Abstract
Staphylococcus aureus is a Gram-positive pathogen responsible for tremendous morbidity and mortality. As with most bacteria, S. aureus requires iron to cause disease, and it can acquire iron from host hemoglobin. The current model for staphylococcal hemoglobin-iron acquisition proposes that S. aureus binds hemoglobin through the surface-exposed hemoglobin receptor IsdB. IsdB removes heme from bound hemoglobin and transfers this cofactor to other proteins of the Isd system, which import and degrade heme to release iron in the cytoplasm. Here we demonstrate that the individual components of the Isd system are required for growth on low nanomolar concentrations of hemoglobin as a sole source of iron. An in-depth study of hemoglobin binding by IsdB revealed key residues that are required for hemoglobin binding. Further, we show that these residues are necessary for heme extraction from hemoglobin and growth on hemoglobin as a sole iron source. These processes are found to contribute to the pathogenicity of S. aureus in a murine model of infection. Together these results build on the model for Isd-mediated hemoglobin binding and heme-iron acquisition during the pathogenesis of S. aureus infection.
Keywords: Staphylococcus aureus, heme, hemoglobin, iron-regulated surface determinant system, infection, iron, pathogenesis
To colonize the host and cause disease Staphylococcus aureus requires iron. The most abundant iron source within vertebrates is hemoglobin. Staphylococci capture hemoglobin and subsequently import and degrade heme using a series of proteins known collectively as the iron-regulated surface determinant (Isd) system [1]. The Isd system consists of 10 proteins, which act in concert to acquire iron from hemoglobin (Figure 1A) [1, 2]. The current model for Isd-mediated heme capture proposes that IsdA and IsdB are anchored to the cell wall by sortase A and are localized to the bacterial surface [1, 3]. IsdC is embedded within the cell wall and is covalently anchored to peptidoglycan by a dedicated sortase B [1, 2]. IsdB binds hemoglobin and removes heme from the protein backbone of hemoglobin [1, 4–7]. IsdB then rapidly passes heme to IsdA, which transfers it to IsdC [5, 6, 8–10]. Multiple IsdC molecules are suggested to pass heme across the cell wall and deliver heme to the membrane transport component IsdE [5, 6, 11, 12]. The kinetics of heme binding and unidirectional transfer have been extensively studied using mass spectrometry, ultraviolet-visible spectrometry, magnetic circular dichroism spectrometry, nuclear magnetic resonance, and X-ray crystallography [5–7, 9, 13–18]. IsdH is another protein that binds hemoglobin and heme in vitro [18, 19]. Despite significant amino acid similarity to IsdB, IsdH does not appear to be required for hemoglobin binding and hemoglobin-derived iron acquisition in vivo [4]. It is possible that IsdH contributes to hemoglobin binding at low concentrations or to binding of hemoglobin-haptoglobin complexes. Membrane-localized IsdE allows for heme passage into the cytoplasm, where heme is degraded by the heme oxygenases IsdG and IsdI to liberate iron and form staphylobilin [15, 20–26]. Consistent with the importance of heme-iron acquisition to the pathogenesis of staphylococcal infections, inactivation of isdA, isdB, isdC, isdG, or isdI reduces bacterial proliferation within the host [3, 4, 21, 27–29].
Figure 1.
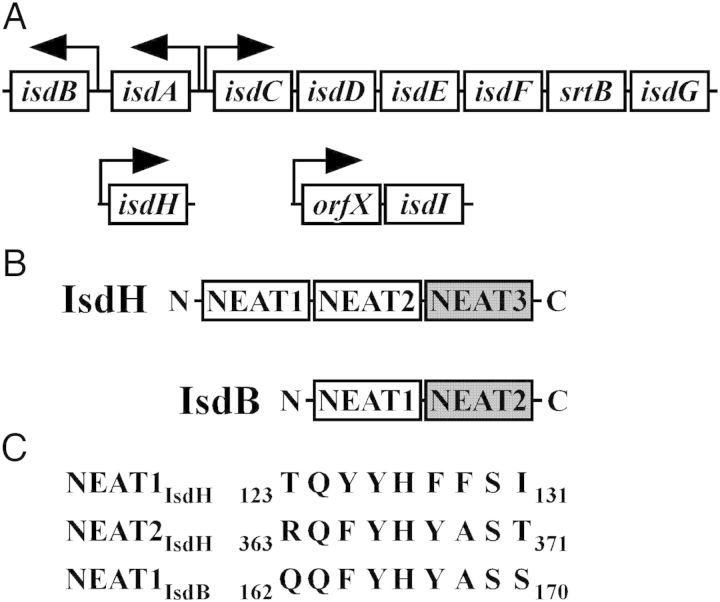
S. aureus iron regulated surface determinant system (Isd). (A) Genomic arrangement of the genes encoding the Isd system. Arrows denote promoter regions and the direction of transcription. (B) IsdB and IsdH hemoglobin-binding (white) and heme-binding (grey) NEAT domains. (C) Alignment of the hemoglobin binding motifs from IsdH and IsdB.
