Abstract
Tandem helical repeats have emerged as an important DNA binding architecture. DNA glycosylase AlkD, which excises N3- and N7-alkylated nucleobases, uses repeating helical motifs to bind duplex DNA and to selectively pause at non-Watson-Crick base pairs. Remodeling of the DNA backbone promotes nucleotide flipping of the lesion and the complementary base into the solvent and toward the protein surface, respectively. The important features of this new DNA binding architecture that allow AlkD to distinguish between damaged and normal DNA without contacting the lesion are poorly understood. Here, we show through extensive mutational analysis that DNA binding and N3-methyladenine (3mA) and N7-methylguanine (7mG) excision are dependent upon each residue lining the DNA binding interface. Disrupting electrostatic or hydrophobic interactions with the DNA backbone substantially reduced binding affinity and catalytic activity. These results demonstrate that residues seemingly only involved in general DNA binding are important for catalytic activity and imply that base excision is driven by binding energy provided by the entire substrate interface of this novel DNA binding architecture.
Keywords: Base excision repair, DNA glycosylase, Protein-DNA interaction, HEAT repeat, ALK motif, Alkylpurine
1. Introduction
Genomic integrity is continuously threatened by chemical modifications caused by endogenous metabolites and environmental toxins. Alkylation, oxidation, and deamination produce a variety of single-base lesions with mutagenic and cytotoxic effects [1]. Each type of chemical alteration, however, is recognized by a DNA glycosylase that cleaves the N-glycosidic bond to liberate the damaged base and initiate base excision repair [2]. Recognition by most DNA glycosylases is achieved by penetration of the protein into the DNA duplex to exploit structural and/or energetic differences between normal and damaged bases pairs, and by flipping the damaged nucleobase into the enzyme active site [3].
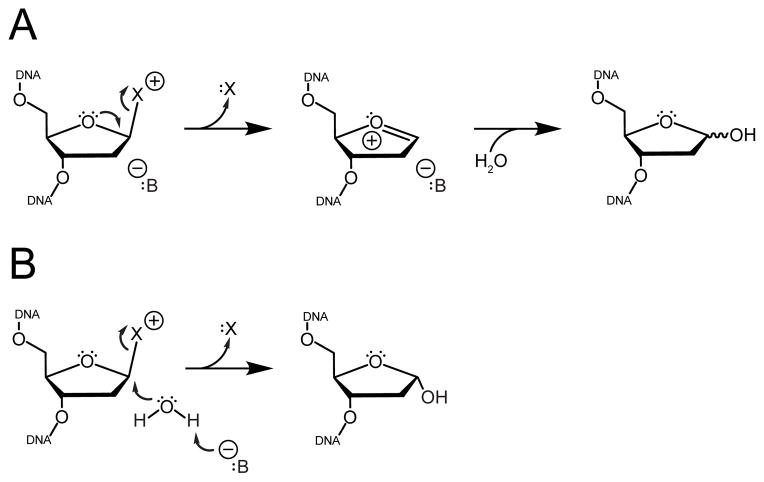
The glycosylase AlkD selectively excises positively charged N3- and N7-methylpurines by a mechanism distinct from other monofunctional glycosylases [4–7]. AlkD is structurally unique, composed of six tandem helical ALK repeats that bear resemblance to HEAT motifs normally associated with protein interactions [8]. Crystal structures of AlkD in complex with DNA containing a 3mA mimetic and an abasic site analogue revealed that, unlike other glycosylases, AlkD does not contact the damaged nucleotide, but rather anchors itself to the DNA duplex through interactions with the strand directly opposite the lesion and with regions of the damaged strand several nucleotides away from the lesion [7]. The enzyme-bound DNA is trapped in a distorted conformation in which the lesion is flipped toward the solvent and away from the protein, and the complementary nucleotide is flipped toward the protein surface. This arrangement is inconsistent with the typical catalytic mechanism of monofunctional glycosylases because it precludes a side-chain carboxylate from stabilizing the oxocarbenium intermediate formed during N-glycosidic bond cleavage or activating a water for nucleophilic attack at the anomeric C1′ carbon (Figure 1) [9–14].
