Abstract
Significance: Functional states of organisms vary rhythmically with a period of about a day (i.e., circadian). This endogenous dynamic is shaped by day–night alternations in light and energy. Mammalian circadian rhythms are orchestrated by the hypothalamic suprachiasmatic nucleus (SCN), a brain region specialized for timekeeping. These autonomous ∼24-h oscillations are cell-based, requiring transcription–translation-based regulation. SCN circadian oscillations include the maintenance of intrinsic rhythms, sensitivities to input signals, and generation of output signals. These change predictably as time proceeds from dawn to day, dusk, and through the night. SCN neuronal excitability, a highly energy-demanding process, also oscillates over ∼24 h. The nature of the relationship of cellular metabolism and excitability had been unknown. Recent Advances: Global SCN redox state was found to undergo an autonomous circadian rhythm. Redox state is relatively reduced in daytime, when neuronal activity is high, and oxidized during nighttime, when neurons are relatively inactive. Redox modulates neuronal excitability via tight coupling: imposed reducing or oxidizing shifts immediately alter membrane excitability. Whereas an intact transcription–translation oscillator is necessary for the redox oscillation, metabolic modulation of excitability is too rapid to be under clockwork control. Critical Issues: Our observations lead to the hypothesis that redox state and neuronal activity are coupled nontranscriptional circadian oscillators in SCN neurons. Critical issues include discovering molecular and cellular substrates and functional consequences of this redox oscillator. Future Directions: Understanding interdependencies between cellular energy metabolism, neuronal activity, and circadian rhythms is critical to developing therapeutic strategies for treating neurodegenerative diseases and brain metabolic syndromes. Antioxid. Redox Signal. 20, 2955–2965.
Introduction
Daily and seasonal cycles of the Earth's relationship to the Sun are critical variables that profoundly shape the conditions for life. These environmental fluctuations determine the availability of light, nutrients, and warmth. Organisms have adapted to these variables by patterning their behaviors, physiology, and internal metabolism to fluctuate with respect to energy availability and need. These rhythms are not driven by the varying environment. Rather, they are generated by an internal timing system that adapts the organism to the changing external world by orchestrating metabolism, physiology, and behavior to anticipate environmental conditions. As a result, organismic functions are coordinated with environmental conditions optimal for their occurrence.
Mammalian circadian and seasonal rhythms are orchestrated by a brain region specialized for ∼24-h timekeeping, the hypothalamic suprachiasmatic nucleus (SCN, Fig. 1). The SCN is named for its position directly above the optic chiasm and lies at the base of the hypothalamus near brain nuclei that control sleep-wake, feeding, drinking, and sexual/reproductive/affiliative behaviors, body temperature, and autonomic functions. These functions oscillate in circadian rhythms coordinated by the SCN. Timekeeping is cell-based, but the ability to pattern behavioral rhythms and appropriately orchestrate oscillations in tissue and organ systems resides in integrated properties of this specialized brain structure.
FIG. 1.
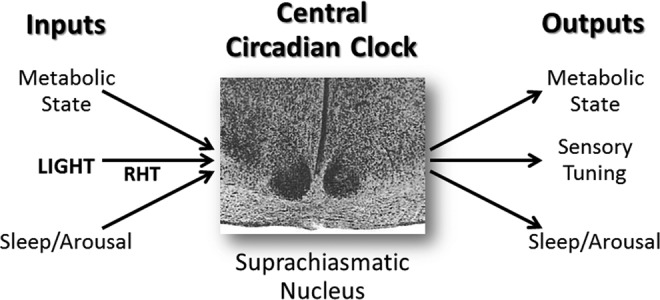
Organization of the circadian timing system of mammals. Circadian and seasonal rhythms are orchestrated by a central circadian clock in the brain, the SCN. The SCN is an endogenous oscillator, generating a near-24-h timebase when isolated in a hypothalamic brain slice (5, 16, 23, 59, 67, 73, 83). The rat SCN is seen here in transverse section as two densely staining clusters of Nissl-positive cells at the base of the third ventricle (the dark vertical line). This image shows each SCN nestled in the optic chiasm (ventral lighter structure) in the ventromedial hypothalamus. Each SCN is ∼500 μm across and the somata of individual cells ranges from 8 to 12 μm in diameter. The SCN produces output signals that coordinate circadian rhythms of physiology and behavior, including metabolic state, sensory tuning, and sleep and arousal. Phasing of the SCN clock can be adjusted by a range of inputs, including those that communicate metabolic state, environmental light (via the RHT), and sleep/arousal. Windows of sensitivity to phase-resetting signals are gated by the SCN clock so that signals communicating loss of desynchronization with day–night or output targets adaptively reset SCN clock phasing (22, 24, 45, 51). RHT, retinohypothalamic tract; SCN, suprachiasmatic nucleus.
SCN circadian dynamics extend from transcription, translation, and post-translational modification of clock genes and proteins to levels of small molecular regulators, temporal gating of signal transduction pathways that respond to incoming signals, neuropeptide release within the SCN, and neuronal activity that transmits output signals. SCN neuronal electrical activity peaks at mid-day and is low at night, as are energy availability and use. Cellular metabolism has been evaluated only recently. Reduction–oxidation and (redox) homeostasis in this tissue are not static, as has been predicted for basal cellular metabolism, but rather they are intrinsically dynamic. Redox state oscillates within a narrowly buffered range with a predictable phase relation to day and night. What is the consequence of this regular rhythm of redox changes and energetics for the physiological state of this brain region?
As in other organs, energy metabolic dysfunction in the brain is often pathogenic (4). Knowledge of the interdependency between cellular metabolic disturbance and deficit in neuronal activity is essential for the development of therapeutic strategies for the treatment of neurodegenerative diseases and metabolic syndromes in the brain. While activity-dependent metabolic state changes have been under intensive study, the inverse pathway, the influence of metabolic state on neuronal activity, is relatively unexplored. Emerging evidence suggests a reciprocal interaction between metabolic state and neuronal excitability in the normal dynamics of the brain's circadian clock (79). Thus, metabolic state could be an important modulator of information processing in the brain. These new findings in the context of a dynamic, near-24-h biological oscillator are the subject of this review.
The Circadian Clock in the SCN
Circadian rhythms are properties of all mammalian cells, but these myriad clocks are controlled by the SCN (42, 61, 88). The SCN comprises bilateral clusters of 10,000 neurons and glia each. The paired brain nuclei are located in the anterior ventral hypothalamus, directly above the optic chiasm, and separated by the third ventricle (Fig. 1) (50). SCN lesion abolishes circadian rhythms of behavior and physiology, indicating that the SCN is necessary for organismic circadian rhythmicity. Transplantation of fetal SCN into a host rendered arrhythmic by an SCN lesion restores that organism's rhythms, except for hormone release from the pituitary. The period of the restored behavioral rhythms is that of the transplanted SCN, rather than the host when genotypes differ. These findings position the SCN as the master clock coordinating a hierarchical clock system (42, 61).
SCN is a coherent circadian oscillator
The SCN is distinguished from other tissue-base clocks by its intrinsic coherence (88). When tissues from various organs or brain regions are cultured, their rhythms are synchronized immediately after removal from the body. As the days ex vivo pass, coordinated tissue-level rhythms gradually diminish and disappear. However, the SCN continues to oscillate coherently for the duration that it survives in vitro, which may be months (83). Thus, the SCN possesses unusual clock properties and serves in a privileged position as the conductor of the peripheral clock orchestra.
