultures, the autocrine inhibition loop is interrupted, freeing cells to transform to their competent sexual forms (Luporini et al., 2005; Vallesi et al., 1995). This system enables Euplotes species to limit sexual differentiation in highly clonal populations where genetic recombination is unlikely to be beneficial, and also allows greater mating reactivity than is observed in dioecious species. Neither the predominance of heterotypic encystment nor the low mating reactivity of A. tamarense isolates refute the presence of a Euplotes-like mating barrier, but its mating system must have additional complexity to explain these characteristics.
A simple modification to the Euplotes model is the addition of a second barrier that prevents adhesion of homotypic pairs (for instance, + and − types like those of C. reinhardtii). This second adhesion barrier is contrasted to the first Euplotes-like barrier that acts as a self-recognition mechanism. Given a system with four self-recognition types and two adhesion types, the probability that a pairwise combination is positive would converge toward 37.5% where each clone is characterized by one each of the self-recognition and adhesion types. This target compares favorably to the 31.8% positive encystment ratio found among Group I combinations but is much less than the 48.4% observed among Group III (Table 1). The higher ratio in the latter case could be explained by an absence of the self-recognition barrier or by a larger number of self-recognition types among Group III clones. As an alternative to the proposed two-step model, a one-step model would require a multiple mating type adhesion system whereby each isolate expresses a factor that is recognizable as non-self by other isolates and in turn promotes adhesion between them. Such a one-step system was also hypothesized for Euplotes before the molecular characterization of its self-recognition system (Heckmann and Kuhlmann, 1986). A weakness of such a model for A. tamarense species is that low positive encystment ratios could only be explained by non-transitive interactions between mating factors, both between ribosomal groups and also within ribosomal groups where gene flow has been less restricted. Given the low positive ratios within the A. tamarense clades, it is unclear what advantage a multiple mating type adhesion system would lend versus a less complex dioecious system. In contrast, a two-step system would both limit gametic differentiation when sexual recombination is unprofitable and also ensure heterotypic conjugation.
The precise system that mediates mating among Alexandrium cells has substantial implications for the interaction of unlike, natural populations, such as the mixing of toxic Group I and nontoxic Group III cells. If one accepts that reduced encystment among out-group co-cultures results from impaired biological signaling, the pre-mating barriers must occur either during gametogenesis or adhesion. If the barrier affects adhesion, conjugation between unlike ribosomal clades would be inhibited under both the one- and two-step models. If instead the barrier affects gametogenesis, greater sexual interaction between unlike clades might be expected under certain natural conditions. This is because differentiated gametes would freely conjugate with those of other clades, particularly where populations commingle and are mutually induced to enter the sexual cycle. It would also follow that the finding of selectivity for isolates of the same ribosomal species should not be taken to represent the behavior of natural polyclonal populations. Instead, the finding of selectivity would be due to the contrivance of co-culturing only two clones. In natural, polyclonal populations such as those occurring in Belfast Lough, both may be self-stimulating so that an individual Group I cell would undergo gametogenesis upon stimulation by its conspecifics, and a Group III cell similarly by its own population. The resulting Group I and III gametes could then intermingle, conjugate, and encyst unimpeded.
5. Summary
Co-culture experiments were first undertaken as an approach to characterize relationships among globally distributed populations, and many of these experiments pre-date our understanding of A. tamarense ribosomal phylogeny. Persistent questions about the nature of sexual interactions among dinoflagellates led to development of the nested PCR method for assessing parentage of hypnozygotes produced in co-cultures. Using this method, the predominance of heterotypic encystment was demonstrated and normal heterothallic sexuality (heterotypic conjugation) between two A. tamarense clades was verified. Substantial copy reduction of rDNA loci was also discovered upon encystment.
