Abstract
Recent genomic profiling of childhood acute lymphoblastic leukemia (ALL) identified a novel high-risk subtype with a gene expression signature resembling Philadelphia chromosome-positive ALL and a poor prognosis (Ph-like ALL). However, the role of inherited genetic variation in Ph-like ALL pathogenesis remains unknown. In a genome-wide association study (GWAS) of 511 ALL cases and 6,661 non-ALL controls, we identified a single susceptibility locus for Ph-like ALL (GATA3, rs3824662, P=2.17×10−14, odds ratio [OR]=3.85, for Ph-like ALL vs. non-ALL; P=1.05×10−8, OR=3.25, for Ph-like ALL vs. non-Ph-like ALL) that was independently validated. The rs3824662 risk allele was associated with somatic lesions underlying Ph-like ALL (i.e., CRLF2 rearrangement, JAK mutation, and IKZF1 deletion) and directly influenced GATA3 transcription. Finally, GATA3 SNP genotype was also associated with early treatment response and the risk of ALL relapse. Our results provide insights into interactions between host and tumor genomes and their importance in ALL pathogenesis and prognosis.
Progressive intensification and risk-adapted chemotherapy have improved the 5-year survival rate of childhood ALL to over 85% in most developed countries1. However, prognosis remains poor for approximately 20% of patients with high-risk features (e.g., older age and higher leukocyte count at diagnosis, Philadelphia chromosome-positive [Ph+] ALL)2-5 .
Recent genomic profiling studies have revealed the remarkable heterogeneity of childhood ALL with more granular classification of molecular subtypes. Up to 15% of childhood B-lineage ALL cases exhibit a gene expression signature similar to that of Ph+ ALL6-9. Defined by this common expression profile, the “Ph-like” ALL subtype has a range of structural genetic alterations in the tumor genome that activate lymphoid development, cytokine receptor, and kinase signaling pathways. Ph-like ALL commonly harbors somatic IKZF1 deletion or mutation6,9. Up to 50% of Ph-like ALL cases carry CRLF2 rearrangements, with concurrent JAK mutations in approximately half of CRLF2-related cases8,10. Ph-like ALL cases without CRLF2 alterations harbor a range of genomic lesions targeting cytokine receptors and tyrosine kinases7. Importantly, Ph-like ALL is associated with a high risk of relapse 6,8,11.
GWAS have identified germline single nucleotide polymorphisms (SNPs) in ARID5B, IKZF1, CEBPE, PIP4K2A, and CDKN2A/CDKN2B that strongly influence susceptibility to childhood ALL12-15. In fact, children carrying the ARID5B variants not only are more likely to develop ALL in general, but are at a particularly high risk of having hyperdiploid ALL14,16,17, implying interactions between inherited and acquired genetic variations during leukemogenesis. Similarly in myeloproliferative neoplasms, germline variation at the JAK2 locus was linked to somatic JAK2V617F mutation18-20. Together, these observations indicate that both germline and somatic genetic variations play critical roles in tumor pathogenesis.
To this end, we conducted a GWAS of Ph-like ALL to identify germline genetic variants related to susceptibility to this ALL subtype, and to evaluate their association with somatic lesions underlying Ph-like ALL and with the risk of relapse.
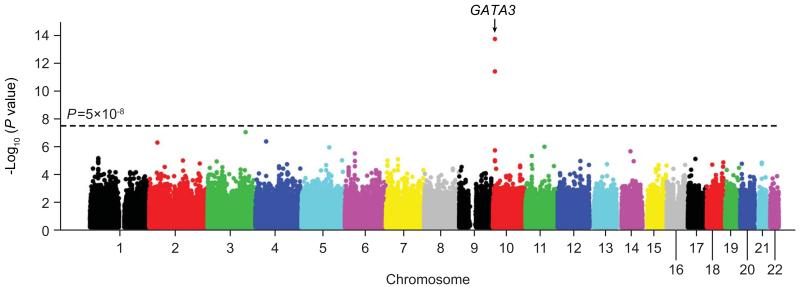
In the discovery GWAS, we compared genotype frequency at 718,890 SNPs between 75 children with Ph-like ALL from the Children’s Oncology Group (COG) AALL0232 cohort and 6,661 non-ALL controls (Supplementary Fig.1). After adjusting for genetic ancestry, two SNPs at 10p14 within the GATA3 gene reached genome-wide significance: rs3824662 (P=2.17×10−14, OR=3.85 [95%CI, 2.71 to 5.47]) and rs3781093 (P=4.94×10−12, OR=3.45 [2.42 to 4.93], Table 1 and Fig. 1). These two SNPs were in strong linkage disequilibrium (LD, r2=0.94, D’=1 in HapMap CEU, Supplementary Fig. 2), representing a single susceptibility locus. The A allele at rs3824662 and the C allele at rs3781093 were over-represented in Ph-like ALL, conferring increased disease risk across ethnicity (Table 1 and Supplementary Fig. 3). We next performed a second GWAS comparing children in the COG AALL0232 cohort who had the Ph-like expression profile (N=75) with those who did not have the Ph-like profile (“non-Ph-like”, N=436). After adjusting for genetic ancestry, the same GATA3 SNPs, rs3824662 and rs3781093, exhibited the strongest association across the genome (P=1.05×10−8, OR=3.25 [2.16 to 4.89], and P=2.62×10−7, OR=2.89 [1.92 to 4.34], respectively, Table 1 and Supplementary Figs. 3 and 4). Imputation of genotypes at 37,493 additional SNPs at this locus (chr10: 60,523 to 10,060,447) did not reveal any variants with a stronger association with Ph-like ALL than the original GWAS hits (Supplementary Fig. 5).
