SUMMARY
Vinculin, a cytoskeletal scaffold protein essential for embryogenesis and cardiovascular function, localizes to focal adhesions and adherens junctions, connecting cell surface receptors to the actin cytoskeleton. While vinculin interacts with many adhesion proteins, its interaction with filamentous actin regulates cell morphology, motility, and mechanotransduction. Disruption of this interaction lowers cell traction forces and enhances actin flow rates. Although a model for the vinculin:actin complex exists, we recently identified actin-binding deficient mutants of vinculin outside sites predicted to bind actin, and developed an alternative model to better define this novel actin-binding surface, using negative-stain EM, discrete molecular dynamics, and mutagenesis. Actin-binding deficient vinculin variants expressed in vinculin knockout fibroblasts fail to rescue cell-spreading defects and reduce cellular response to external force. These findings highlight the importance of this new actin-binding surface and provide the molecular basis for elucidating additional roles of this interaction, including actin-induced conformational changes which promote actin bundling.
INTRODUCTION
Vinculin (Vcn) is a highly conserved, abundant protein that localizes to focal adhesions (FAs), focal complexes, and adherens junctions (Geiger et al., 2001; Geiger et al., 2009). Vcn plays an essential role in embryogenesis, as knockout mice show defects in heart and nerve formation and do not survive past E10 (Xu et al., 1998). Cells deficient in Vcn exhibit rounded morphology, increased motility (Xu et al., 1998), and resistance to apoptosis and anoikis (Subauste et al., 2004). Consistent with these observations, Vcn regulates FA turnover (Saunders et al., 2006), adhesion dynamics at the leading edge of cells (Thievessen et al., 2013), and force transduction (Grashoff et al., 2010). However, the mechanisms by which Vcn regulates these functions are poorly understood.
Vcn is a molecular scaffold protein comprised of three domains: a 91 kDa head (Vh), a proline-rich linker, and a 22 kDa tail (Vt) (Ziegler et al., 2006). Cytosolic Vcn exists in an inactive, autoinhibited conformation mediated by a Vh:Vt interaction that obscures binding to many ligands (Johnson and Craig, 1994, 1995). Disruption of tight autoinhibitory contacts is required for Vcn activation, and is mediated by multiple mechanisms, including ligand binding to both Vh and Vt, mechanical force, and phosphorylation (Peng et al., 2011).
Vcn binds to F-actin through Vt and subsequently crosslinks F-actin filaments into fibers (Huttelmaier et al., 1997; Johnson and Craig, 1995). This interaction links the actin cytoskeleton to integrins and the extracellular matrix, and is believed to be critical for FA maturation (Humphries et al., 2007; Thievessen et al., 2013), cell movement (Hu et al., 2007), and force transduction (Grashoff et al., 2010; Ji et al., 2008; Shen et al., 2011). In addition to binding F-actin, Vt also binds raver1 (Lee et al., 2009), paxillin (Wood et al., 1994), and phosphatidylinositol 4,5-bisphosphate (PIP2) (Palmer et al., 2009). Vt contains a five-helix bundle fold, with an N-terminal strap (residues 879–892, NT) and C-terminal arm (residues 1046–1066, CT) that interact to bring the termini in close proximity to each other (Bakolitsa et al., 2004; Bakolitsa et al., 1999).
A structural model (J-model) of the Vt:F-actin complex, derived from low resolution electron microscopy (EM) data, places helices 2 and 3 (H2 and H3) of Vt in a hydrophobic cleft at the junction between two of the actin subunits (Janssen et al., 2006). However, specific Vt sites that interact with actin have not been verified by targeted mutagenesis. Although Vcn variants deficient in F-actin binding have been employed to probe the functional consequences of this interaction, results from these studies are complicated, as the variants possess multiple mutations or large deletions in Vt that disrupt Vcn structure and/or interactions with other tail ligands (Palmer et al., 2009). A computational model has since been published, but lacks supporting experimental evidence (Golji and Mofrad, 2013).
Herein, we employ mutagenesis, negative-stain EM, and molecular modeling to identify a novel actin binding surface. We also identify a conservative Vcn point mutant that retains Vt structure and PIP2 binding, yet disrupts binding to F-actin. Interestingly, the mutation site (V1001) is outside the reported actin-binding interface (Janssen et al., 2006). While this hydrophobic site is distinct from the surface identified in the J-model, it is consistent with current mutagenesis data, known ligand interactions, and occlusion of the site in the full length protein (Johnson and Craig, 1995; Lee et al., 2009; Shen et al., 2011). To examine the consequences of disrupting the Vcn:F-actin interaction, we transfected F-actin binding deficient variants into Vcn knockout murine embryonic fibroblasts (Vin−/− MEFs), and find that loss of actin binding by Vcn alters cell and FA size and limits the ability of cells to respond to external force.
RESULTS
Identification of Vt variants deficient in actin binding
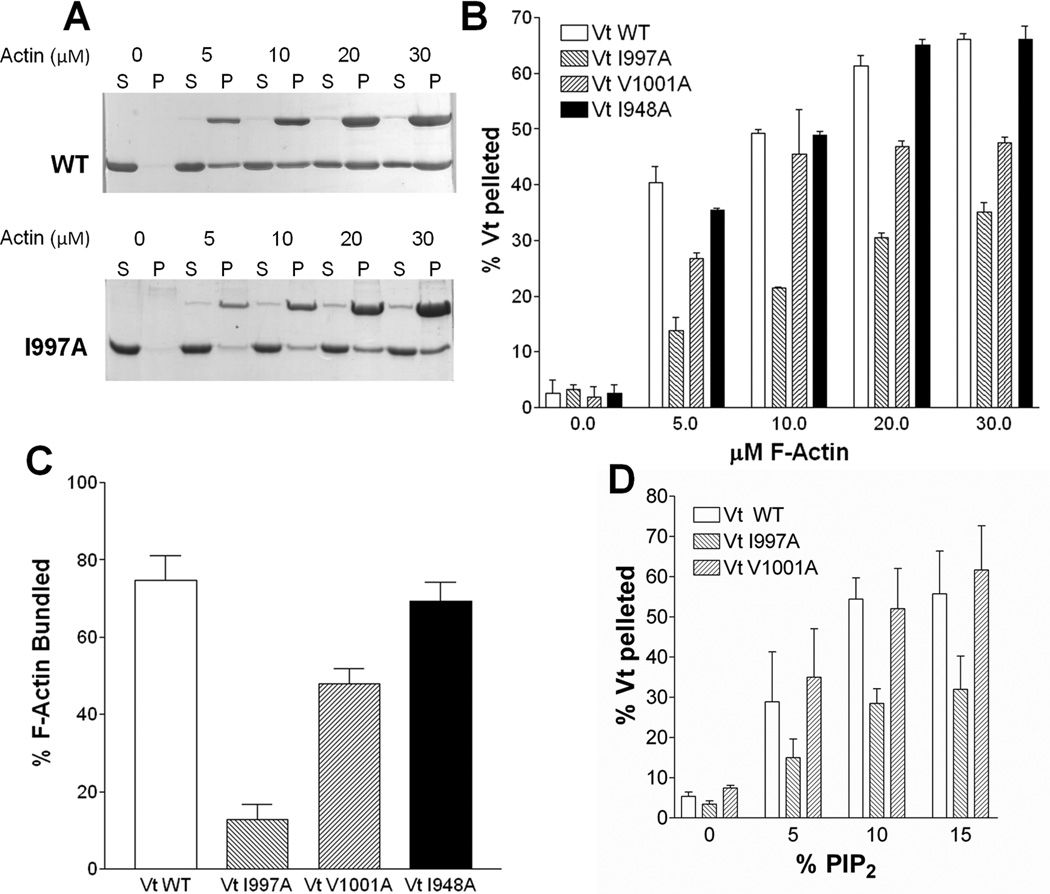
Although Vcn variants impaired in actin binding have been identified, they contain multiple point mutations (Cohen et al., 2005) or deletions (Huttelmaier et al., 1997; Marg et al., 2010; Menkel et al., 1994) and have not been fully characterized to determine if Vt structure or other ligand binding interactions are altered. We generated two Vt variants, VtI997A and VtV1001A, which exhibit a significantly decreased affinity for F-actin (Fig. 1A, B). This was unexpected, as the surfaces associated with the J-model for the Vt:F-actin interface do not include residues I997 or V1001 (Janssen et al., 2006), and actin-deficient variants at these sites have not been reported.
