Abstract
Objective
To characterize the pharmacokinetics and pharmacogenetics of nifedipine in pregnancy.
Study Design
Pregnant women receiving oral nifedipine underwent steady-state pharmacokinetic testing over one dosing interval. DNA was obtained and genotyped for cytochrome P450 (CYP) 3A5, and CYP3A4*1B. Nifedipine and oxidized nifedipine concentrations were measured in plasma and pharmacokinetic parameters were compared between those women who expressed a CYP3A5*1 allele and those who expressed only variant CYP3A5 alleles (*3,*6, or *7).
Results
Fourteen women had complete data to analyze. Four women (29%) expressed variant CYP3A5; three of these women were also CYP3A4*1B allele carriers. The mean half-life of nifedipine was 1.68±1.56 hours. AUC0–6 for the women receiving nifedipine every 6 hours was 207±138μg·h/L. Oral clearance was different between high expressers and low expressers (232.0±37.8 μg/mL vs. 85.6±45.0 μg/mL, respectively; p=0.007).
Conclusion
CYP3A5 genotype influences the oral clearance of nifedipine in pregnant women.
Keywords: nifedipine, pharmacokinetics, pharmacogenetics, pregnancy, tocolysis
Introduction
Nifedipine is a calcium channel blocker commonly used for preterm labor tocolysis. It has been demonstrated to be effective in delaying preterm delivery for at least 48 hours.1 A decision analysis and expert opinions propose that nifedipine may be considered a first line tocolytic drug.2,3 However, there is significant variability in tocolytic response to nifedipine, with some women experiencing rapid contraction cessation while others continue contracting and deliver despite the medication. Additionally, some women develop adverse effects, such as profound hypotension, after nifedipine administration while others have no significant adverse effects.4,5 Pharmacokinetic analyses of nifedipine used as a tocolytic reveal great interindividual variability in plasma concentrations even when women are given the same dose.6–8 In such situations where drug response and disposition are highly variable, genetic differences between individuals in drug metabolizing enzymes and transporters that act on the drug may be responsible.9,10 Understanding the pharmacogenetic relationships may lead to improved tocolysis and fewer drug-related adverse events with use of this important medication.
Nifedipine is metabolized primarily by the cytochrome P450 (CYP) 3A sub-family of enzymes.11 Unlike most other CYP3A substrates, there is no evidence of significant nifedipine transport by p-glycoprotein or other transporters.12 Genetic variations in CYP3A enzymes impact the metabolism and activity of several medications.13–15 However, pharmacogenetic influence on nifedipine disposition remains unclear.8,16
The objective of this Obstetric-Fetal Pharmacology Research Unit (OPRU) Network study was to characterize the pharmacokinetics and pharmacogenetics of nifedipine in pregnancy. We hypothesized that differences in CYP3A genotype would influence the pharmacokinetic disposition characteristics of nifedipine. Improved understanding of the pharmacogenomic biomarkers of interindividual variability in drug metabolism could then lead to optimized pharmacotherapy for pregnant women.
Materials and Methods
Subjects
This protocol was conducted at the clinical sites of the NICHD-funded OPRU Network (Georgetown University, University of Pittsburgh, University of Texas Medical Branch-Galveston, and University of Washington). This study was part of the Under-studied Drugs in Pregnancy Protocol in which pregnant women who were administered a physician-prescribed drug as part of medically-indicated clinical care were approached for a pharmacokinetic (PK) study of that drug. The primary objective of the Under-studied Drugs in Pregnancy Protocol was to estimate drug PK parameters in pregnant women for medications they were already receiving. This was not a treatment protocol and all clinical decisions about a woman’s care were made by the treating provider. The underlying hypothesis of the Under-studied Drugs in Pregnancy Protocol was that drug disposition would change significantly throughout pregnancy. Neither clinical outcomes nor maternal vital signs while taking the drugs were recorded as part of the protocol so no pharmacodynamic measures are reported. Inclusion criteria included women at least 18 years of age, ability to give written informed consent, and receiving nifedipine as prescribed by the provider for preterm labor tocolysis. Women were excluded if they had a hematocrit <28%.
