Abstract
Since the discovery of tubulin as the major component of microtubules over 40 years ago, its diversity of forms has raised a continuum of fundamental questions about its regulation and functions in a variety of organisms across phyla. Its high abundance in the brain or in specialized organelles such as cilia has allowed early characterization of this important target for anticancer drugs. However, it was only when matrix-assisted laser desorption ionization and electrospray ionization mass spectrometry technologies became available in the late 1980's that the full complexity of tubulin expression patterns became more obvious. This contributed in a major way to the idea that due to increasing and conserved tubulin heterogeneity during evolution, a tubulin code read by microtubule associated proteins might exist and be of functional significance. We review here the merging of recent genetic and cell biology studies with proteomics to decipher this code and illustrate some of the tubulin proteomic approaches with new data generated in our laboratories.
Keywords: tubulin, microtubule, proteomics, mass spectrometry
Introduction
Microtubules (MTs3) are dynamic polymers of α/β tubulin heterodimers that are involved in numerous processes such as cell division and migration. MT dynamics is characterized by stochastic transitions between phases of growth and phases of disassembly (catastrophes) and vice versa (pauses and rescues) occurring at the tips of MTs [1]. The resulting basal dynamics is precisely tuned by the expression profile of tubulin isotypes [2], the transient association of proteins to MTs and by the crosstalk between MTs, chromosomes, actin filaments and adhesion sites [3]. MTs also function as a scaffold for a large number of signaling proteins and may serve to localize and regulate their activities. Protein-protein interactions occurring at the surface of MTs have been shown to depend on the extent and nature of tubulin posttranslational modifications [4–6]. Progress in deciphering the enzymatic complexes involved in these modifications of tubulin has enabled the development of in vivo models that are key to obtaining insight into the functional significance of this diversity of tubulin species [7; 8]. Besides the physiological role of the spatio-temporal expression of different tubulin isotypes, some forms of tubulin have been associated with disease states and therefore represent useful biomarkers [9–13]. These findings underscore the need for advanced qualitative and quantitative analytical approaches allowing the detection of α- and β-tubulin isotype expression in human cells and tissues. This review will focus on the contributions of high resolution protein separation and mass spectrometry either directly towards this goal or indirectly through the validation of antibodies directed against specific α- or β-tubulin isotypes. The other members of the tubulin family of proteins (γ, δ, ε, ζ) are not included in the review, but some of the cited principles may be applicable to the analysis of their expression.
Tubulin isotypes and functional significance: first hints of a tubulin code
Tubulin is a 100 kDa heterodimer formed by an α- and a β-subunit that are equivalent in size and structure [14]. In mammals, tubulin represents about 3–4% of the total proteins in cells and up to 10% in brain. In humans, eight α-tubulin and seven β-tubulin genes have been identified [15; 16]. These tubulin isotypes have been detected at the mRNA and/or protein level and are differentially distributed in tissues [16]. Additional complexity is generated by the fact that each isotype can undergo extensive posttranslational modifications, including polyglutamylation, polyglycylation, reversible tyrosination, phosphorylation and acetylation [16]. Consequently, a great number of combinations of tubulin subunits can form tubulin heterodimers. Nevertheless, only a subset of these possible combinations are expressed in a given cell population. So far, it appears that the degree of complexity in posttranslational modifications is related to the stability of MTs, with highly dynamic MTs being relatively unmodified [17–19]. Heavily modified stable MTs are concentrated in specialized organelles such as centrioles and cilia or in the axons of neurons. More broadly, the proportion of stable vs dynamic MTs tends to increase and the tubulin isotype expression profile changes during cell differentiation. One exciting possibility is that the extensive tubulin diversity represents a complex code that can be deciphered and acted upon by interacting proteins [17; 20; 21].
Alignment of the amino acid sequences of the α- or β-tubulin isotypes reveals that most of the divergence is contained in the last 20 amino acids [16]. Tubulin nomenclature based on these divergent sequences is still confusing (see Table 1 for new and old names of human α and β-tubulin isotypes). For β-tubulin, we favor the concept of class I, II, III, IV, V, VI that is used for human β-tubulins, and we applied in the text this nomenclature to mouse β-tubulin for simplicity. This is possible because either the C-termini of human and mouse tubulins are identical or they display minimal divergence. For non mammalian organisms, this is not applicable therefore we used the names as they appear in literature.
Table 1.
Identity of human tubulin isotypes: NCBI accession number, isoelectric point and mass.
| Tubulin isotypea | Accession number NCBI | Protein mass (Da) | pI | C-terminal sequenceb | Mass (Da) CNBr C-terminal peptide |
|---|---|---|---|---|---|
| α1A (α brain specific, hum-a-tub1, hum-a-tub2, α3) | NP_006000 | 50,135.6 | 4.94 | MAALEKDYEEVGVDSVEGEGEEEGEEY | 2860.87 |
| α1B (Kα1, α-tub isoform 1, α1, α-tubulin ubiquitous) | AAC31959 | 50,151.6 | 4.94 | MAALEKDYEEVGVDSVEGEGEEEGEEY | 2860.87 |
| α1C (α6) | Q9BQE3 | 49,895.3 | 4.96 | MAALEKDYEEVGADSADGEDEGEEY | 2590.55 |
| α3Cd (α2) | Q13748 | 49,959.5 | 4.98 | LAALEKDYEEVGVDSVEAEAEEGEEY | 4152.21 |
| α3E | NP_997195 | 49,916.6 | 4.97 | LAALEKDCEEVGVDSVEAEAEEGEEY | 4093.17 |
| α4A (α1 testis specific) | NP_005991 | 49,924.4 | 4.95 | MAALEKDYEEVG I D SYEDEDEGEE | 2633.61 |
| α8 (α-like 2) | Q9NY65 | 50,093.5 | 4.94 | LAALEKDYEEVGTDSFEEENEGEEF | 4158.17 |
| α-like 3 | NP_079079 | 49,908.7 | 5.68 | LAALERDYEEVAQSF | 3060.23 |
| Bα1e (α3) | CAA25855 | 50,157.7 | 5.02 | MAALEKDYEEVGVHSVEGEGEEEGEEY | 2882.93 |
|
| |||||
| βI (β5, β4, Ib) | AAD33873 | 49,670.8 | 4.78 | YQDATAEEEEDFGEEAEEEA | 3367.30 |
| βIIf (class II β-tubulin, β2A / β2B) | NP 001060 / AAH01352 | 49,953.1 / 49,907.0 | 4.78 / 4.78 | YQDATADEQGEFEEEEGEDEA | 3467.38 |
| βIII (β4) | AAH00748 | 50,432.7 | 4.83 | YQDATAEEEGEMYEDDEEESEAQGPK | 1624.58 |
| βIVag (β4, β5) | CAA25318 / NP_006078 | 49,630.9 / 49,585.8 | 4.81 / 4.78 | YQDATAEQGEFEEEAEEEVA YQDATAEEGEFEEEAEEEVA |
3350.36 / 3351.35 |
| βIVb (β2C) | CAA26203 | 49,831.0 | 4.79 | YQDATAEEEGEFEEEAEEEVA | 3480.46 |
| βV (β6) | NP_115914 | 49,857.1 | 4.77 | YQDATANDGEEAFEDEEEE I DG | 3552.49 |
| βVI (β1) | NP_110400 | 50,326.9 | 5.05 | FQDAKAVLEEDEEVTEEAEMEPEDKGH | 809.82 |
Nomenclature of α-tubulin is based on recent revision [15]. Old names used for human tubulin isotypes are indicated in parenthesis.
First amino acid residue has been arbitrarily chosen.
Calculated average [M-H]− mass of the CNBr C-terminal peptides when analyzed by MALDI-TOF in the negative mode.
Another gene coding for the same sequence has been cloned (TUBA3D) [15].
This isotype was not considered in the new α-tubulin nomenclature
Two different sequences were found in the NCBI protein database.
Two βIVa-tubulin sequences with distinct C-termini were found in the NCBI protein database. The upper C-terminal sequence was found in human brain and lower sequence was found in a human oligodendroglioma and in mouse brain.
With the exception of acetylation of Lys40, most known posttranslational modifications of tubulin occur within this highly acidic and unstructured region that lies as a flexible arm at the surface of the MT lattice [14]; this location makes the C-termini of tubulin accessible to the enzymes responsible for the various posttranslational modifications and for interactions with regulatory, structural and motor proteins. In addition, in vitro studies have indicated that the ability of certain structural and motor microtubule associated proteins (MAPs) to interact with tubulin varies with the length of the polyglutamyl side chain [4–6].