Figure 1. VtI997A and VtV1001A are deficient in F-actin binding and bundling yet retain association with PIP2.
(A) SDS-polyacrylamide gel electrophoresis of supernatant (S) and pellet (P) fractions after co-sedimentation of Vt with F-actin. Actin concentrations and Vt variants are noted. (B) Quantification of F-actin co-sedimentation assays identifies Vt variants in H4 deficient in F-actin binding. (C) Vt variants deficient in binding to F-actin are also defective in F-actin bundling. (D) VtV1001A, while deficient in actin binding, retains PIP2 binding comparable to VtWT. VtI997A is impaired in PIP2 binding. Error bars are standard deviation, n= 3. See also Figure S1.
To quantify actin binding of these Vt variants, we performed F-actin co-sedimentation assays (Figure 1A, B). At 5 µM F-actin (concentration at approximately half-saturation), 40% VtWT pellets with F-actin. Three-fold less VtI997A and two-fold less VtV1001A bind actin at this concentration. As both I997 and V1001 lie outside the actin binding site in the J-model, we generated another variant, VtI948A, which lies within the reported Vt:F-actin interface (Janssen et al., 2006). However, VtI948A did not significantly decrease the affinity of Vt for F-actin (Figure 1B). Moreover, we performed actin co-sedimentation experiments on additional variants (summarized in Table S1). We also assessed actin binding properties of a subset of these mutations in the full-length protein. While the actin-binding site of Vcn resides in Vt, it is partially masked in the full-length protein due to autoinhibitory contacts between Vh and Vt (Cohen et al., 2005). Consistent with this, binding of F-actin by VcnWT is significantly reduced compared to isolated Vt (Figure S1A). As the interaction between Vh and Vt can be disrupted by an IpaA peptide from Shigella (Hamiaux et al., 2006; Izard et al., 2006), we added IpaA to Vcn, and observed significantly enhanced F-actin binding (Figure S1A). As previously reported, VcnI997A has a 10-fold weaker Kd for F-actin than VcnWT (Thievessen et al., 2013). At 30 µM F-actin, both VcnI997A and VcnV1001A bind roughly half as much F-actin as VcnWT (Figure S1A). Actin binding profiles observed for VcnI948A are similar to those observed for VcnWT, with enhanced F-actin binding observed in the presence of the IpaA peptide (Figure S1A). These results suggest that actin-binding deficient mutations in Vt similarly impair actin-binding in Vcn.
Binding of F-actin to Vcn facilitates bundling of F-actin filaments. This occurs through a conformational change that promotes Vt dimerization and crosslinking of actin filaments (Janssen et al., 2006; Johnson and Craig, 2000). We showed that the Vcn CT hairpin is required for generation of this actin-induced dimer and for F-actin bundling (Shen et al., 2011). Vt variants deficient in F-actin binding are also notably impaired in their ability to bundle F-actin (Figure 1C), with deficiencies in F-actin binding correlated with deficiencies in bundling. The Vt variant most impaired in F-actin binding, VtI997A, is most impaired in F-actin bundling and possesses a bundling defect similar to our previously characterized CT hairpin deletion variant (VtΔC5) (Shen et al., 2011). VtV1001A is able to partially bind and bundle F-actin, whereas VtI948A retains F-actin binding (VtI948A) and is fully capable of bundling F-actin.
We also employed circular dichroism (CD) and NMR spectroscopy to assess the structural integrity of our Vt variants. Far-UV CD spectra for all variants are similar, with characteristic minima at 208 and 222 nm (Figure S1B, C), indicating that the α-helical secondary structure of the Vt variants is preserved. For Vt, a distinct near-UV CD signal is observed between 270 and 300 nm and reflects tertiary packing of W912 in the H1/H2 loop with W1058 in the CT (Palmer et al., 2009), which is preserved in VtI997A, VtV1001A, and VtI948A (Figure S1D, E). To further confirm the structural integrity of VtI997A and VtV1001A, we acquired Heteronuclear Single Quantum Coherence (HSQC) 2D NMR spectra on 15N-enriched VtWT and the actin-binding deficient Vt variants. The peaks in the 1H-15N HSQC spectra for both VtI997A and VtV1001A remain dispersed, indicative of well-folded protein, and overlap well with the peaks of VtWT (Figure S1F, G). Peaks that shift correspond to residues near the site of mutation (Figure S1H, I). The amide (NH) line widths and intensity are also unchanged from those of VtWT, suggesting that the mutations do not significantly alter dynamic properties of Vt. These data, taken together, suggest that the actin-binding deficiencies of VtI997A and VtV1001A are not the result of structural defects (Figure S1).
To determine if these actin-binding deficient Vt variants are altered in their interactions with PIP2, we performed lipid co-sedimentation experiments. While the F-actin deficient variants VtI997A and VtV1001A retain specificity for PIP2 over PS, VtI997A exhibits a 50% decrease in binding to PIP2 (Figure 1D). To discriminate consequences of the common actin-binding defect, given differences in PIP2 affinity, we evaluated the cellular properties of these variants.
Deficiencies in Actin Binding by Vcn Alter Cellular Properties
Vcn variants containing multiple mutations (Cohen et al., 2005) or deletions that remove helix 2 and 3 (Marg et al., 2010) or the entire tail domain (Humphries et al., 2007) have been generated to prevent the interaction of Vcn with F-actin. However, these variants likely display phenotypes resulting from disruption of multiple ligand interactions in addition to the actin defect. While the Vcn:F-actin interaction is thought to play a critical role in adhesion turnover, cell motility and force transduction, it remains to be determined if phenotypes associated with these deletion variants are attributed to the Vcn:F-actin interaction alone. Given our well-characterized actin-binding deficient Vcn variants, we explored the role of actin-binding by expressing VcnI997A and VcnV1001A in Vin−/− MEFs.