In contrast to the significant amount of information regarding heme relay between the Isd proteins, hemoglobin capture has not been studied in detail. Binding of hemoglobin and removal of heme are carried out in vitro by distinct domains within IsdB called NEAT1IsdB and NEAT2IsdB, respectively (Figure 1B) [5, 7, 18, 28, 30, 31]. NEAT1IsdB is required and sufficient to bind hemoglobin, whereas NEAT2IsdB is required and sufficient for heme binding [7, 28]. Biochemical and crystallographic analyses of NEAT1 from IsdH (NEAT1IsdH) have identified a sequence of consecutive aromatic residues within IsdH that is required for hemoglobin binding in vitro [18, 31]. However, differences exist between the residues that are predicted to bind hemoglobin within NEAT1IsdH and those found within NEAT1IsdB (Figure 1C).
We have previously found that IsdB binds human hemoglobin with a higher affinity than hemoglobin from other species and that mice expressing human hemoglobin (αHβA) are more susceptible to S. aureus infection [29, 32]. These results suggest that specific hemoglobin binding by IsdB is required for iron acquisition. To directly test this hypothesis, we first interrogated the role of NEAT1IsdB in hemoglobin binding to S. aureus in vivo and identified the residues that are required for hemoglobin capture. Using a strain expressing an IsdB point mutant that is deficient in hemoglobin capture, we demonstrate that hemoglobin binding activity of IsdB is required for heme extraction, iron acquisition, and virulence in αHβA mice. Together, these results establish the Isd system as a physiologically relevant heme-iron acquisition system within S. aureus and elucidate the role of hemoglobin binding by IsdB in the function of the Isd system.
MATERIALS AND METHODS
Bacterial Strains and Construction of isd Gene Knockouts
All experiments were performed using the S. aureus strain Newman or mutants generated in its background. Newman ΔisdA::tetM (H734) and Newman ΔisdB::ermC were constructed previously [1, 16], whereas Newman ΔisdC:km (H833) was constructed as described here. Briefly, isdC was polymerase chain reaction (PCR)-amplified and cloned into the plasmid pBC SK(+). A kanamycin resistance determinant was subsequently inserted into isdC, and the Pwo PCR-amplified fragment was inserted into pAUL-A. Recombination with the S. aureus chromosome was performed as described elsewhere [33]. Strains mutated for isdA, isdB, or isdC were complemented in trans using a plasmid expressing isdABC from their native promoters (Figure 1C). The resulting plasmid was named pJS019 and maintained through addition of 5 µg/mL chloramphenicol. Point mutations were generated by Pfu mutagenesis and confirmed by sequencing.
Hemoglobin-dependent Growth
The utilization of hemoglobin as an iron source by iron-restricted S. aureus Newman, and its isogenic isd mutants, described above, was assessed using freshly prepared human hemoglobin. In brief, erythrocytes were pelleted by centrifugation and washed 3 times in 3 cell pellet volumes of sterile saline. Erythrocytes were then lysed in 50 mM Tris pH 8.6, 2 mM EDTA. The supernatant from the lysate was collected following centrifugation. Stroma were precipitated through the addition of 50 mg/mL of NaCl and removed via centrifugation. The resulting supernatant was removed and dialyzed overnight against buffer A (50 mM Tris pH 8.6, 1 mM EDTA). Following dialysis, the hemoglobin solution was filtered and purified through anion exchange on a Mono Q HR 16/10 column (GE Healthcare) by running a gradient where buffer B consisted of 50 mM Tris pH 8.6, 1 mM EDTA, 0.5 M NaCl. The resulting fractions were dialyzed twice into 50 mM Tris pH 8.0 and filter-sterilized. Aliquots were stored at −80°C.
Single, isolated colonies of S. aureus were resuspended in 120 µL Roswell Park Memorial Institute (RPMI) medium supplemented with 1 µM of the iron chelator ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA, LGC Standards GmbH). One hundred microliters of the colony resuspension were used to inoculate 2 mL of the same media, and cultures were grown for 8–10 hours, until OD600 was approximately 1. The precultures were normalized to an OD600 of 1 and subcultured (1:400) into 2 mL of the same media with hemoglobin supplied as a sole iron source. Cultures were incubated at 37°C in 14-mL round-bottom polypropylene tubes, shaken at 200–220 rpm. A path length correction included in the Gen5 microtiter plate software (Bio-Tek) was applied such that the absorbance values are comparable to the 1-cm horizontal path length of standard spectrophotometers. See supplement for method B.
Mapping isdB Genetic Variation Across S. aureus Genome Projects
Three thousand two hundred and seventy-seven raw sequencing project submissions isolated primarily from clinical specimens were downloaded from the public National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) on 10 January 2013. These were mapped against the S. aureus strain N315 reference genome (GenBank ID NC_002745) using BWA short read aligner [34], and single-nucleotide polymorphisms (SNPs) were called using Genome Analysis Toolkit (GATK) [35]. The SNPs within the isdB genes of all these strains were extracted using a custom Perl script.