The endogenous clock can maintain circadian rhythms without external environmental cues. Mice housed in constant conditions in a dark room continue to exhibit strong circadian rhythms similar to under day/night conditions. However, the intrinsic period, measured by the pattern of wheel-running activity, is not equivalent to the precise 24-h solar cycle. The endogenous free-running circadian period (τ) of these mice is <24 h, while in rats τ is >24 h. In humans, τ also is a little longer than 24 h for most individuals. Thus, these rhythms are not driven by the solar cycle.
Phase-resetting the SCN clock
To align with the changing day and night length of the seasons, animals must readjust their intrinsic clocks to the day–night cycle in nature, a process called entrainment (24). Light is the most important environmental time cue for entraining the circadian clock (22). Photic information is conveyed directly from intrinsically photosensitive, melanopsin-bearing retinal ganglion cells to the SCN via the retinohypothalamatic tract (Fig. 1) (9, 30). The effect of light on SCN phasing depends on the time of day, such that sensitivity and response will report an error with respect to anticipated environmental light (15, 22). In early night, a light pulse signals an extended day; the phase of the SCN clock delays to adapt to this change. On the other hand, in late night, premature light signals an early day; the clock responds by advancing its phase. Clock phase is not altered by light experienced in constant dark conditions during the subjective daytime, when light is normally present; this is compatible with synchronization.
Nonphotic stimuli also entrain the circadian clock (72). Among the most salient of these cues is food, when availability is restricted to only a few hours/day (31, 80). After entrainment to restricted food availability, motor activity increases significantly in anticipation of feeding. This anticipatory behavior is evidence that a time-of-day signature has been encoded in the entrainment process. Notably, sensitivity to both photic and nonphotic entrainment cues is circadian time (CT)-dependent (24).
Transcription–Translation Oscillator: The Molecular Clockwork Core
At the most fundamental level, circadian rhythms emerge from interacting feedback loops of core clock genes and proteins (44). Clock genes, which are necessary to circadian timekeeping, were discovered first in Drosophila and then in mouse, man, and virtually every species, including cyanobacteria (64, 65, 78). These genes are highly conserved across species, so much so that human clock genes can substitute for fly homologs to generate behavioral rhythms (70). The transcriptional, translational, and post-translational regulation of clock genes and their products forms the core clockwork mechanism, a transcription–translation oscillator (TTO, Fig. 2).
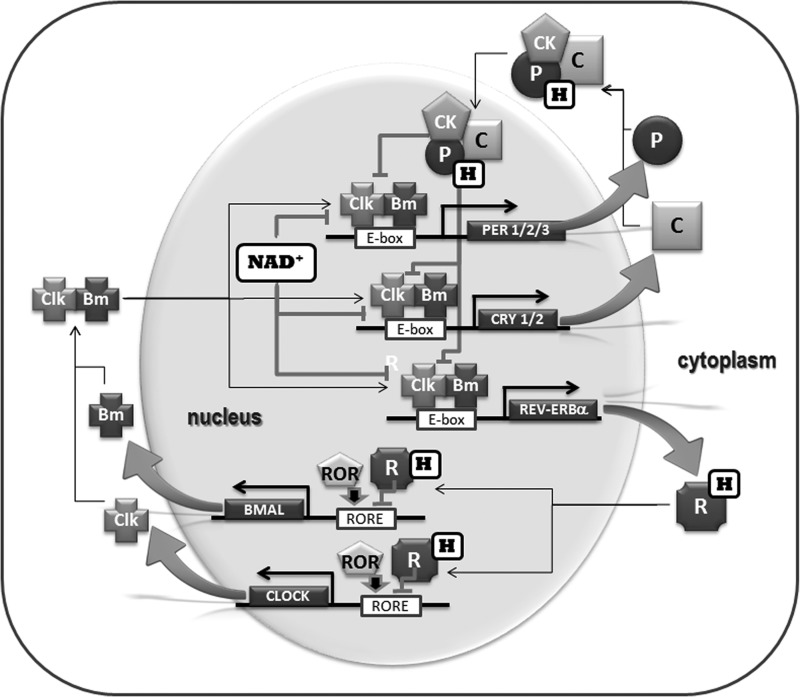
FIG. 2.
Model of the circadian TTO with nodes of redox regulation. The critical components sustaining cell-based circadian rhythms have been thought to be clock genes and their proteins. These essential elements interact as a ∼24-h regenerative oscillator, the TTO. Heterodimers of positive transcription factors CLOCK/BMAL (or CLOCK/NPAS2 in some brain regions) bind to E-box motifs in the promoters of Per 1/2/3, Cry 1/2 and Rev-Erbα, activating transcription. The protein products undergo complex post-translational modifications in the cytoplasm, including phosphorylation (by casein kinase 1δ or ɛ [CK], glycogen synthase kinase 3β, protein kinase C, and other kinases not included in this model), sumoylation, acetylation, and ubiquitination. PER (P) and CRY (C) proteins heterodimerize, translocate to the nucleus, and repress transcriptional activation by CLOCK/BMAL. An additional regulatory feedback loop involves REV-ERBα (R, nuclear heme receptor, a negative regulator) and ROR (a positive regulator), which compete for binding to and regulation of ROREs within the Bmal1 promoter. REV-ERBα also can bind to ROREs in the Per and Cry promoters, modulating transcription. Binding of transcription factors is a dynamic process, so their action is subject to relative stoichiometries. Among the many elements in this complex set of pathways, rates of both synthesis and proteosomal degradation are important to rhythm generation (22, 24, 45, 51, 76). Elements within these molecular loops are sensitive to redox state, which thereby can modulate both binding and transcriptional activity. NAD+ and heme (H) denote redox-sensitive nodes on clock proteins. CRY-bound FAD oxidizes CLOCK-bound NADPH to NAD(P)+, inhibiting CLK/BMAL activation of transcription at E-box elements (22, 24, 45, 51). The PERs and REV-ERBα contain heme-binding moieties (H) in their ligand-binding domains (16, 67, 86, 87). Heme binding interferes with the ability of PER2 to interact with CRY proteins (84), whereas it stabilizes REV-ERBα binding to a nuclear corepressor of transcription. Temporal relationships between the RXO in the SCN and these modulatory effects on the TTO have not been evaluated fully. CRY, CRYPTOCHROME; FAD, flavin adenine dinucleotide; NAD+, nicotinamide adenine dinucleotide; PER, PERIOD; ROR, retinoic acid-related orphan receptor; ROREs, retinoic acid-related orphan receptor response elements; RXO, redox oscillator; TTO, transcription–translation oscillator.