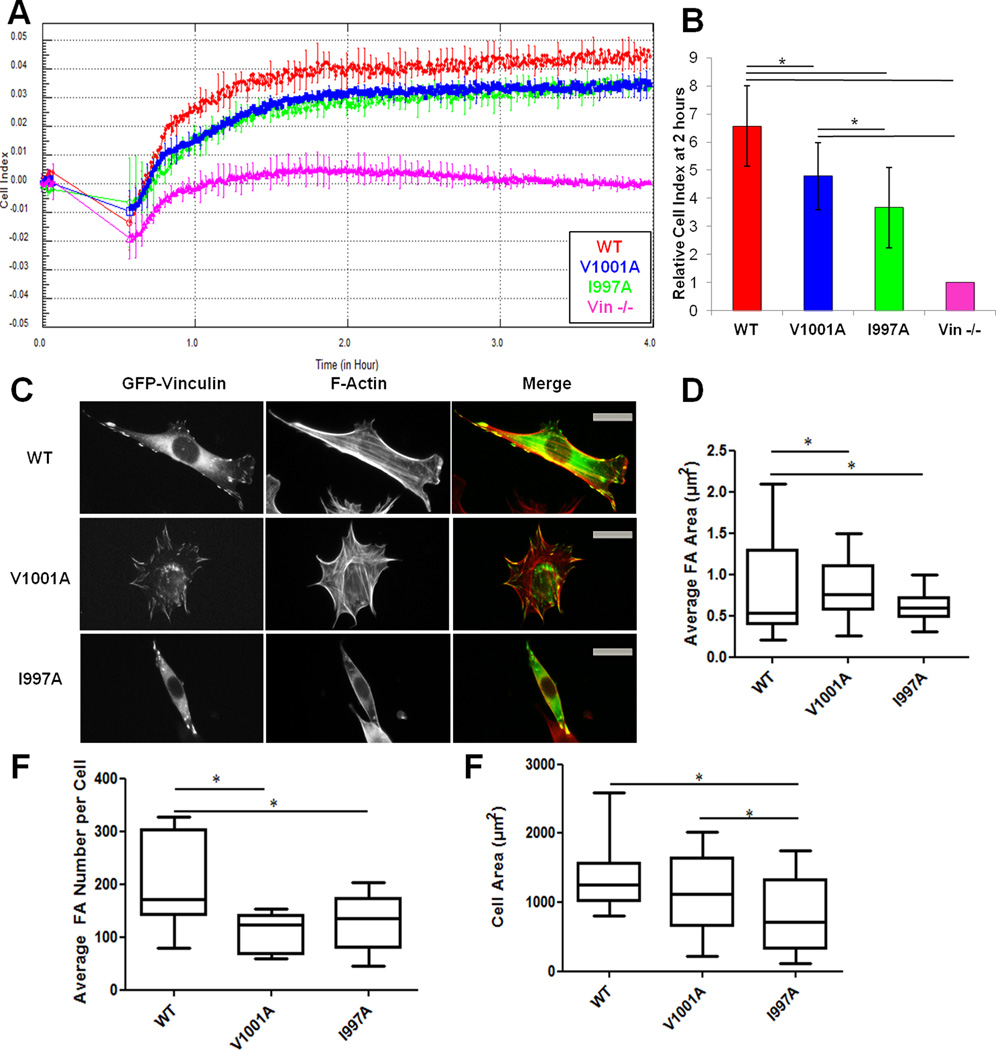
We reported a Vcn variant that retains actin binding but is deficient in F-actin bundling (Shen et al., 2011). Expression of this variant (VcnΔC5) in Vin−/− MEFs resulted in larger FAs and smaller cell area when cells adhered and spread on fibronectin (FN). We anticipated similar results for a loss of actin-binding by Vcn, as a deficiency in binding necessitates a deficiency in bundling. First, we expressed the GFP-vinculin in Vin−/− MEFs and verified their expression level by western blot (data not shown). Initially, we examined the ability of the cells to attach and spread over time on FN using the real-time cell analyzer (RTCA) xCELLigence system, an impedance-based system (given in arbitrary units as cell index, CI) that monitors changes in electrical resistance as cells adhere to the microelectrode in the dish (Figure 2A, B) (Atienza et al., 2005). Cells expressing VcnWT have 6.6-fold higher CI (6.57 ± 1.43 CI) than Vin−/− MEFs and readily spread. These findings support previous observations that Vin−/− MEFs have difficulties in adhering and spreading on substrates (Coll et al., 1995). MEFs expressing VcnI997A (3.67 ± 1.43 CI) and VcnV1001A (4.78 ± 1.19 CI) showed reduced spreading compared to cells expressing VcnWT, suggesting that Vcn binding to F-actin plays an integral role in cell spreading. VcnV1001A impairs spreading less than VcnI997A, in agreement with its increased affinity for F-actin. We also performed immunofluorescence studies on VcnWT, VcnI997A, and VcnV1001A to verify that our variants retain localization to FAs upon expression in Vin−/− MEFs (Figure 2C). Our finding that Vcn variants localize properly is expected, as Vh is sufficient to localize Vcn to FAs (Humphries et al., 2007). We also find that cells expressing VcnI997A and VcnV1001A have significantly larger FAs and 35% and 46% fewer FAs, respectively, in comparison to cells expressing VcnWT (Figure 2D, 3). However, cells expressing VcnV1001A did not show a significant change in cell area (only 20% smaller), while cells expressing VcnI997A were significantly smaller (42%) than those expressing VcnWT (Figure 2F). While there is slightly higher CI with VcnV1001A over VcnI997A, and cells expressing VcnV1001A do not exhibit a change in cell area, the observed CI could be attributed to the number and size of FAs found in these cells, as the system is sensitive enough to detect cytoskeletal changes and an increase in adhesion to the substrate (Atienza et al., 2005) These results suggest that the number and average size of FAs during spreading events are directly influenced by Vcn's interactions with F-actin.
Figure 2. Vcn variants deficient in actin binding affect spreading and cell adhesion in MEFs.
(A) RTCA using the xCELLigence system shows that Vin−/− cells expressing VcnWT have higher cell impedance, hence more spread, than cells expressing VcnV1001A, VcnI997A or Vin−/−.MEFs. A representative trace of cell impedance (graphed as cell index (CI)) taken every 15 seconds for 13 hours; lower impedance indicates less contact with the sensor. Each data point represents an average CI of at least triplicate wells for each condition. (B) A graph showing the relative CI of cells spread on FN two hours following plating, which corresponds to the same time as the pictures shown in (C). Data is the average ± SEM combined from four independent experiments. *p≤ 0.05, in comparison to VcnWT. (C) Vin−/− MEFs transfected with GFP-tagged VcnWT, VcnI997A, or VcnV1001A and plated on FN for two hours. VcnI997A and VcnV1001A exhibit the same localization as VcnWT. (D,E,F) Box and whisker plots of FA area (D), FA number (E) and cell area (F). Areas were calculated using Matlab (Methods) (n=25). Cells expressing VcnI997A and VcnV1001A had fewer and larger FAs, *p-value≤0.05. Cells expressing VcnI997A were significantly smaller, but those expressing VcnV1001A were not. Scale bar is 25 µm.