Women who were receiving oral immediate release nifedipine to treat preterm labor contractions were recruited. Nifedipine was being administered and dosing determined by the subject’s treating clinical team. At the time of the study, subjects were receiving steady-state doses of 10–20 mg every 4–8 hours, per the clinician’s preference. On each study day, plasma was obtained at predefined times: 0 (trough), 0.5, 1, 1.5, 2, 3, 4, 6, and 8 hours post-dose, truncated to the dosing interval (i.e. at 4, 6, or 8 hours). Subjects were not allowed to eat for at least five hours before until at least one hour after the study dose was given. Blood was centrifuged at high speed for 10 minutes and the plasma aliquots stored at −80° until assayed. Whole blood was obtained from all subjects for DNA extraction and genotyping. While women recruited to the Under-studied Drugs in Pregnancy Protocol could be at any gestational age, women recruited on nifedipine were all between the gestational ages of 24 to 36 weeks. Women were also receiving standard clinical care, including antenatal corticosteroid therapy (beginning the day of admission or several days before the study dose) and antibiotic prophylaxis for Group B beta streptococcus when indicated. Provider-ordered intravenous fluid administration depended on clinically-assessed volume status. All subjects provided written informed consent for this protocol, which was approved by all governing Institutional Review Boards. Subjects were recruited between October 2007 and January 2010.
Nifedipine Assays
Nifedipine and oxidized nifedipine (2,6-Dimethyl-4-(2′-nitrophenyl)-3,5-pyridinecarboxylic acid dimethyl ester) were quantified by High-Performance Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (API 3200, AB Sciex, Foster City, CA). Nitrenpidine was added to 100 μL of plasma as internal standard. Following the addition of 200 μL of 2 M NaOH/1M glycine, pH 11.3, compounds were extracted from plasma with ethyl acetate. The organic phase was evaporated to dryness and compounds reconstituted in 10:90 acetonitrile:5mM ammonium acetate (mobile phase A) The compounds were injected onto a C8 50x4.6 mm, 5 micron column. A gradient elution methods was employed as follows: 100% mobile phase A linearly increased to 100% B (80:20 acetonitrile:5mM ammonium acetate) over 6 minutes; 100% B maintained for 1 minute before return to 100% A. The compounds were detected at Q1/Q3 mass spectrometer settings of: 347/315 for nifedipine, oxidized nifedipine 345/284, and 361/315 for nitrendipine. The assay was linear between 0.01 and 1000 ng/mL. Inter- and intra-day coefficients of variation were <15%.
Pharmacokinetic Analysis
Nifedipine and oxidized nifedipine pharmacokinetic parameters were determined using standard noncompartmental pharmacokinetic methods in Excel (Microsoft Corp., Redmond, WA.) Peak concentration (Cmax) and time to peak concentration (tmax) were determined by visual inspection of the concentration-time curve. Elimination rate constant (kel) was estimated by linear regression of the log concentration-time curve, and half-life estimated as ln2/kel. The area under the concentration time curve from time 0 to next subsequent dose ( ) was estimated using the trapezoidal rule. Average plasma concentration (Cave) was estimated as divided by the dosing interval (τ). Apparent oral clearance (Cl/F) was calculated as Dose/ and apparent volume of distribution (Vd/F) was estimated from apparent oral clearance divided by kel.
DNA Extraction and CYP3A Genotyping
Whole blood was stored and the DNA extracted using the QIAamp® DNA mini kit (Qiagen Inc., Valencia, CA). Manufacturer centrifuge and processing protocol instructions were followed for all kits. Isolated DNA was transferred into cryovials and all samples were stored at −80 ° C until analysis.
CYP3A5 genotyping was performed using real-time PCR and commercially available assays. CYP3A5 *3, *6, *7, and CYP3A4*1B genotypes were analyzed using SYBR green assays (rs776746, rs10264272, rs41303343, and rs2740574 respectively). Since these are variants most likely to result in reduced activity in the population studied, individuals who did not display variants for these SNPs were considered CYP3A high expressers (CYP3A5 *1 or CYP3A4*1, respectively). Those who expressed any CYP3A5 variant (*3,*6, or *7) were classified as CYP3A5 low expressers (CYP3A5*0).17
Statistical Analysis
Subjects with at least one CYP3A5*1 allele were categorized as CYP3A5 high expressers and subjects with two variant alleles (CYP3A5*0) were classified as low expressers. Pharmacokinetic parameters for nifedipine and oxidized nifedipine were compared between genotype groups using linear regression and ANOVA. As the majority of subjects were receiving 20 mg doses, all concentrations were normalized to that dose and corrected for subject weight. Continuous variables were compared using the Wilcoxon rank sum tests for nonparametric data, and categorical variables were compared using the chi-square test. All analyses were performed using R v. 2.13.1.18 Due to the clinical nature of the study and the expected difficulty in participant recruitment, sample size calculations were not performed a priori.