Table 1. Association of GATA3 SNPs with Ph-like ALL in the discovery GWAS and replication cohorts1.
| Chr | Position2 | SNP | Alleles3 | Cohort4 | Risk allele frequency (total number of subjects) |
Ph-like ALL vs. non-ALL | Ph-like ALL vs. non-Ph-like ALL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ph-like ALL | non-Ph-like ALL | non-ALL5 | P-value5 | OR (95%, CI)5 | P-value5 | OR (95%, CI)5 | |||||
| 10 | 8144214 | rs3824662 | A/C | Discovery | 58% (75) | 29% (436) | 20% (6,661)−14 | 2.17×10 | 3.85 (2.71-5.47) | 1.05×10−8 | 3.25 (2.16-4.89) |
| Replication | 50% (32) | 30% (139) | 18% (5,755) | 3.69×10−5 | 3.14 (1.18-5.44) | 0.01 | 2.16 (1.18-3.97) | ||||
| 10 | 8141933 | rs3781093 | C/T | Discovery | 52% (75) | 26% (436) | 22% (6,661) | 4.94×10−12 | 3.45 (2.42-4.93) | 2.62×10−7 | 2.89 (1.92-4.34) |
| Replication | 46% (32) | 25% (139) | 18% (5,755) | 0.0001 | 2.95 (1.68-5.16) | 0.004 | 2.55 (1.33-4.88) | ||||
Abbreviations: Chr, chromosome; OR, odds ratio; CI, confidence interval.
Association of SNP genotype and Ph-like ALL was evaluated by comparing allele frequency between Ph-like ALL and non-ALL, also between Ph-like ALL and non-Ph-like ALL, after adjusting for genetic ancestry.
Chromosomal locations are based on hg18.
Bold indicates risk allele for Ph-like ALL.
Discovery cohort: COG AALL0232, Replication cohort: COG P9906.
P-values were estimated by the logistic regression test and OR represents the increase in risk of developing Ph-like ALL for each copy of the risk allele compared with subjects who don't carry the risk allele.
Figure 1. Genome wide association study (GWAS) of the susceptibility of Ph-like ALL.
The association between genotype and Ph-like ALL was evaluated using logistic regression model for 718,890 SNPs in 75 Ph-like ALL and 6,661 non-ALL controls. P-values (−log 10 P, y axis) were plotted against respective chromosomal position of each SNP (x axis). The blue horizontal line indicates the genome-wide significant threshold (P<5×10−8). Gene symbol was indicated for the GATA3 locus at 10p14.
To validate the association of GATA3 SNPs with Ph-like ALL, we then genotyped rs3824662 and rs3781093 in 171 children with B-ALL enrolled in the COG P9906 study and in an independent cohort of 5,755 non-ALL controls. In this replication analysis, risk alleles at both GATA3 SNPs were consistently over-represented in Ph-like ALL (N=32) compared to non-ALL controls: rs3824662 (P=3.69×10−5, OR=3.14, [1.18 to 5.44]), and rs3781093 (P=0.0001, OR=2.95 [1.68 to 5.16]), or compared to non-Ph-like ALL (N=139): rs3824662 (P=0.01, OR=2.16 [1.18 to 3.97]) and rs3781093 (P=0.004, OR=2.55 [1.33 to 4.88], Table 1).