Until recently, strong evidence of the existence of a tubulin code which translates into specific functions in vivo has been lacking. The first insights came from invertebrates. Drosophila testis specific β2-tubulin cannot be substituted by the more ubiquitous β3 isotype during spermatogenesis [22]. Surprisingly, Hvβt the closest ortholog of Drosophila β2-tubulin from another insect, Heliothis virescens, with equivalent function in the formation of axonemal MTs in spermatides, could not substitute for Drosophila β2-tubulin, despite the presence in both C-terminal sequences of the EGEF motif common to axonemal β-tubulin conserved across evolution [22; 23]. Similar results were obtained with the Drosophila α-84B testis specific tubulin [24]. In the worm Caenorhabditis elegans, the β-tubulin mec-7 is associated with the 15 protofilament MTs of touch neurons [25]. Similar functional genetic studies of a tubulin isotype in vertebrates comes from a knock-out mouse model for β1-tubulin which is normally specifically expressed in the blood cell lineage, like its avian β6- and human βVI-tubulin orthologs [26]. βVI-tubulin is the most divergent and in mouse it represents the major β-tubulin isotype expressed in megakaryocytes (MKs) and concentrates in the marginal bands of protoplatelets and platelets. βVI−/− -tubulin mice harbor MKs that fail to form protoplatelets in culture and they present severe thrombocytopenia. Also, platelets from these mice have their marginal bands depleted of microtubules and total tubulin content, and their response to thrombin-induced activation is attenuated [26]. Interestingly, two other β-tubulin isotypes, β2 (human βII) and β5 (human βI), are overexpressed in MKs from βVI−/− mice which obviously does not compensate for the lack of βVI-tubulin. Moreover, in a search for MAPs specifically interacting with βVI-tubulin, a yeast two hybrid system using the C-terminal domain of βVI-tubulin as a bait identified the secretory leukocyte protease inhibitor (SLPI) [27]. When the bait was replaced by the βI-tubulin equivalent no such interaction was found. SLPI colocalizes with platelet microtubules in wild type mice and is released from platelets upon their activation by thrombin, however not in βVI−/− mice. Collectively, these observations underscore the existence of specific functions of βVI-tubulin related to platelet biogenesis, structure and function.
The most recent progress in determining how the tubulin isotype composition of microtubules can specify function in vivo comes from the identification of some of the enzymes responsible for tubulin-specific posttranslational modifications. The C-terminal tyrosine of α-tubulin can be removed by a still unidentified carboxypeptidase. This tyrosine residue can be added again to detyrosinated α-tubulin (glu-tubulin) by tubulin tyrosine ligase (TTL) but cannot be added when the penultimate glutamate residue of glutubulin has been removed (Δ2-tubulin) by another unknown carboxypeptidase [28; 29]. This detyrosination-tyrosination cycle operates in various eukaryotes except in yeast, where the C-terminal tyrosine is replaced by a phenylalanine and no TTL is expressed. In order to decipher the role of this cycle, a budding yeast strain, lacking the last phenylalanine residue of its unique α-tubulin, was generated and displayed a defect in nuclear oscillations during cell division [30]. This was due to a lack of interaction between glu-tubulin and Bik1p, the budding yeast homolog of mammalian CLIP-170, at the plus ends of microtubules. This delocalization of CLIP-170 from microtubule plus ends due to the absence of the C-terminal tyrosine on α-tubulin was confirmed in TTL−/− neurons and embryonic fibroblasts, in cultures isolated from knock-out mice [7]. TTL−/− neurons exhibit unstable neurite extensions and premature axon differentiation that are associated with the death, soon after birth, of TTL−/− mice whose brain presents neuronal disorganization. TTL−/− fibroblasts present defects in mitotic spindle positioning and in polarity during migration [31]. The structural bases for this requirement for tyrosinated α-tubulin in the interaction between microtubules and CLIP-170 were recently uncovered (see ref [32] for review). Addition of side chains of glutamate residues on tubulin C-termini is performed by TTL-like enzymes (TTLL), comprised of different subunits [33]. In mammals TTLL1 and TTLL7 preferentially catalyze the addition of glutamate residues to α- and β-tubulin, respectively [34]. TTLL1 activity requires the presence of a PGs1 subunit whose function is lost in ROSA22 mice [8]. These mice have a loss of α-tubulin polyglutamylation in neurons and a decreased affinity of MAPs and motors for their microtubules. In particular, an impairment of the targeting of the kinesin KIF1A to neurites in vivo correlates with a decreased density of its cargo synaptic vesicles that modulates continuous synaptic transmission. Mutations in polyglycylation sites of Tetrahymena thermophyla in β-tubulin, but not α-tubulin, induce death, but axonemes from heterokarions lack the central pair of microtubules and arrest in cytokinesis [35].
We can expect further insights into the functional translation of the tubulin code with the knock-out of minor tubulin isotypes or of isotypes more specifically expressed in certain organs, and with the identification of other enzymes modifying specifically or preferentially tubulin (polyglycylases, carboxypeptidases, acetyl transferase, kinases …). Mass spectrometry and other proteomic methods have been essential for the detection of tubulin posttranslational modifications (see below), the validation of knock-out models and the identification of TTL and TTLLs.
Analysis of tubulin expression profiles: key contributions from mass spectrometry
Tubulin isotype expression profiling has been frequently assessed by RT-PCR of mRNA and at the protein level by antibody-based approaches [2]. These approaches are useful in a high-throughput setting, but results must be cautiously interpreted. This is particularly relevant to β-tubulin mRNA because it appears to be autoregulated by the level of the free β-tubulin in cells [36], implying that levels of mRNA may not reflect levels of the corresponding tubulin. Antibodies directed against the C-terminal peptides of most of the β-tubulin, but not α-tubulin, isotypes are readily available and have been extensively used to evaluate their respective isotype expression levels. However, such studies are semi-quantitative at best, due to the differing binding affinities of the antibodies employed. Specificity of some of these antibodies needs to be further verified. Antibodies recognizing polyglutamylated tubulin are also available, but they provide no information as to the exact length of the appended side chain. Moreover, multiple entries for some human tubulin isotypes can be found in the databases and determination of which of these are effectively expressed at the protein level is necessary in order to detect mutations or polymorphisms.
Mass spectrometry (MS)-based analyses were thus instrumental in first detecting and characterizing in detail the complexity of tubulin protein species and also in the validation of tubulin sequences and of antibody specificity. The methodologies leading to these goals and the emergence of MS based quantitation of tubulin isotype expression are presented in the following paragraphs.
Focus on C-terminal tubulin peptides
Tubulin analysis by MS was pioneered using brain and axonemal/cilia material because of their high content of tubulin [37; 38]. Since the carboxyl-terminal peptides are both characteristic of a specific tubulin isotype and are also the main site for posttranslational modifications, generation and purification of these peptides from tubulin has been the basis for most mass spectrometry-based characterization of tubulin across phyla [39–55]. These carboxyl-terminal peptides can be generated from purified tubulin by chemical cleavage with cyanogen bromide (CNBr) or enzymatic digestion with an endoprotease. In most studies, anion exchange enrichment of these acidic peptides was performed prior to reverse phase HPLC separation and MS. Antibodies have also been used to capture specific populations of carboxyl-terminal peptides on immuno-affinity columns thereby allowing the analysis of discrete tubulin isoforms. Frankfürter and colleagues have combined CNBr cleavage and antibody capture of C-terminal peptides to demonstrate the glutamylation and phosphorylation of βIII-tubulin from bovine brain [56].
Determination of tubulin composition is more difficult to achieve from material where tubulin is less concentrated such as cells in culture or in non-neuronal tissues. Sackett used rapid high yield purification of tubulin from cell and tissue extracts by solid-phase ion exchange chromatography [57] but the cartridges used are no longer available from the manufacturer. This method needs to be adapted to spin-columns or mini columns that are easily available. As an alternative, Taxol, which decreases the critical concentration of tubulin for polymerization, can be used to isolate assembly-competent tubulin from cell extracts obtained by sonication and ultracentrifugation [58]. We have selected the most direct analytical protocols, from sample preparation to MS, to study tubulin isotype content in cells in culture.
CNBr released C-terminal tubulin peptides
In our initial analysis of human cell lines we used CNBr to release the C-terminal tubulin fragments [59], a method that was first used to detect posttranslational modifications of βIII-tubulin from bovine brain [56] and polyglycylated tubulin from bull sperm [49]. CNBr efficiently cleaves C-terminal to methionine residues and releases peptides with a C-terminal homoserine lactone, except when a serine or threonine residue follows methionine in the amino acid sequence. Direct MS analysis of the human α- and β-tubulin CNBr-derived C-terminal peptides is facilitated because their masses are all unique and are in the 2000–4000 mass range (Table 1), and their acidic nature makes them amenable to selective MALDI-TOF-MS detection in the negative ion mode [60]. For MS analysis, total cell extracts are resolved by SDS-PAGE, transferred to nitrocellulose, and the region of the blot corresponding to tubulin (~ 50 kDa) excised and cleaved with CNBr/formic acid. A rabbit polyclonal antibody prepared against a synthetic peptide corresponding to the final 12 amino acids of human α1B-tubulin (Kα1 in old nomenclature [15], see Table1) was used to follow the release of the CNBr C-terminal fragment. Under our cleavage conditions [59], i.e. CNBr (150 mg/ml) in 70 % formic acid for 3.5 hr at room temperature, no immunoreactivity remained on the nitrocellulose membrane after the incubation period indicating a complete CNBr cleavage/release of the C- terminal peptides.