Results
Fourteen women were enrolled and studied. Mean maternal age was 29.2±8.2 years (range 16–39) and the gestational ages (EGA) at time of study were 32±4 weeks (range 24–336). The mean BMI of subjects was 31.1±5.8 kg/m2 (range 22.3–44.6). There were 4 African-American, 8 Caucasian, and 2 Hispanic women enrolled. Three African-American women and one Hispanic woman were heterozygous (high) expressers of CYP3A5*1 (minor allele frequency = 14.3%). Of these, one woman was a CYP3A4*1B homozygote and two were heterozygous for CYP3A4*1B. No other individuals carried the CYP3A4*1B allele. No woman was receiving a strong CYP3A inhibitor or inducer. Twelve women received nifedipine every 6 hours. One woman received nifedipine every 8 hours and one received nifedipine every 4 hours; both were Caucasian and CYP3A5 low expressers. All subjects had normal renal and hepatic function.
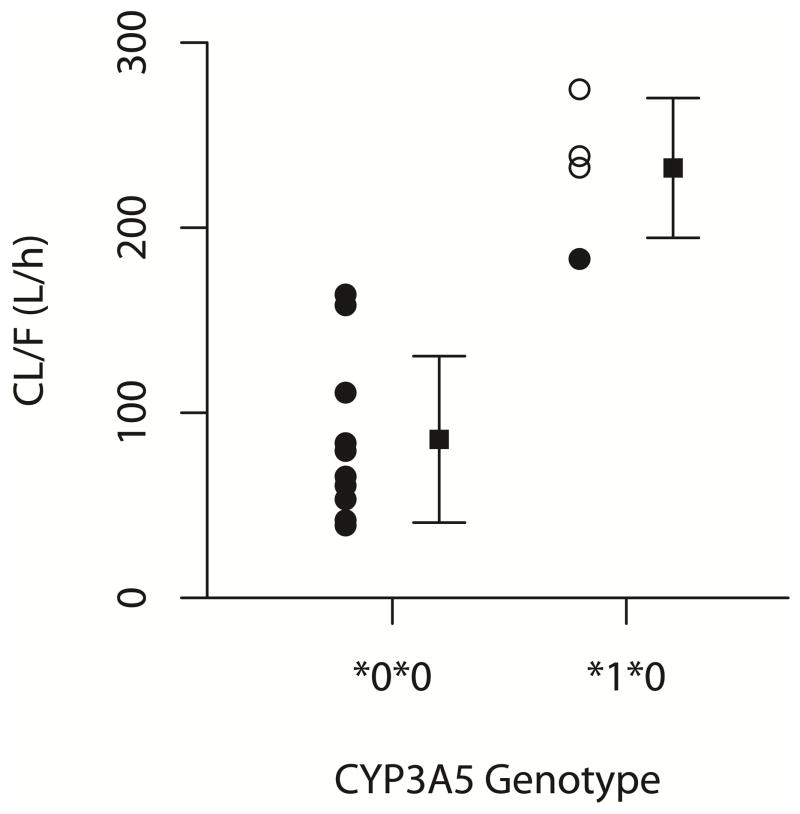
The pharmacokinetic parameters for nifedipine and oxidized nifedipine are presented in Table 1. One patient (CYP3A5 low expresser) had an extended half-life of 6.91 hrs. Exclusion of that individual resulted in a mean half-life of 1.28±0.42 hours. Mean oral clearance of nifedipine was 129±80.4 L/h. AUC0–6 for the women receiving nifedipine every 6 hours was 207±139 μg·h/L. Figure 1 displays the concentration-time curves for CYP3A5 high expressers compared to low expressers. The differences in the average plasma concentrations, CL/F, and Vd/F of nifedipine were all significant when comparing CYP3A5 high expressers and low expressers (Table 2, Figure 2, CL/F: high expressers 232±37.8L/h; low expressers 85.6±45 L/h, p=0.000095). The association between CYP3A4*1B and CL/F was also significant (249±22.9 L/h for CYP3A4*1B allele carriers vs. 94.5±51.8 L/h for CYP3A4 wild type, p=0.0004). The haplotype combination (CYP3A4*1B + CYP3A5*1) was significantly associated with increased CL/F (p = 0.0005).
Table 1.
Steady-state pharmacokinetic parameters of nifedipine and its oxidized metabolite during pregnancy
| Parameter | Nifedipine | Oxidized Nifedipine |
|---|---|---|
| Cmax (μg/L) | 165±112 (40.6 – 397) | 89.9±80.9 (14.4 – 236) |
| Cave (μg/L) | 41.2±31.0 (12.2–119) | 20.4±15.0 (4.48 – 56.1) |
| kel (h−1) | 0.55±0.20 (0.10 – 0.87) | 0.59±0.34 (0.15 – 1.3) |
| t1/2 (h) | 1.68±1.56 (0.79 – 6.91) | 1.67±1.19 (0.54 – 4.7) |
| CL/F (L/h) | 128±80.4 (39.1–274.8) | |
| Vd/F(L) | 295±262 (60.6–832) |
Data are presented as mean ± standard deviation (range).
Figure 1.