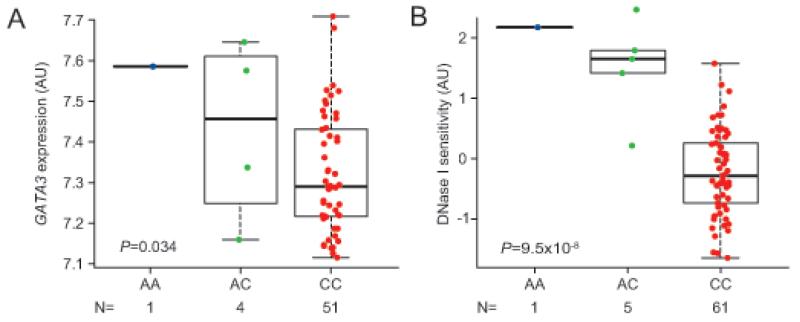
To explore the functions of these germline GATA3 variants, we first examined the relationships between rs3824662 SNP genotype and GATA3 mRNA expression. In lymphoblastoid cell lines, rs3824662 A allele was associated with significantly increased GATA3 mRNA level (HapMap YRI, N=56, P=0.034, Fig. 2A; CEU and MEX, Supplementary Fig. 6). Consistently, the A allele was also linked to higher levels of DNase hypersensitivity at this locus (HapMap YRI, N=67, P=9.5×10−8, Fig. 2B), indicating its influence on local chromatin accessibility and transcriptional activity. Association of germline GATA3 SNP genotype and GATA3 expression was confirmed in ALL blasts in both COG AALL0232 and COG P9906 cohorts (N=511, P=9.2×10−8 and N=173, P=3.6×10−6, respectively, Supplementary Fig. 7). Interestingly, ectopic overexpression of GATA3 in ALL cell lines consistently led to global changes in gene expression pattern, with a highly significant enrichment of genes within the Ph-like ALL expression signature (UOCB1 cell line, P=0.0004; Nalm6 cell line, P=0.001, Supplementary Fig. 8).
Figure 2. rs3824662 as a cis-acting regulatory element of GATA3 transcription.
GATA3 SNP rs3824662 risk allele (the A allele) was associated with higher GATA3 mRNA in 56 unrelated lymphoblastoid cell lines from HapMap population (YRI) (A), and was related to increased DNase hypersensitivity (higher transcription activity) in 67 unrelated HapMap cell lines (YRI). GATA3 expression and DNase hypersensitivity at this locus were obtained from previously published datasets30,31. (B). Genotype-expression association and genotype-DNase hypersensitivity association was evaluated using a linear regression model, adjusting genetic ancestry as appropriate. AU, arbitrary unit. Boxes include data between the twenty-fifth and the seventy-fifth percentiles.
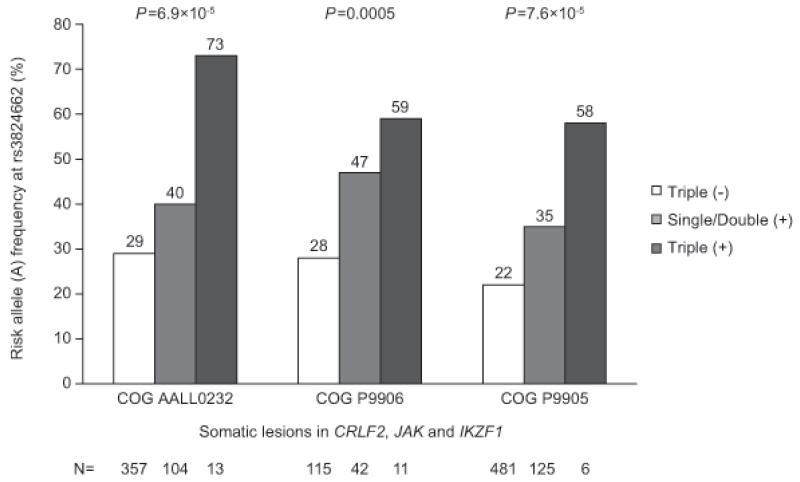
Recurrent genomic lesions targeting lymphoid development, cytokine receptor, and tyrosine kinase signaling are a hallmark of Ph-like ALL. In both COG AALL0232 and COG P9906, the GATA3 SNP rs3824662 was associated with CRLF2 lesion, JAK mutation, and IKZF1 deletion, which was also validated in a third cohort of 781 children enrolled on the COG P9905 protocol (Table 2). The A risk allele at rs3824662 was further enriched among patients with multiple “Ph-like ALL related” somatic lesions. In COG AALL0232, the frequency of the rs3824662 A allele was highest (73%) in ALL cases with CRLF2 lesion, JAK mutation, and IKZF1 deletion simultaneously, followed by patients with one or two of lesions (40%), and lowest (29%) among patients without any of the three lesions (P=6.09×10−5, Fig. 3). This correlation was also validated in the COG P9906 cohort (P=0.0005) and in the COG P9905 cohort (P=7.6×10−5, Fig. 3). Within Ph-like ALL, there was a trend that rs3824662 A allele was over-represented in cases with CRLF2 lesions (P=0.05, Supplementary Fig. 9). However, the association of rs3824662 with Ph-likeness remained significant within ALL cases that were negative for CRLF2 alterations (P=8.8×10−5, Supplementary Fig. 9), JAK mutation (P=2.1×10−5), or IKZF1 deletion (P=0.001), and in a multivariate model after adjusting for all three lesions (P=0.001).