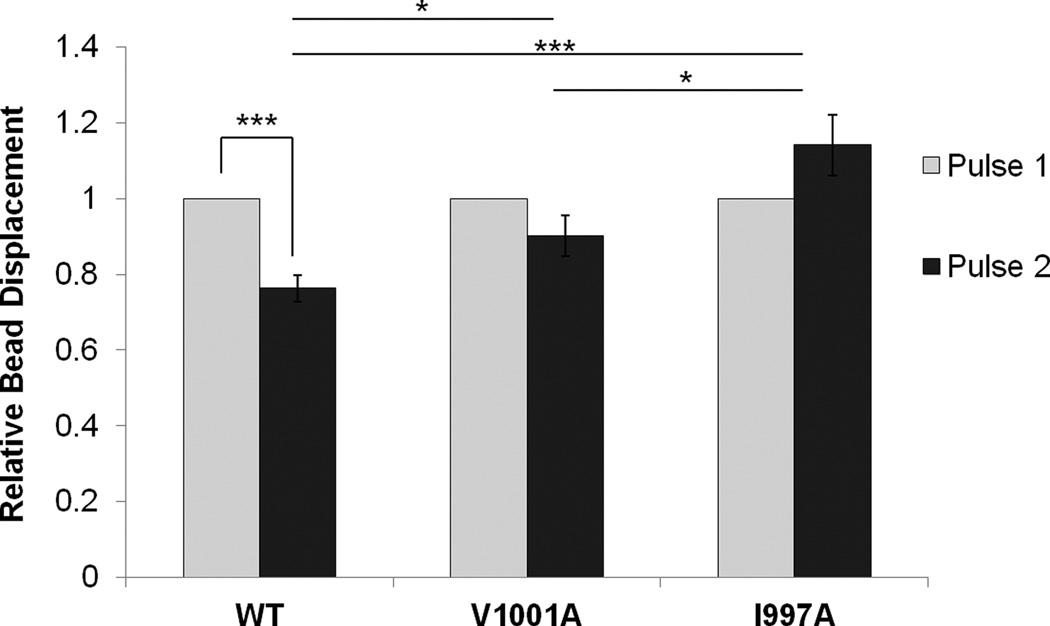
Figure 3. Actin binding to Vcn is necessary for the mechanical response to force on integrins.
Upon applying pulses of constant force, a decrease in the relative bead displacement of Vin−/− MEFs transfected with VcnWT is observed, in contrast to the increase observed for Vin−/− MEFs transfected with VcnI997A. Two force pulses were applied to FN-coated beads bound to Vin−/− MEFs transfected with either VcnWT (n=20), VcnV1001A (n=19) or VcnI997A (n = 26) and displacement was measured. * indicates a p-value< 0.05. *** indicates a p-value<0.001. Error bars are ± SEM. These results indicate that actin binding to Vcn plays a role in Vcn’s ability to respond to force.
Vcn plays a critical role in regulation of the cellular response to force (Grashoff et al., 2010; Ji et al., 2008). We recently reported that bundling of F-actin by Vcn is essential for reinforcement, the process by which cells locally stiffen when force is applied to FAs (Shen et al., 2011). Given these observations, we predicted that disruption of Vcn binding to F-actin (which is necessary for bundling) will also prevent reinforcement in cells. To test this, we exposed Vin−/− MEFs expressing VcnWT, VcnI997A, or VcnV1001A with FN-coated magnetic beads. Using the three dimensional force microscope (3DFM), we applied pulses of constant force to cells transfected with the various GFP-Vcn variants (O'Brien et al., 2008). The relative displacement of the bead was determined for the first and second pulses. Cells transfected with VcnWT showed a 23% decrease in bead displacement upon application of the second pulse (p-value < 0.001; Figure 3), indicating stiffening in response to force. Cells transfected with VcnV1001A exhibited a slight stiffening response (10% decrease), though it was not significant (p-value 0.07). Vin−/− MEFs transfected with VcnI997A showed a striking 14% increase in the relative displacement of the bead upon application of a second pulse (p-value < 0.001; Figure 3). The failure of VcnI997A- and VcnV1001A-transfected MEFs to exhibit reinforcement further supports the role of the Vcn:actin interaction in force transduction.
Identification of an alternative actin binding surface
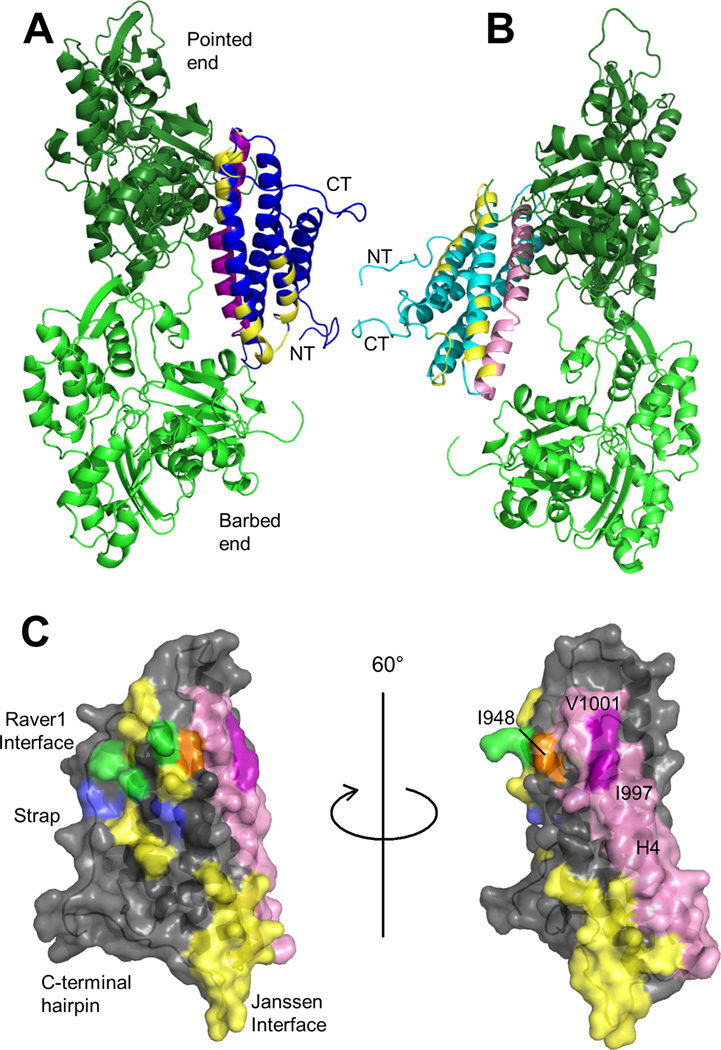
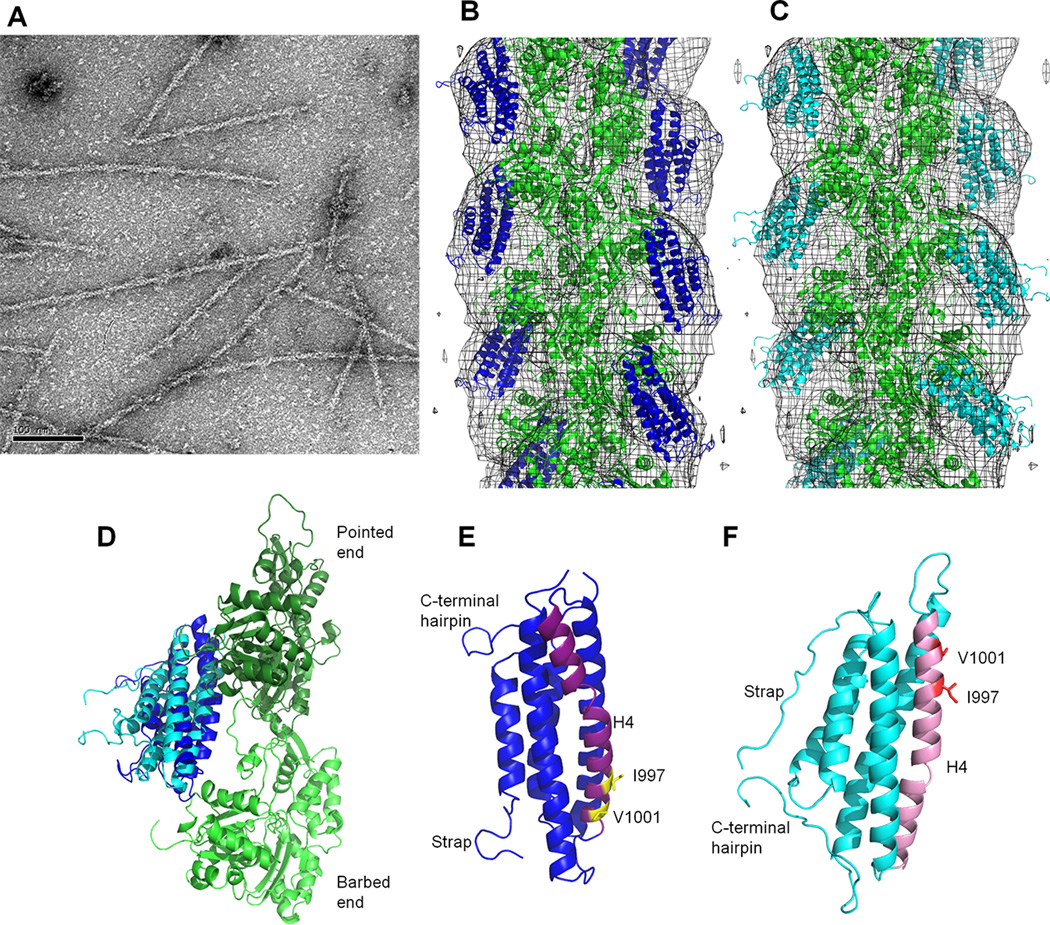
We identified conservative Vcn variants that retain Vt structure but disrupt F-actin binding. The mutations that most impair actin binding are located on helix 4 (H4), outside of the actin binding surfaces identified in the J-model (Janssen et al., 2006) (Figure 5A). Given the discrepancy between the sites we identified as being critical for actin binding, and the Vt surfaces postulated to bind actin, we collected electron micrographs of F-actin filaments decorated with Vt and generated a reconstruction with a resolution of ~20 Å (Figure 4A, B). An atomic model of the actin filament (PDB 3MFP) and the Vt crystal structure (PDB 1QKR) were docked manually into the 3D-reconstruction of F-actin-Vt complex using Chimera (Pettersen et al., 2004). As we find multiple plausible orientations of Vt in agreement with the EM map, we conclude that the orientation of Vt cannot be uniquely defined by the EM reconstruction at this resolution. However, an orientation that fits our experimental data but is distinct from the J-model is plausible, and is shown in Figure 4B. Cryo-EM attempts were unsuccessful, as Vt decoration on F-actin was lost upon blotting and freezing, and bundling activity created sample heterogeneity. In addition to manual docking of Vt into the EM reconstruction, we applied computational refinement approaches using discrete molecular dynamics (DMD) to fit the EM map and generated an alternative model of the Vt:F-actin complex (Figure 4C). While manual docking and DMD yielded different orientations of Vt with respect to F-actin, in both models the surface identified by mutagenesis faces F-actin to mediate binding (Figure 4D).
Figure 5. The proposed binding surface is not accounted for in the J-model.