Figure 1. Mechanisms of cationic lesion excision by monofunctional DNA glycosylases.
Cleavage of the N-glycosidic bond liberates the damaged nucleobase (“X”) and generates an abasic site. (A) Dissociative mechanisms utilize anionic protein side chains or DNA phosphate groups (“B”) to directly assist bond breakage and to stabilize the resulting oxocarbenium intermediate. (B) Associative mechanisms employ a general base (“B”) to activate a water for nucleophilic attack at the anomeric C1′ carbon.
Alternatively, inhibition of AlkD catalyzed 7mG excision by methylphosphonate substitution suggests that the distorted DNA backbone conformation may allow a phosphate in close proximity to the lesion to provide electrostatic stabilization or nucleophilic activation through a substrate-assisted mechanism of catalysis [15], as previously observed for uracil excision from DNA by UDG [11, 16, 17]. Such a mechanism would suggest that without contacts to the lesion, AlkD must rely on enzyme-substrate binding energy to distort the DNA backbone into an autocatalytic conformation to promote base excision. Here, we demonstrate through mutational analysis that residues throughout the concave DNA binding surface, including residues distant from the lesion, affect binding and catalysis. These required, concerted interactions with multiple sites on both damaged and undamaged strands are consistent with a mechanism in which the DNA backbone must be remodeled into a catalytic conformation.
2. Materials and methods
2.1. Protein purification
AlkD was purified by immobilized metal affinity, heparin affinity, and size exclusion chromatography steps as previously described [6]. AlkD mutants were generated by site-directed mutagenesis using a Quik-Change kit (Stratagene) and purified in the same manner as wild-type AlkD (Figure S1).
2.2. Thermal melting
Structural integrity of AlkD mutants was verified by far-UV circular dichroism spectroscopy using a J-810 spectropolarimeter (Jasco) and a Peltier temperature controller (Jasco). Molar ellipticity was monitored at 222 nm as mixtures containing 10 μM enzyme, 20 mM Bis-Tris propane (pH 6.5), 100 mM NaCl, and 0.1 mM EDTA were heated at 2°C/min in a 0.1-cm cell. Melting temperatures (Tm) were derived by fitting the data to the equation θ = 1/(1+e(Tm−T)/k), where θ is molar ellipticity, T is temperature, Tm corresponds to the temperature at 50% denaturation, and k describes the cooperativity of the transition (Figure S2).
2.3. DNA binding
DNA binding was monitored by changes in fluorescence anisotropy as enzyme was added to a 25-mer oligonucleotide duplex [d(GACCACTACACCXATTCCTAACAAC)/d(GTTGTTAGGAATYGGTGTAGTGGTC)-FAM] labeled with 6-carboxyfluorescein (FAM). Unmodified G•C-DNA contained X=G and Y=C, mismatched G•T DNA contained X=G and Y=T, and abasic THF•C-DNA contained X=THF and Y=C. Enzyme (0–30 μM AlkD-D113N and AlkD-R148A; 0–75 μM wild-type AlkD and all other mutants) was added to 50 nM FAM-DNA, 20 mM Bis-Tris propane (pH 6.5), 100 mM NaCl, 2 mM DTT, and 0.1 mM EDTA and incubated at 25°C for 10 min. Fluorescence anisotropy measurements were recorded as previously described [6]. Equilibrium dissociation constants (Kd) were derived by fitting a two-state binding model to the data (Figure S3).
2.4. Base excision from oligonucleotide substrate
Excision of 7mG from a 25-mer oligonucleotide duplex [d(GACCACTACACC(7mG)ATTCCTTACAAC)/d(GTTGTAAGGAATCGGTGTAGTGGTC)] was measured by autoradiography as previously described [6]. Reactions were performed at 37°C and contained 20 μM enzyme (5 μM AlkD-D113N and AlkD-R148A), 2 nM DNA, and glycosylase buffer [50 mM HEPES (pH 7.5), 100 mM KCl, 10 mM DTT, and 2 mM EDTA]. Due to thermal instability, reactions containing AlkD-D113N (Tm = 30.7°C) were also carried out at 25°C to rule out inactivity due to protein unfolding. Second-order rate constants (kobs) were obtained from single-exponential fits to the data (Figure S4).