S. aureus Hemoglobin Binding Assay
Hemoglobin purification and coprecipitation was performed as described elsewhere [29]. ΔisdB-pOS1-isdB was grown in the presence of 10 µg/mL chloramphenicol to maintain the plasmid. Solubilized hemoglobin was subjected to 15% SDS-PAGE. Proteins were transferred from the gels onto nitrocellulose membranes, which were immunoblotted with a rabbit α-hemoglobin antibody at 1:1000 dilution (Santa Cruz, SC-21005) followed by an α-rabbit secondary antibody conjugated to a fluorophore at 1:25 000 dilution (Alexa Fluor 680 goat anti-rabbit immunoglobulin G [IgG](H + L)). Fluorescent signal was detected using an Odyssey infrared imaging system (LI-COR). IsdB was detected as described elsewhere [1].
Heme-transfer Experiments
Recombinant IsdB (rIsdB) was expressed and purified as described elsewhere [1]. Prior to measurements, rIsdB was subject to a final purification step by gel filtration (Superose 12, GE Healthcare). Human hemoglobin was purified from blood as described elsewhere [31]. Conversion to met-hemoglobin for heme-transfer assays was achieved by addition of 5-fold molar excess of potassium ferricyanide followed by buffer exchange into 20 mM sodium phosphate pH 7.0 over Sephadex G25 (GE Healthcare). To measure heme-transfer, 1.5 µM met-hemoglobin was mixed with 6 µM IsdB in 150 mM sodium phosphate pH 7.0, and spectra were obtained in the range of 700–350 nm every 40 seconds at 4°C (Jasco V630 spectrophotometer). Heme transfer rates were calculated from spectral change at 406 nm fit to single or double-exponential expressions. Absorbance was normalized to the starting condition.
Systemic Mouse Infections
Infections were carried out as described elsewhere [29]. Animal experiments were approved by the Institutional Animal Care and Use Committee of Vanderbilt University.
RESULTS
Components of the Isd System Are Required for Growth on Nanomolar Concentrations of Human Hemoglobin
To determine whether individual components of the Isd system are required for growth in the presence of hemoglobin, growth assays were performed in media that were supplemented with human hemoglobin as the sole iron source. Wild-type S. aureus and its isogenic isd knockout mutants were grown under iron-restriction and subcultured into media containing human hemoglobin at concentrations ranging from 5 to 100 nM. Five nM hemoglobin was found to be insufficient to support growth of any strain, with the exception of ΔisdA (H734) complemented with pJS019, which exhibited minimal growth at 36 hours (Figure 2). The addition of 10 nM hemoglobin promoted growth of the wild-type and complemented mutant strains but did not promote growth of ΔisdA (H734), ΔisdB, or ΔisdC (H833). Increasing the hemoglobin concentration to 20 nM facilitated more rapid and robust growth, where ΔisdA (H734), ΔisdB, and ΔisdC (H833) were still strongly deficient in hemoglobin-dependent growth relative to the wild-type. The addition of hemoglobin at concentrations at or above 100 nM promoted the growth of all strains and reduced discernible differences between the wild-type and isd knockout strains (Figure 2). Using an alternative method, it was shown that in contrast to wild-type S. aureus, ΔisdB, ΔisdC, ΔisdG, and ΔisdGΔisdI mutants are unable to proliferate in hemoglobin as an iron source (Supplementary Figure 1A). Similar results were obtained when free heme was added as a sole iron source (Supplementary Figure 1B).
Figure 2.
Hemoglobin-dependent growth of Staphylococcus aureus Newman WT, its isogenic isd mutants, and corresponding complemented strains. Strains were cultured in RPMI chelated with 1 μM EDDHA and 5, 10, 20, or 100 nM of human hemoglobin was supplied as the sole source of iron. Culture densities were assessed at 12, 24, and 36 h by determining OD600. Data shown are representative of at least 3 experiments, where samples were performed in quintuplicate and error bars represent standard error of the mean. Statistical analysis was performed using a 2-way analysis of variance coupled with Bonferroni post hoc tests where the wild type was set as the comparator; a, P < .001; b, P < .01, and c, P < .05.
In summary, we have demonstrated that isd mutants are defective in iron uptake using heme or freshly prepared human hemoglobin as the iron source, and that this requirement for Isd components is only markedly demonstrable at nanomolar concentrations of heme/hemoglobin.