Clock genes and the molecular clockwork mechanism
Chronobiologists dissected the molecular clockwork (Fig. 2) from studies of mutations or gene deletions in mouse that alter the circadian patterning of locomotor activity. Takahashi and colleagues first cloned the mouse gene Clock, a positive transcriptional regulator (40, 77). After decades of exploration, the crystal structure of CLOCK bound to its heterodimeric partner, BMAL1, which together form the transcriptional activator complex, has been resolved (32). PERIOD and CRYPTOCHROME (PER and CRY), the negative regulatory elements, repress their own transcription by binding to E-box sequences in their gene promoters. The CLOCK:BMAL complex binds to PER:CRY, relieving the transcriptional repression, so Per and Cry can again be transcribed, reinitiating the cycle. This cycle of transcription, translation, cytoplasmic post-translational modification, and cytoplasmic–nuclear translocations takes ∼24-h (39, 56). The core molecular clockwork also encompasses a loop in which a nuclear heme receptor, REV-ERBα, is a key player (43, 86, 87). REV-ERBα can bind to the retinoic acid-related orphan receptor response elements (ROREs) in the promoters of Clock and Bmal1 to initiate transcription (6). This interacting oscillatory loop is proposed to add stability to the rhythm-generating loop; it also provides points of TTO modulation by metabolic state (see further). This model of the TTO (Fig. 2) as the driving force of circadian rhythms is now well established [see ref. (44) for a comprehensive review].
The clockwork machinery has expanded to include not only the transcriptional and translational elements of these clock genes but also post-translational modulators. Central clock elements in the SCN include CLOCK, BMAL, CRY, PER2, and the kinases/phosphatases that post-translationally modify them (e.g., casein kinase 1δ, ɛ, glycogen synthase kinase) (39, 56). Genetic alterations in specific kinases and phosphatases that modify clock proteins can alter the generation or period of circadian rhythms (2, 63, 74).
Health consequences of altered clock genes
Can the circadian TTO contribute to health or disease? Evidence supporting roles for altered molecular clockworks in disease generation is substantial. A hereditary human sleep disorder was identified in a single-base mutation in hPer2 that changes coding of serine662, the substrate for casein kinase 1ɛ (Fig. 2), to a glycine, causing this phosphorylation site to be lost. This one-base change in the DNA causes lifetime shortening of the circadian period of carriers, resulting in familial advanced sleep phase disorder (74, 89) in which their circadian cycles complete in ∼20 h every day.
Clues that clock genes also may act beyond the SCN came from studies of mice in which clock genes had been mutagenized, deleted, or genetically engineered. Takahashi and colleagues generated a gene knock-in mouse with the luminescent protein, luciferase (LUC), encoded in tandem with a clock protein, PER2. PER2::LUC bioluminescence oscillates with a circadian beat in the SCN and, indeed, within all the cells of all tissues examined (88). Animals harboring altered or missing clock genes not only exhibit profound rhythms deficits, they have other abnormalities, as well. These may include weight gain or loss, impaired glucose tolerance, diminished fecundity, arthritis, cardiovascular disease, tumors, susceptibility to addiction, sleep–wake cycle disorders, and shortened lifespan (14, 29, 33, 37, 46–48, 71, 89).
The Rise of Nontranscriptional Circadian Oscillators
The dominion of the TTO over sustained circadian rhythms has been challenged by studies of animals with deficits in clock gene transcription that unexpectedly exhibit minimal effects on rhythmic behavior or even clock protein expression (38, 41, 52, 85). These findings suggest that the rhythmic transcriptional regulation of clock genes may not be fully necessary. It follows that the TTO model may not be a comprehensive interpretation of the circadian clock.
Circadian oscillations in small regulatory molecules such as cAMP (53, 60) and Ca2+ (10, 27, 34) have been observed in SCN tissue. Analysis of interdependence between oscillations in these cytosolic versus nuclear activities suggests that the TTO is not sufficient to generate the circadian rhythm (Fig. 3) (10, 28). The concept of cytosolic oscillators as core components of circadian clocks emerged from the study of cAMP oscillations in SCN tissue (53): (i) pharmacological manipulation of cellular cAMP concentration affects the daily-rhythmic expression of clock genes, (ii) suppression of the level of cAMP compromises the free-running oscillation of clock genes, and drug washout causes convergence to a common phase in the cycle, and (iii) the rate of cAMP synthesis influences the length of the intrinsic period generated by the central pacemaker. Similar properties have been described for Ca2+ in Drosophila (27) and cyclic ADP-ribose in the plant Arabodopsis (17). In the SCN, circadian oscillations of intracellular Ca2+ concentration ([Ca2+]i) drive transcription of Per and Cry genes (10). The membrane receptor-associated GTP-binding protein, Gq, can reprogram these circadian rhythms via intercellular signals activating the receptor (10). These findings suggest that critical interplay between cytoplasmic small molecules and the TTO, rather than the TTO alone, drives intrinsic circadian rhythms (Fig. 3).
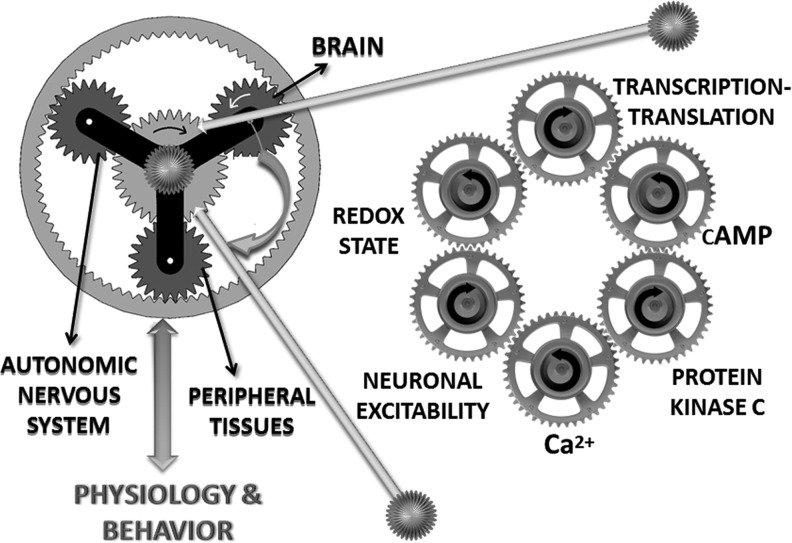
FIG. 3.
Multiple oscillators of the core circadian clockwork. Circadian timekeeping is a cellular property, key elements of which are represented here. In cells of the SCN, the circadian rhythm is generated on molecular level by a negative feedback loop of a group of clock genes and proteins that generate a TTO. The transcriptional–translational post-translational regulation of clock genes and their products forms the molecular clockwork machinery (44). Proteins that mediate post-translational modification, such as protein kinases/phosphatases (63), and the small cytoplasmic molecules cAMP (53, 60) and Ca2+ (10, 27) also participate in generating the intrinsic TTO. Redox state is emerging as a parallel cellular oscillator. The RXO is tightly integrated with the TTO, as well as with cellular physiology. At the tissue level, signals between cells synchronize and shape the multitude of cellular oscillators (10). Coherent oscillations of the SCN lead to coordinated timekeeping in the brain, autonomic nervous system, and peripheral tissues. These oscillations give rise to integrated circadian rhythms of physiology and behavior.
Neuronal Excitability as a Nontranscriptional Oscillator
Spontaneous neuronal activity undergoes circadian oscillation
Each SCN is a neural network of thousands of units that undergo daily oscillation in electrical activity. This electrical oscillation is essential for the functionality of the central pacemaker in synchronizing brain and body clocks to each other and to changing environmental time cues (11, 13). One level at which neuronal excitability oscillates is a daily fluctuation of spontaneous action potentials (SAP) generated by neurons of the SCN (Fig. 4, top) (49, 60). Frequencies of the SAP activity are significantly higher in daytime than during nighttime in both nocturnal and diurnal species. This pattern can be detected both in vivo and in vitro, by single- or multiunit recording (26, 35, 81). Thus, circadian changes in SCN neuronal excitability may comprise another nontranslational oscillatory system, one that encompasses the cell membrane.