2.5. Base excision from genomic DNA substrate
Excision of 3mA and 7mG from methylated calf thymus DNA was measured by HPLC-MS/MS as previously described [18]. Reactions were performed at 37°C for 1 h and contained 5 μM enzyme, 10 μg DNA, glycosylase buffer, and 0.1 mg/mL BSA. Due to thermal instability, reactions containing AlkD-D113N (Tm = 30.7°C) were also carried out at 25°C to rule out inactivity due to protein unfolding. In lieu of enzyme, controls contained 5 N HCl or 2 mM Bis-Tris propane (pH 6.5), 10 mM NaCl, and 0.01 mM EDTA.
3. Results and discussion
3.1. DNA binding architecture
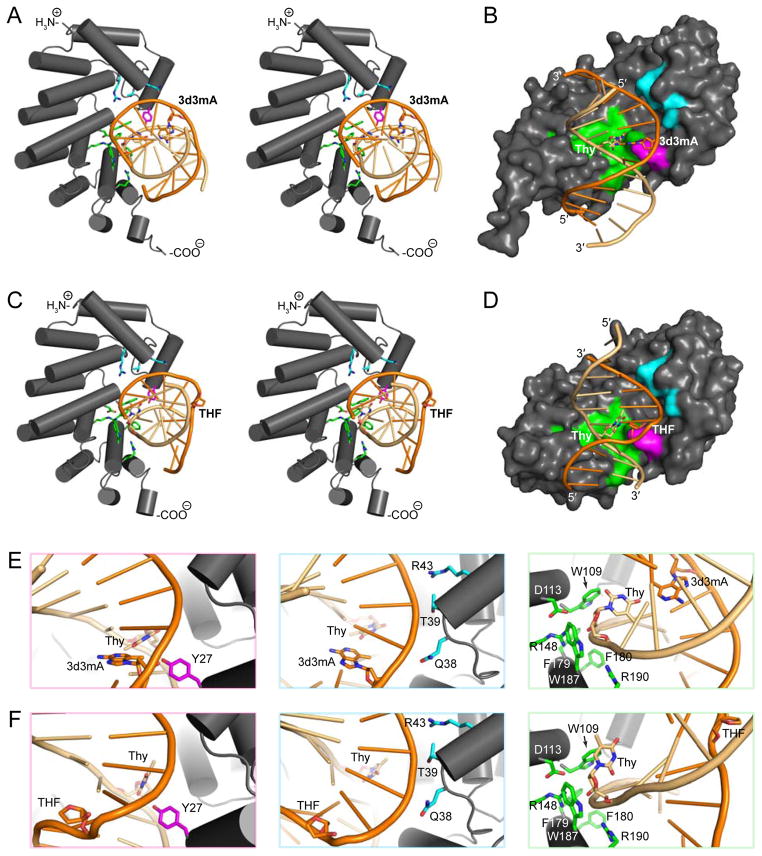
Tandem helical repeats have emerged as an important and widespread structural feature among DNA binding proteins [8]. AlkD is composed of six antiparallel two-helix ALK motifs that stack into a short left-handed solenoid with a positively charged concave binding surface created by basic residues on each C-terminal helix (Figure 2). Unlike other tandem helical repeats that bind nucleic acids, ALK motifs contact the backbone but not the nucleobases [8]. Sixteen residues on the concave surface of AlkD form electrostatic or hydrophobic contacts with phosphate or deoxyribose groups in substrate- and product-like complexes with DNA containing 3-deaza-N3-methyladenine (3d3mA) and tetrahydrofuran (THF), respectively (Figure 2). The DNA in both complexes is markedly distorted. In the substrate-like complex, the 3d3mA•T base pair is sheared due to rotation of the thymine into the minor groove and toward the protein surface (Figure 2). A nearly identical conformation is present in a complex containing DNA with a mismatched G•T base pair (PDB: 3JXY) [7]. In the product-like complex, both the thymine and the THF are fully extruded from the duplex, creating a single-base bulge in which base stacking is maintained by the flanking bases (Figure 2).