We applied this methodology to bovine brain tubulin and tubulin from total extracts from MDA-MD-231 and A549, breast and lung cancer cell lines, respectively [59]. In these human cells, three major, and several minor, molecular ions were clearly resolved. To confirm that these ions were derived from tubulin peptides, the three major ions were subjected to MS/MS. The fragmentation patterns obtained confirmed that the molecular ions with m/z values of 2860.5 and 3367.0 corresponded to the C-terminal CNBr fragments of α1B- and βI tubulin, respectively (Table 1). However, the molecular ion with a m/z value of 2590.4 did not correspond to the mass of any cloned α/β-tubulin C-terminal fragment in the databases at that time. A partial fragmentation pattern for this molecular ion was observed by MS/MS and yielded ions corresponding to the first several N-terminal amino acids of a peptide that was clearly derived from an α-tubulin. The remaining sequence of this peptide was determined by a combination of tandem MS and MALDI-TOF MS analysis of partial carboxypeptidase Y digests. The amino acid sequence of this peptide shared similarities with the corresponding region of the mouse α6 isotype, but was clearly distinct. Subsequently, the gene encoding the human α1C-tubulin (α6 in old nomenclature [15], see Table 1) was identified [61] and the deduced sequence of its C-terminal peptide was identical to the sequence we had obtained [59]. The minor ion with a m/z value of 3478.9 was tentatively identified as the βIVb isotype [59].
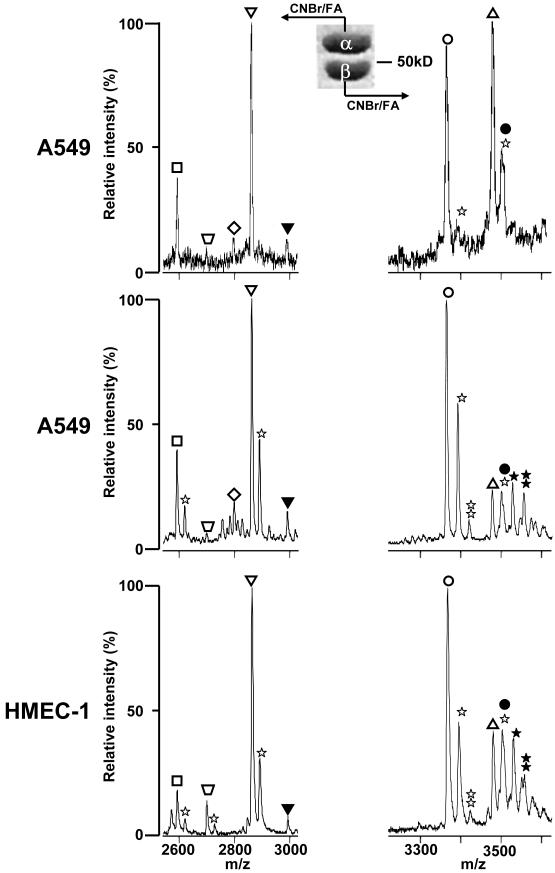
Because of ion suppression effects in MALDI-TOF MS, comparison of m/z peak intensities from two different peptides does not reflect the relative expression of the proteins from which they were derived. Ions from minor tubulin species could be suppressed by the presence of more highly abundant C-terminal tubulin peptides. In addition, peptides may have differential ionization efficiencies. Consequently, analysis of the C-terminal tubulin peptides derived from total cell extracts may underrepresent specific isotypes [59]. Therefore, we reduced the complexity of the sample prior to CNBr digestion [62] by performing a Taxol-based purification of tubulin from cell extracts followed by the separation of α-tubulin and β-tubulin by SDS-PAGE before transfer to a nitrocellulose membrane. Tubulin can be resolved into two distinct bands by 1D SDS-PAGE using SDS containing large aliphatic chain alkyl sulfate contaminants [63]. This separation of α- and β-tubulin can be improved by raising the pH of the electrophoresis running buffer to 9.5 and adding up to 6M urea to a 10% acrylamide resolving gel [49]. With mammalian tubulin, the band with the slowest electrophoretic mobility contains only α-tubulin and the fastest β-tubulin (Fig. 1). The region of the blot containing either α-tubulin or β-tubulin was treated with CNBr and MALDI-TOF MS analysis of CNBr peptides was performed in the negative ion mode (Fig. 1). These modifications were used with A549, HMEC-1 vascular endothelial cells (Fig.1) and HeLa cell extracts [64], which resulted in a major increase of the m/z peak intensity, corresponding to the βIVb-tubulin C-terminal peptide compared to that obtained from tubulin analyzed from unfractionated cell extracts [59]. Also, very small peaks of the C-terminal peptide of tyrosinated α4-tubulin were detected. Formic acid diluted to 70% was used to dissolve CNBr, and in most experiments, only minor or undetectable m/z peaks, corresponding to formylated peptides, were observed in mass spectra (Fig. 1, top panels). When formylation of serine and threonine residues was extensive (Fig. 1, middle and bottom panels), the profiles of m/z peaks of β-tubulins were more complex and resulted in a strong decrease in intensity of the CNBr C-terminal peptide from βIVb-tubulin (Fig. 1). Because of this variability, it is recommended that trifluoroacetic acid (TFA) be used instead of formic acid.
Figure 1. Mass spectrometry analysis of CNBr C-terminal peptides from α- or β-tubulins-separated by SDS-PAGE.
Tubulin was isolated from A549 or HMEC-1 cells and separated on a 10 % acrylamide gel containing 2 M urea at pH 9.5. After Coomassie staining, the gel band containing either α-tubulin (left panels) or β-tubulin (right panels) was incubated with CNBr and formic acid (FA) and analyzed by MALDI-TOF MS in the negative mode. Top and middle panels, two independent preparations of CNBr peptides from tubulin isolated from A549 cells; bottom panels, CNBr peptides from tubulin isolated from HMEC-1 cells; mono- and bi- formylated peptides are labeled by one star (+28 Da) and two stars (+56 Da), respectively; monoglutamylated peptides are labeled with filled symbols (+129 Da); peptides from the different tubulin isotypes are indicated as follow: α1C (□), detyrosinated α1B ( , −163 Da), tyrosinated α4A (◇, +163 Da), α1B (∇), βI (◯), βIVb (△). The assignment of each m/z peak for β-tubulin is an interpretation based on the CNBr C-terminal peptides of βI- and βIVb-tubulin and their monoformylated derivatives.
, −163 Da), tyrosinated α4A (◇, +163 Da), α1B (∇), βI (◯), βIVb (△). The assignment of each m/z peak for β-tubulin is an interpretation based on the CNBr C-terminal peptides of βI- and βIVb-tubulin and their monoformylated derivatives.
However, the βII, βIII and βIVa-tubulin C-terminal peptides are still undetectable with this method, whereas RT-PCR–based analysis of mRNA isolated from A549 cells, for instance, reveals the expression of these isotypes [2]. Since we could detect βII tubulin in bovine brain microtubule preparations [59], where it is the major isotype, we presumed that our inability to detect the βII-tubulin in human cancer and vascular endothelial cell lines was not a problem inherent to our MS-based method. In an attempt to detect them at the protein level, we implemented further separation of each isotype in combination with trypsin digestion or CNBr cleavage and MALDI-TOF MS.