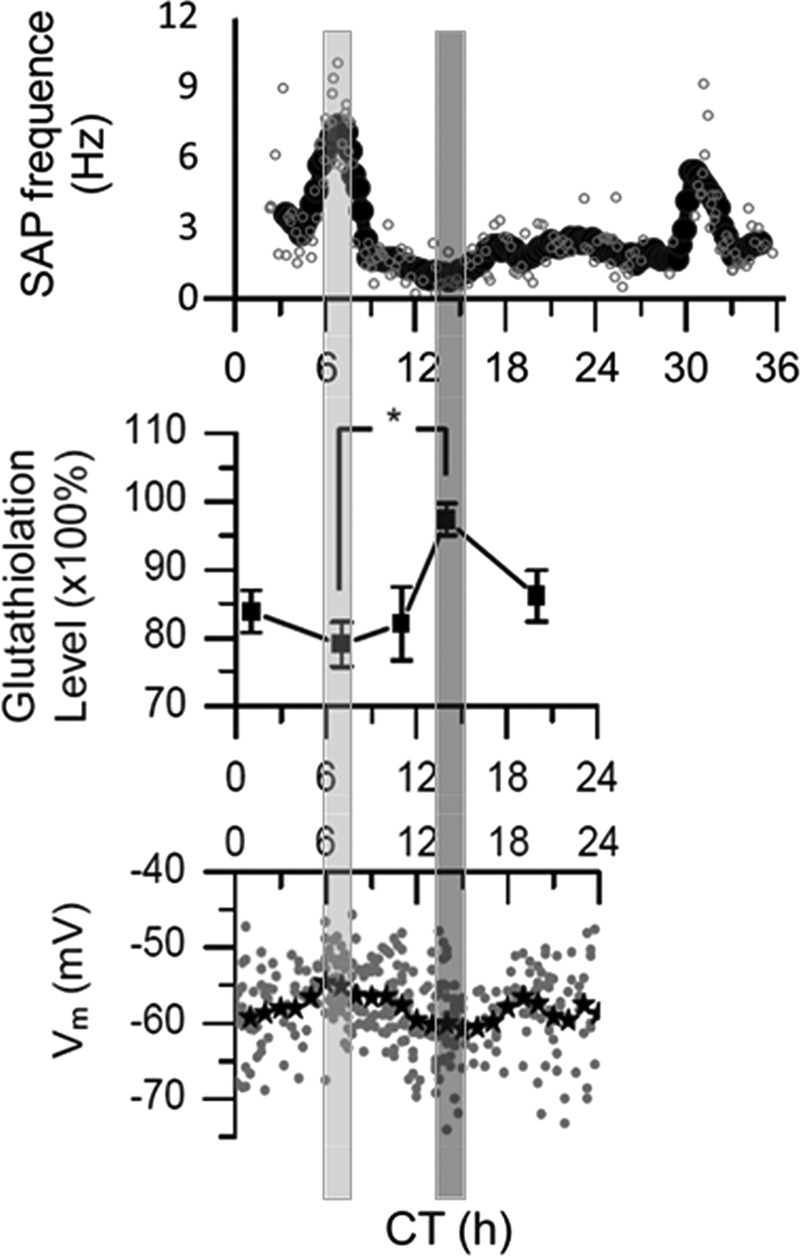
FIG. 4.
Circadian rhythms of SCN excitability and redox state are aligned. The SCN generates a ∼24-h rhythm of SAP that peaks ∼CT 7, mid-subjective day in a hypothalamic brain slice (top). SAP activity is lowest at CT 14, early during subjective night. Redox state, measured via protein glutathiolation of SCN ex vivo, oscillates in a circadian rhythm. SCN tissue is most reduced at CT 7 and most oxidized at CT 14 (*p<0.05, ANOVA, Tukey HSD, n=6, middle). Vm, assessed by patch clamp of 337 neurons over each of 24 CTs, varies significantly (p<001, ANOVA, Tukey HSD, bottom). Vm is most depolarized at CT 7 and most hyperpolarized at CT 14. Adapted from Wang et al. (79). ANOVA, analysis of variance; CT, circadian time; SAP, spontaneous action potentials.
Ionic mechanisms underlying oscillatory electrical activity
Dissection of underlying ionic mechanisms by patch-clamp recordings of membrane properties of mouse (8) and rat (79) neurons reveals very similar patterns of rhythmic oscillations in the excitatory state of neuronal membranes (membrane potential, Vm; Fig. 4, bottom). At least three types of ionic factors, K+ currents, Ca2+ currents, and [Ca2+]i, contribute to the oscillations in membrane excitability (8). These ionic fluxes are of three functional types: (i) currents generating the excitatory drive that elevates Vm to the threshold for action potential generation, (ii) currents responding to the excitatory drive and generating action potentials, and (iii) currents contributing to the nightly silencing of firing through hyperpolarizing Vm (13). Modulation of these currents could be on the levels of channel expression, localization, and/or post-translational modification of conductance and/or gating properties of specific ion channels.
The circadian oscillation of membrane excitability and electrical activity had been thought to be driven by the TTO, but a study in Drosophila suggests that the rhythmic electrical activity is necessary for the clock gene expression. Electrical silencing of pacemaker neurons in the fly brain by the genetic manipulation of K+ channels results in arrhythmic behavior and dampens the circadian pattern clock gene expression (51). Experimental evidence for this relationship in the mammalian SCN has yet not been gathered, but a similar mechanism is predicted. Thus, neuronal excitability may be another type of nontranscriptional oscillator, like the cytosolic oscillators cAMP and Ca2+, that acts as a core component of the clockwork machinery. However, it is not a cytosolic oscillator because neuronal excitability is not based on specific molecules in cytoplasm; rather it describes a cellular state (Fig. 3; Table 1).
Table 1.
Nontranscriptional Circadian Oscillators
| Oscillator | Oscillatory system | Species |
|---|---|---|
| Cytosolic small molecules and proteins | cAMP | Mouse53 |
| Ca2+ | Drosophila27, mouse10 | |
| cADPR | Arabidopsis17 | |
| PKC | Mouse63 | |
| Cellular state | Redox state | Human54 |
| Ostreococcus55 | ||
| Mouse/fly/fungus/bacteria/Archaea19 | ||
| Rat79, mouse79 | ||
| Electrical activity | Drosophila51 | |
| Membrane excitability | Rat51, mouse8 |
A Circadian Redox Oscillator
Circadian rhythms and metabolism
The organism's metabolic state, including the levels of liver enzymes and energy production and utilization, oscillates with a circadian rhythm. Superimposed upon circadian rhythms of metabolism are near 24-h oscillations due to the contingencies of life, such as metabolites in circulation and intracellular microenvironments, hormones related to feeding and fasting, and ingestive behaviors (6, 25). Genomic analysis found that 20% of mRNAs that exhibit circadian rhythms of expression are related to metabolism (1). These include enzymes critical for energy metabolism, such as glycogen phosphorylase, cytochrome oxidase, lactate dehydrogenase, and the monocarboxylate transporter, MCT2 (66). Disruption in mice of transcription of the core clock genes, Clock and Bmal 1, causes obesity, diabetes, and other metabolic syndromes (46, 75). These data support the contention that daily patterning of metabolism and the body systems it engages is one of the most important evolutionary drivers for an internal circadian clock.