Table 2. Association of GATA3 SNP rs3824662 with somatic lesions in CRLF2, JAK and IKZF11.
| COG AALL0232 | COG P9906 | COG P9905 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Genomic lesions |
Subjects with lesion (RAF) |
Subjects without lesion (RAF) |
P-value2 | OR (95% CI)2 | Subjects with lesion (RAF) |
Subjects without lesion (RAF) |
P-value2 | OR (95% CI)2 | Subjects with lesion (RAF) |
Subjects without lesion (RAF) |
P-value2 | OR (95% CI)2 |
|
| ||||||||||||
|
CRLF2 genomic
lesion |
39 (62%) | 472 (30%) | 9.64×10−7 | 3.68 (2.18-6.23) | 23 (50%) | 150 (32%) | 0.08 | 1.87 (0.99-3.87) | 66 (34%) | 711 (24%) | 0.009 | 1.70 (1.13-2.57) |
| JAK mutation | 23 (67%) | 488 (31%) | 2.06×10−5 | 4.31 (2.18-8.52) | 18 (58%) | 155 (32%) | 0.008 | 2.89 (1.29-6.44) | 21 (38%) | 610 (25%) | 0.05 | 1.89 (0.96-3.71) |
| IKZF1 deletion | 84 (45%) | 371 (30%) | 0.00026 | 1.97 (1.36-2.86) | 45 (50%) | 122 (30%) | 0.002 | 2.30 (1.23-3.96) | 89 (37%) | 519 (23%) | 0.0002 | 1.94 (1.35-2.78) |
Abbreviations; RAF, risk allele frequency (i.e., allele A at rs3824662); OR, odds ratio; CI, confidence interval.
Association of SNP genotype with CRLF2 genomic lesions (either IGH@-CRLF2 or P2RY8-CRLF2), JAK mutation (JAK1, JAK2 or JAK3) or IKZF1 deletion was evaluated by comparing allele frequency between ALL cases with vs. without the specific lesion or mutation, after adjusting for genetic ancestry.
P-values were estimated by logistic regression test and OR represents the increase in the likelihood of CRLF2, JAK or IKZF1 somatic lesions/mutations for each copy of the A allele compared with subjects who don't carry the A allele at this SNP.
Figure 3. GATA3 SNP rs3824662 risk allele frequency and the constellation of multiple “Ph-like ALL related” genomic lesions (CRLF2 lesion, JAK mutation, and IKZF1 deletion).
Patients in COG AALL0232, COG P9906, and COG P9905 cohorts were grouped as triple positive, double positive, single positive and triple negative based on their status for somatic CRLF2 lesion, JAK mutation, and IKZF1 deletion. Risk (A) allele frequency at rs3824662 was highest in patients carrying all three lesions and lowest in patients carrying no lesions at these three genes, with a positive correlation between A allele frequency and the cumulative number of lesions in three cohorts (P=6.9×10−5, P=0.0005 and P=7.6×10−5, respectively), as determined by the ordinal regression test adjusting genetic ancestry.