Figure 2. Protein-DNA interactions on the concave surface of AlkD.
(A,C) Stereodiagrams of AlkD-DNA interactions in substrate- (A, PDB: 3JX7) and product-like (C, PDB: 3JXZ) complexes containing 3-deaza-N3-methyladenine (3d3mA) and tetrahydrofuran (THF), respectively. (B,D) Surface representations of contacts in the substrate- (B) and product-like (D) complexes shown in panels A and C. (E,F) Close-up views of interactions in the substrate- (E) and product-like (F) complexes shown in panels A and C. In all panels, 3d3mA, THF, and the complementary thymidine are shown as sticks. Minor groove contacts (Tyr27), damaged strand contacts (Gln38, Thr39, and Arg43), and undamaged strand contacts (Trp109, Asp113, Arg148, Phe179, Phe180, Trp187, and Arg190) are colored magenta, cyan, and green, respectively. The damaged strand and the undamaged strand are colored bright orange and light orange, respectively.
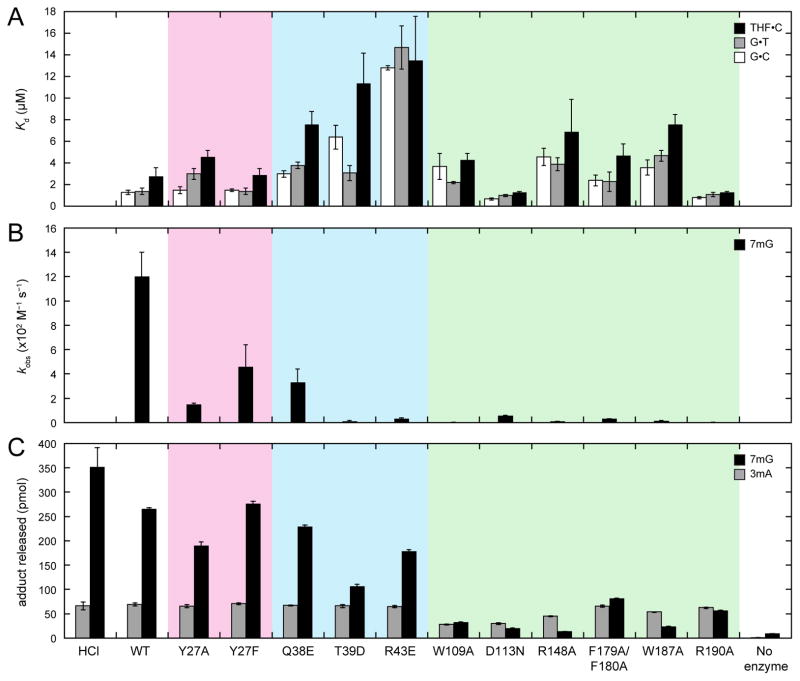
In order to understand how this unique nucleic acid binding surface recognizes DNA damage, we mutated 10 of the 16 residues that contact the DNA in the crystal structures and measured DNA binding to 25-mer oligonucleotides containing a centrally located Watson-Crick G•C base pair, a G•T mismatch, or a THF•C abasic site; 7mG excision from the same 25-mer oligonucleotide; and 3mA and 7mG release from methylated genomic DNA. Wild-type AlkD binds G•C-, G•T-, and THF•C-DNA with weak (low micromolar) affinity typical of protein-DNA complexes involving only nonspecific backbone contacts (Figure 3A and Table S1) [6].
Figure 3. DNA binding affinities and base excision activities of wild-type and mutant AlkD.