Benefits from high resolution isoelectrofocusing of proteins
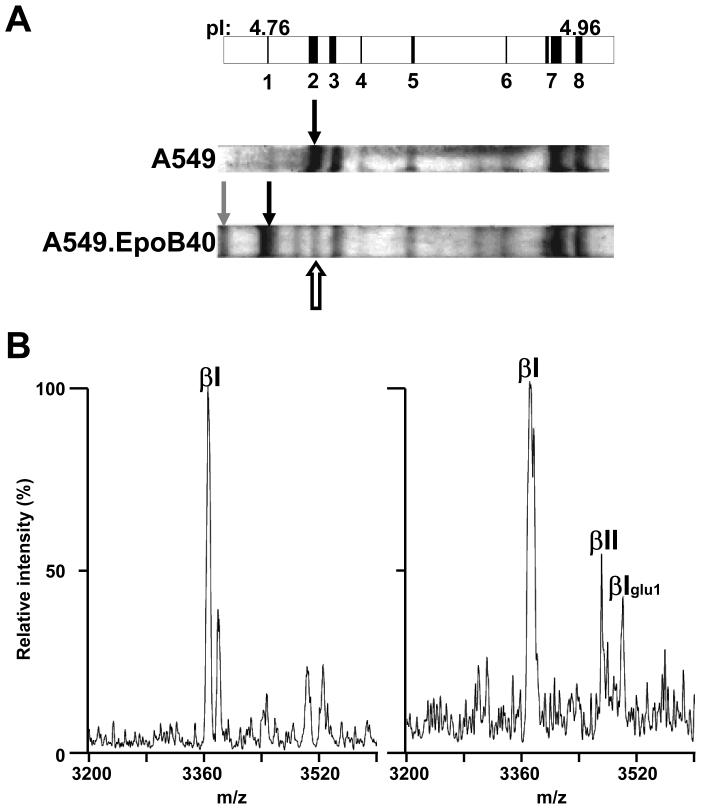
The recent technological improvements in isoelectrofocusing (IEF) and the commercial availability of precast immobilized pH gradient (IPG) gel strips were instrumental in separating tubulin isotypes. These IPG gels, up to 24 cm in length and containing a narrow pH gradient from 4.5 to 5.5, allows the resolution of tubulins that differ in pI by only 0.01 pH units. (Table 1). After polymerization in cell extracts, the Taxol-stabilized microtubules are pelleted and solubilized in IEF sample buffer prior to isoelectrofocusing. Proteins can be stained in IEF gels with imidazole and zinc salts [62] with subsequent direct processing by enzymes or chemicals. For practical reasons, we also used Coomassie staining which produces low background on IPG strips [62]. Protein bands are cut out of the strips with the plastic backing, processed for trypsin digestion [62] or CNBr cleavage [65] and analyzed by MALDI-TOF MS analysis in the positive and the negative ion mode, respectively. Mass analysis of each tryptic digest detected several isotype specific peptides that confirmed the predicted position of the individual tubulin isotypes expressed in the MDA-MB 231 and A549 cell lines [62]. In addition, the expression of βIII-tubulin was clearly demonstrated. When we combined high resolution IEF separation of tubulin isotypes and CNBr cleavage, the tubulin C-terminal peptide from each protein band corresponded to the expected tubulin isotype based on respective pI and previous tryptic peptide-based identification [65]. Moreover, the presence of monoglutamylated tubulins was also established, and human βV-tubulin, for which no antibody was available, was detected for the first time at the protein level in human cells [65]. Nevertheless, no C-terminal βIII-tubulin peptides were detected from CNBr cleavage of the band containing βIII-tubulin. Thus, we examined the MALDI-TOF MS behavior of an identical synthetic C-terminal βIII-tubulin peptide in the negative ion mode and found no problem of ionization/desorption and retention of a sufficient signal to noise ratio even at high dilutions (unpublished personal observations). We concluded that this peptide might be particularly sensitive to ion suppression. CNBr C-terminal peptides from βIVa-tubulin or tryptic peptides specific for this isotype were not detected at either of the two predicted focusing positions (Table 1), thereby calling into question previous results (2). Because βI- and βII-tubulin have the same pI, we used an A549 epothilone-resistant cell line (A549.EpoB40) that bears a mutation, Q292E, in βI-tubulin [66] that shifts its position on IEF gels to a more acidic pI (Fig 1A). A band was still visible at the position of the wild type βI-tubulin and CNBr cleavage of this band yielded a C-terminal βI-tubulin peptide and a small m/z peak that matched the (M-H) calculated value for C-terminal βII-tubulin peptide (Fig.2B, Table 1), indicating that βII-tubulin was expressed at low levels.
Figure 2. Mass spectrometry analysis of CNBr C-terminal peptides from IEF-separated tubulin isotypes from A549 and A549.EpoB40 cells.
A, Schematic representation of a portion of an IPG strip (pH 4.5–5.5) with predicted positions of tubulin isotypes depicted as vertical black bands starting with monoglutamylated βI-tubulin (band 1, pI: 4.76) and finishing with α1C-tubulin (band 8, pI: 4.96). Bands 2 to 7 represent βI-, βIVb-, monoglutamylated βIII-, βIII, monoglutamylated α1B-, tyrosinated α4A/α1B- and α1C-tubulin, respectively. On Coomassie-stained IEF gels, the black indicate the position of wild type and mutated βI-tubulin from A549 and A549.EpoB40, respectively. The white arrow and the grey arrow indicate the focalization position of βII-and monoglutamylated mutated βI-tubulin, respectively. B, the bands corresponding to mutated βI-tubulin (left panel) and βII tubulin (right panel)were cut out and the protein was cleaved by CNBr. Peptides were analyzed by MALDI-TOF MS in the negative mode and only the band expected to contain βII-tubulin produced a small m/z peak (3467.71) corresponding to the C-terminus of βII-tubulin. Another m/z peak at 3496.50 corresponded to contaminating monoglutamylated βI tubulin (βIglu1). Maximal intensity is 114 for left spectrum vs 30 for right spectrum.
In principle, the combination of IEF and MS for the analysis of tubulin isotypes from cells in culture can be applied to tissue samples. Nevertheless, there are more uncontrolled variables in tissue proteomic studies such as physiological conditions prior to death, lag-time between death and tissue processing, methods of preservation and storage, and the presence of associated fat tissue and blood vessels. We initiated a tubulin isotyping study in mouse tissues and encountered difficulties in obtaining tubulin from liver. By raising ionic strength, high concentrations of glutamate are known to promote tubulin stabilization and polymerization [67], while preventing co-purification of MAPs. Therefore, glutamate was added to the usual MES-based extraction buffer. This modification in combination with Taxol-induced polymerization increased significantly the yield of tubulin isolated from mouse liver. This modification was used in a study of tubulin isotype expression during induced tumorigenesis in rat liver by the IEF-MS approach [13]. After CNBr cleavage of an IEF band with a slightly more basic pI than βIVb-tubulin, a peak was detected at m/z of 3308.31. MS/MS analysis allowed for the assignment of this peak to the βIVb isotype with two residues proteolytically removed from the C-terminus.
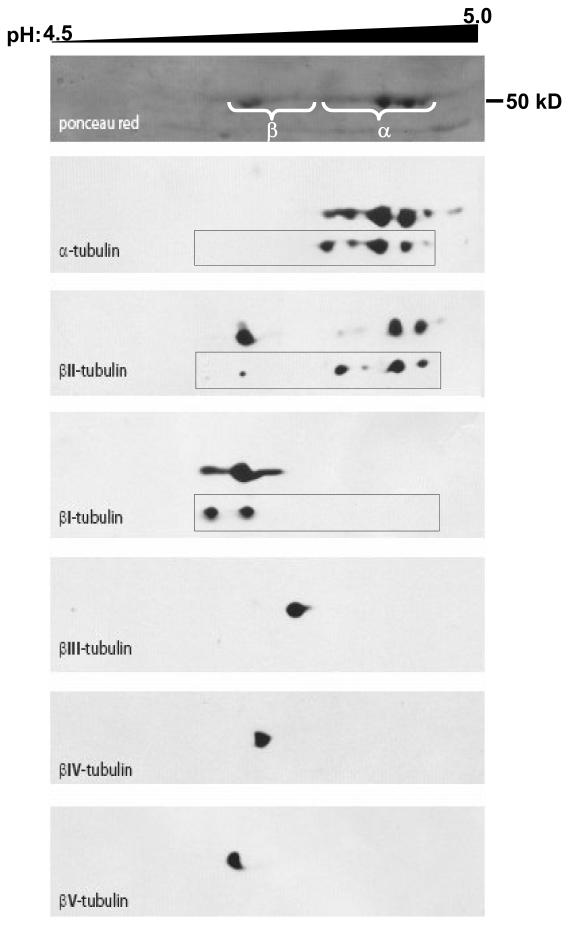
The ability to resolve and identify each tubulin isotype on high-resolution IEF gels also allowed us to evaluate the specificity of a panel of tubulin isotype-specific antibodies [62]. The anti-β-tubulin isotype monoclonal antibodies produced and characterized by R.F. Ludueña and coworkers [16] represent valuable tools that have helped to unravel some of the functional significance of the different tubulin isotypes. Based on the combination of IEF and MS described above, and after transfer of tubulin from IEF gels onto nitrocellulose, no cross-reactivity of the anti-βI, βIII-, and βIV-tubulin antibodies was observed [62]. However, the anti-βII-tubulin antibody, JDR.3B8 (antigen: EGEEDEA; [68]), and the anti-βI-tubulin antibody had identical Western blot patterns of tubulin isotypes isolated from A549.EpoB40 expressing Q292E βI-tubulin [62], demonstrating extensive cross-reactivity of this anti-βII-tubulin antibody with βI-tubulin. In order to evaluate the βII content in human immortalized endothelial cells (HMEC-1), we performed 2D electrophoresis and used another monoclonal anti-βII antibody, 7B9 (antigen: EEEEGEDEA; [69]). Although there was no cross-reactivity with βI (Fig 3) a spot was labeled with the expected pI (Table 1) and additional spots located between α-tubulin spots were labeled that may represent basic forms of β-tubulin. Such uncharacterized basic forms of β-tubulin have been described previously [70]. Note that the βII-tubulin C-terminal sequence was used as an antigen to produce JDR.3B8 and 7B9 antibodies from chicken and human βII-tubulin sequences, respectively. Similarly, C-terminal avian, rodent and human C-terminal sequences for βV-tubulin are distinct [65].We produced a polyclonal antibody that specifically binds to human βV-tubulin (Fig 3), based on our previous combined IEF-MS analysis [62], whereas R.F. Ludueña and co-workers produced a monoclonal anti-βV-tubulin, SHM.12G11, recognizing mouse but not human βV-tubulin [71]. We checked the specificities of anti-α1C- and anti-α1B-tubulin antibodies using this approach and showed that the former was specific for α1C and the latter cross-reacted with α1C [72].