SCN energetics
The brain is the most metabolically active organ, consuming about 20% of the O2 and 25% of all glucose in the human body, despite occupying only 2% of body mass (7). Most of the energy is spent on maintaining and restoring ionic gradients across the membrane (3), the basis of electrical signals that process information. Changes of Vm in the form of action potentials and synaptic potentials rely on the translocation of ions across the plasma membrane, leading to changes of energetic state. Indeed, task-dependent neuronal activity in the brain is accompanied by increased local blood flow and glucose consumption. This property constitutes the basis of functional imaging and mapping of the brain, such as positron emission tomography and functional magnetic resonance imaging (20). The intense energetics of brain functional dynamics requires an efficient machinery to coordinate energy production, delivery, storage, and utilization. It is widely accepted that activity-induced changes of brain energy metabolism are the result rather than the cause of neural activity (7).
Early functional measures of SCN circadian rhythmicity evaluated energy metabolism based on uptake of tracer amounts of 2-deoxy[1-14C]glucose (21, 69). This approach enables the determination of glucose consumption of individual brain structures in vivo and reports the functional activity. Morning glucose utilization in rat SCN is 2×higher than in the evening (p<0.01). Mitochondrial cytochrome C oxidase activity, measured by the changes in light absorption at 550 nm, mitochondrial aggregation, and glucose concentration, is higher during the light period, as well (36). These measures of energetics find high-energy availability when SCN electrical activity also is high (82). However, the ATP level in the SCN and anterior hypothalamic area is negatively correlated with electrical activity (82).
Redox state as a circadian oscillator
Redox state, the potential of molecular substrates of cellular metabolism to receive or donate electrons, is the manifestation of cellular metabolic state (18, 68). Redox state is determined by multiple interacting molecular electron couples (Fig. 5). The biochemical balance of interconversions of several sets of metabolites, such as lactate and pyruvate, depends on reducer/oxidizer homeostasis. Reactive free radicals, such as nicotinamide adenine dinucleotide (NAD+ and flavin adenine dinucleotide (FAD), originate from the redox reactions in metabolism and occur universally in cells. Redox molecules integral to intermediary metabolism also contribute to cell physiology and intracellular/intercellular signaling (18).
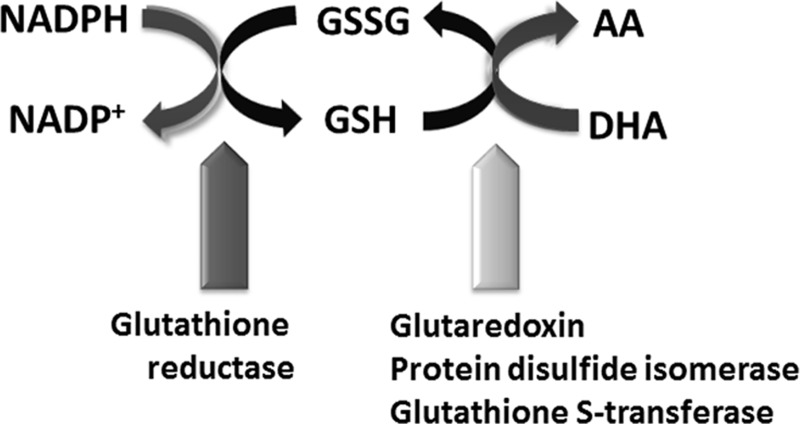
FIG. 5.
Cellular redox state is determined by a network of molecular interrelationships between electron couples. Electron couples are paired states of molecules that exchange electrons based on the intracellular reduction–oxidation environment. These reversible reactions are enzyme-mediated, dynamic, and have consequences for molecular conformation, physical interactions, and function. Presented are relationships between redox pairs, NADPH/NADP+, GSSG/GSH, and AA/DHA, and associated enzymes. AA, ascorbic acid (reduced form, also known as vitamin C); DHA, dehydroascorbic acid (oxidized form); GSH, glutathione (reduced form); GSSG, glutathione disulfide (oxidized form); NADP+, nicotinamide adenine dinucleotide phosphate (oxidized form); NADPH, reduced form of NADP+.
It follows that the redox state can be evaluated through the ratio of redox-molecule pairs, such as NAD(P)+/NAD(P)H, glutathione disulfide/glutathione (GSH), or dehydroascorbic acid (DHA)/ascorbic acid (AA). Ratios of these redox couples in the SCN were measured at points around the 24-h cycle in vivo using three different methods (79). Noninvasive redox analysis by real-time imaging of FAD/NAD(P)H in organotypic brain slices from both rat and mouse found highly significant circadian oscillations in redox state (p<0.00.1, 23.7±0.3 h). When analyzed in SCN from a Bmal1−/− mouse, which lacks a functional circadian clock (12, 41), the FAD/NAD(P)H ratios do not oscillate. This indicates some form of coupling of transcriptional and redox oscillators (RXOs) in the SCN.
Two different redox couples were analyzed in SCN either ex vivo or maintained as brain slices for several hours. Glutathiolation, the capacity of proteins to incorporate reduced GSH, of SCN ex vivo shows a significant time-of-day effect (p<0.5), with daily minimum 7 h into the day and maximum 2 h into the night (Fig. 4, middle). Furthermore, the AA system, among the most important antioxidant buffers in the brain (62), was analyzed using capillary electrophoresis with laser-induced fluorescence detection. We directly measured concentrations of DHA versus AA, its reduced counterpart, in acute SCN brain slices. Ratios of DHA/AA also oscillate significantly with a circadian rhythm (p<0.5). Like glutathiolation, levels are lowest midday and highest in early night. Discovery of parallel changes in different redox couples using distinct analytical methods reveals that global redox state oscillates with a circadian rhythm. SCN global redox state oscillates both in vivo and in vitro, such that conditions are significantly reduced in midday and oxidized in early night (Fig. 4, middle).
Redox oscillation is not limited to mammals' central circadian clock. A circadian rhythm in redox state based on peroxiredoxin oligomerization has been reported in human red blood cells (54), green algae (55), and many other species (19). This suggests that changing peroxiredoxin state is a conserved element of metabolic circadian rhythms in all organisms. The physiological relevance of peroxiredoxins to cellular function and the molecular clockwork remains to be established. Rhythmic activity of peroxiredoxin is independent of transcription/translation in the anucleate RBC but is disrupted in embryonic fibroblasts from clock-mutant mice (54). This also suggests that in nucleated cells, the transcriptional and RXOs are closely coupled.
Support for the RXO as an oscillator independent of the TTO is building. As in the SCN, the circadian oscillation of redox state in both central (79) and peripheral (54) tissues is disrupted when clock genes are deleted or mutated. While these clock-disrupted animals exhibit metabolic syndromes (46, 75), questions remain as to whether the disrupted redox oscillation results directly from the malfunction of TTO or is consequence of the metabolic disorder. To distinguish these two possibilities, we need advanced techniques for monitoring redox state in live specimens with genetically manipulated clock genes. Real-time redox imaging (79) combined with acute clock-gene knock-down has the potential to achieve this goal. Nevertheless, redox state can be regarded as a nontranscriptional circadian oscillator. Similar to membrane excitability, the RXO belongs to the category of cell-state oscillator (Fig. 3; Table 1).