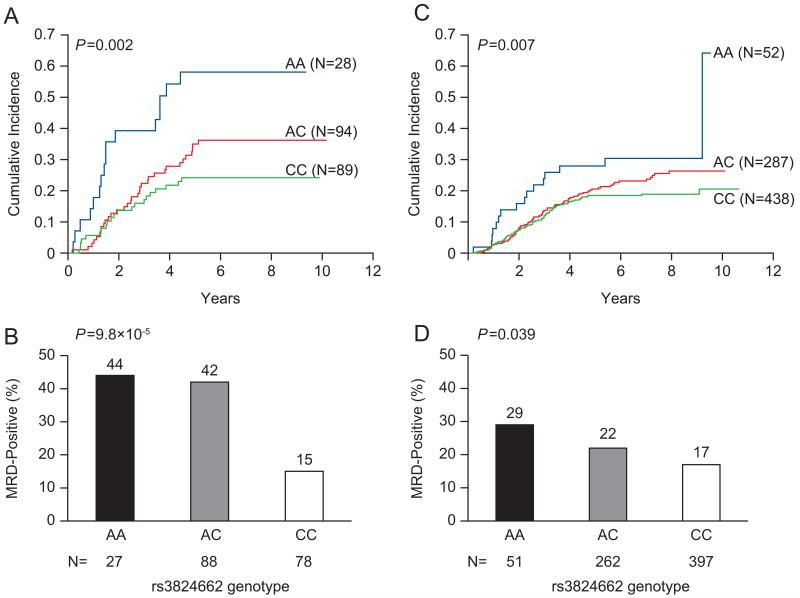
Given the poor prognosis of Ph-like ALL, we next examined the relationships between GATA3 SNP genotypes and ALL relapse. In the COG P9906 cohort, the GATA3 allele linked to Ph-like ALL was also associated with a higher risk of relapse after adjusting for genetic ancestry (rs3824662, N=215, P=0.002, Fig. 4A). While rs3824662 was strongly related to early treatment response (i.e., minimal residual disease [MRD] at the end of induction therapy, N=193, P=9.8×10−5, Fig. 4B), it remained prognostic even within patients who were MRD negative (N=132, P=0.028). To further define the prognostic value of the GATA3 SNP, we tested the association of rs3824662 with relapse in the COG P9905 protocol. In this cohort, genotype at rs3824662 was significantly associated with relapse, with each copy of A allele linked to 1.43-fold increase (95% CI, 1.10 to 1.86) in the risk of disease recurrence (N=781, P=0.007, Fig. 4C). Also, the A allele at rs3824662 was associated with a higher MRD level at the end of induction therapy (N=710, P=0.039, Fig. 4D), and there was a trend for it to be linked to higher relapse risk within patients negative for MRD in the COG P9905 cohort (N=566, P=0.094).
Figure 4. Genotype at GATA3 SNP rs3824662 and ALL treatment response.
The cumulative incidence of relapse was compared by genotype at rs3824662 in the COG P9906 (A) and COG P9905 (B), with P value estimated by hazard regression test including ancestry as covariate. Early treatment response measured by minimal residual disease (MRD) at the end of induction was also related to genotype at rs3824662 in both COG P9906 (C) and COG P9905 (D), with P value estimated by Spearman Rank test. For both relapse and MRD, the allele linked to Ph-like ALL was also associated with worse treatment response.
While association of rs3824662 with Ph-like ALL was consistent across ethnicity (Supplementary Fig. 3), the risk allele frequency varied significantly among different ethnic groups. Among worldwide populations, the rs3824662 allele related to Ph-like ALL and relapse was markedly more common in Guatemalans with high Native American (NA) genetic ancestry and US Hispanics than individuals of European descent (52%, 40%, and 14%, respectively, Supplementary Fig. 10), consistent with the racial disparities in ALL treatment outcomes21.
The majority of children with ALL can be cured with individualized combination chemotherapy, and treatment outcome continues to improve as new molecular prognostic markers are incorporated to achieve more precise risk classification2. Until recently, little is known about why a child develops a specific subtype of ALL in the first place and whether inherited genetic variations that predispose to a subtype also influence prognosis12,14,16. Therefore, the goal of this GWAS was to discover the genetic basis of the susceptibility to Ph-like ALL and to better understand the biology of this important high-risk subtype. The discovery of GATA3 variants associated with Ph-like ALL and related genomic lesions points to potentially novel mechanisms of ALL etiology and also previously unrecognized function of GATA3 in leukemogenesis. GATA3 belongs to a group of transcription factors characterized by 2 highly-conserved zinc fingers that mediate binding to the (A/G)GATA(A/G) sequence and protein-protein interactions22. Stage-specific transcription of GATA3 has been extensively characterized during T cell development and differentiation23. GATA3 is critical for the generation of early T-lineage progenitor cells24 and somatic loss-of-function mutations in GATA3 are enriched in early T-cell precursor ALL25. Inherited genetic variation in GATA3 has also been linked to the susceptibility to Hodgkin lymphoma26, although they are not related to rs3824662 or rs3781093 (r2<0.1 in HapMap CEU). Other members of GATA family are critical for different stages of hematopoietic development, and germline or somatic mutations in these genes can lead to a variety of hematologic disorders27,28.