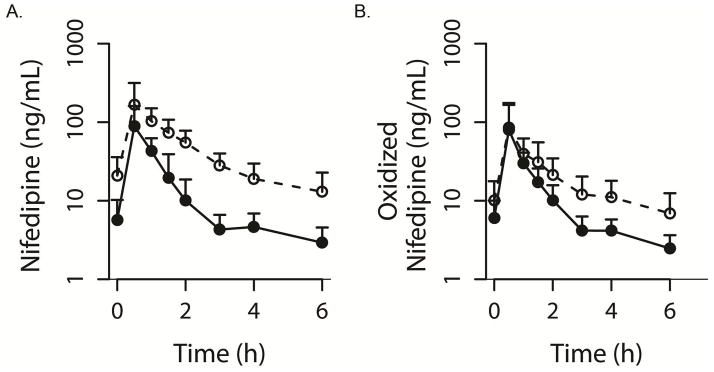
Concentration-time curves for nifedipine (A) and oxidized nifedipine (B) in women who are CYP3A5 high expressers (solid line) versus CYP3A5 low expressers (dashed line).
Table 2.
Comparison of pharmacokinetic parameters of nifedipine for CYP3A5 high expressers vs. low expressers
| CYP3A5 high expressers (n=3) | CYP3A5 low expressers (n=10) | P-value | |
|---|---|---|---|
| Cmax (μg/L) | 102±56.5 (60.8–184) | 190±121 (40.6–397) | 0.2 |
| Cave (μg/L) | 14.7±2.5 (12.2–18.2) | 51.8±30.8 (20.3–119) | 0.002 |
| kel (h−1) | 0.49±0.18 (0.29–0.71) | 0.58±0.21 (0.10–0.88) | 0.5 |
| t1/2 (h) | 1.6±0.6 (0.98–2.35) | 1.7±1.8 (0.8–6.9) | 0.5 |
| Cl/F (L/h) | 232±37.8 (183–275) | 85.6±45.0 (39.1–164) | 0.000095 |
| Vd/F (L) | 524±207 (331–788) | 204±228 (60.6–832) | 0.02 |
Data are presented as mean±sd (range)
CYP3A5 high expressers carry at least one *1 allele whereas nonexpressers all carried the *3/*3 alleles.
Figure 2.
Effect of CYP3A genotype on oral clearance of nifedipine. CYP3A4*1B allele carriers are indicated by open circles.
There were no statistically significant associations of oxidized nifedipine concentration or half-life with any genotypes studied. There were also no significant correlations between any PK parameters and any other demographic variables such as caffeine intake, maternal age, or body mass index (data not shown). Exclusion of the two subjects with 4 or 8 hour dosing intervals did not alter the overall results (data not shown).
Comment
This study adds to the developing literature documenting that the pharmacokinetics of commonly used drugs in pregnancy. The OPRU has documented such differences for clonidine, glyburide, metformin, and amoxicillin among others.19–22. The current study supports previous findings of a shorter half-life and lower concentrations for nifedipine in pregnancy.23,24 Nonpregnant subjects have a reported half-life of 2–3.4 hours or more.25–27 The half-life in the current study was under 2 hours. Mean nifedipine clearance in young healthy nonpregnant females is 35–38 L/h28, significantly lower than that observed in our pregnant population, 128 L/h. Interestingly, the Cmax noted in the current study is higher than in several other studies.
Nifedipine is prescribed for the treatment of preterm labor; however, optimal dosing regimens are unclear. Previous studies describing loading-dose nifedipine PK in pregnant women have been small.6,7 Two studies examined nifedipine during the initial loading dose, administered as 10 mg tablets sublingually. Ferguson et al. estimated the half-life following the final sublingual loading dose to be 81±26 minutes (range = 49–137) 6, similar to our observations. By contrast, Papatsonis et al. employed a nifedipine regimen of 10 mg capsules every 15 minutes for four doses, followed by a 20 mg slow release tablet 60 minutes after the last 10 mg dose 7. Mean plasma concentrations were 67±28 ng/mL. In fifteen women taking 10 mg nifedipine every 6 hours for gestational hypertension, oral clearance was 146±63 L/hr, consistent with our results (128±80 L/h). 24. Similarly, in twenty women being treated for preterm labor with 10 mg nifedipine every 6 hours the mean steady-state dose-interval AUC was 86.1±61.1 μg·h/L 8, which is comparable to our results, dose-adjusted to 10mg, for the 12 women receiving nifedipine every 6 hours (104 μg·h/L).