RXO–TTO Engagement
Direct RXO–TTO engagement via transcriptional regulation
As a potential core component of the central pacemaker, the RXO has two pathways that can engage the clockwork machinery: one transcriptional and one nontranscriptional (Figs. 2 and 6). The transcriptional pathway originally was identified in cultured cells and is well characterized in peripheral tissues, such as liver (6, 25). Redox species, including FAD, NAD(P)H, and CO, can regulate the functions of clock proteins, CLOCK, BMAL1, and CRY. One mechanism involves CRY-bound FAD oxidizing CLOCK/NPAS2-bound reduced form of NADP+ (NADPH), thus transforming the activating effect of the reduced nicotinamide cofactor [NAD(P)H] to the inhibiting action of the oxidized cofactor [NAD(P)+] (Fig. 2) (16, 67).
FIG. 6.
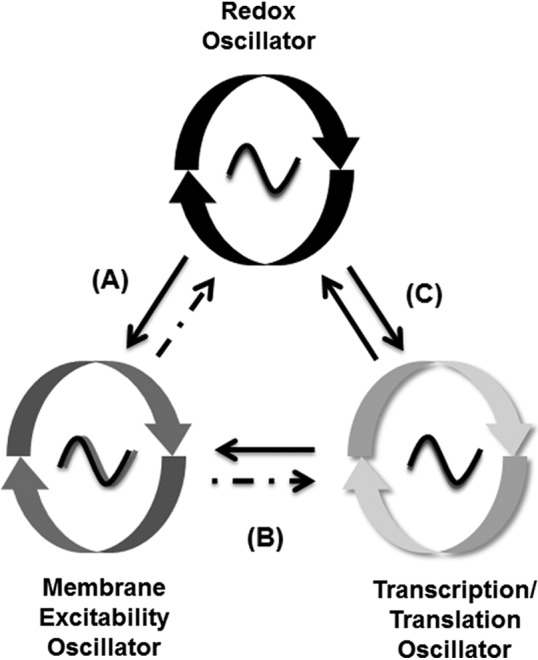
Reciprocal interactions of the RXO, membrane excitability oscillator, and transcriptional–translational oscillator. (A) The RXO regulates the circadian rhythm of SCN neuronal activity by modulating opening of K+ channels (solid arrow) (79). Concomitantly, increased neuronal activity can increase blood flow, glucose uptake by astrocytes, and energy availability, which feeds back, modulating neuronal metabolic state (dashed arrow) (66). (B) Some ion channels that underlie membrane excitability are rhythmically expressed under the control of clock genes (solid arrow), although acute shifts in redox state can immediately alter membrane excitability (11, 13). At the same time, membrane excitability of SCN neurons gates signal input to the TTO, which in turn affects clock gene expression (dashed arrow) (22, 24, 45, 51). (C) The circadian oscillation of SCN redox state depends on a functional TTO (79). Redox state feeds back to the TTO, modulating clock gene expression (solid arrows) (5, 16, 59, 67). Adapted from Wang et al. (79).
Binding of heme, an iron-containing prosthetic group, adds another mode of redox sensitivity to clock proteins. Redox state determines the binding of reactive gases, such as CO and NO, to the heme moiety, and thus the protein's conformational state. PER and CLOCK/NPAS2 bind heme via PAS domains (Fig. 2) (16, 67). Heme-dependent binding of CO to neuronal PAS-domain protein 2 inhibits the DNA binding activity (16, 67). Metabolic signals regulate rhythmic expression of the nuclear receptors REV-ERBα and RORα that engage the core clockwork (5, 59). Furthermore, heme is the ligand for REV-ERBα, stabilizing its binding to a nuclear corepressor of transcription. Depletion of intracellular heme abolishes the interaction between REV-ERBα and the nuclear corepressor protein. These conditions favor RORα binding to ROREs and transcriptional activation (86, 87).
Transcriptional regulation of clock genes by redox state is likely to occur in the SCN, but is not yet explored. Based on reports of redox regulation of clock gene transcription (66) and redox oscillation in SCN tissue (79), it is reasonable to speculate that the redox state could modulate the amplitude, period, and/or phase of the transcriptional oscillation of clock genes in the SCN. Differences in local redox states of microdomains of the cytoplasm or nucleus could fine-tune this modulation.
Indirect RXO–TTO engagement via modulating membrane excitability
Recent evidence indicates that cellular metabolic state at the level of redox molecules oscillates and contributes importantly to circadian timekeeping in the SCN (79). Acute exposure of the SCN to reducing or oxidizing reagents rapidly reverses the polarity of Vm, which determines the state of excitability. The directionality and amplitude of this effect changes with CT (time reckoned from the endogenous circadian clock of the SCN free-running in constant conditions). The effect of nontranscriptional regulation of redox state on neuronal excitability in SCN neurons is a novel pathway for integrating the metabolic cycle into the core clockwork machinery.
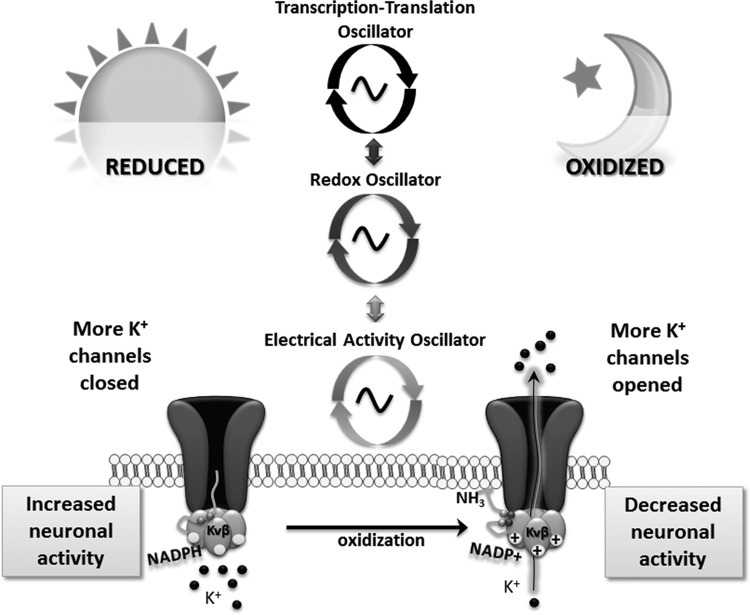
The circadian redox oscillation directly modulates the neuronal membrane, driving circadian changes in SCN electrical activity via post-translational modification of K+ channels (79). Sensitivity to specific blockers suggests that leak and A-type K+ channels mediate the redox-sensitive changes in excitability. The reduced redox state of daytime (Fig. 4) closes these K+ channels, diminishing the hyperpolarizing influences on the membrane and permitting depolarization and increased neuronal electrical activity (Fig. 7). Conversely, the oxidized state of nighttime (Fig. 4) increases the activity of these channels and reduces neuronal excitability (Fig. 7). Expression of leak K+ channels (KcnK1) in the SCN exhibits robust circadian rhythm (58), supporting our finding that the redox modulation of membrane excitability depends on the CT (79). This emphasizes the complexity of the ionic mechanisms underlying the circadian oscillation of electrical activity in the SCN.
FIG. 7.