(A) Equilibrium dissociation constants (Kd) for binding of FAM-labeled 25-mer oligonucleotide duplexes containing a centrally located G•C (white bars), G•T (gray bars), or THF•C (black bars). (B) Second-order rate constants (kobs) for excision of 7mG (black bars) from a 25-mer oligonucleotide duplex. (C) 3mA (gray bars) and 7mG (black bars) released from methylated genomic DNA. HCl and no-enzyme controls represent the upper and lower limits for adduct removal. All panels show the averages and standard deviations from three experiments. Values are provided in Tables S1–S3. Mutants are colored in accordance with residues in Figure 2.
Cleavage of 7mG from the same 25-mer oligonucleotide occurs at 1.2 × 103 M−1 s−1 (Figure 3B and Table S2), while excision of 7mG and 3mA from methylated genomic DNA occurs 5-fold more slowly (2.2 × 102 M−1 s−1) and 7-fold more rapidly (8.0 × 103 M−1 s−1), respectively [18]. These rates of lesion removal are comparable to some 3mA glycosylases that extrude the damaged base into a nucleobase binding pocket during catalysis [19–22].
3.2. Damaged strand interactions
Six basic or neutral hydrophilic residues interact with the modified DNA strand. Three of these residues (Gln38, Thr39, and Arg43), clustered on the 3′ side of the damaged base, were mutated to aspartate or glutamate. As expected, electrostatic repulsion between the carboxylate side chains and the DNA backbone reduced binding affinity for all three oligonucleotide constructs by 2.2–11-fold (Figure 3A and Table S1). Correspondingly, excision of 7mG from oligonucleotide DNA was slowed by 3.6–110-fold (Figure 3B and Table S2), and release of 7mG from methylated genomic DNA was reduced by ≤2.5-fold (Figure 3C and Table S3). Like wild-type AlkD, all mutants completely excised 3mA from genomic DNA. Despite the lack of contacts with the lesion, there is a correlation between reduction in DNA binding and loss of base excision activity as a result of mutating residues contacting the damaged DNA strand.
3.3. Undamaged strand interactions
Ten basic or hydrophobic residues contact the unmodified DNA strand. Six of these residues (Trp109, Arg148, Phe179, Phe180, Trp187, and Arg190), which are located in or around a shallow, aromatic-rich cleft, were mutated to alanine. With the exception of the Arg190→Ala mutation, all alanine substitutions decreased affinity for the three DNA substrates by 1.5–3.5-fold, while alanine substitution of Arg190 slightly increased affinity, most notably for THF•C-DNA (Figure 3A and Table S1). This modest increase in binding affinity by AlkD-R190A is consistent with the THF-DNA complex structure, which suggests that removal of the Arg190 side chain would allow for optimized van der Waals and stacking interactions from Trp109 and Trp187 (Figure S5). Irrespective of the type of interaction, the binding data confirms the importance of each of these residues in the concave surface. Likewise, excision of 7mG from oligonucleotide DNA by all mutants, including AlkD-R190A, was retarded by 37–360-fold (Figure 3B and Table S2), and release of 3mA and 7mG from methylated genomic DNA was decreased by ≤2.4-fold and 3.2–20-fold, respectively (Figure 3C and Table S3). Thus, the coordinated reduction of DNA binding and catalytic activity resulting from disrupting electrostatic interactions with the damaged strand extends to hydrophobic contacts with the undamaged strand. The increased DNA binding affinity and decreased lesion excision activity of AlkD-R190A are consistent with the use of binding energy to distort the DNA into a disfavored but catalytically competent conformation.
Asp113 does not contact the DNA in either crystal structure but does form a salt bridge with Arg148 at the center of the aromatic-rich cleft near the base opposite the lesion. Mutation of Asp113 has been shown previously to modestly strengthen DNA binding yet substantially slow lesion excision [5, 6, 18]. Relative to wild-type AlkD, AlkD-D113N binds all three oligonucleotide constructs 1.4–2.0-fold more tightly (Figure 3A and Table S1), but excises 7mG from oligonucleotide substrate 21-fold more slowly (Figure 3B and Table S2). Correspondingly, removal of 3mA and 7mG from methylated genomic DNA is decreased by 2.3-fold and 13-fold, respectively (Figure 3C and Table S3). These seemingly contradictory phenomena have been attributed to reduced electrostatic repulsion between the carboxylate side chain and the DNA backbone but decreased thermal stability (Figure S2) due to disruption of the salt bridge [6, 7]. The latter could correlate with loss of the structural rigidity necessary to appropriately remodel the DNA backbone.