Figure 3. High resolution 2D-electrophoresis tubulin isotypes in Taxol-stabilized microtubules from HMEC-1, endothelial cells.
Tubulin was isolated from HMEC-1 cells as described in the text. Tubulin isotypes were separated in the first dimension on a 24 cm IPG-strip pH 4.5–5.5 (pH 4.5–5.0 portion shown). The second dimension was run on a 10% acrylamide gel and proteins were blotted on a nitrocellulose membrane. The membrane was probed with antibodies against the indicated isotypes and stripped between each antibody. Alignment of blots was performed using the actin spots (not displayed). An anti-human βV-tubulin rabbit polyclonal antibody was produced and specifically labeled a spot on the acidic side of βI-tubulin as predicted. Insets show equivalent blots for α-, βI- and βII-tubulins in A549 cells; the most acidic spot for βI-tubulin corresponds to its monoglutamylated form.
Determination of the mass of expressed tubulin isotypes by LC-MS
Although some tubulins have identical pIs, their presence in Taxol-stabilized microtubules can be detected by electrospray ionization (ESI)–mass spectrometry [64; 65; 73]. Tubulins were first separated by reversed-phase HPLC on C4-columns using linear aqueous acetonitrile gradients containing TFA prior to in-line MS. If necessary, samples can be reduced and alkylated prior to LC and poorly soluble tubulins can be solubilized in 6 M guanidine-HCl with no change in retention time. As expected, all the masses in the range of human tubulins elute from the HPLC column at retention times corresponding to the largest total ion current peak [64; 65; 73]. After averaging the scans, we obtained overall profiles of the deconvoluted mass spectra that matched closely those of βI-, βIVb-, α1C-, tyrα4-, α1B-, glu-α1B-, βV- and βIII-tubulin (Table 1, [64; 65; 73]) from human cancer cell lines. The identity of the A549 tubulin isotypes was confirmed by tryptic mass mapping of the isotypes resolved on the C4 column. In these experiments, the column effluent was split for fraction collection and for delivery to the ion trap mass spectrometer. The total ion current was deconvoluted in one min segments across the gradient and fractions that contained the same protein masses were pooled. After removal of solvents, tryptic digestion was performed and the resulting peptides were analyzed by MALDI-TOF MS and tubulin isotype-specific tryptic peptides were identified [64; 65; 73]. No βII or βIVa isotype-specific tryptic peaks were observed during these mass mapping experiments, but βIII tubulin and its monoglutamylated isoform were detected.
In addition, the search for tubulin mutations in cell lines selected for resistance to microtubule-stabilizing drugs, such as Taxol and the epothilones, has been largely restricted to βI-tubulin. However, mutations in α1B-tubulin in some drug resistant cell lines also occur [72; 73]. Moreover, βIVb- and α1C-tubulins are present at significant levels in many of the cell lines examined [59; 62; 64; 65; 73]. Although technically feasible, the complete sequencing of the multiple tubulin transcripts in human cell lines would be a lengthy and difficult task partly because they are so highly conserved. Knowing whether mutations that occur to a single tubulin allele are expressed, and to what levels, is important since the ratio of wild-type to mutant tubulin should influence the level of resistance. A major advantage of the LC/ESI-MS method is that it can be used directly to analyze mutations that occur to tubulin isotypes in drug-resistant cell lines [64; 73]. In a series of Taxol- and epothilone-resistant human cell lines that harbor heterozygous mutations in either βI and/or α1B-tubulins, the expression of the mutant tubulins was clearly observed at levels that appeared higher than the wild-type isotype [64; 73]. Using an ion trap mass spectrometer in these studies, we were able to discriminate wild-type and mutant βI-tubulins that differ in mass by 26 daltons. Higher resolution mass spectrometers should be better able to discriminate expression of mutant tubulins.
Mass spectrometry-based quantitation of tubulin
Differential expression of α and β-tubulin isotypes has been proposed as a potential mechanism of resistance towards microtubule-targeting drugs. The level of β-tubulin mRNA is regulated by the size of the pool of free tubulin dimers [36], itself dependent on the levels of expression of associated proteins such as stathmin [2]. Therefore, quantitation of tubulin protein expression is also important to assess the status of the microtubule cytoskeleton in cells. Because of different ionization potentials of peptides, ion suppression effects and variation in response by detectors, mass spectra cannot be used directly to determine the amount of tubulin between different samples or the relative expression of tubulin isotypes in a given sample. Nevertheless, in the past few years several labeling methods have been introduced that allow investigators to quantify relative protein levels between biological samples and absolute levels in individual samples (see ref [74] for review).
In our tubulin proteomics program, we have quantified the relative tubulin expression levels in human cell lines using stable isotope labeling by amino acids in cell culture (SILAC) [75; 76] to differentially label proteins in Taxol-sensitive and resistant cell lines or Taxol-treated vs non-treated cells (Fig 4). In the SILAC method, two cell lines are cultured for several generations in media containing either a “heavy” essential amino acid (e. g., 2H n=3-Leu or 13C n=6-Arg) or its normal “light” counterpart. After five doublings in the presence of the “heavy” medium, the majority of the “light” tubulin has been replaced by its “heavy” counterpart. For relative quantitation of protein expression levels, equal amounts of total cytosolic proteins from cell lines grown in “light” medium and “heavy” medium are mixed, separated by electrophoresis and digested by trypsin, prior to MALDI-TOF MS. In the case of the leucine labeling protocol, for example, leucine-containing tryptic peptides will be represented by two isotopic envelopes separated by a multiple of 3 Da in the mass spectra, depending on the number of leucine residues in the peptide. Since both heavy and light tryptic peptides are chemically equivalent, the ratio of signal intensities for each peptide pair is a reflection of the relative protein levels in the two samples.
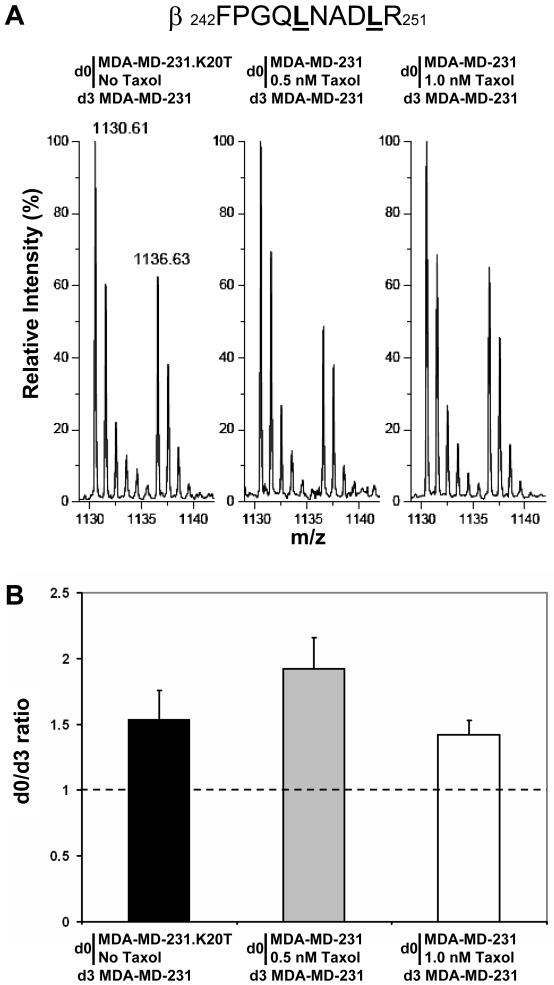
Figure 4. Relative quantitation of total tubulin expression by SILAC.