In strong LD in European, Hispanic, and Asian populations (r2=0.94, 0.90, and 0.97 in HapMap CEU, MEX, and CHB/JPT, respectively), rs3824662 and rs3781093 both achieved genome-wide significance in the discovery GWAS with similar association with Ph-like ALL (Supplementary Figs. 2, 3, and 11). However, rs3781093 became non-significant in multivariate analysis conditioning on rs3824662 (Supplementary Table 1). In African subjects in which these 2 SNPs are poorly linked (r2=0.006 in the HapMap YRI), the A allele at rs3824662 remained over-represented in Ph-like ALL whereas rs3781093 no longer showed any evidence of association with Ph-like ALL (Supplementary Fig. 11). Also, rs3781093 was not associated with GATA3 expression nor with local DNase hypersensitivity in HapMap YRI samples, whereas consistent evidence points to rs3824662 as a potential expression quantitative trait locus across ancestry in HapMap populations (Supplementary Figs. 6 and 12). In fact, rs3842662 was the top SNP influencing DNase hypersensitivity at this locus in the YRI population (Supplementary Fig. 12). Further examination of the ENCODE data suggested possible enhancer activities within the region encompassing rs3824662 in lymphoblastoid cell lines, based on histone methylation marks and PU.1 and P300 binding (Supplementary Fig. 13). Although functional studies are warranted to determine the exact causal variant(s) at this locus and molecular mechanisms by which GATA3 variants influence Ph-like ALL leukemogenesis, these lines of evidence consistently point to rs3824662 as a potentially functional variation with possibly direct contribution to the GWAS signal.
The GATA3 allele linked to Ph-like ALL was also associated with an increased risk of relapse in the COG P9906 cohort, which was validated in the COG P9905 cohort (Fig. 4). However, in the COG P9906 cohort, the GATA3 SNP was not prognostic after adjusting for Ph-likeness, arguing that the association with relapse might be largely driven by its relationship with Ph-like ALL. GATA3 SNP genotype was also related to CRLF2 rearrangement, JAK mutation, and IKZF1 deletion, but remained associated with Ph-like ALL after adjusting for these genomic lesions. To explore this further, we attempted to build a classification model for Ph-like ALL on the basis of GATA3 germline SNPs, somatic lesions in CRLF2, JAK, and IKZF1, and genetic ancestry in 682 patients in COG AALL0232 and COG P9906, using classification and regression tree methods (CART29). In this analysis (Supplementary Fig. 14), CRLF2, IKZF1, rs3824662, and NA genetic ancestry were independent predictors of Ph-like ALL, and rs3824662 was associated with Ph-likeness regardless of CRLF2 status. Interestingly, NA genetic ancestry remained significant after stratifying on the GATA3 SNP, indicative of additional ancestry-related germline variants that are associated with Ph-like ALL. There was also significant over-representation of the rs3824662 risk alleles in non-Ph-like ALL compared with non-ALL control (P=0.0008 and 0.00035 in the discovery GWAS and replication cohorts, respectively), suggesting effects of this variant on ALL susceptibility in general.
In conclusion, our genome-wide germline SNP analysis identified genetic variations in the GATA3 gene that influence susceptibility to Ph-like ALL and the risk of relapse. These findings highlight the intricate interactions between host and tumor genomes and their importance in the pathogenesis and prognosis of cancer in general.
ONLINE METHODS
Subjects and genotyping
The ALL cases investigated comprised children with newly-diagnosed B-precursor ALL who were treated on the Children’s Oncology Group (COG) trials AALL0232, P990532 and P990610 (Supplementary Table 2), and non-ALL controls included 12,416 subjects14,33-35. The number of subjects included in each analysis was described in Supplementary Figs 15, 16, and 17, and in the text as appropriate. This study was approved by the Institutional Review Boards with proper informed consent.
Germline genomic DNA was extracted from peripheral blood or bone marrow samples obtained during clinical remission for children with ALL. Genotyping was done for COG AALL0232 and COG P9905 cohorts and for non-ALL controls using the Affymetrix Human SNP Array 6.0. Quality control was performed for samples and SNPs according to call rate and minor allele frequency (Supplementary Fig. 1). Theta (allele signal intensity) plots were constructed using Affymetrix Genotyping Console for rs3824662 and rs3781093 (Supplementary Fig. 18). GATA3 SNPs (rs3824662 and rs3781093) were genotyped in the COG P9906 cohort and in the Guatemalan samples by Sanger sequencing (Supplementary Table 3).