Conserved Residues Are Required for Hemoglobin Binding to S. aureus
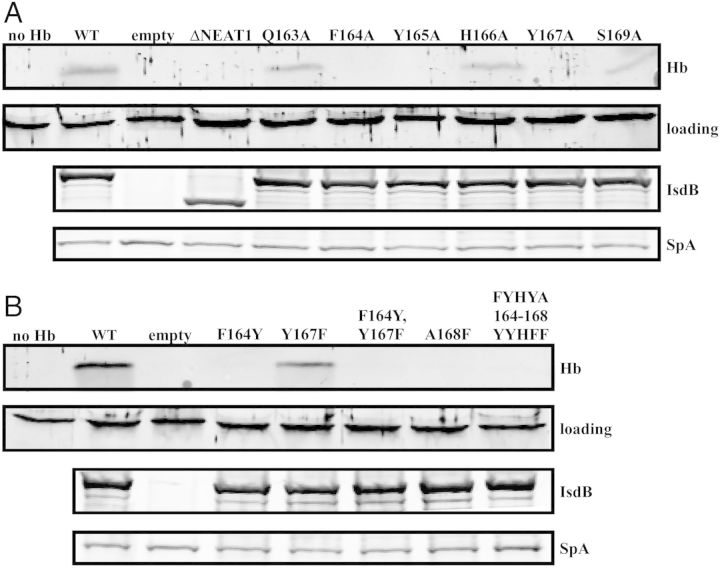
IsdB is the primary hemoglobin receptor of S. aureus; however, the residues required for IsdB-mediated hemoglobin capture have not been defined. To test the role of NEAT1IsdB in hemoglobin binding, we transformed a mutated version of isdB, which lacks the NEAT1 domain (isdB ΔNEAT1), in trans into S. aureus ΔisdB. This resulted in a strain that exclusively expresses IsdB ΔNEAT1 (Figure 3A, third panel). Using a similar approach, we created strains of S. aureus that express a series of alanine substitution mutants within the predicted hemoglobin-binding motif of NEAT1 (Figure 3A). Hemoglobin binding assays revealed that IsdB ΔNEAT1 is unable to capture hemoglobin at the surface of S. aureus (Figure 3A). This is consistent with a previous report demonstrating the requirement for recombinant NEAT1IsdB for hemoglobin binding in vitro [28]. In addition, alanine substitutions of phenylalanine 164 (F164A) and tyrosine 167 (Y167A) result in complete abrogation of hemoglobin binding (Figure 3A). In contrast, substitutions of glutamine 163 (Q163A), histidine 166 (H166A), and serine 169 (S169A) result in milder defects in hemoglobin binding. The observed reductions in hemoglobin binding were not due to uneven gel loading, differences in the expression level of IsdB, or reduced surface expression of IsdB (Figure 3A, lower panels and Supplementary Figure 2). These results demonstrate the requirement for the NEAT1IsdB domain in hemoglobin capture by S. aureus and identify specific residues that are critical to this process.
Figure 3.
Identification of residues within NEAT1IsdB required for hemoglobin binding to Staphylococcus aureus. A, Hemoglobin bound by S. aureus IsdB mutants containing alanine substitutions of individual residues of the hemoglobin-binding motif. B, Hemoglobin bound by S. aureus IsdB mutants containing a chimeric IsdB/IsdH hemoglobin-binding motif. For both (A) and (B), IsdB was expressed in S. aureus ΔisdB from pOS1 under iron-limiting conditions. S. aureus was incubated with 15 nM hemoglobin and washed to remove unbound hemoglobin. Cell wall-associated proteins were eluted and analyzed by SDS-PAGE. Bound hemoglobin was detected by immunoblotting (top panel). Equal loading of the hemoglobin-containing samples was controlled with a cross-reactive protein (second panel). To detect IsdB the cell wall proteins were liberated through cell wall digestion, resolved with SDS-PAGE, and immunoblotted (third panel). The bottom panel depicts protein A (Spa), which nonspecifically binds antibodies and is a loading control for cell wall proteins.
The specific residues within NEAT1IsdB and NEAT1IsdH that bind hemoglobin are similar but not identical (Figure 1C). To test if the residues involved in hemoglobin recognition by IsdB and IsdH are interchangeable, we substituted the individual residues within IsdB for corresponding amino acids of IsdH (F164Y; Y167F; A168F). Residues 164 and 167 were also swapped (F164Y, Y167F), and the hemoglobin-binding motif of IsdB was substituted for the corresponding motif of IsdH (FYHYA164–168YYHFF). Binding experiments demonstrated that all of the above substitutions, with the exception of Y167F mutation, resulted in abrogation of hemoglobin binding by S. aureus (Figure 3B). These data indicate that the unique sequence within NEAT1IsdB is required for hemoglobin binding by S. aureus.
To investigate the pattern of natural genetic variation in the isdB gene we mapped data from 3277 public S. aureus whole genome shotgun projects downloaded from the NCBI SRA database (Figure 4). Nonsynonymous mutations were highly nonrandom in their distribution, with clustering in 2 regions (approximately between residues 36–127 and 459–564). Most importantly, the critical residues in the NEAT1IsdB region between 164 and 168 did not contain mutations in any of the 3277 strains. This suggests that there is selection across S. aureus to maintain hemoglobin binding proficiency.
Figure 4.

IsdB polymorphisms across Staphylococcus aureus strains. A, Hotspots of polymorphisms in the isdB gene as revealed by the analysis of 3,277 S. aureus strains from the NCBI SRA database compared to N315 reference genome. B, Frequency distribution of all the SNPs across 3,277 strains. The horizontal blue line above the x-axis indicates 645-amino acids long IsdB protein, vertical lines being the polymorphic positions shown in Figure 4A. The NEATIsdB region (aa 162-170) is indicated. Abbreviation: SNP, single-nucleotide polymorphism.