A, relative intensities of the m/z peaks of β-tubulin peptide β242-251 were obtained from SILAC experiments using d0- or d3-Leu in cell culture medium. The two leucine residues present in the β242-251 peptide are underlined and a difference of 6 Da is observed between the d0-β242-251 and d3-β242-251 m/z peaks. B, relative quantitation of total β-tubulin expression obtained after deisotoping of m/z peaks and averaging of relative intensities of m/z peaks in spectra from different β-tubulin peptides containing Leu. Tubulin was isolated and analyzed by MALDI-TOF MS from the following experiments as indicated on the top of mass spectra in A and under each bar of the graph in B: MDA-MD-231.K20T cells were cultured in the presence of d0-Leu after removal of normal culture medium containing 20 nM Taxol and one passage in Taxol-free medium. MDA-MD-231 cells were cultured in the presence of d3-Leu; MDA-MD-231 cells were maintained in the presence of d0-Leu and treated with 0.5 nM Taxol for 24 h, and untreated MDA-MD-231 cells were maintained in the presence of d3-Leu; MDA-MD-231 cells were maintained in the presence of d0-Leu and treated with 1.0 nM Taxol for 24 h, and untreated MDA-MD-231 cells were maintained in the presence of d3-Leu.
After labeling cells with either d3- or d0-Leu, equivalent amounts of total protein from each corresponding cytosolic extract are mixed and tubulins are isolated by Taxol-induced polymerization and resolved by 1D SDS-PAGE for relative quantitation of total α- and β-tubulin levels (Fig 4). For relative quantitation of individual tubulin isotype levels, samples are separated by IEF and gel bands containing a particular isotype analyzed by tryptic mass mapping [65]. This approach allowed us to demonstrate that βV-tubulin is specifically and significantly overexpressed in Hey, an ovarian cancer cell line, compared to A549 cells [65]. It showed also that low concentrations of Taxol were able to increase the level of total tubulin and that this increase in expression was conserved in resistant cells selected with Taxol. This trend was confirmed by SILAC experiments with two other Taxol-resistant cell lines, A549.AT12 and A549.AT24, which express respectively 1.98 ± 0.21 and 2.11 ± 0.35 fold more tubulin compared to A549 cells.
For relative specific quantitation of a tubulin species in a tissue such as rat liver, we used 15N-labeled tubulin synthetic peptides as standards [13]. An internal CNBr peptide, common to all rat β-tubulins, was synthesized with two additional C-terminal residues following the methionine such that CNBr fragmentation of this standard peptide would lead to formation of homoserine lactone identical to that resulting from the cleavage of endogenous β-tubulin in the sample. The other standard was the CNBr C-terminal peptide corresponding to βIVb-tubulin lacking the last two residues; a modification of βIVb-tubulin found in rat liver tumors [13]. Standard peptides were added to the SDS-PAGE gel piece containing tubulin prior to CNBr cleavage. The relative intensities between m/z peaks from light sample peptides and corresponding heavy standard peptides in MALDI-TOF MS spectra in the negative ion mode were calculated. The use of the internal β-tubulin peptide for normalization of total β-tubulin levels among samples determined that the C-terminal dipeptidic deletion form of βIVb-tubulin was increased by 3-fold in precancerous and tumor bearing livers compared to livers from control rats unchallenged by carcinogens [13].
Despite the undeniable advantage of highthroughput flexibility offered by antibody- based quantitation of tubulin isotype expression, the methodology is contingent on proper validation, limited multiplexing, compatibility with fixation procedure and careful determination of the signal dynamic range that depends on the antibody itself and the detection method used. Indeed, detection of differences in expression between 1- and 2-fold as revealed by SILAC (Fig 4) would be difficult to assess by Western blotting. Mass spectrometry-based relative quantitation is useful when no specific antibody is yet available for a newly discovered tubulin species [13; 65].
Use and development of tubulin proteomics
Collectively, high resolution IEF-MS and LC-MS determination of pI and MW of human tubulin isotypes, respectively, constitute a bi-dimensional analysis that validates which sequences are expressed at the protein level. Nevertheless, each of these approaches has its limitations and some tubulin species may be below the detection level. Taxol-dependent polymerization does not selectively enrich for specific β-tubulin isotypes [62], but it may exclude from the analysis a non polymerizable, but not necessarily denatured, pool of tubulin in the cell material used. Even at the highest available resolution of tubulin isotypes by IEF, incomplete focusing of tubulin isotypes cannot be avoided (Fig. 2 and 3). This can be viewed as and advantage as the major C-terminal peptide can provide internal calibration. Pursuit of functional studies on tubulin posttranslational modifications such as polyglutamylation [8] and detection of less well characterized modifications such as phosphorylations [56; 77], oxidation [78], nitrosylation [79], sumoylation [80] and proteolysis [81] will require the use of mass spectrometry. Importantly, statistical methods for the analysis of complex mass spectra [82] and the development of relative or absolute quantitation using isotopically labeled synthetic peptides as standards will help in the detection of differential expression of tubulins in tissues.
The impact of tubulin proteomics in the clinic is still limited but it may be important for the discovery and validation of tubulin-based biomarkers. Changes in tubulin isotype expression profiles, including posttranslational modifications may be signs of physiological alterations associated with disease progression or an indicator of a risk for an illness that could be prevented [9–13]. Based on immunohistochemistry, the increase in βIII-tubulin expression in tumors has been associated with resistance to drug treatment and with poor prognosis [83]. Mass spectrometry has qualitatively confirmed the specificity of antibodies against βIII-tubulin, but quantitative mass spectrometry-based approaches could be used also to demonstrate that the observed increase in βIII-tubulin is specific for this isotype. It is likely that the association between antibody-based isolation of a particular isotype, such as βIII, and mass spectrometry-based analysis of posttranslational modifications will reveal the full potential of tubulin as a useful biomarker in cancer, neurological disorders or other diseases.
Acknowledgment of financial support
Supported by ANR-05-BLAN-SPV00551 (to D.L) and INCa-Cancéropôle PACA 2003 (Plateforme Protéomique Timone; to D.B) and NIH Grants, CA077263, CA124898, and the National Foundation for Cancer Research (to S.B.H.) and CA110150 (to R.H.A). E. P. is supported by a Cancer Institute New South Wales “Early Career Development” Fellowship. L.M.M. is supported by an NIH postdoctoral fellowship (CA125923).
Footnotes
This manuscript is dedicated to the memory of our esteemed colleague, Professor George A. Orr, whose ideas formed the foundation of this research. His creativity and dedication continue to inspire our work in tubulin proteomics.
Abbreviations. CID/PSD: collision induced dissociation/post source decay; CNBr: cyanogen bromide; d3- or d0-Leu: 2H n=3 Leu or H n=3 Leu; 1D or 2D: mono- or bi-dimensional; ESI: electrospray ionization; FA: formic acid; FT ICR: Fourier transform ion cyclotron resonance; IEF: isoelectrofocusing; IPG: immobilized pH gradients; LC: liquid chromatography; MALDI-TOF: matrix-assisted laser desorption ionization-time of flight; MAPs: microtubule associated proteins; MES: 4-morpholineethanesulfonate; MKs: megakaryotypes; MT: microtubule; pI: pH at isoelectric point; QTOF: quadripole-time of flight; SILAC: stable isotope labeling with amino acid in culture; SLPI : secretory leukocyte protease inhibitor; TFA: trifluoroacetic acid; TTL : tubulin tyrosine ligase; TTLL: TTL-like.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Honoré S, Pasquier E, Braguer D. Understanding microtubule dynamics for improved cancer therapy. Cell Mol Life Sci. 2005;62:3039–56. doi: 10.1007/s00018-005-5330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–95. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- [4].Bonnet C, Boucher D, Lazereg S, Pedrotti B, Islam K, Denoulet P, Larcher JC. Differential binding regulation of microtubule-associated proteins MAP1A, MAP1B, and MAP2 by tubulin polyglutamylation. J Biol Chem. 2001;276:12839–48. doi: 10.1074/jbc.M011380200. [DOI] [PubMed] [Google Scholar]