Genetic ancestry was determined by using STRUCTURE21,36 and was used to define ethnicity (Supplementary Note).
Ph-like ALL and GWAS
Ph-like ALL was identified in the COG ALL0232 cohort and in the COG P9906 cohort on the basis of unsupervised hierarchical clustering analysis of global gene expression profile, as described previously7,9,37 .
The discovery GWAS of Ph-like ALL comprised 511 ALL cases enrolled on the COG AALL0232 protocol and 6,661 non-ALL controls from the dbGaP MESA dataset. We performed two association tests to identify germline SNPs related to Ph-like ALL: we compared the genotype frequency at each SNP 1) in Ph-like ALL (N=75) vs. non-ALL controls (N=6,661) and 2) in Ph-like ALL (N=75) vs. ALL cases without Ph-like profile (“non-Ph-like ALL”, N=436). Association was evaluated with logistic regression under an additive model with genetic ancestry as covariates. Population stratification was assessed by the construction of a quantile-quantile (Q-Q) plot (Supplementary Fig. 19). SNPs that reached P≤5×10−8 in the discovery GWAS were tested in an independent replication cohort: 171 ALL cases from the COG P9906 protocol and 5,755 non-ALL controls. Association with Ph-like ALL was evaluated by logistic regression with genetic ancestries as covariates by comparing 1) Ph-like ALL (N=32) vs. non-ALL controls (N=5,755) and 2) Ph-like ALL (N=32) vs. non-Ph-like ALL (N=139). Independently, the Ph-like phenotype was also identified by the recognition of outliers by sampling ends (ROSE) algorithm (Supplementary Fig. 20). GATA3 SNPs (rs3824662 and rs3781093) and expression were also evaluated in a separate cohort of patients with Ph+ ALL (Supplementary Note and Supplementary Fig. 21).
Functional characterization of GATA3 SNPs was performed by examining the association of SNP genotype with GATA3 expression, local DNase hypersensitivity, and global gene expression in ALL (Supplementary Note, Supplementary Table 4, Fig. 2, Supplementary Figs. 6,7, 12, 22, and 23), partly using previously published data sets30,31,38. Associations of GATA3 SNPs with CRLF2, JAK, and IKZF1 somatic lesions were evaluated in the COG AALL0232, COG P9906, and COG P9905 cohorts, and with relapse in the COG P9906 and COG P9905 cohorts (Supplementary Note). Germline SNPs within the JAK2 gene were tested for association with somatic JAK2 mutation in ALL (Supplementary Table 5). R 2.15.1 statistical software was used for all analyses unless indicated otherwise (Supplementary Note). Statistical tests were chosen as appropriate and according to the phenotype distribution (e.g., normally or binomially distributed for continuous or categorical variables, respectively).
Supplementary Material
Acknowledgements
We thank the patients and parents who participated in the COG protocols included in this study, the clinicians and research staff at COG institutions and J. Pullen (University of Mississippi at Jackson) for assistance in classification of patients with ALL. Genome-wide genotyping of COG P9905 samples was performed by the Center for Molecular Medicine with the generous financial support from the Jeffrey Pride Foundation and the National Childhood Cancer Foundation. V.P.A is supported by the Spanish Ministry of Education Fellowship Grant and by the St. Jude Children’s Research Hospital Academic Programs Special Fellowship. J.J.Y. is supported by the American Society of Hematology Scholar Award, Alex Lemonade Stand Foundation for Childhood Cancer Young Investigator Grant, and by the Order of St. Francis Foundation. K.G.R. is supported by a National Health and Medical Research Council (Australia) Overseas Training Fellowship and a Haematology Society of Australia and New Zealand Novartis New Investigator Scholarship. C.G.M. is a Pew Scholar in the Biomedical Sciences and a St. Baldrick’s Scholar. We thank M. Shriver (Pennsylvania State University) for sharing SNP genotype data of the Native American references, Jonathan Pritchard and Jacob Degner (University of Chicago) for sharing DNase hypersensitivity data of HapMap Yoruba cell lines, Raul C. Ribeiro (St. Jude Children’s Research Hospital) and Pedro De Alarcon (University of Illinois College of Medicine at Peoria) for coordinating collaborations in Guatemala. This work was supported by the National Institutes of Health (grant numbers CA156449, CA21765, CA36401, CA98543, CA114766, CA98413, CA140729 and GM92666), in part by the intramural Program of the National Cancer Institute, and by the American Lebanese Syrian Associated Charities (ALSAC). The study sponsors were not directly involved in the design of the study, the collection, analysis, and interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript.