(A) Manual fit model of the Vt:F-actin complex with the J-model surface mapped on Vt. The actin protomers are green. Vt is blue, with H4 in purple. The N- and C-termini are labeled. Yellow residues were identified in the J-model as mediating the Vt:F-actin interaction (Janssen et al., 2006). (B) Vt:F-actin complex from DMD model with the J-model surface mapped on Vt. The actin protomers are green. Vt is cyan, with H4 in pink. The Vt termini are labeled. Yellow residues were identified in the J-model as part of the Vt:F-actin interaction surface. (C) The J-model surface, raver1 interface, and H4 on Vt. Vt is shown with a semi-transparent surface in gray. Residues in the J-model interface are yellow. Those in the raver1 interface are blue (Lee et al., 2009). Those shared between the J-model interface and the raver1 interface are green. H4 is in pink. I997 and V1001 are purple and labeled, while I948 is orange. Two views are shown, rotated 60°. Note that I997 and V1001 are distal from the J-model actin binding interface.
Figure 4. The proposed actin binding surface on Vt is consistent with EM reconstruction.
(A) Negative-stain EM image of F-actin decorated with VtWT. Scale bar is 100 nm. (B) Manual fit model of Vt bound to F-actin. Crystal structure of the Vt domain (blue ribbon, PDB 1QKR) and the atomic model of F-actin (green ribbon, PDB 3MFP) are manually docked into the 3D-reconstruction (gray mesh). (C) DMD model of Vt bound to F-actin. F-actin is in green, Vt in cyan. (D) Comparison of Vt domain orientation from B and C with respect to the two adjacent actin protomers (long-pitch helix F-actin dimer). The color scheme is maintained. (E,F) Comparison of Vt H4 orientation in the manual fit and DMD models from B and C, respectively. The orientation and color scheme of the models has been maintained from D. The manual fit and DMD models are related to each other by an approximately 180° rotation, with H4 at the F-actin interface. H4 is purple and pink in the manual fit and DMD model, respectively. Residues I997 and V1001 are labeled and shown as yellow and red sticks in the respective models. See also Figure S2, Table S1.
The DMD model was further evaluated by comparing F-actin binding properties of VtWT and several Vt variants (Table S1) with the predicted change in binding energy (ΔΔG) for the variants (Figure S2A). These values are listed in Table S1, with the mutation sites mapped onto the Vt structure in Figure S2B. These ΔΔG values show agreement with our experimental data (correlation coefficient of 0.68). Our model is distinct from that proposed by Golji and Mofrad, as it highlights a surface comprising H4 and H5 and the importance of hydrophobic residues on H4 instead of an electrostatic surface on H3 and H4 (Golji and Mofrad, 2013).
In both the manual fit and DMD models, the orientation of Vt within the EM reconstruction places the NT and CT outside of the reconstructed volume (Figure 4B, C). These regions are absent or have larger B-factors relative to the helix bundle in the Vt crystal structure (Bakolitsa et al., 1999), suggesting conformational heterogeneity. To evaluate whether regions fit to low density in the averaged reconstruction are conformationally mobile, we collected NMR heteronuclear NOE data (Farrow et al., 1994) to evaluate the fast dynamics of VtQ1018K, a Vt variant with a decreased propensity to form the non-physiological Vt dimer at NMR concentrations (Bakolitsa et al., 1999) (Figure S2D). Low heteronuclear NOE values were observed for the Vt CT hairpin, suggesting that these residues are mobile in solution and unlikely to contribute a unified signal by EM. We also used fast-HSQC (Hwang et al., 1998) and CLEANEX (Hwang et al., 1997) NMR to measure solvent exchange. These results reveal that backbone amides associated with the NT and CT for both VtWT and VtQ1018K possess high rates of solvent exchange, further suggesting that these regions are intrinsically disordered and conformationally variable (Figure S2C). Taken together, the NMR data suggest that the NT and CT are unlikely to be observed by EM as they do not have a single defined orientation, consistent with our inability to fit these regions in the micrograph.