Hemoglobin Binding Is Required for Heme Transfer to IsdB
To investigate the effect of mutations in the hemoglobin-binding NEAT1IsdB domain on heme transfer, UV-visible absorbance spectroscopy was employed (Figure 5A). Mixing wild-type IsdB with met-Hb leads to rapid changes in absorbance (Figure 5), characteristic of heme transfer from met-hemoglobin to IsdB. All tested IsdB mutations cause a reduction in the rate of spectral change, indicating reduced heme transfer (Figure 5B and Table 1). The Q163A, H166A, and FYHYA164–168YYHFF mutations result in 15-20-fold reductions in heme transfer rate compared to wild-type rIsdB. Transfer rates to rIsdBY165A are approximately 50-fold slower than for wild-type rIsdB. As this mutant shows no detectable interaction with hemoglobin, the slow heme transfer is likely to be the result of simple heme dissociation from Hb, followed by uptake of heme from solution by rIsdBY165A [6]. Because NEAT1 does not contribute to heme binding to IsdB, this effect is not due to a reduction in heme binding [28]. Overall, these results indicate that mutations in NEAT1IsdB that reduce hemoglobin binding also slow down heme transfer from hemoglobin to NEAT2IsdB, suggesting that direct interaction between hemoglobin and the hemoglobin-binding domain of IsdB is essential for efficient heme extraction.
Figure 5.
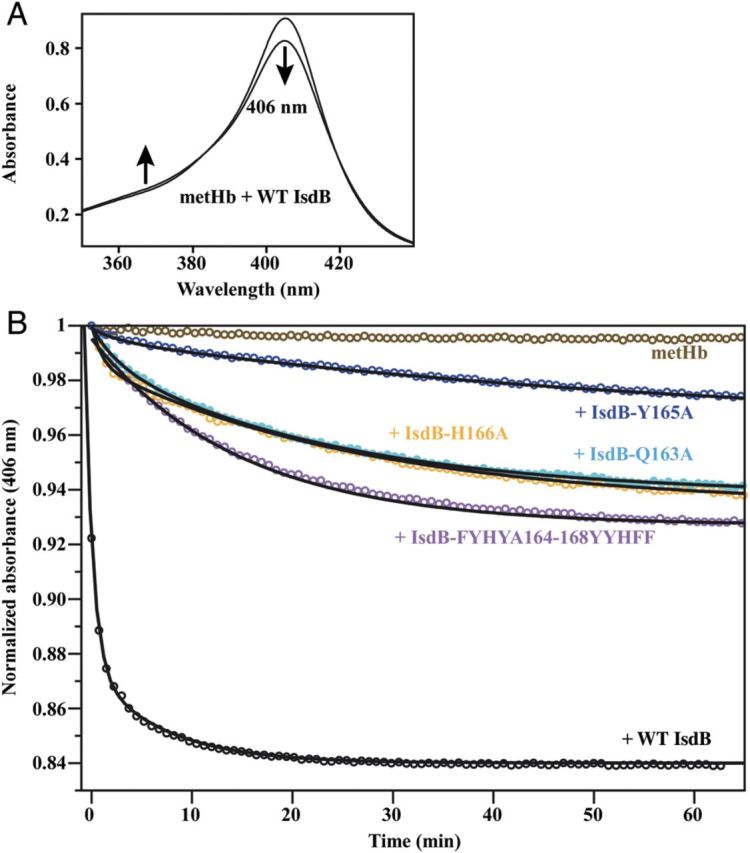
Effect of mutations in the hemoglobin-binding motif of NEAT1IsdB on heme extraction from hemoglobin. A, A change in the UV-visible absorption spectrum of rIsdB-hemoglobin mixtures indicates heme transfer to rIsdB. B, Spectral change at 406 nm was monitored for mixtures of hemoglobin with rIsdB.
Table 1.
Rates of Heme Transfer From Hemoglobin to rIsdB at 4°C
| IsdB Construct | k1 min−1 | k2 min−1 |
|---|---|---|
| WT | 1.08 ± 0.24 | 0.114 ± 0.024 |
| FYHYA164–168YYHFF | 0.066 ± 0.001 | |
| H166A | 0.042 ± 0.001 | |
| Q163A | 0.051 ± 0.002 | |
| Y165A | 0.020 ± 0.004 |
UV-vis data were fit to single or double-exponential expressions. Rate measurements are average ± 1 SD from 2 to 3 experiments. Abbreviations: IsdB, iron-regulated surface determinant B; rIsdB, recombinant IsdB; WT, wild type.