Nifedipine is oxidized primarily by CYP3A enzymes.11 In humans, the CYP3A isoforms are CYP3A4, CYP3A5, and CYP3A7. CYP3A5 is highly polymorphic, with active CYP3A5 enzyme expressed in only 10 to 20% of Caucasians but in 75% of African-Americans.29 CYP3A7 is primarily expressed in fetal liver. CYP3A4 is 5–20-fold more efficient at metabolizing nifedipine than CYP3A5 in recombinant enzyme systems and CYP3A7 has very low activity toward nifedipine.30 The CYP3A4*1B variant possesses a SNP in the nifedipine-specific response element of the 5-prime promoter region of the CYP3A4 gene 31,32, that has been associated with increased promoter activity.33 However, functional effects of this variation on drug metabolism are controversial.34–36 As observed in this study, strong linkage disequilibrium exists between CYP3A4*1B and CYP3A5, making it difficult to distinguish between the effects of these individual SNPs.37,38
The CYP3A4*1B/CYP3A5 haplotype was significantly associated with average steady-state nifedipine plasma concentrations in our study of pregnant women. Women expressing CYP3A5*1/CYP3A4*1B had much lower concentrations of nifedipine than the CYP3A5*0/CYP3A4*1 expressers. This may have an impact on the likelihood of successful tocolysis following nifedipine. CYP3A5*1 is more frequently expressed in African-Americans than in Caucasians31 (PharmGKB.org), as was the case in this study, with three out of four African-Americans carrying the functional allele. Due to the small sample size, race was not significantly associated with nifedipine clearance (p>0.05). However, it may be expected that African-Americans are more likely to express SYP3A5*1 than Caucasians. In the one previous study examining the impact of genotype on nifedipine tocolysis, women with genotypes expected to result in high expression of CYP3A5 had less improvement in contraction frequency compared to low expressers.8 In that study, CYP3A4 genotype was not associated with either PK or tocolytic outcomes. CYP3A5 genotypes associated with high enzyme expression are associated with lower medication concentrations in the case of a few agents such as midazolam.39 Our data support a similar association between CYP3A5 genotype and nifedipine plasma concentrations. While the current study did not monitor pharmacodynamic outcomes such as contraction frequency or blood pressure, we hypothesize that women who are low expressers of CYP3A5 will have higher nifedipine concentrations, resulting in both fewer contractions and greater vasodilation.
The effect of CYP3A5 polymorphisms on nifedipine pharmacokinetics is unclear. In vitro studies indicate that nifedipine has lower intrinsic clearance in CYP3A5 variants.15 However, clinical studies examining the effect of CYP3A5 expression are contradictory. In a previous study of nifedipine for preterm labor tocolysis (n=20), CYP3A5 genotype high expressers had higher exposure to nifedipine than did low expressers.8 However, firm conclusions of CYP3A5 effect in that study were limited due to the presence of several women in both groups who were receiving CYP3A inhibitors. Our study excluded women taking any known CYP3A inhibitors. Fukuda et al.16 examined the pharmacokinetics of nifedipine following a single 10 mg oral dose in 16 healthy male Japanese volunteers genotyped as CYP3A5*1*3 (n=8) or CYP3A5*3*3 (n=8). No significant difference was observed in AUC0 →∞ (219±80.9 vs. 178±92.8 ng/ mL/h for CYP3A5*1*3 and CYP3A5*3*3, respectively). In contrast, we found a significant difference in average steady-state concentrations between CYP3A5 high and low expressers. This may be due to differences in metabolism between pregnant women and healthy males or the effect of CYP3A4*1B expression on nifedipine metabolism, as this variant was not examined by Fukuda et al. Additional CYP3A5 variants, other than those tested for (CYP3A5*3, *6, and *7), may impact nifedipine metabolism as well.15
Our study was limited by its small sample size. However, meticulous sampling and characterization of the samples, sensitive and specific analytical methods, and study of the drug at steady-state aid in the interpretation of our results and their extrapolation to clinical pregnant populations. In addition, post hoc analyses indicate that our sample size provided 90% power to detect a 2-fold difference in oral clearance between CYP3A5 genotypes, with α=0.05. Due to the high linkage disequilibrium between CYP3A5 and CYP3A4*1B genotypes, separating out the effect of the individual SNPs would require a much larger sample size. We did not analyze the concentration of nifedipine in cord blood as most of the women were no longer receiving the drug at the time of delivery. Other studies have reported umbilical cord plasma nifedipine concentrations at delivery to be 57–80% of maternal concentrations 40,41, suggesting roles for placental and fetal metabolism or transport. Thus, there is potential that in addition to maternal genotype, fetal genotype may also play a role in determining fetal drug exposure to nifedipine.42 Since the role of placental CYP3A remains controversial, we plan to study placental transfer and metabolism of nifedipine in future studies. Additionally, our results suggest the utility of future protocols to simultaneously model tocolytic and hypotensive pharmacodynamic responses along with nifedipine PK in order to better individualize therapy for pregnant women in preterm labor. Research combining strict entry criteria similar to those in this study, with the robust clinical pharmacodynamic data is currently being planned.