While both pseudo-atomic models yielded a reasonable match with the EM density, the resolution limits interpretation of the Vt:F-actin complex on a per-residue basis. Despite the ambiguity in positioning the Vt domain onto the actin filament, the actin surface that interacts with Vt is similar to the J-model (Janssen et al., 2006). However, the Vt surface in our manual and DMD models is significantly different from the J-model. The manual fit places the bottom of the helix bundle at the pointed end of the actin filament instead of the barbed end, while the DMD model is roughly flipped (Figure 4 C, D, and E). In both of our models, H3 and H4 are oriented towards the F-actin filament, as opposed to the strap, H2, and H3 (Janssen et al., 2006) (Figure 5A, B). The DMD model is rotated roughly 70° and the manual fit model is rotated roughly 70° in one axis and 180° degrees in another with respect to the J-model. Again, due to resolution limitations, we cannot advocate one of our models over the other; however, both models are supported by mutagenesis data and contain a similar actin-binding surface that contains residues (I997, V1001) critical for actin binding. Notably, this novel actin binding surface is distinct from that previously proposed by Janssen et al (Figure 5C) and is the first report of specific hydrophobic residues driving the interaction between F-actin and Vcn.
DISCUSSION
Vcn is an essential scaffolding protein that plays key roles in regulating FA assembly and disassembly. While recent studies have begun to unravel multi-component functions of Vcn (Carisey et al., 2013; Thievessen et al., 2013), the challenge of generating Vcn variants deficient in specific interactions has limited the ability to link a specific interaction with specific roles at FAs. Here, we report characterization of two Vcn variants, I997A and V1001A, which retain Vcn structure but are deficient in actin binding.
Previously, studies on Vcn variants deficient in actin binding used deletions that removed part of the helix bundle and disrupted the domain structure (Huttelmaier et al., 1997; Johnson and Craig, 2000; Menkel et al., 1994). Although a Vcn variant (T10) containing three point mutations showed decreased actin binding, only a modest 20% drop was observed (2 µM Vt, 5 µM F-actin) (Cohen et al., 2005). In contrast, our I997A and V1001A point mutations result in 50% and 30% reductions, respectively, in actin binding (10 µM Vt, 5 µM F-actin, Figure 1B). Importantly, both variants maintain Vt structure and PS binding. While VtI997A shows reduced affinity for PIP2, VtV1001A retains PIP2 binding, making these variants useful tools for studying the Vt:F-actin interaction.
The significant decrease in actin binding by VtI997A and VtV1001A is intriguing, as both mutations are outside of the binding sites reported by Janssen et al (Janssen et al., 2006) (Figure 5). This suggests that the J-model is incomplete in identifying the actin-binding interface. The J-model places the F-actin binding interface on H2 and H3 of Vt, split between two sites (Janssen et al., 2006), supported by previously reported mutagenesis data (Cohen et al., 2005; Janssen et al., 2006). The variants most deficient in F-actin binding, identified in this earlier study (T9, T10, and T19, though defects in binding are small, <20%), all support the lower site in the J-model, which resides primarily on H3 and at the N-terminus of H4. However, less evidence exists for the upper site. While MD simulations by Golji and Mofrad support the lower site identified in the J-model, their upper site contains part of the surface we identify here (Golji and Mofrad, 2013). Both Janssen et al. and Golji and Mofrad predicted the importance of hydrophobic interactions at the upper interface, but we identify some of these residues (I997 and V1001) and reject others identified as part of the actin-binding surface (L928 and I948). Our results also conflict with previous findings that removal of residues 979–1066 retains acting binding to Vt (Le Clainche et al., 2010). However, this construct removes half of Vt and disrupts the helix bundle.
While VtI997A retains PS binding, a reduction in PIP2 association is observed, suggesting that the actin and lipid binding interfaces on Vt may overlap. This observation is supported by data that Vcn binding to F-actin and PIP2 are mutually exclusive events (Steimle et al., 1999). To understand the implications of these binding interactions and their interplay, an improved understanding of how Vt binds PIP2 is required. We are currently pursuing a structural model for this interaction and generating Vcn variants that will allow us to probe the function of the Vcn:PIP2 interaction in cells.
As demonstrated in Figures 2 and 3, cells expressing these variants display defects in cell spreading and have abrogated responses to pulses of force, a phenotype similar to that observed for an actin-bundling deficient mutant (Shen et al., 2011). These results are expected given that actin binding is required for filament bundling. While cells transfected with either VcnI997A or VcnV1001A show a loss of reinforcement, the effect is more dramatic for VcnI997A, likely due to its weaker affinity for F-actin, though a reduced PIP2 affinity may also play a role.
The cellular phenotypes associated with these actin-binding deficient Vcn variants closely match and support findings published by Thievessen et al (Thievessen et al., 2013). Cells expressing actin-binding deficient Vcn variants show a decrease in cell spreading, in FA number, and an increase in FA size. Similarly, Thievessen et al. found that average FA size increased in cells expressing VcnI997A, likely due to an increase in FA growth rate. Additionally, they reported an increase in F-actin flow rates in the lamellipodium and at FAs and a decrease in FA formation density. Similar phenotypes were observed when activating mutations in Vh were introduced in the context of the VcnI997A mutation, indicating that alterations in cellular phenotype are due to the actin binding defect (Thievessen et al., 2013) instead of an activation defect. This finding raises a new question regarding whether F-actin binding to Vcn is required to initiate Vcn activation.
Our findings also elucidate factors influencing FA growth and maturation. The size of FAs is influenced by multiple factors such as rate of assembly and disassembly, density of the matrix to which the cells adhere, mechanical tension and other undetermined factors. The role of mechanical tension in the assembly and growth of FAs is controversial (Lessey et al., 2012). Initial studies implicated tension as a critical factor (Chrzanowska-Wodnicka and Burridge, 1996; Riveline et al., 2001). However, the role of tension in FA maturation has been questioned, as tension alone cannot drive FA maturation in the absence of stress fibers (Oakes et al., 2012). Interestingly, we find that cells expressing actin-binding deficient Vcn variants have larger and fewer FAs than cells expressing VcnWT. These cells also exhibit a decreased mechanotransduction response and fail to stiffen when external tension is applied to FN-coated beads that are attached to the cells. The observation that these cells have larger FAs argues that mechanical tension mediated by the Vcn:F-actin interaction is not required for FA maturation and stabilization (Thievessen et al., 2013).
A factor that limits structural analysis of the Vt:actin interaction is that both Vt and F-actin likely undergo conformational changes upon binding (Johnson and Craig, 2000; Wen et al., 2009), which places limitations on fitting isolated structures of Vt and actin into the complex, especially given the low resolution of negative-stain EM. Additional data supporting model selection or elimination is required. For example, we have generated Vcn mutants that provide support for this novel actin-binding surface. Additionally, Vt is able to simultaneously bind F-actin and the RNA binding protein raver1 (Lee et al., 2009). It is therefore unlikely that these interfaces overlap. The binding site for raver1, identified through x-ray crystallography and supported by mutagenesis, overlaps the upper site in the J-model (Figure 5C), suggesting that the upper site of the J-model is incomplete.
Based upon these concerns and the new data presented herein, we propose a new F-actin binding surface on Vt. This surface on H4, located at the Vt:F-actin interface in both models generated here (Figure 5A, B), is obscured in full length Vcn due to autoinhibitory interactions with Vh, consistent with previous work showing that the Vh:Vt interaction impairs binding to F-actin (Johnson and Craig, 1995). Additionally, this surface is not involved in binding raver1, allowing for simultaneous interactions with both ligands. The hydrophobic nature of the new surface, as shown by the importance of the isoleucine and valine sidechains, is congruent with reports that many actin-binding proteins recognize a hydrophobic cleft in actin (Dominguez, 2004, 2009). While we are unable to uniquely determine the orientation of Vt with respect to F-actin or identify specific residue-residue contacts given the resolution of our EM data (~20 Å), we have identified a novel surface of Vcn important for actin binding, supported not only by our mutagenesis and cellular data, but also by the current literature.