3.4. Minor groove interactions
A single bulky hydrophilic residue (Tyr27) inserted into the minor groove forms a hydrogen bond with the nucleobase on the unmodified strand immediately 3′ to the thymine opposite the damaged base. Tyr27 was mutated to alanine and phenylalanine to differentiate effects due to hydrogen-bonding and steric interactions. Phenylalanine substitution had no appreciable influence on binding affinity for any of the three DNA constructs (Figure 3A and Table S1), and likewise failed to alter levels of 3mA and 7mG released from genomic DNA (Figure 3C and Table S3). Excision of 7mG from the oligonucleotide substrate was reduced by 2.6-fold (Figure 3B and Table S2). Conversely, alanine substitution modestly decreased affinity for the oligonucleotide constructs containing a mismatched G•T base pair or an abasic THF•C site by 2.1-fold and 1.6-fold, respectively, but did not significantly affect binding of G•C-DNA (Figure 3A and Table S1). Release of 3mA and 7mG from genomic DNA was decreased by 1.1-fold and 1.4-fold, respectively (Figure 3C and Table S3), and excision of 7mG from oligonucleotide DNA was slowed by 7.7-fold (Figure 3B and Table S2). While the small influences of the Tyr27→Phe mutation are consistent with loss of the hydrogen bond observed in the crystal structures, the greater, albeit small, effects of the Tyr27→Ala mutation suggest that the steric bulk of the aromatic ring also plays a role in Tyr27-DNA interactions.
4. Conclusions
We mutated ten residues on the DNA binding surface of AlkD and found that every mutation, regardless of location relative to the lesion, reduced the rate of 7mG excision from an oligonucleotide substrate. Most mutations also decreased cleavage of 7mG from a genomic DNA substrate, but few mutations altered cleavage of 3mA. Apparent changes in 3mA excision, however, were attenuated by substrate depletion, masking small to moderate changes in the rate of 3mA cleavage and obscuring possible alterations of substrate specificity. While aspartate or glutamate substitution of residues contacting the damaged strand at sites flanking the lesion had the greatest impact on DNA binding affinity, alanine substitution of residues interacting with the undamaged strand directly across from the lesion had the largest effect on catalysis. Mutation of Tyr27, the only residue that contacts a nucleobase, had only very modest influences on DNA affinity and lesion excision. These results support a mechanism of catalysis in which electrostatic interactions throughout the binding interface distort the DNA into a catalytically competent conformation. In the previously proposed autocatalytic single-base bulge [7], the positively charged and inherently labile alkylated lesion would be flipped out of the duplex through the major groove and into the proximity of a phosphate in the kinked DNA backbone, conceivably providing stabilization for the oxocarbenium intermediate formed upon cleavage of the N-glycosidic bond or facilitating activation of a water for nucleophilic attack at the anomeric C1′ carbon. Concomitantly, the base opposite the lesion would be flipped through the minor groove and retained in an aromatic-rich cleft, stabilizing the altered conformation of the DNA and extending the lifetime of the damaged base near the catalytic phosphate. We suggest that transient interrogating interactions between Tyr27 and nucleobase edges may provide additional sensitivity as AlkD scans double-stranded DNA for energetically unstable base pairs. Analogously, glycosylases that flip the lesion into a nucleobase binding pocket utilize DNA intercalating “plug” and “wedge” residues to prolong extrusion of the damaged base and to disrupt irregular base pairs, respectively [3].