- [5].Boucher D, Larcher JC, Gros F, Denoulet P. Polyglutamylation of tubulin as a progressive regulator of in vitro interactions between the microtubule-associated protein Tau and tubulin. Biochemistry. 1994;33:12471–7. doi: 10.1021/bi00207a014. [DOI] [PubMed] [Google Scholar]
- [6].Larcher JC, Boucher D, Lazereg S, Gros F, Denoulet P. Interaction of kinesin motor domains with alpha- and beta-tubulin subunits at a tau-independent binding site. Regulation by polyglutamylation. J Biol Chem. 1996;271:22117–24. doi: 10.1074/jbc.271.36.22117. [DOI] [PubMed] [Google Scholar]
- [7].Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, Schweitzer A, Saoudi Y, Pointu H, Bosc C, Salin PA, Job D, Wehland J. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc Natl Acad Sci U S A. 2005;102:7853–8. doi: 10.1073/pnas.0409626102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, Campbell PK, Yuasa S, Janke C, Macgregor GR, Setou M. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci U S A. 2007;104:3213–8. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Katsetos CD, Herman MM, Mork SJ. Class III beta-tubulin in human development and cancer. Cell Motil Cytoskeleton. 2003;55:77–96. doi: 10.1002/cm.10116. [DOI] [PubMed] [Google Scholar]
- [10].Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mialhe A, Lafanechère L, Treilleux I, Peloux N, Dumontet C, Bremond A, Panh MH, Payan R, Wehland J, Margolis RL, Job D. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 2001;61:5024–7. [PubMed] [Google Scholar]
- [12].Freson K, De Vos R, Wittevrongel C, Thys C, Defoor J, Vanhees L, Vermylen J, Peerlinck K, Van Geet C. The TUBB1 Q43P functional polymorphism reduces the risk of cardiovascular disease in men by modulating platelet function and structure. Blood. 2005;106:2356–62. doi: 10.1182/blood-2005-02-0723. [DOI] [PubMed] [Google Scholar]
- [13].Miller LM, Menthena A, Chatterjee C, Verdier-Pinard P, Novikoff PM, Horwitz SB, Angeletti RH. Increased levels of a unique post-translationally modified βIVb-tubulin isotype in liver cancer. Biochemistry. 2008;47:7572–7582. doi: 10.1021/bi8005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nogales E. Structural insight into microtubule function. Annu Rev Biophys Biomol Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- [15].Khodiyar VK, Maltais LJ, Sneddon KM, Smith JR, Shimoyama M, Cabral F, Dumontet C, Dutcher SK, Harvey RJ, Lafanechere L, Murray JM, Nogales E, Piquemal D, Stanchi F, Povey S, Lovering RC. A revised nomenclature for the human and rodent alpha-tubulin gene family. Genomics. 2007;90:285–9. doi: 10.1016/j.ygeno.2007.04.008. [DOI] [PubMed] [Google Scholar]
- [16].Luduena RF. Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol. 1998;178:207–75. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- [17].Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–47. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- [18].Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol. 1987;105:251–64. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gundersen GG, Khawaja S, Bulinski JC. Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol. 1989;109:2275–88. doi: 10.1083/jcb.109.5.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bloom K. Microtubule composition: cryptography of dynamic polymers. Proc Natl Acad Sci U S A. 2004;101:6839–40. doi: 10.1073/pnas.0401266101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–60. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- [22].Hoyle HD, Raff EC. Two Drosophila beta tubulin isoforms are not functionally equivalent. J Cell Biol. 1990;111:1009–26. doi: 10.1083/jcb.111.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Raff EC, Hutchens JA, Hoyle HD, Nielsen MG, Turner FR. Conserved axoneme symmetry altered by a component beta-tubulin. Curr Biol. 2000;10:1391–4. doi: 10.1016/s0960-9822(00)00784-3. [DOI] [PubMed] [Google Scholar]
- [24].Hutchens JA, Hoyle HD, Turner FR, Raff EC. Structurally similar Drosophila alpha-tubulins are functionally distinct in vivo. Mol Biol Cell. 1997;8:481–500. doi: 10.1091/mbc.8.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Savage C, Hamelin M, Culotti JG, Coulson A, Albertson DG, Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3:870–81. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- [26].Schwer HD, Lecine P, Tiwari S, Italiano JE, Jr., Hartwig JH, Shivdasani RA. A lineage-restricted and divergent beta-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr Biol. 2001;11:579–86. doi: 10.1016/s0960-9822(01)00153-1. [DOI] [PubMed] [Google Scholar]
- [27].Schulze H, Korpal M, Bergmeier W, Italiano JE, Jr., Wahl SM, Shivdasani RA. Interactions between the megakaryocyte/platelet-specific beta1 tubulin and the secretory leukocyte protease inhibitor SLPI suggest a role for regulated proteolysis in platelet functions. Blood. 2004;104:3949–57. doi: 10.1182/blood-2004-03-1179. [DOI] [PubMed] [Google Scholar]
- [28].Lafanechere L, Job D. The third tubulin pool. Neurochem Res. 2000;25:11–8. doi: 10.1023/a:1007575012904. [DOI] [PubMed] [Google Scholar]
- [29].Erck C, MacLeod RA, Wehland J. Cloning and genomic organization of the TTL gene on mouse chromosome 2 and human chromosome 2q13. Cytogenet Genome Res. 2003;101:47–53. doi: 10.1159/000073418. [DOI] [PubMed] [Google Scholar]
- [30].Badin-Larcon AC, Boscheron C, Soleilhac JM, Piel M, Mann C, Denarier E, Fourest-Lieuvin A, Lafanechere L, Bornens M, Job D. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc Natl Acad Sci U S A. 2004;101:5577–82. doi: 10.1073/pnas.0307917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, Wehland J, Andrieux A, Job D. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174:839–49. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008 doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- [33].Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Edde B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–62. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- [34].van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–48. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- [35].Thazhath R, Liu C, Gaertig J. Polyglycylation domain of beta-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat Cell Biol. 2002;4:256–9. doi: 10.1038/ncb764. [DOI] [PubMed] [Google Scholar]
- [36].Cleveland DW, Sullivan KF. Molecular biology and genetics of tubulin. Annu Rev Biochem. 1985;54:331–65. doi: 10.1146/annurev.bi.54.070185.001555. [DOI] [PubMed] [Google Scholar]
- [37].Stephens RE. Preparation of ciliary and flagellar remnants. Methods Cell Biol. 1995;47:361–4. doi: 10.1016/s0091-679x(08)60830-4. [DOI] [PubMed] [Google Scholar]
- [38].Shelanski ML, Gaskin F, Cantor CR. Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci U S A. 1973;70:765–8. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Redeker V, Frankfurter A, Parker SK, Rossier J, Detrich HW., 3rd Posttranslational modification of brain tubulins from the Antarctic fish Notothenia coriiceps: reduced C-terminal glutamylation correlates with efficient microtubule assembly at low temperature. Biochemistry. 2004;43:12265–74. doi: 10.1021/bi049070z. [DOI] [PubMed] [Google Scholar]
- [40].Redeker V, Le Caer JP, Rossier J, Prome JC. Structure of the polyglutamyl side chain posttranslationally added to alpha-tubulin. J Biol Chem. 1991;266:23461–6. [PubMed] [Google Scholar]
- [41].Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bre MH. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 1994;266:1688–91. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- [42].Redeker V, Levilliers N, Vinolo E, Rossier J, Jaillard D, Burnette D, Gaertig J, Bre MH. Mutations of tubulin glycylation sites reveal cross-talk between the C termini of alpha- and beta-tubulin and affect the ciliary matrix in Tetrahymena. J Biol Chem. 2005;280:596–606. doi: 10.1074/jbc.M408324200. [DOI] [PubMed] [Google Scholar]
- [43].Redeker V, Melki R, Prome D, Le Caer JP, Rossier J. Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett. 1992;313:185–92. doi: 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- [44].Redeker V, Rossier J, Frankfurter A. Posttranslational modifications of the C-terminus of alpha-tubulin in adult rat brain: alpha 4 is glutamylated at two residues. Biochemistry. 1998;37:14838–44. doi: 10.1021/bi981335k. [DOI] [PubMed] [Google Scholar]
- [45].Redeker V, Rusconi F, Mary J, Prome D, Rossier J. Structure of the C-terminal tail of alpha-tubulin: increase of heterogeneity from newborn to adult. J Neurochem. 1996;67:2104–14. doi: 10.1046/j.1471-4159.1996.67052104.x. [DOI] [PubMed] [Google Scholar]
- [46].Rudiger A, Rudiger M, Weber K, Schomburg D. Characterization of post-translational modifications of brain tubulin by matrix-assisted laser desorption/ionization mass spectrometry: direct one-step analysis of a limited subtilisin digest. Anal Biochem. 1995;224:532–7. doi: 10.1006/abio.1995.1083. [DOI] [PubMed] [Google Scholar]