EXPERIMENTAL PROCEDURES
Vcn expression and purification
Expression of the tail domain of chicken Vcn (Vt, residues 879–1066) was performed as previously described (Palmer et al., 2009). Briefly, E. coli BL21-DE3 RIPL cells were transfected with a pET15b (Novagen) vector containing the cDNA for chicken Vcn residues 879–1066. Cells were grown at 37°C until an OD of 0.6 and were then induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) (0.25 mM) and the temperature was dropped to 18°C. Cells were grown for an additional 18 hours, centrifuged, and resuspended in lysis buffer. Cells were lysed by sonication and the lysate cleared by centrifugation. Vt was purified using Ni-NTA-agarose beads (Qiagen) and cation-exchange chromatography. Vt variants were generated by QuikChange site-directed mutagenesis (Stratagene) and sequences verified by DNA sequencing (Genewiz).
Full-length chicken Vcn and its variants were expressed and purified (Thievessen et al., 2013). The final product was evaluated by SDS-PAGE for purity. PMSF, benzamidine, antipain, and leupeptin were used to limit protease activity during purification.
Actin co-sedimentation assays
Actin binding and bundling by Vt were measured with a co-sedimentation assay as previously described (Shen et al., 2011). Actin binding by Vcn was measured in the same way, using 10 µM Vcn in the place of Vt. IpaA peptide was used at a concentration of 100 µM, in ten-fold excess to Vcn. The percent Vcn pelleted was determined in the same way as before. Briefly, the supernatant and pellet fractions were run on a gel, and the band intensity was calculated using ImageJ (Abramoff et al., 2004). Percent binding was determined by dividing the intensity of the pellet by the sum of the intensities of the pellet and supernatant and multiplying by 100%.
Lipid co-sedimentation assays
Vcn tail binding to phosphatidylinositol 4,5-bisphosphate (PIP2) was evaluated by lipid co-sedimentation assays using small, unilamellar vesicles (SUVs) as reported (Palmer et al., 2009). SUVs were generated using 250 µg lipid per reaction, with the reported PIP2% and a 3:1:1 ratio of phosphatidylethanolamine to phosphatidylcholine to phosphatidylserine and/or PIP2. The lipids were resuspended (40 mM MES pH 6.0, 150 mM NaCl, and 2 mM DTT) and subsequently extruded in a mini-extruder (Avanti Polar Lipids). Relative protein amounts were quantified using ImageJ (Abramoff et al., 2004).
EM sample preparation and analysis
G-actin was prepared from rabbit skeletal muscle (Strzelecka-Golaszewska et al., 1980) and clarified by chromatography over a Superdex-200 column. G-actin in complex with calcium was polymerized (20 mM imidazole-HCl, pH 7.2, 50 mM KCl, 2 mM MgCl2, and 1 mM EGTA) for 2–3 hours at 23°C. Decoration of actin filaments was performed on carbon-covered EM-grids. One drop of 1.5–2 µM F-actin was applied to the glow-discharged grid, blotted and then washed with 1–3 drops of 2.5 µM VtWT or Vt variants. The last drop was incubated up to 1 min, the grid was blotted and negatively stained with a 2% (w/v) solution of uranyl acetate.
A Tecnai-12 electron microscope at an accelerating voltage of 80 keV and a nominal magnification of 30X was employed. BSOFT package (Heymann and Belnap, 2007) was used to determine defocus values to correct for the contrast transfer function in the images. Images were digitized at a raster of 4.28Å/pixel, and 6416 segments (100 pixels long) were processed using the SPIDER (Frank et al., 1996) and IHRSR (Egelman, 2000) packages. Cross-correlation approach was used to extract segments of filaments fully decorated with Vt. The first of two models created was a model of actin filament (PDB 3MFP) (Fujii et al., 2010), while the second contained the actin filament model with Vt (PDB 1QKR) (Bakolitsa et al., 1999) attached to each of the actin protomers as suggested by Janssen et al (Janssen et al., 2006). Segments that yielded the best correlation with the second model (n=1716) converged to a helical solution of −167° rotation and 27.8 Å translation. The resolution of the resultant 3D-reconstruction was judged to be ~ 20 Å using the Fourier shell correlation equal to 0.5 criterion. UCSF Chimera software (Pettersen et al., 2004) was used to fit the model of the actin filament (PDB 3MFP) and the crystal structure of Vt (PDB 1QKR) into the experimental map. Atomic coordinates from crystal structures were converted to density maps, filtered to the resolution of the experimental map, and docked manually.
DMD model generation
The 6.6 Å electron cryo-microscopy map for F-actin was used to reconstruct the long-pitch helix F-actin dimer (Fujii et al., 2010). Parameters for accurate rotation and rise per subunit were obtained from the header of the corresponding protein databank deposition (PDB 3MFP). The actin dimer generated was used in the EM-fitting, done with Situs 2.5 (Wriggers, 2010). Prior to the fitting process, the 4-methyl histidine (HIC) at position 73 (PDB 3MFP) was replaced with a canonical histidine (HIS) for compatibility. Coordinates for Vt (residues 879–1065) were obtained from the crystal structure of Vcn (PDB 1ST6) (Bakolitsa et al., 2004). Density for Vt alone was obtained upon subtraction of the density corresponding to actin using Chimera (Pettersen et al., 2004). The crystal structure of Vt was fit into the isolated density using Situs 2.5 (Wriggers, 2010).
Steric clashes resulting from 3D reconstruction using EM constraints were resolved using Chiron (Ramachandran et al., 2011). Various orientations of Vt with respect to the actin dimer were sampled using DMD simulations (Ding et al., 2008; Dokholyan et al., 1998). The backbone of the actin dimer was maintained static during the simulations, while the side chains were allowed to freely sample different rotameric states. Rigid body movement of Vt was allowed to sample different orientations of Vt with respect to the actin dimer. In order to maintain Vt in the vicinity of the actin dimer for enhanced sampling, a distance constraint of 5 Å was imposed between the center of mass of the actin dimer and that of Vt. One thousand snapshots from the simulations were retrieved at regular time intervals and were clustered based on root mean square deviation. The centroid structure from the largest cluster was chosen for prediction of binding free energy change upon mutation using Eris (Ding and Dokholyan, 2006; Yin et al., 2007). Details pertaining to the force field used for simulations are presented elsewhere (Ding and Dokholyan, 2006; Ding et al., 2008; Dokholyan et al., 1998).
Cell culture
Vin−/− MEFs were obtained from Dr. Eileen Adamson (Burnham Institute) and grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 5% fetal bovine serum and antibiotic-antimycotic solution.
DNA constructs and transfection
DNA constructs were generated for cell culture as previously reported (Shen et al., 2011). Cells were transfected with Vcn expression constructs using Lipofectamine (Invitrogen) and Plus Reagent (Invitrogen) according to the manufacturer’s protocol and examined 48–72 hours following transfection.
Cell Resuspension and Spreading Assay
Prior to plating, cells were serum-starved in DMEM media supplemented with 0.5% delipidated BSA and antibiotic-antimycotic solution. Cells were then resuspended in the serum-free de-lipidated BSA media for approximately two hours. For the RTCA xCELLigence System (Acea Biosciences), 2500 cells per well were seeded into the E-plate 16 that were coated with 50 µg/mL FN. Attachment and spreading, monitored by impedance and reported as cell index (CI), was recorded with the RTCA apparatus every 15 seconds over 13 hours. For the adhesion site analysis, cells were prepared as described above prior to seeding onto glass coverslips containing FN (50 µg/mL).