Footnotes
URLs
dbGaP database: http://www.ncbi.nlm.nih.gov/gap
HapMap: http://hapmap.ncbi.nlm.nih.gov
STRUCTURE: http://pritch.bsd.uchicago.edu/structure_software/release_versions/v2.3.3/html/structure.html
Zoom Locus: http://csg.sph.umich.edu/locuszoom
GSEA: http://www.broadinstitute.org/gsea/index.jsp
Epigenome Browser: http://epigenomegateway.wustl.edu/browser
1000 Genomes: http://www.1000genomes.org
Database accession numbers
NCBI dbGAP: phs000638, phs000209, phs000021, phs000017
NCBI GEO: GSE11877, GSE7851, GSE5859
NCI caArray: EXP-578
Author Contribution
Jointly supervised research: J.J.Y; Conceived and designed the experiments: V.P.A, S.P.H., C.L.W., C.G.M and J.J.Y.; Performed the experiments: V.P.A, K.G.R, R.C.H., J.G.F., S.E., I-M.C., G.N., E.G.B., D.G.T. and C.N.V.; Performed statistical analysis: V.P.A, J.J.Y., R.C.H., W.Y., C.C., D.P., Y.F., M.D., C.S. and G.N.; Analyzed the data: V.P.A, K.G.R., R.C.H., W.Y., H.X., S.E., J.Y.S.L., I-M.C., Y.F., M.J.B., C.S., G.N., E.G.B., D.T., F.A.K., C.N.V., M-L.L., M.D., D.B., C-H.P., W.E.E., M.V.R., S.P.H., C.L.W. and C.G.M.; Contributed to reagents/materials/analysis tools: R.C.H., J.G.F., J.Y.S.L.,Y.F., E.G.B., F.A.K., C.N.V., N.J.W., B.M.C., E.R., B.W., F.Y., W.L.C., E.L., W.P.B., M-L.L., M.D., S.P.H., C.L.W. and C.G.M.; Wrote the paper: V.P.A and J.J.Y.
Competing Interest and Financial Disclosures
The authors declare do not have any relevant competing interest and full disclosures are provided in the
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–78. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Mullighan CG, Evans WE, Relling MV. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood. 2012;120:1165–74. doi: 10.1182/blood-2012-05-378943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunger SP, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol. 2012;30:1663–9. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanulla M, et al. Integrating molecular information into treatment of childhood acute lymphoblastic leukemia--a perspective from the BFM Study Group. Blood Cells Mol Dis. 2007;39:160–3. doi: 10.1016/j.bcmd.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Biondi A, et al. Imatinib after induction for treatment of children and adolescents with Philadelphia-chromosome-positive acute lymphoblastic leukaemia (EsPhALL): a randomised, open-label, intergroup study. Lancet Oncol. 2012;13:936–45. doi: 10.1016/S1470-2045(12)70377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Den Boer ML, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts KG, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey RC, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116:4874–84. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey RC, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–21. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh ML, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood. 2013;121:485–8. doi: 10.1182/blood-2012-04-422691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–10. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherborne AL, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42:492–4. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trevino LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–5. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105:733–42. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30:751–7. doi: 10.1200/JCO.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsson K, et al. Genetic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2010;107:21719–24. doi: 10.1073/pnas.1006981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones AV, et al. JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat Genet. 2009;41:446–9. doi: 10.1038/ng.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olcaydu D, et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet. 2009;41:450–4. doi: 10.1038/ng.341. [DOI] [PubMed] [Google Scholar]
- 20.Kilpivaara O, et al. A germline JAK2 SNP is associated with predisposition to the development of JAK2(V617F)-positive myeloproliferative neoplasms. Nat Genet. 2009;41:455–9. doi: 10.1038/ng.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang JJ, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43:237–41. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara T, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36:667–81. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei G, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yagi R, Zhu J, Paul WE. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int Immunol. 2011;23:415–20. doi: 10.1093/intimm/dxr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enciso-Mora V, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010;42:1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasquet M, et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121:822–9. doi: 10.1182/blood-2012-08-447367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn CN, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–7. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies SM, et al. Pharmacogenetics of minimal residual disease response in children with B-precursor acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood. 2008;111:2984–90. doi: 10.1182/blood-2007-09-114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang RS, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–63. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borowitz MJ, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: a Children’s Oncology Group study. Blood. 2008;111:5477–85. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–7. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burchard EG, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–92. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 36.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.