The discovery that clones from widely dispersed populations will encyst is similar to reports of gametic compatibility among geographically dispersed isolates of G. catenatum, but hybrids of genetically isolated A. tamarense Group I and III species failed to yield viable progeny. This discovery has immediate practical applications because Group I cells produce PSP toxins but Group III cells do not (Scholin et al., 1994; Lilly et al., 2007). Further, the ranges of the these two species only overlap at their shared boundary. Taken together, post-zygotic lethality and the species’ minimal biogeographic overlap implicate interbreeding between them as a mechanism hindering expansion of one type into the range of the other, and vice versa. It also follows that deliberate introduction of the nontoxic Group III species should be considered a promising approach for mitigation of recurrent toxic Group I blooms.
Supplementary Material
Acknowledgments
We thank K. Norton, B. Keafer, J. Kleindinst and other members of the Anderson laboratory for technical support, and are also grateful to S. Bickel and R. Sloboda at Dartmouth College (Hanover, NH) for hosting MLB during completion of the laser catapult experiments. We also thank a large number of colleagues who shared their Alexandrium cultures for these experiments. Work by MLB, DLE, and DMA was supported by NSF Grant nos. OCE-0402707 and OCE-9808173 and by the Woods Hole Center for Oceans and Human Health through NSF Grant no. OCE-0430724 and NIEHS Grant no. P50ES012742-0. Research support has also been provided through NOAA Grant no. NA06-NOS4780245, an EU SEED Grant no. GOCE-CT-2005-003875 (JL, LP), and a STAR graduate fellowship to MLB (FP-91688601) from the US Environmental Protection Agency. The EPA has not formally reviewed this publication, and the EPA does not endorse any of the products mentioned in it. The views expressed are solely those of the authors. This is ECOHAB Contribution no. 309.
Abbreviations
- LSU rDNA
large subunit ribosomal DNA sequence
- PCR
polymerase chain reaction
- PSP
paralytic shellfish poisoning
- qPCR
real-time, quantitative PCR
Footnotes
Appendix A. Supporting information Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dsr2.2009.09.005.
References
- Adair WS, Hwang C, Goodenough UW. Identification and visualization of the sexual agglutinin from the mating-type plus flagellar membrane of chlamydomonas. Cell. 1983;33(1):183–193. doi: 10.1016/0092-8674(83)90347-1. [DOI] [PubMed] [Google Scholar]
- Anderson D, Lindquist N. Time-course measurements of phosphorus depletion and cyst formation in the dinoflagellate Gonyaulax tamarensis Lebour. Journal of Experimental Marine Biology and Ecology. 1985;86(1):1–13. [Google Scholar]
- Anderson DM. Toxic algal blooms and red tides: a global perspective. In: Okaichi T, Anderson DM, Nemoto T, editors. Red Tides: Biology, Environmental Science, and Toxicology. Elsevier Science Publishing Co. Inc.; 1989. [Google Scholar]
- Anderson DM, Fukuyo Y, Matsuoka K. Cyst methodologies. In: Hallegraeff GM, Anderson DM, Cembella AD, editors. Manual on Harmful Marine Microalgae. United Nations Educational; Paris, France: 2003. pp. 165–190. [Google Scholar]
- Anderson DM, Keafer BA. An endogenous annual clock in the toxic marine dinoflagellate Gonyaulax tamarensis. Nature. 1987;325(6105):616–617. doi: 10.1038/325616a0. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Kulis DM, Binder BJ. Sexuality and cyst formation in the dinoflagellate Gonyaulax tamarensis: cyst yield in batch cultures. Journal of Phycology. 1984;20(3):418–425. [Google Scholar]