Drug metabolism can be impacted by genetic and environmental factors. As observed for nifedipine, CYP3A5 genotype may impact drug PK parameters. Other drugs commonly prescribed to pregnant women that are metabolized by the polymorphic CYP3A enzymes include erythromycin, fentanyl, and ondansetron. Other enzymes are also highly polymorphic, including CYP2C9 and CYP2D6. CYP2D6 is responsible for activating metabolism of opioid analgesics. In addition to genetic variations, several medications have the potential to inhibit drug metabolizing enzymes. For instance, CYP3A is inhibited by commonly prescribed drugs such as erythromycin, cimetidine, and drinking grapefruit juice. The environmental and genetic factors affecting drug pharmacokinetics, and thereby activity, are important considerations for optimal drug utilization.
Individualized pharmacotherapy in pregnancy has only recently been applied to the care of pregnant women. With several options available for treatment of preterm labor43, and with the variability in response to any of these agents, this area of obstetrical therapy is ideal for individualized therapy. While many factors may affect drug metabolism and response, especially in the complex state of pregnancy, our results suggest that, as in cardiovascular and oncologic therapeutics,44–46 pharmacogenetic analyses are crucial to design of clinical trials as they may contribute to both therapeutic and adverse drug effects.
In conclusion, CYP3A5 genotype is associated with changes in the steady-state average concentration of nifedipine when used as a tocolytic. Incorporating pharmacogenetic data into therapeutic models may lead to improved pharmacotherapy in pregnancy.
Acknowledgments
This research was supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development to the Obstetric-fetal Pharmacology Research Units Network (#5U10HD063094, 5U10HD047892, 5U10HD047890, 2U10HD04789107, 1U10HD057753) and the NIH National Center for Research Resources grant # UL1RR025014 and UL1RR031975. The views expressed in this manuscript represent the authors and not those of the National Institutes of Health.
Footnotes
Reprints not available from authors
Disclosure: None of the authors have a conflict of interest.
References
- 1.King JF, Flenady VJ, Papatsonis DN, Dekker GA, Carbonne B. Calcium channel blockers for inhibiting preterm labour. Cochrane Database Syst Rev. 2003;(1):CD002255. doi: 10.1002/14651858.CD002255. [DOI] [PubMed] [Google Scholar]
- 2.Grimes DA, Nanda K. Magnesium sulfate tocolysis: time to quit. Obstet Gynecol. 2006;108(4):986–989. doi: 10.1097/01.AOG.0000236445.18265.93. [DOI] [PubMed] [Google Scholar]
- 3.Haas DM, Imperiale TF, Kirkpatrick PR, et al. Tocolytic Therapy: A Meta-Analysis and Decision Analysis. Obstetrics & Gynecology. 2009;113(3):585–594. doi: 10.1097/AOG.0b013e318199924a. [DOI] [PubMed] [Google Scholar]
- 4.Goldenberg RL. The management of preterm labor. Obstet Gynecol. 2002;100(5 Pt 1):1020–1037. doi: 10.1016/s0029-7844(02)02212-3. [DOI] [PubMed] [Google Scholar]
- 5.Hearne AE, Nagey DA. Therapeutic agents in preterm labor: tocolytic agents. Clin Obstet Gynecol. 2000;43(4):787–801. doi: 10.1097/00003081-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson JE, 2nd, Schutz T, Pershe R, Stevenson DK, Blaschke T. Nifedipine pharmacokinetics during preterm labor tocolysis. American Journal of Obstetrics & Gynecology. 1989;161(6 Pt 1):1485–1490. doi: 10.1016/0002-9378(89)90909-5. [DOI] [PubMed] [Google Scholar]
- 7.Papatsonis DNM, Bos JM, Van Geijn HP, Lok CAR, Dekker GA. Nifedipine pharmacokinetics and plasma levels in the management of preterm labor. Am J Ther. 2007;14:346–350. doi: 10.1097/01.mjt.0000209679.76335.df. [DOI] [PubMed] [Google Scholar]
- 8.Haas DM, Quinney SK, McCormick CL, Jones DR, Renbarger JL. A pilot study of the impact of genotype on nifedipine pharmacokinetics when used as a tocolytic. J Matern Fetal Neonatal Med. 2011 doi: 10.3109/14767058.2011.583700. in press. [DOI] [PubMed] [Google Scholar]