- [47].Rudiger AH, Rudiger M, Carl UD, Chakraborty T, Roepstorff P, Wehland J. Affinity mass spectrometry-based approaches for the analysis of protein-protein interaction and complex mixtures of peptide-ligands. Anal Biochem. 1999;275:162–70. doi: 10.1006/abio.1999.4319. [DOI] [PubMed] [Google Scholar]
- [48].Rudiger M, Plessman U, Kloppel KD, Wehland J, Weber K. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992;308:101–5. doi: 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- [49].Rudiger M, Plessmann U, Rudiger AH, Weber K. Beta tubulin of bull sperm is polyglycylated. FEBS Lett. 1995;364:147–51. doi: 10.1016/0014-5793(95)00373-h. [DOI] [PubMed] [Google Scholar]
- [50].Rudiger M, Weber K. Characterization of the post-translational modifications in tubulin from the marginal band of avian erythrocytes. Eur J Biochem. 1993;218:107–16. doi: 10.1111/j.1432-1033.1993.tb18357.x. [DOI] [PubMed] [Google Scholar]
- [51].Edde B, Rossier J, Le Caer JP, Prome JC, Desbruyeres E, Gros F, Denoulet P. Polyglutamylated alpha-tubulin can enter the tyrosination/detyrosination cycle. Biochemistry. 1992;31:403–10. doi: 10.1021/bi00117a014. [DOI] [PubMed] [Google Scholar]
- [52].Mary J, Redeker V, Le Caer JP, Prome JC, Rossier J. Class I and IVa beta-tubulin isotypes expressed in adult mouse brain are glutamylated. FEBS Lett. 1994;353:89–94. doi: 10.1016/0014-5793(94)01018-8. [DOI] [PubMed] [Google Scholar]
- [53].Mary J, Redeker V, Le Caer JP, Rossier J, Schmitter JM. Posttranslational modifications in the C-terminal tail of axonemal tubulin from sea urchin sperm. J Biol Chem. 1996;271:9928–33. doi: 10.1074/jbc.271.17.9928. [DOI] [PubMed] [Google Scholar]
- [54].Vinh J, Langridge JI, Bre MH, Levilliers N, Redeker V, Loyaux D, Rossier J. Structural characterization by tandem mass spectrometry of the posttranslational polyglycylation of tubulin. Biochemistry. 1999;38:3133–9. doi: 10.1021/bi982304s. [DOI] [PubMed] [Google Scholar]
- [55].Vinh J, Loyaux D, Redeker V, Rossier J. Sequencing branched peptides with CID/PSD MALDI-TOF in the low-picomole range: application to the structural study of the posttranslational polyglycylation of tubulin. Anal Chem. 1997;69:3979–85. doi: 10.1021/ac970449j. [DOI] [PubMed] [Google Scholar]
- [56].Alexander JE, Hunt DF, Lee MK, Shabanowitz J, Michel H, Berlin SC, MacDonald TL, Sundberg RJ, Rebhun LI, Frankfurter A. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc Natl Acad Sci U S A. 1991;88:4685–9. doi: 10.1073/pnas.88.11.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sackett DL. Rapid purification of tubulin from tissue and tissue culture cells using solid-phase ion exchange. Anal Biochem. 1995;228:343–8. doi: 10.1006/abio.1995.1361. [DOI] [PubMed] [Google Scholar]
- [58].Vallee RB. A taxol-dependent procedure for the isolation of microtubules and microtubule-associated proteins (MAPs) J Cell Biol. 1982;92:435–42. doi: 10.1083/jcb.92.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rao S, Aberg F, Nieves E, Band Horwitz S, Orr GA. Identification by mass spectrometry of a new alpha-tubulin isotype expressed in human breast and lung carcinoma cell lines. Biochemistry. 2001;40:2096–103. doi: 10.1021/bi002323d. [DOI] [PubMed] [Google Scholar]
- [60].Jai-nhuknan J, Cassady CJ. Negative ion postsource decay time-of-flight mass spectrometry of peptides containing acidic amino acid residues. Anal Chem. 1998;70:5122–8. doi: 10.1021/ac980577n. [DOI] [PubMed] [Google Scholar]
- [61].Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Verdier-Pinard P, Wang F, Martello L, Burd B, Orr GA, Horwitz SB. Analysis of tubulin isotypes and mutations from taxol-resistant cells by combined isoelectrofocusing and mass spectrometry. Biochemistry. 2003;42:5349–57. doi: 10.1021/bi027293o. [DOI] [PubMed] [Google Scholar]
- [63].Stephens RE. Electrophoretic resolution of tubulin and tektin subunits by differential interaction with long-chain alkyl sulfates. Anal Biochem. 1998;265:356–60. doi: 10.1006/abio.1998.2909. [DOI] [PubMed] [Google Scholar]
- [64].Verdier-Pinard P, Wang F, Burd B, Angeletti RH, Horwitz SB, Orr GA. Direct analysis of tubulin expression in cancer cell lines by electrospray ionization mass spectrometry. Biochemistry. 2003;42:12019–27. doi: 10.1021/bi0350147. [DOI] [PubMed] [Google Scholar]
- [65].Verdier-Pinard P, Shahabi S, Wang F, Burd B, Xiao H, Goldberg GL, Orr GA, Horwitz SB. Detection of human betaV-tubulin expression in epithelial cancer cell lines by tubulin proteomics. Biochemistry. 2005;44:15858–70. doi: 10.1021/bi051004p. [DOI] [PubMed] [Google Scholar]
- [66].He L, Yang CP, Horwitz SB. Mutations in beta-tubulin map to domains involved in regulation of microtubule stability in epothilone-resistant cell lines. Mol Cancer Ther. 2001;1:3–10. [PubMed] [Google Scholar]
- [67].Hamel E, Lin CM. Glutamate-induced polymerization of tubulin: characteristics of the reaction and application to the large-scale purification of tubulin. Arch Biochem Biophys. 1981;209:29–40. doi: 10.1016/0003-9861(81)90253-8. [DOI] [PubMed] [Google Scholar]
- [68].Banerjee A, Roach MC, Wall KA, Lopata MA, Cleveland DW, Luduena RF. A monoclonal antibody against the type II isotype of beta-tubulin. Preparation of isotypically altered tubulin. J Biol Chem. 1988;263:3029–34. [PubMed] [Google Scholar]
- [69].Moody SA, Miller V, Spanos A, Frankfurter A. Developmental expression of a neuron-specific beta-tubulin in frog (Xenopus laevis): a marker for growing axons during the embryonic period. J Comp Neurol. 1996;364:219–30. doi: 10.1002/(SICI)1096-9861(19960108)364:2<219::AID-CNE3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- [70].Verrills NM, Walsh BJ, Cobon GS, Hains PG, Kavallaris M. Proteome analysis of vinca alkaloid response and resistance in acute lymphoblastic leukemia reveals novel cytoskeletal alterations. J Biol Chem. 2003;278:45082–93. doi: 10.1074/jbc.M303378200. [DOI] [PubMed] [Google Scholar]
- [71].Banerjee A, Jensen-Smith H, Lazzell A, Prasad V, Elguezabal G, Hallworth R, Luduena RF. Localization of betav tubulin in the cochlea and cultured cells with a novel monoclonal antibody. Cell Motil Cytoskeleton. 2008;65:505–14. doi: 10.1002/cm.20280. [DOI] [PubMed] [Google Scholar]
- [72].Martello LA, Verdier-Pinard P, Shen HJ, He L, Torres K, Orr GA, Horwitz SB. Elevated levels of microtubule destabilizing factors in a Taxol-resistant/dependent A549 cell line with an alpha-tubulin mutation. Cancer Res. 2003;63:1207–13. [PubMed] [Google Scholar]
- [73].Yang CP, Verdier-Pinard P, Wang F, Lippaine-Horvath E, He L, Li D, Hofle G, Ojima I, Orr GA, Horwitz SB. A highly epothilone B-resistant A549 cell line with mutations in tubulin that confer drug dependence. Mol Cancer Ther. 2005;4:987–95. doi: 10.1158/1535-7163.MCT-05-0024. [DOI] [PubMed] [Google Scholar]
- [74].Fenselau C. A review of quantitative methods for proteomic studies. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:14–20. doi: 10.1016/j.jchromb.2006.10.071. [DOI] [PubMed] [Google Scholar]
- [75].Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–8. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- [76].Ong SE, Mann M. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol Biol. 2007;359:37–52. doi: 10.1007/978-1-59745-255-7_3. [DOI] [PubMed] [Google Scholar]
- [77].Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Mol Biol Cell. 2006;17:1041–50. doi: 10.1091/mbc.E05-07-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pamplona R, Dalfo E, Ayala V, Bellmunt MJ, Prat J, Ferrer I, Portero-Otin M. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J Biol Chem. 2005;280:21522–30. doi: 10.1074/jbc.M502255200. [DOI] [PubMed] [Google Scholar]
- [79].Lefievre L, Chen Y, Conner SJ, Scott JL, Publicover SJ, Ford WC, Barratt CL. Human spermatozoa contain multiple targets for protein S-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics. 2007;7:3066–84. doi: 10.1002/pmic.200700254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Adrain C, Duriez PJ, Brumatti G, Delivani P, Martin SJ. The cytotoxic lymphocyte protease, granzyme B, targets the cytoskeleton and perturbs microtubule polymerization dynamics. J Biol Chem. 2006;281:8118–25. doi: 10.1074/jbc.M509361200. [DOI] [PubMed] [Google Scholar]
- [82].Olson MT, Blank PS, Sackett DL, Yergey AL. Evaluating reproducibility and similarity of mass and intensity data in complex spectra--applications to tubulin. J Am Soc Mass Spectrom. 2008;19:367–74. doi: 10.1016/j.jasms.2007.11.012. [DOI] [PubMed] [Google Scholar]
- [83].Seve P, Dumontet C. Is class III beta-tubulin a predictive factor in patients receiving tubulin-binding agents? Lancet Oncol. 2008;9:168–75. doi: 10.1016/S1470-2045(08)70029-9. [DOI] [PubMed] [Google Scholar]