- Anderson DM, Wall D. Potential importance of benthic cysts of Gonyaulax tamarensis and G. excavata in initiating toxin dinoflagellate blooms. Journal of Phycology. 1978;14(2):224–234. [Google Scholar]
- Binder BJ, Anderson DM. Green light-mediated photomorphogenesis in a dinoflagellate resting cyst. Nature. 1986;322(6080):659–661. [Google Scholar]
- Blackburn SI, Bolch CJS, Haskard KA, Hallegraeff GM. Reproductive compatibility among four global populations of the toxic dinoflagellate Gymnodinium catenatum (Dinophyceae) Phycologia. 2001;40(1):78–87. [Google Scholar]
- Bull JJ, Pease CM. Combinatorics and variety of mating-type systems. Evolution. 1989;43(3):667–671. doi: 10.1111/j.1558-5646.1989.tb04263.x. [DOI] [PubMed] [Google Scholar]
- Destombe C, Cembella A. Mating-type determination, gametic recognition and reproductive success in Alexandrium excavatum (Gonyaulacales, Dinophyta): a toxic red-tide dinoflagellate. Phycologia. 1990;29(3):316–325. [Google Scholar]
- Dini F, Bleyman LK, Giubbilini P. Non-Mendelian inheritance of early maturity in Euplotes crassus. Journal of Eurkaryotic Microbiology. 1990;37(6):475–478. [Google Scholar]
- Dini F, Nyberg D. Sex in ciliates. Advances in Microbial Ecology. 1993;13:85–153. doi: 10.1007/s002489900126. [DOI] [PubMed] [Google Scholar]
- Erdner D, Percy L, Keafer BA, Lewis J, Anderson DM. A quantitative real-time PCR assay for the identification and enumeration of Alexandrium cysts in marine sediments. Deep-Sea Research II. 2010;57(3–4):279–287. doi: 10.1016/j.dsr2.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa RI, Bravo I. Sexual reproduction and two different encystment strategies of Lingulodinium polyedrum (Dinophyceae) in culture. Journal of Phycology. 2005;41(2):370–379. [Google Scholar]
- Figueroa RI, Bravo I, Garcés E. Effects of nutritional factors and different parental crosses on the encystment of Alexandrium catenella (Dinophyceae) in culture. Phycologia. 2005;44(6):658–670. [Google Scholar]
- Figueroa RI, Bravo I, Garcés E. Multiple routes of sexuality in Alexandrium taylori (Dinophyceae) in culture. Journal of Phycology. 2006;42(5):1028–1039. [Google Scholar]
- Galluzzi L, Bertozzini E, Penna A, Perini F, Garcés E, Magnani M. Analysis of rRNA gene content in the mediterranean dinoflagellate Alexandrium catenella and Alexandrium taylori: implications for the quantitative real-time PCR-based monitoring methods. Journal of Applied Phycology. 2009 10.1007/s10811-009-9411-3. [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine plankton diatoms I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Canadian Journal of Microbiology. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Hallegraeff GM. A review of harmful algal blooms and their apparent global increase. Phycologia. 1993;32(2):79–99. [Google Scholar]
- Heckmann K, Kuhlmann HW. Mating types and mating inducing substances in Euplotes octocarinatus. Journal of Experimental Zoology. 1986;237:87–96. [Google Scholar]
- Hoekstra RF. The evolution of sexes. In: Stearns SC, editor. The Evolution of Sex and its Consequences. Birkhäuser Verlag; Boston: 1987. pp. 59–91. [Google Scholar]
- John U, Fensome RA, Medlin LK. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense ‘‘Species Complex’’ (Dinophyceae) Molecular Biology and Evolution. 2003;20(7):1015–1027. doi: 10.1093/molbev/msg105. [DOI] [PubMed] [Google Scholar]
- Leaw CP, Lim PT, Ng BK, Cheah MY, Ahmad A, Usup G. Phylogenetic analysis of Alexandrium species and Pyrodinium bahamense (Dinophyceae) based on theca morphology and nuclear ribosomal gene sequence. Phycologia. 2005;44(5):550–565. [Google Scholar]
- Lilly EL, Halanych KM, Anderson DM. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae) Journal of Phycology. 2007;43(6):1329–1338. [Google Scholar]