Supplementary Material
Acknowledgments
This work was funded by the National Science Foundation (MCB-1122098). Additional support for facilities was provided by the Vanderbilt Center in Molecular Toxicology (P30 ES000267) and the Vanderbilt-Ingram Cancer Center (P30 CA068485). E.A.M. and E.H.R. were supported by the Vanderbilt Training Program in Molecular Toxicology (NIH T32 ES07028).
Abbreviations
- 3d3mA
3-deaza-N3-methyladenine
- 3mA
N3-methyladenine
- 7mG
N7-methylguanine
- FAM
6-carboxyfluorescein
- THF
tetrahydrofuran
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Author contributions
B.F.E. oversaw the project; E.H.R. generated and purified AlkD mutants and performed oligonucleotide-based base excision assays, DNA binding assays, and thermal melting experiments; E.A.M. performed genomic DNA-based base excision assays; all authors analyzed data and wrote the paper.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2. ASM Press; Washimgton, DC: 2006. [Google Scholar]
- 2.Lindahl T. New class of enzymes acting on damaged DNA. Nature. 1976;259:64–66. doi: 10.1038/259064a0. [DOI] [PubMed] [Google Scholar]
- 3.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alseth I, Rognes T, Lindback T, Solberg I, Robertsen K, Kristiansen KI, Mainieri D, Lillehagen L, Kolsto AB, Bjoras M. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol Microbiol. 2006;59:1602–1609. doi: 10.1111/j.1365-2958.2006.05044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalhus B, Helle IH, Backe PH, Alseth I, Rognes T, Bjoras M, Laerdahl JK. Structural insight into repair of alkylated DNA by a new superfamily of DNA glycosylases comprising HEAT-like repeats. Nucleic Acids Res. 2007;35:2451–2459. doi: 10.1093/nar/gkm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinson EH, Metz AH, O’Quin J, Eichman BF. A new protein architecture for processing alkylation damaged DNA: the crystal structure of DNA glycosylase AlkD. J Mol Biol. 2008;381:13–23. doi: 10.1016/j.jmb.2008.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinson EH, Gowda AS, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubinson EH, Eichman BF. Nucleic acid recognition by tandem helical repeats. Curr Opin Struct Biol. 2011;22:101–109. doi: 10.1016/j.sbi.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Berti PJ, Schramm VL. Transition-state analysis for depurination of DNA by ricin A-chain. J Am Chem Soc. 2000;122:6527–6534. [Google Scholar]
- 10.Werner RM, Stivers JT. Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion-uracil anion intermediate. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 11.Dinner AR, Blackburn GM, Karplus M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature. 2001;413:752–755. doi: 10.1038/35099587. [DOI] [PubMed] [Google Scholar]
- 12.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 13.Berti PJ, McCann JA. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 14.McCann JA, Berti PJ. Transition-state analysis of the DNA repair enzyme MutY. J Am Chem Soc. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 15.Rubinson EH, Christov PP, Eichman BF. Depurination of N7-methylguanine by DNA glycosylase AlkD Is dependent on the DNA backbone. Biochemistry. 2013;52:7363–7365. doi: 10.1021/bi401195r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang YL, Ichikawa Y, Song F, Stivers JT. Powering DNA repair through substrate electrostatic interactions. Biochemistry. 2003;42:1922–1929. doi: 10.1021/bi027014x. [DOI] [PubMed] [Google Scholar]
- 17.Parker JB, Stivers JT. Uracil DNA glycosylase: revisiting substrate-assisted catalysis by DNA phosphate anions. Biochemistry. 2008;47:8614–8622. doi: 10.1021/bi800854g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins EA, Rubinson EH, Pereira KN, Calcutt MW, Christov PP, Eichman BF. An HPLC-tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods. 2013;64:59–66. doi: 10.1016/j.ymeth.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metz AH, Hollis T, Eichman BF. DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG) EMBO J. 2007;26:2411–2420. doi: 10.1038/sj.emboj.7601649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J. 2003;22:4898–4909. doi: 10.1093/emboj/cdg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien PJ, Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem. 2004;279:26876–26884. doi: 10.1074/jbc.M403860200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.