To assess the contribution of hemoglobin binding by IsdB to iron acquisition in vivo, we generated a chromosomal mutation of isdB, resulting in a tyrosine 165 substitution for alanine (isdB Y165A). IsdBY165A is expressed from the chromosome at the same level as wild-type IsdB; however, it does not bind hemoglobin to the surface of S. aureus (Supplementary Figure 1C). To assess the impact of this mutation on iron acquisition, the growth of wild-type, isdBY165A, and ΔisdB was compared in the presence of hemoglobin (Supplementary Figure 1D). Although the wild-type S. aureus proliferated in the presence of hemoglobin, both ΔisdB and isdBY165A were deficient in growth. These data indicate that hemoglobin binding by IsdB is required for heme extraction from hemoglobin and growth on hemoglobin as a sole iron source.
Hemoglobin Binding Is Required for Virulence
S. aureus strains lacking isdB are reduced for colonization in murine models of infection [3, 4, 27–29]. Due to the increased specificity of S. aureus for human hemoglobin compared to mouse hemoglobin, we have established transgenic mice expressing human hemoglobin (αHβA) as an enhanced model of S. aureus infection [29, 32]. To test whether IsdB-mediated hemoglobin capture is required for proliferation within the host, we systemically infected αHβA mice with wild-type, ΔisdB, or isdBY165A. Infection with ΔisdB and isdBY165A resulted in a nearly 100-fold decrease in the number of colony-forming units (CFUs) isolated from the kidneys and hearts of infected mice (Figure 6A and 6B). Consistent with a previous report, we did not observe a difference in the number of CFUs isolated from the livers of mice infected with either of the strains (Figure 6C) [3]. Further, mice infected with isdBY165A or ΔisdB lose significantly less weight than mice infected with wild-type bacteria (Figure 6D). Taken together, these data indicate that hemoglobin binding by IsdB is required for hemoglobin-derived iron acquisition and virulence.
Figure 6.
isdBY165A pathogenicity in a systemic murine model of infection. Seven week-old female αHβA mice were retro-orbitally infected with approximately 1 × 107 CFUs of Staphylococcus aureus. Ninety-six hours postinoculation the mice were killed. The number of CFUs in (A) kidneys, (B) heart, and (C) liver was quantified by serial dilutions followed by plating on growth media. D, Weight loss was quantified by weighing the mice before inoculation and after death. Error bars denote standard deviation. Asterisks indicate statistically significant differences as determined by Student t test (P ≤ .05). Abbreviation: CFU, colony-forming unit.
DISCUSSION
Due to the requirement for iron by virtually all forms of life, this element often plays a decisive role during host-pathogen interactions. Within vertebrates, most iron is intracellular, bound to host proteins, and enclosed within the protoporphyrin ring of heme. S. aureus acquires heme-iron from hemoglobin in a series of steps that involve binding of hemoglobin to the bacterial surface, followed by removal and import of heme into the bacterial cytoplasm whereupon heme is either degraded to release free iron or incorporated intact into bacterial hemoproteins [1]. The process of iron acquisition from hemoglobin in S. aureus is carried out by the iron-regulated surface determinant system [1].
Numerous studies have assigned heme import and degradation functions to the individual Isd components [1–14, 16–26, 28–31]. However, the wealth of literature on the contribution of the Isd system to hemoglobin/heme-derived iron acquisition was challenged by 2 recent reports [36, 37]. Hurd et al reported that the IsdA, IsdB, and IsdH are not required for hemoglobin and heme-derived iron. Wright and Nair argued that IsdE is dispensable for heme acquisition. We suggest that the discrepancies between these 2 reports and the abundant data implicating the Isd system in heme-iron acquisition stem from the utility of the Isd system at low extracellular heme/hemoglobin concentrations not used in these 2 studies. Also, both of the aforementioned studies used hemoglobin stored in the lyophilized form, which is structurally compromised and contains significant free heme [36–38]. In contrast to hemoglobin purified from fresh blood, lyophilized hemoglobin appears as a diffuse smear when resolved on a native gel (Supplementary Figure 3). Our data suggest that Isd is a high-affinity heme-iron acquisition system that has maximal utility at the low nanomolar concentrations of heme/hemoglobin, consistent with the predicted physiological concentrations of free heme and cell-free plasma hemoglobin of 30 and 150 nM, respectively [19, 39]. The role of isd in heme-iron acquisition may therefore be masked when heme-iron sources are applied in excess. The assays reported here were repeated in separate laboratories using independently generated mutants and reagents, each yielding similar results (Figure 2 and Supplementary Figure 1). Therefore, these experiments have demonstrated the requirement for the S. aureus Isd components during hemoglobin-derived iron acquisition.
Previous studies have identified the motif within IsdH that is required for hemoglobin binding by this protein [18, 31]. Here we define a similar but not identical motif within IsdB that is required for hemoglobin binding to S. aureus in vivo (Figure 3). Notably, this motif is absolutely conserved among 3277 S. aureus strains (Figure 4), establishing it as a potential target for therapeutic inhibitors of hemoglobin binding. Further, we demonstrate that heme extraction and iron import are dependent on hemoglobin binding because IsdBY165A, which is deficient in hemoglobin binding, fails to efficiently extract heme from hemoglobin in vitro and fails to provide S. aureus with iron required for growth (Figure 5 and Supplementary Figure 1C and 1D). This is consistent with a recent report that has demonstrated the requirement for a hemoglobin-binding domain of IsdH for heme extraction by IsdH [40]. Finally, it is demonstrated that hemoglobin-binding activity of IsdB is required for S. aureus pathogenesis in a murine model of systemic infection (Figure 6).