- Lilly EL, Kulis DM, Gentien P, Anderson DM. Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: evidence from DNA and toxin analysis. Journal of Plankton Research. 2002;24(5):443–452. [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434(7036):1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Luporini P, Alimenti C, Ortenzi C, Vallesi A. Ciliate mating types and their specific protein pheremones. ACTA Protozoologica. 2005;44:89–101. [Google Scholar]
- MacKenzie L, de Salas M, Adamson J, Beuzenberg V. The dinoflagellate genus Alexandrium (Halim) in New Zealand coastal waters: comparative morphology, toxicity and molecular genetics. Harmful Algae. 2004;3(1):71–92. [Google Scholar]
- Matrai P, Thompson B, Keller M. Circannual excystment of resting cysts of Alexandrium spp. from eastern Gulf of Maine populations. Deep-Sea Research II. 2005;52(19–21):2560–2568. [Google Scholar]
- Musgrave A, Eijk E, Welscher R, Broekman R, Lens P. Sexual agglutination factor from Chlamydomonas eugametos. Planta. 1981;153(4):362–369. doi: 10.1007/BF00384255. [DOI] [PubMed] [Google Scholar]
- Oshima Y, Itakura H, Lee KC, Yasumoto T, Blackburn S, Hallegraeff G. Toxin production by the dinoflagellate Gymnodinium catenatum. In: Smayda TJ, Shimizu Y, editors. Toxic Phytoplankton Blooms in the Sea; Proceedings of the Fifth International Conference on Toxic Marine Phytoplankton; Newport, RI, USA. 28 October 1991; The Netherlands: Elsevier, Amsterdam; 1993. pp. 907–912. [Google Scholar]
- Paterson HEH. Evolution and the Recognition Concept of Species. The Johns Hopkins University Press; Baltimore, MD: 1993. [Google Scholar]
- Pfiester LA, Anderson DM. Dinoflagellate reproduction. In: Taylor FJR, editor. The Biology of Dinoflagellates. Blackwell Scientific Publications; Oxford: 1987. pp. 611–648. [Google Scholar]
- Scholin CA, Anderson DM. Identification of Group- and Strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). I. RFLP analysis of SSU rRNA genes. Journal of Phycology. 1994;30(4):744–754. [Google Scholar]
- Scholin CA, Hallegraeff GM, Anderson DM. Molecular evolution of the Alexandrium tamarense ‘species complex’ (Dinophyceae): dispersal in the North American and west Pacific regions. Phycologia. 1995;34(6):472–485. [Google Scholar]
- Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of Group- and Strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. Journal of Phycology. 1994;30(6):999–1011. [Google Scholar]
- Schwinghamer P, Anderson DM, Kulis DM. Separation and concentration of living dinoflagellate resting cysts from marine sediments via density-gradient centrifugation. Limnology and Oceanography. 1991;36(3):588–592. [Google Scholar]
- Silverman BW. Density Estimation for Statistics and Data Analysis. Chapman & Hall; New York: 1986. [Google Scholar]
- Solow AR. A randomization test for misclassification probability in discriminant analysis. Ecology. 1990;71(6):2379–2382. [Google Scholar]
- Tower J. Developmental gene amplification and origin regulation. Annual Review of Genetics. 2004;38(1):273–304. doi: 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- Turpin DH, Dobell PER, Taylor FJR. Sexuality and cyst formation in Pacific strains of the toxic dinoflagellate Gonyaulax tamarensis. Journal of Phycology. 1978;14(2):235–238. [Google Scholar]
- Uchida T. The role of cell contact in the life cycle of some dinoflagellate species. Journal Plankton Research. 2001;23(8):889–891. [Google Scholar]
- Vallesi A, Giuli G, Bradshaw RA, Luporini P. Autocrine mitogenic activity of pheromones produced by the protozoan ciliate Euplotes raikovi. Nature. 1995;376(6540):522. doi: 10.1038/376522a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.