Day–night differences in SCN neuronal activity depend on redox state but are the consequence of multiple interacting cellular oscillatory systems. Most proximally, redox species bind to subunits of K+ channels within the neuronal membrane, altering the open–closed states of the channel (bottom) (57). Changes in the open–closed state of K+ channels alter K+ fluxes across the membrane. When redox state is reduced in daytime, K+ channels close and the membrane is biased toward depolarization, facilitating the activation of channels that contribute to excitation and action potentials. During nighttime, the oxidized state of the subunits facilitates K+-channel opening, leading to membrane hyperpolarization and lowering the probability of achieving threshold for action potentials (79). Thus, oscillations in membrane excitability are regulated most proximally by the RXO, which is coupled to the TTO.
A recent biophysical study described a mechanism by which N-type inactivation of a voltage-sensitive K+ channel, Kvβ1, is modulated by redox state in an NADPH-dependent manner (57). Each Kvβ subunit binds one molecule of NADPH cofactor and has an N terminus that closes the channel by N-type inactivation; oxidation of the NADPH suppresses inactivation and increases channel current.
The redox-based post-translational modulation of neuronal excitability in the SCN links metabolic state to circadian physiology at multiple levels. By driving the daily oscillation of membrane excitability, the metabolic oscillator can modulate coupling between the central pacemaker and both input signals and output targets (22, 66).
Conclusion
The SCN is the central component of an organism-wide circadian system that coordinates daily rhythms in metabolism, physiology, and behavior. Circadian timekeeping emerges from a cell-based molecular TTO. Recent findings reviewed here report an autonomous circadian rhythm of global SCN redox state, which is reduced in daytime and oxidized at night. This RXO modulates circadian rhythms of neuronal excitability via specific K+ channels. SCN neurons are both metabolically and electrically more active in daytime when SCN redox state is reduced and K+ conductances are diminished. During nighttime, SCN redox state is reversed: neurons are more oxidized, K+ conductances are enhanced, and neurons are relatively inactive. Energetic fluctuation in the central nervous system has been considered to be a consequence of the neuronal activity. However, these new findings suggest that changes in cellular metabolic state can be the cause, rather than the result, of the neuronal activity. Thus, cross talk between energetic and neuronal states can bridge cellular state and physiology with important functional consequences. These findings suggest regulatory features for future experimental investigation.
Abbreviations Used
- [Ca2+]i
intracellular Ca2+ concentration
- AA
ascorbic acid
- ANOVA
analysis of variance
- CRY
CRYPTOCHROME, a clock protein
- CT
circadian time
- DHA
dehydroascorbic acid
- FAD
flavin adenine dinucleotide
- GSH
glutathione
- GSSG
glutathione disulfide
- LUC
luciferase
- NAD+
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate (oxidized form)
- NADPH
reduced form of NADP+
- PER
PERIOD, a clock protein
- RHT
retinohypothalamatic tract
- ROR
retinoic acid-related orphan receptor
- ROREs
retinoic acid-related orphan receptor response elements
- RXO
redox oscillator
- SAP
spontaneous action potentials
- SCN
suprachiasmatic nucleus
- TTO
transcription–translation oscillator
- Vm
membrane potential
- τ
tau, circadian period
Acknowledgments
The authors thank Jennifer Mitchell for provocative discussions and contributions to the article. Support of the National Institutes of Health (RO1HL086870 and R21MH101655) is gratefully acknowledged. Content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, and Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Allada R, Emery P, Takahashi JS, and Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24: 1091–1119, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Alle H, Roth A, and Geiger JR. Energy-efficient action potentials in hippocampal mossy fibers. Science 325: 1405–1408, 2009 [DOI] [PubMed] [Google Scholar]
- 4.Aon MA, Cortassa S, and O'Rourke B. Mitochondrial oscillations in physiology and pathophysiology. Adv Exp Med Biol 641: 98–117, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsalobre A, Damiola F, and Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93: 929–937, 1998 [DOI] [PubMed] [Google Scholar]
- 6.Bass J. and Takahashi JS. Circadian integration of metabolism and energetics. Science 330: 1349–1354, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belanger M, Allaman I, and Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14: 724–738, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Belle MD, Diekman CO, Forger DB, and Piggins HD. Daily electrical silencing in the mammalian circadian clock. Science 326: 281–284, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Berson DM, Dunn FA, and Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295: 1070–1073, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Brancaccio M, Maywood ES, Chesham JE, Loudon AS, and Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78: 714–728, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown TM. and Piggins HD. Electrophysiology of the suprachiasmatic circadian clock. Prog Neurobiol 82: 229–255, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, and Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12: 553–569, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson AJ, Sellix MT, Daniel J, Yamazaki S, Menaker M, and Block GD. Chronic jet-lag increases mortality in aged mice. Curr Biol 16: R914–R916, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Coursey PJ. Daily light sensitivity rhythm in a rodent. Science 131: 33–35, 1960 [DOI] [PubMed] [Google Scholar]
- 16.Dioum EM, Rutter J, Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA, and McKnight SL. NPAS2: a gas-responsive transcription factor. Science 298: 2385–2387, 2002 [DOI] [PubMed] [Google Scholar]
- 17.Dodd AN, Gardner MJ, Hotta CT, Hubbard KE, Dalchau N, Love J, Assie JM, Robertson FC, Jakobsen MK, Goncalves J, Sanders D, and Webb AA. The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318: 1789–1792, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Droge W. Free radicals in the physiological control of cell function. Physiol Rev 82: 47–95, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, and Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figley CR. and Stroman PW. The role(s) of astrocytes and astrocyte activity in neurometabolism, neurovascular coupling, and the production of functional neuroimaging signals. Eur J Neurosci 33: 577–588, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Fuchs JL. and Moore RY. Development of circadian rhythmicity and light responsiveness in the rat suprachiasmatic nucleus: a study using the 2-deoxy[1-14C]glucose method. Proc Natl Acad Sci U S A 77: 1204–1208, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillette MU. and Mitchell JW. Signaling in the suprachiasmatic nucleus: selectively responsive and integrative. Cell Tissue Res 309: 99–107, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Gillette MU. and Reppert SM. The hypothalamic suprachiasmatic nuclei: circadian patterns of vasopressin secretion and neuronal activity in vitro. Brain Res Bull 19: 135–139, 1987 [DOI] [PubMed] [Google Scholar]
- 24.Golombek DA. and Rosenstein RE. Physiology of circadian entrainment. Physiol Rev 90: 1063–1102, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Green CB, Takahashi JS, and Bass J. The meter of metabolism. Cell 134: 728–742, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green DJ. and Gillette R. Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res 245: 198–200, 1982 [DOI] [PubMed] [Google Scholar]
- 27.Harrisingh MC, Wu Y, Lnenicka GA, and Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci 27: 12489–12499, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastings MH, Maywood ES, and O'Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18: R805–R815, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, and Panda S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15: 848–860, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattar S, Liao HW, Takao M, Berson DM, and Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295: 1065–1070, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honma K. and Honma S. The SCN-independent clocks, methamphetamine and food restriction. Eur J Neurosci 30: 1707–1717, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, and Takahashi JS. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 337: 189–194, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber R, Ghilardi MF, Massimini M, and Tononi G. Local sleep and learning. Nature 430: 78–81, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, and Allen CN. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38: 253–263, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Inouye ST. and Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 76: 5962–5966, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isobe Y, Hida H, and Nishino H. Circadian rhythm of metabolic oscillation in suprachiasmatic nucleus depends on the mitochondrial oxidation state, reflected by cytochrome C oxidase and lactate dehydrogenase. J Neurosci Res 89: 929–935, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Kennaway DJ, Boden MJ, and Varcoe TJ. Circadian rhythms and fertility. Mol Cell Endocrinol 349: 56–61, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Kim EY, Bae K, Ng FS, Glossop NR, Hardin PE, and Edery I. Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron 34: 69–81, 2002 [DOI] [PubMed] [Google Scholar]