Adhesion site analysis
Adhesion sites were analyzed as previously reported (Shen et al., 2011), except that cells were permeabilized in 0.5% Triton X-100 instead of 0.3%.
3D force microscopy
Three-dimensional force microscopy (3DFM) was used to apply controlled and precise 60–100 pN local force to focal adhesions. Tosyl-activated magnetic dynabeads (2.8 µm, Invitrogen) were washed with PBS and incubated for 24 hours with FN at 37°C. After three washes with PBS and incubation with 5% de-lipidated BSA (Sigma) for 1 hour at 37°C, the beads were sonicated and incubated with cells for 30 min. Force application and bead displacement were performed as previously described (Shen et al., 2011). The tracked displacements are reported as mean ± S.E.M. Two-tailed Student’s t test for p values were performed.
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this work was provided, in whole or in part, by the National Institutes of Health [GM081764 and GM080568 (S.L.C.), GM029860 (K.B.), GM081303 (E.H.E.), R01GM080742 (N.V.D.)], the National Science Foundation Graduate Research Fellowship [2008072760 (P.M.T)], and the American Heart Association [12PRE11820012 (P.M.T.)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.L.C. conceived of the project, with support from K.B., E.H.E., and N.V.K. P.M.T., K.S., S.M.P., and K.M.P. performed actin co-sedimentation experiments. P.M.T. performed the lipid co-sedimentation, CD, and NMR experiments. A.O., V.E.G, and E.H.E. collected and manually fit the EM data. P.K. and N.V.D. performed the DMD modeling and binding calculations. C.E.T. performed the cellular experiments and force measurements. R.S. designed and provides oversight for the 3DFM equipment. P.M.T., C.E.T., and S.L.C. wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with Image. J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Atienza JM, Zhu J, Wang X, Xu X, Abassi Y. Dynamic monitoring of cell adhesion and spreading on microelectronic sensor arrays. J Biomol Screen. 2005;10:795–805. doi: 10.1177/1087057105279635. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, Cohen DM, Bankston LA, Bobkov AA, Cadwell GW, Jennings L, Critchley DR, Craig SW, Liddington RC. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C, de Pereda JM, Bagshaw CR, Critchley DR, Liddington RC. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. Journal of Cell Biology. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. The Journal of biological chemistry. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- Coll JL, Ben-Ze'ev A, Ezzell RM, Rodriguez Fernandez JL, Baribault H, Oshima RG, Adamson ED. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc Natl Acad Sci U S A. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Computional Biology. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- Golji J, Mofrad MR. The interaction of vinculin with actin. PLoS Comput Biol. 2013;9:e1002995. doi: 10.1371/journal.pcbi.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C, van Eerde A, Parsot C, Broos J, Dijkstra BW. Structural mimicry for vinculin activation by IpaA, a virulence factor of Shigella flexneri. EMBO Rep. 2006;7:794–799. doi: 10.1038/sj.embor.7400753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur J Biochem. 1997;247:1136–1142. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- Hwang TL, Mori S, Shaka A, Van Zijl PCM. Application of phase-modulated CLEAN chemical EXchange spectroscopy (CLEANEX-PM) to detect water-protein proton exchange and intermolecular NOEs. Journal of the American Chemical Society. 1997;119:6203–6204. [Google Scholar]
- Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- Izard T, Tran Van Nhieu G, Bois PR. Shigella applies molecular mimicry to subvert vinculin and invade host cells. The Journal of cell biology. 2006;175:465–475. doi: 10.1083/jcb.200605091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen ME, Kim E, Liu H, Fujimoto LM, Bobkov A, Volkmann N, Hanein D. Three-dimensional structure of vinculin bound to actin filaments. Mol Cell. 2006;21:271–281. doi: 10.1016/j.molcel.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. The Journal of biological chemistry. 1994;269:12611–12619. [PubMed] [Google Scholar]
- Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–264. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. Actin activates a cryptic dimerization potential of the vinculin tail domain. The Journal of biological chemistry. 2000;275:95–105. doi: 10.1074/jbc.275.1.95. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Dwivedi SP, Didry D, Carlier MF. Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. The Journal of biological chemistry. 2010;285:23420–23432. doi: 10.1074/jbc.M110.102830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessey EC, Guilluy C, Burridge K. From Mechanical Force to RhoA Activation. Biochemistry. 2012;51:7420–7432. doi: 10.1021/bi300758e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marg S, Winkler U, Sestu M, Himmel M, Schonherr M, Bar J, Mann A, Moser M, Mierke CT, Rottner K, et al. The vinculin-DeltaIn20/21 mouse: characteristics of a constitutive, actin-binding deficient splice variant of vinculin. PLoS One. 2010;5:e11530. doi: 10.1371/journal.pone.0011530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkel AR, Kroemker M, Bubeck P, Ronsiek M, Nikolai G, Jockusch BM. Characterization of an F-actin-binding domain in the cytoskeletal protein vinculin. The Journal of cell biology. 1994;126:1231–1240. doi: 10.1083/jcb.126.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien ET, Cribb J, Marshburn D, Taylor RM, Superfine R. Magnetic Manipulation for Force Measurements in Cell Biology. Method Cell Biol. 2008;89 doi: 10.1016/S0091-679X(08)00616-X. 433-+. [DOI] [PubMed] [Google Scholar]
- Oakes PW, Beckham Y, Stricker J, Gardel ML. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. The Journal of cell biology. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SM, Playford MP, Craig SW, Schaller MD, Campbell SL. Lipid binding to the tail domain of vinculin: specificity and the role of the N and C termini. The Journal of biological chemistry. 2009;284:7223–7231. doi: 10.1074/jbc.M807842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Kota P, Ding F, Dokholyan NV. Automated minimization of steric clashes in protein structures. Proteins. 2011;79:261–270. doi: 10.1002/prot.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. Journal of Cell Biology. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RM, Holt MR, Jennings L, Sutton DH, Barsukov IL, Bobkov A, Liddington RC, Adamson EA, Dunn GA, Critchley DR. Role of vinculin in regulating focal adhesion turnover. Eur J Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Shen K, Tolbert CE, Guilluy C, Swaminathan VS, Berginski ME, Burridge K, Superfine R, Campbell SL. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. The Journal of biological chemistry. 2011;286:45103–45115. doi: 10.1074/jbc.M111.244293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle PA, Hoffert JD, Adey NB, Craig SW. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J Biol Chem. 1999;274:18414–18420. doi: 10.1074/jbc.274.26.18414. [DOI] [PubMed] [Google Scholar]
- Strzelecka-Golaszewska H, Prochniewicz E, Nowak E, Zmorzynski S, Drabikowski W. Chicken-gizzard actin: polymerization and stability. Eur J Biochem. 1980;104:41–52. doi: 10.1111/j.1432-1033.1980.tb04397.x. [DOI] [PubMed] [Google Scholar]
- Subauste MC, Pertz O, Adamson ED, Turner CE, Junger S, Hahn KM. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. The Journal of cell biology. 2004;165:371–381. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. The Journal of cell biology. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KK, Rubenstein PA, DeMali KA. Vinculin nucleates actin polymerization and modifies actin filament structure. The Journal of biological chemistry. 2009;284:30463–30473. doi: 10.1074/jbc.M109.021295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CK, Turner CE, Jackson P, Critchley DR. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J Cell Sci. 1994;107(Pt 2):709–717. [PubMed] [Google Scholar]
- Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.