- 9.Andersson T, Flockhart DA, Goldstein DB, et al. Drug-metabolizing enzymes: evidence for clinical utility of pharmacogenomic tests. Clin Pharmacol Ther. 2005;78(6):559–581. doi: 10.1016/j.clpt.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Swen JJ, Huizinga TW, Gelderblom H, et al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 2007;4(8):e209. doi: 10.1371/journal.pmed.0040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: role of cyp3a4 and cyp3a5. Drug Metabolism & Disposition. 2003;31(7):938–944. doi: 10.1124/dmd.31.7.938. [DOI] [PubMed] [Google Scholar]
- 12.Kim RB, Wandel C, Leake B, Cvetkovic M. Interrelationship Between Substrates and Inhibitors of Human CYP3A and P-Glycoprotein. Pharmaceutical Research. 1999;16(3):408–408. 414. doi: 10.1023/a:1018877803319. [DOI] [PubMed] [Google Scholar]
- 13.Cholerton S, Daly AK, Idle JR. The role of individual human cytochromes P450 in drug metabolism and clinical response. Trends Pharmacol Sci. 1992;13(12):434–439. doi: 10.1016/0165-6147(92)90140-2. [DOI] [PubMed] [Google Scholar]
- 14.Lee S-J, Bell DA, Coulter SJ, Ghanayem B, Goldstein JA. Recombinant CYP3A4*17 is defective in metabolizing the hypertensive drug nifedipine, and the CYP3A4*17 allele may occur on the same chromosome as CYP3A5*3, representing a new putative defective CYP3A haplotype. J Pharmacol Exp Ther. 2005;313(1):302–309. doi: 10.1124/jpet.104.078758. [DOI] [PubMed] [Google Scholar]
- 15.Lee S-J, Usmani KA, Chanas B, et al. Genetic findings and functional studies of human CYP3A5 single nucleotide polymorphisms in different ethnic groups. Pharmacogenetics. 2003;13(8):461–472. doi: 10.1097/00008571-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Onishi S, Fukuen S, et al. CYP3A5 genotype did not impact on nifedipine disposition in healthy volunteers. Pharmacogenomics Journal. 2004;4(1):34–39. doi: 10.1038/sj.tpj.6500218. [DOI] [PubMed] [Google Scholar]
- 17.Egbelakin A, Ferguson MJ, MacGill EA, et al. Increased risk of vincristine neurotoxicity associated with low CYP3A5 expression genotype in children with acute lymphoblastic leukemia. Pediatr Blood Cancer. 2011;56(3):361–367. doi: 10.1002/pbc.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Accessed 7-8-10]. Available at http://www.R-project.org. [Google Scholar]
- 19.Andrew MA, Easterling TR, Carr DB, et al. Amoxicillin pharmacokinetics in pregnant women: modeling and simulations of dosage strategies. Clin Pharmacol Ther. 2007;81(4):547–556. doi: 10.1038/sj.clpt.6100126. [DOI] [PubMed] [Google Scholar]
- 20.Buchanan ML, Easterling TR, Carr DB, et al. Clonidine pharmacokinetics in pregnancy. Drug Metab Dispos. 2009;37(4):702–705. doi: 10.1124/dmd.108.024984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyal S, Easterling TR, Carr D, et al. Pharmacokinetics of metformin during pregnancy. Drug Metab Dispos. 2010;38(5):833–840. doi: 10.1124/dmd.109.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert MF, Ma X, Naraharisetti SB, et al. Are we optimizing gestational diabetes treatment with glyburide? The pharmacologic basis for better clinical practice. Clin Pharmacol Ther. 2009;85(6):607–614. doi: 10.1038/clpt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton JR, Prevost RR, Wilson DA, Whybrew WD, Sibai BM. Nifedipine pharmacokinetics and pharmacodynamics during the immediate postpartum period in patients with preeclampsia. American Journal of Obstetrics & Gynecology. 1991;165(4 Pt 1):951–954. doi: 10.1016/0002-9378(91)90446-x. [DOI] [PubMed] [Google Scholar]
- 24.Prevost RR, Akl SA, Whybrew WD, Sibai BM. Oral nifedipine pharmacokinetics in pregnancy-induced hypertension. Pharmacotherapy. 1992;12(3):174–177. [PubMed] [Google Scholar]
- 25.Sorkin EM, Clissold SP, Brogden RN. Nifedipine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy, in ischaemic heart disease, hypertension and related cardiovascular disorders. Drugs. 1985;30(3):182–274. doi: 10.2165/00003495-198530030-00002. [DOI] [PubMed] [Google Scholar]
- 26.Taburet AM, Singlas E, Colin JN, et al. Pharmacokinetic studies of nifedipine tablet. Correlation with antihypertensive effects. Hypertension. 1983;5(4 Pt 2):II29–33. doi: 10.1161/01.hyp.5.4_pt_2.ii29. [DOI] [PubMed] [Google Scholar]
- 27.Product Information: Procardia(R), nifedipine capsules. Pfizer labs; Clearwater, FL: 2000. [Google Scholar]