- 39.King DP. and Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci 23: 713–742, 2000 [DOI] [PubMed] [Google Scholar]
- 40.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, and Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell 89: 641–653, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, and Takahashi JS. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8: e1000513, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, and Bittman EL. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci 7: 1626–1638, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowrey PL. and Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet 5: 407–441, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowrey PL. and Takahashi JS. Genetics of circadian rhythms in mammalian model organisms. Adv Genet 74: 175–230, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundkvist GB, Kwak Y, Davis EK, Tei H, and Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci 25: 7682–7686, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, and Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F, Backx PH, Ralph MR, and Sole MJ. Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension 49: 1104–1113, 2007 [DOI] [PubMed] [Google Scholar]
- 48.McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, and Nestler EJ. Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc Natl Acad Sci U S A 102: 9377–9381, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meijer JH, Schaap J, Watanabe K, and Albus H. Multiunit activity recordings in the suprachiasmatic nuclei: in vivo versus in vitro models. Brain Res 753: 322–327, 1997 [DOI] [PubMed] [Google Scholar]
- 50.Moore RY, Speh JC, and Leak RK. Suprachiasmatic nucleus organization. Cell Tissue Res 309: 89–98, 2002 [DOI] [PubMed] [Google Scholar]
- 51.Nitabach MN, Blau J, and Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Numano R, Yamazaki S, Umeda N, Samura T, Sujino M, Takahashi R, Ueda M, Mori A, Yamada K, Sakaki Y, Inouye ST, Menaker M, and Tei H. Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc Natl Acad Sci U S A 103: 3716–3721, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, and Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320: 949–953, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Neill JS. and Reddy AB. Circadian clocks in human red blood cells. Nature 469: 498–503, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, and Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature 469: 554–558, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okamura H, Yamaguchi S, and Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res 309: 47–56, 2002 [DOI] [PubMed] [Google Scholar]
- 57.Pan Y, Weng J, Levin EJ, and Zhou M. Oxidation of NADPH on Kvbeta1 inhibits ball-and-chain type inactivation by restraining the chain. Proc Natl Acad Sci U S A 108: 5885–5890, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, and Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002 [DOI] [PubMed] [Google Scholar]
- 59.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, and Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260, 2002 [DOI] [PubMed] [Google Scholar]
- 60.Prosser RA. and Gillette MU. Cyclic changes in cAMP concentration and phosphodiesterase activity in a mammalian circadian clock studied in vitro. Brain Res 568: 185–192, 1991 [DOI] [PubMed] [Google Scholar]
- 61.Ralph MR, Foster RG, Davis FC, and Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science 247: 975–978, 1990 [DOI] [PubMed] [Google Scholar]
- 62.Rice ME. Ascorbate regulation and its neuroprotective role in the brain. Trends Neurosci 23: 209–216, 2000 [DOI] [PubMed] [Google Scholar]
- 63.Robles MS, Boyault C, Knutti D, Padmanabhan K, and Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science 327: 463–466, 2010 [DOI] [PubMed] [Google Scholar]
- 64.Roenneberg T. and Merrow M. The network of time: understanding the molecular circadian system. Curr Biol 13: R198–R207, 2003 [DOI] [PubMed] [Google Scholar]
- 65.Rosbash M. The implications of multiple circadian clock origins. PLoS Biol 7: e62, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rutter J, Reick M, and McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem 71: 307–331, 2002 [DOI] [PubMed] [Google Scholar]
- 67.Rutter J, Reick M, Wu LC, and McKnight SL. Regulation of CLOCK and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293: 510–514, 2001 [DOI] [PubMed] [Google Scholar]
- 68.Satoh T. and Lipton SA. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci 30: 37–45, 2007 [DOI] [PubMed] [Google Scholar]
- 69.Schwartz WJ. and Gainer H. Suprachiasmatic nucleus: use of 14C-labeled deoxyglucose uptake as a functional marker. Science 197: 1089–1091, 1977 [DOI] [PubMed] [Google Scholar]
- 70.Shigeyoshi Y, Meyer-Bernstein E, Yagita K, Fu W, Chen Y, Takumi T, Schotland P, Sehgal A, and Okamura H. Restoration of circadian behavioural rhythms in a Period null Drosophila mutant (Per01) by mammalian period homologues mPer1 and mPer2. Genes Cells 7: 163–171, 2002 [DOI] [PubMed] [Google Scholar]
- 71.Silver AC, Arjona A, Walker WE, and Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36: 251–261, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, and Menaker M. Entrainment of the circadian clock in the liver by feeding. Science 291: 490–493, 2001 [DOI] [PubMed] [Google Scholar]
- 73.Tischkau SA, Mitchell JW, Pace LA, Barnes JW, Barnes JA, and Gillette MU. Protein kinase G type II is required for night-to-day progression of the mammalian circadian clock. Neuron 43: 539–549, 2004 [DOI] [PubMed] [Google Scholar]
- 74.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, and Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291: 1040–1043, 2001 [DOI] [PubMed] [Google Scholar]
- 75.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, and Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Ooijen G, Dixon LE, Troein C, and Millar AJ. Proteasome function is required for biological timing throughout the twenty-four hour cycle. Curr Biol 21: 869–875, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, and Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264: 719–725, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wager-Smith K. and Kay SA. Circadian rhythm genetics: from flies to mice to humans. Nat Genet 26: 23–27, 2000 [DOI] [PubMed] [Google Scholar]
- 79.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, and Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337: 839–842, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Webb IC, Baltazar RM, Lehman MN, and Coolen LM. Bidirectional interactions between the circadian and reward systems: is restricted food access a unique zeitgeber? Eur J Neurosci 30: 1739–1748, 2009 [DOI] [PubMed] [Google Scholar]
- 81.Welsh DK, Logothetis DE, Meister M, and Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14: 697–706, 1995 [DOI] [PubMed] [Google Scholar]
- 82.Yamazaki S, Ishida Y, and Inouye S. Circadian rhythms of adenosine triphosphate contents in the suprachiasmatic nucleus, anterior hypothalamic area and caudate putamen of the rat—negative correlation with electrical activity. Brain Res 664: 237–240, 1994 [DOI] [PubMed] [Google Scholar]
- 83.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, and Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685, 2000 [DOI] [PubMed] [Google Scholar]
- 84.Yang J, Kim KD, Lucas A, Drahos KE, Santos CS, Mury SP, Capelluto DG, and Finkielstein CV. A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol Cell Biol 28: 4697–4711, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Z. and Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467, 2001 [DOI] [PubMed] [Google Scholar]
- 86.Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, and Lazar MA. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318: 1786–1789, 2007 [DOI] [PubMed] [Google Scholar]
- 87.Yin L, Wu N, and Lazar MA. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal 8: e001, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, and Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101: 5339–5346, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang L, Ptáček LJ, and Fu YH. Diversity of human clock genotypes and consequences. In: Chronobiology: Biological Timing in Health and Disease, edited by Gillette MU. London: Academic Press; 119: pp. 51–81, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]