Another staphylococcal pathogen, Staphylococcus lugdunensis, encodes an Isd system that is highly similar to that of S. aureus [41, 42]. In particular, S. lugdunensis IsdB contains an aromatic motif that is identical to the one described here. IsdB from both species displays a preference for human hemoglobin over mouse hemoglobin, which has important implications for the evolution of these organisms to infect their primary host [29, 42]. Hemoglobin binding activities of IsdX1 and IsdX2 of Bacillus anthracis have also been investigated [43, 44]. Interestingly, the residues that are responsible for the interaction of IsdX1 and IsdX2 with hemoglobin are not present in IsdB [43, 44]. Heme-iron acquisition systems have been identified in a number of evolutionarily distant Gram-positive pathogens including Bacillus cereus, Listeria monocytogenes, Corynebacterium diphtheria, and Streptococcus pyogenes [45–48]. Therefore, the data presented may be broadly applicable across a variety of infectious diseases.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the members of the Skaar laboratory for critical reading of the manuscript. Some of the data in this article were generated by J. R. S. and D. E. H. during a sabbatical in the laboratory of Dr Cécile Wandersman (Institut Pasteur). They are indebted to her for her generosity and many stimulating scientific conversations.
Financial support. This work was supported by US Public Health Service from the National Institute of Allergy and Infection Diseases (AI69233 and AI073843 to E. P. S.); and American Heart Association Greater Southeast Affiliate Predoctoral Fellowship (09PRE2140221 to G. P.). E. P. S. holds an Investigator in Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Mazmanian SK, Skaar EP, Gaspar AH, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–9. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 2.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A. 2002;99:2293–8. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect Immun. 2009;77:2624–34. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421–9. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muryoi N, Tiedemann MT, Pluym M, Cheung J, Heinrichs DE, Stillman MJ. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J Biol Chem. 2008;283:28125–36. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu H, Xie G, Liu M, et al. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J Biol Chem. 2008;283:18450–60. doi: 10.1074/jbc.M801466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudin CF, Grigg JC, Arrieta AL, Murphy ME. Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry (Mosc) 2011;50:5443–52. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M, Tanaka WN, Zhu H, Xie G, Dooley DM, Lei B. Direct hemin transfer from IsdA to IsdC in the Isd heme acquisition system of Staphylococcus aureus. J Biol Chem. 2008;283:6668–76. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villareal VA, Spirig T, Robson SA, Liu M, Lei B, Clubb RT. Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus. J Am Chem Soc. 2011;133:14176–9. doi: 10.1021/ja203805b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigg JC, Mao CX, Murphy ME. Iron-coordinating tyrosine is a key determinant of NEAT domain heme transfer. J Mol Biol. 2011;413:684–98. doi: 10.1016/j.jmb.2011.08.047. [DOI] [PubMed] [Google Scholar]
- 11.Abe R, Caaveiro JM, Kozuka-Hata H, Oyama M, Tsumoto K. Mapping ultra-weak protein-protein interactions between heme transporters of Staphylococcus aureus. J Biol Chem. 2012;287:16477–87. doi: 10.1074/jbc.M112.346700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiedemann MT, Heinrichs DE, Stillman MJ. Multiprotein heme shuttle pathway in Staphylococcus aureus: Iron-regulated surface determinant cog-wheel kinetics. J Am Chem Soc. 2012;134:16578–85. doi: 10.1021/ja305115y. [DOI] [PubMed] [Google Scholar]
- 13.Mack J, Vermeiren C, Heinrichs DE, Stillman MJ. In vivo heme scavenging by Staphylococcus aureus IsdC and IsdE proteins. Biochem Biophys Res Commun. 2004;320:781–8. doi: 10.1016/j.bbrc.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 14.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J Biol Chem. 2007;282:10625–31. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 15.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Heme coordination by Staphylococcus aureus IsdE. J Biol Chem. 2007;282:28815–22. doi: 10.1074/jbc.M704602200. [DOI] [PubMed] [Google Scholar]
- 16.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol Microbiol. 2007;63:139–49. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 17.Villareal VA, Pilpa RM, Robson SA, Fadeev EA, Clubb RT. The IsdC protein from Staphylococcus aureus uses a flexible binding pocket to capture heme. J Biol Chem. 2008;283:31591–600. doi: 10.1074/jbc.M801126200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilpa RM, Robson SA, Villareal VA, Wong ML, Phillips M, Clubb RT. Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J Biol Chem. 2009;284:1166–76. doi: 10.1074/jbc.M806007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dryla A, Gelbmann D, Von Gabain A, Nagy E. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol. 2003;49:37–53. doi: 10.1046/j.1365-2958.2003.03542.x. [DOI] [PubMed] [Google Scholar]