- 28.Balogh A, Gessinger S, Svarovsky U, et al. Can oral contraceptive steroids influence the elimination of nifedipine and its primary pryidine metabolite in humans? Eur J Clin Pharmacol. 1998;54(9–10):729–734. doi: 10.1007/s002280050543. [DOI] [PubMed] [Google Scholar]
- 29.Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR. Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics. 2004;5(3):243–272. doi: 10.1517/phgs.5.3.243.29833. [DOI] [PubMed] [Google Scholar]
- 30.Williams JA, Ring BJ, Cantrell VE, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30(8):883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 31.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 32.Rebbeck TR, Jaffe JM, Walker AH, Wein AJ, Malkowicz SB. Modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 1998;90(16):1225–1229. doi: 10.1093/jnci/90.16.1225. [DOI] [PubMed] [Google Scholar]
- 33.Amirimani B, Ning B, Deitz AC, et al. Increased transcriptional activity of the CYP3A4*1B promoter variant. Environ Mol Mutagen. 2003;42(4):299–305. doi: 10.1002/em.10199. [DOI] [PubMed] [Google Scholar]
- 34.Wandel C, Witte JS, Hall JM, et al. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5′-promoter region polymorphism. Clin Pharmacol Ther. 2000;68(1):82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 35.Spurdle AB, Goodwin B, Hodgson E, et al. The CYP3A4*1B polymorphism has no functional significance and is not associated with risk of breast or ovarian cancer. Pharmacogenetics. 2002;12(5):355–366. doi: 10.1097/00008571-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 36.Stamer UM, Lee EH, Rauers NI, et al. CYP2D6- and CYP3A-dependent enantioselective plasma concentrations of ondansetron in postanesthesia care. Anesth Analg. 2011;113(1):48–54. doi: 10.1213/ANE.0b013e31821d01bc. [DOI] [PubMed] [Google Scholar]
- 37.Wojnowski L, Hustert E, Klein K, et al. Re: modification of clinical presentation of prostate tumors by a novel genetic variant in CYP3A4. J Natl Cancer Inst. 2002;94(8):630–631. doi: 10.1093/jnci/94.8.630. author reply 631–632. [DOI] [PubMed] [Google Scholar]
- 38.Miao J, Jin Y, Marunde RL, et al. Association of genotypes of the CYP3A cluster with midazolam disposition in vivo. Pharmacogenomics J. 2009;9(5):319–326. doi: 10.1038/tpj.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinney SK, Miao J, Jin Y, et al. CYP3A gene cluster analysis and midazolam pharmacokinetics. Clin Pharmacol Ther. 2009;85(S1):PI-54. [Google Scholar]
- 40.Manninen AK, Juhakoski A. Nifedipine concentrations in maternal and umbilical serum, amniotic fluid, breast milk and urine of mothers and offspring. Int J Clin Pharmacol Res. 1991;11(5):231–236. [PubMed] [Google Scholar]
- 41.Pirhonen JP, Erkkola RU, Ekblad UU, Nyman L. Single dose of nifedipine in normotensive pregnancy: nifedipine concentrations, hemodynamic responses, and uterine and fetal flow velocity waveforms. Obstetrics & Gynecology. 1990;76(5 Pt 1):807–811. doi: 10.1097/00006250-199011000-00016. [DOI] [PubMed] [Google Scholar]
- 42.Atkinson DE, Brice-Bennett S, D’Souza SW. Antiepileptic medication during pregnancy: does fetal genotype affect outcome? Pediatr Res. 2007;62(2):120–127. doi: 10.1203/PDR.0b013e3180a02e50. [DOI] [PubMed] [Google Scholar]
- 43.ACOG practice bulletin. Management of preterm labor Number 43, May 2003. Obstet Gynecol. 2003;101:1039–1047. [Google Scholar]
- 44.Scott SA, Sangkuhl K, Gardner EE, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450-2C19 (CYP2C19) Genotype and Clopidogrel Therapy. Clin Pharmacol Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asseburg C, Frank M, Kohne CH, et al. Cost-effectiveness of targeted therapy with cetuximab in patients with K-ras wild-type colorectal cancer presenting with initially unresectable metastases limited to the liver in a German setting. Clin Ther. 2011;33(4):482–497. doi: 10.1016/j.clinthera.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Ibrahim EM, Zekri JM, Bin Sadiq BM. Cetuximab-based therapy for metastatic colorectal cancer: a meta-analysis of the effect of K-ras mutations. Int J Colorectal Dis. 2010;25(6):713–721. doi: 10.1007/s00384-010-0927-4. [DOI] [PubMed] [Google Scholar]