- 20.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–43. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 21.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–15. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee WC, Reniere ML, Skaar EP, Murphy MEP. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–63. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu R, Skaar EP, Zhang R, et al. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–6. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reniere ML, Ukpabi GN, Harry SR, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol. 2010;75:1529–38. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reniere ML, Haley KP, Skaar EP. The flexible loop of Staphylococcus aureus IsdG is required for its degradation in the absence of heme. Biochemistry (Mosc) 2011;50:6730–7. doi: 10.1021/bi200999q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ukpabi G, Takayama SI, Mauk AG, Murphy ME. Inactivation of IsdI heme oxidation by an active site substitution that diminishes heme ruffling. J Biol Chem. 2012;287:34179–88. doi: 10.1074/jbc.M112.393249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HK, DeDent A, Cheng AG, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28:6382–92. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pishchany G, McCoy AL, Torres VJ, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–50. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilpa RM, Fadeev EA, Villareal VA, Wong ML, Phillips M, Clubb RT. Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J Mol Biol. 2006;360:435–47. doi: 10.1016/j.jmb.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Krishna Kumar K, Jacques DA, Pishchany G, et al. Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH. J Biol Chem. 2011;286:38439–47. doi: 10.1074/jbc.M111.287300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero JR, Suzuka SM, Nagel RL, Fabry ME. Expression of HbC and HbS, but not HbA, results in activation of K-Cl cotransport activity in transgenic mouse red cells. Blood. 2004;103:2384–90. doi: 10.1182/blood-2003-01-0237. [DOI] [PubMed] [Google Scholar]
- 33.Beasley FC, Vines ED, Grigg JC, et al. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol Microbiol. 2009;72:947–63. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurd AF, Garcia-Lara J, Rauter Y, Cartron M, Mohamed R, Foster SJ. The iron-regulated surface proteins IsdA, IsdB, and IsdH are not required for heme iron utilization in Staphylococcus aureus. FEMS Microbiol Lett. 2012;329:93–100. doi: 10.1111/j.1574-6968.2012.02502.x. [DOI] [PubMed] [Google Scholar]
- 37.Wright JA, Nair SP. The lipoprotein components of the Isd and Hts transport systems are dispensable for acquisition of heme by Staphylococcus aureus. FEMS Microbiol Lett. 2012;329:177–85. doi: 10.1111/j.1574-6968.2012.02519.x. [DOI] [PubMed] [Google Scholar]
- 38.Boys BL, Kuprowski MC, Konermann L. Symmetric behavior of hemoglobin alpha- and beta- subunits during acid-induced denaturation observed by electrospray mass spectrometry. Biochemistry (Mosc) 2007;46:10675–84. doi: 10.1021/bi701076q. [DOI] [PubMed] [Google Scholar]
- 39.Sassa S. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide? Antioxid Redox Signal. 2004;6:819–24. doi: 10.1089/ars.2004.6.819. [DOI] [PubMed] [Google Scholar]
- 40.Spirig T, Malmirchegini GR, Zhang J, et al. Staphylococcus aureus uses a novel multidomain receptor to break apart human hemoglobin and steal its heme. J Biol Chem. 2013;288:1065–78. doi: 10.1074/jbc.M112.419119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haley KP, Janson EM, Heilbronner S, Foster TJ, Skaar EP. Staphylococcus lugdunensis IsdG liberates iron from host heme. J Bacteriol. 2011;193:4749–57. doi: 10.1128/JB.00436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapotoczna M, Heilbronner S, Speziale P, Foster TJ. Iron regulated surface determinant (Isd) proteins of Staphylococcus lugdunensis. J Bacteriol. 2012;194:6453–67. doi: 10.1128/JB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekworomadu MT, Poor CB, Owens CP, et al. Differential function of lip residues in the mechanism and biology of an anthrax hemophore. PLoS Pathogens. 2012;8:e1002559. doi: 10.1371/journal.ppat.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honsa ES, Owens CP, Goulding CW, Maresso AW. The near-iron transporter (NEAT) domains of the anthrax hemophore IsdX2 require a critical glutamine to extract heme from methemoglobin. J Biol Chem. 2013;288:8479–90. doi: 10.1074/jbc.M112.430009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei B, Smoot LM, Menning HM, et al. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect Immun. 2002;70:4494–500. doi: 10.1128/IAI.70.8.4494-4500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daou N, Buisson C, Gohar M, et al. IlsA, a unique surface protein of Bacillus cereus required for iron acquisition from heme, hemoglobin and ferritin. PLoS Pathogens. 2009;5:e1000675. doi: 10.1371/journal.ppat.1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Q, Jiang X, Moore KJ, et al. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol. 2011;80:1581–97. doi: 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen CE, Schmitt MP. HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae. J Bacteriol. 2009;191:2638–48. doi: 10.1128/JB.01784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.