Abstract
Measurements of the DNA content of different protist populations can shed light on a variety of processes, including cell division, sex, prey ingestion, and parasite invasion. Here, we modified an Imaging FlowCytobot (IFCB), a custom-built flow cytometer that records images of microplankton, to measure the DNA content of large dinoflagellates and other high-DNA content species. The IFCB was also configured to measure fluorescence from Cy3-labeled rRNA probes, aiding the identification of Alexandrium fundyense (syn. A. tamarense Group I), a photosynthetic dinoflagellate that causes paralytic shellfish poisoning (PSP). The modified IFCB was used to analyze samples from the development, peak and termination phases of an inshore A. fundyense bloom (Salt Pond, Eastham, MA USA), and from a rare A. fundyense ‘red tide’ that occurred in the western Gulf of Maine, offshore of Portsmouth, NH (USA). Diploid or G2 phase (‘2C’) A. fundyense cells were frequently enriched at the near-surface, suggesting an important role for aggregation at the air-sea interface during sexual events. Also, our analysis showed that large proportions of A. fundyense cells in both the Salt Pond and red tide blooms were planozygotes during bloom decline, highlighting the importance of sexual fusion to bloom termination. At Salt Pond, bloom decline also coincided with a dramatic rise in infections by the parasite genus Amoebophrya. The samples that were most heavily infected contained many large cells with higher DNA-associated fluorescence than 2C vegetative cells, but these cells’ nuclei were also frequently consumed by Amoebophrya trophonts. Neither large cell size nor increased DNA-associated fluorescence could be replicated by infecting an A. fundyense culture of vegetative cells. Therefore we attribute these characteristics of the large Salt Pond cells to planozygote maturation rather than Amoebophrya infection, though an interaction between infection and planozygote maturation may also have contributed. The modified IFCB is a valuable tool for exploring the conditions that promote sexual transitions by dinoflagellate blooms but care is needed when interpreting results from samples in which parasitism is prevalent.
Keywords: Alexandrium fundyense, A. tamarense Group I, algal bloom dynamics, imaging flow cytometry, microalgal life cycles
1. Introduction
The dinoflagellates are a morphologically and metabolically diverse protist group that includes several species that endanger human health through the production of neurological and gastrointestinal toxins. They are among the most abundant eukaryotic microbes globally and can dominate planktonic ecosystems as ‘red tides’, dense monospecific blooms that dramatically alter the color of the sea surface. Comprehensive knowledge of their ecology is needed to understand plankton dynamics and also to inform mitigation strategies against harmful species. About half are photosynthetic (autotrophic or mixotrophic) and they include planktonic and benthic species as well as coral and animal endosymbionts. Heterotrophic forms include free-swimming grazers, phagotrophs, and parasites. The majority are motile and many species migrate vertically in the water column (e.g., Hasle, 1950). Especially important to this study, the dinoflagellates share a number of unique nuclear characteristics and have among the largest genomes known: DNA mass per cell is comparable to the multinucleated ciliates and two to three orders of magnitude greater than other planktonic algae (Prescott, 1994; Hou and Lin, 2009).
Changes in DNA content within a dinoflagellate cell may be caused by any of several ecologically important processes: vegetative cell division (mitosis), sexual recombination (zygosis/meiosis), ingestion of other cells as prey, or infection by parasites. Of these, changes due to mitosis and sex are most recognizable because DNA mass changes are large and quantized: 2-fold with each replication, division or fusion. Such quantized patterns in a natural population may be obscured by incorporation of prey cells or parasites, particularly if many prey are ingested or if parasites proliferate intracellularly.
In this study, we have focused on the harmful algal species Alexandrium tamarense Group I, which we refer to as A. fundyense, the renaming proposed by John et al. (in prep). The proposed renaming of this species follows the findings of several studies that have shown polyphyletic relationships between morphospecies in the A. tamarense species complex (A. tamarense, A. fundyense, and A. catenella; see Scholin et al., 1994; Scholin et al., 1995; John et al., 2003; and Lilly et al., 2007). Species boundaries between ribosomal clades were proposed by Lilly et al. and are supported by the discovery of post-zygotic mating incompatibility between two of the most closely related clades, Group I and Group III (Brosnahan et al., 2010).
A. fundyense is a causative organism of paralytic shellfish poisoning (PSP). Consumption of shellfish that are contaminated with PSP toxins can cause paralysis or death, making PSP one of the most dangerous seafood-poisoning syndromes worldwide. Consequently, A. fundyense is the focus of ongoing studies in the Gulf of Maine (GOM) and within the Nauset Marsh system (NMS; Cape Cod, MA). Microplankton samples analyzed in this study were taken in conjunction with these efforts and with the foremost goal of characterizing their A. fundyense populations.
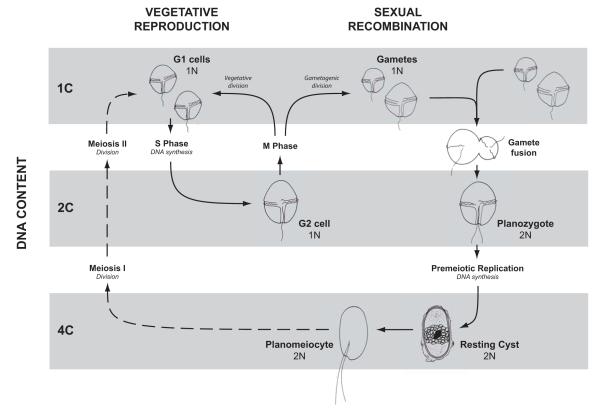
Both the initiation and termination of A. fundyense blooms depend on transitions between the vegetative and sexual phases of the cells’ life cycle (Fig. 1; Anderson et al., 1983). Therefore the detection of sexual transitions is important for predicting bloom decline. Under certain conditions, A. fundyense vegetative cells cease division and transform into gametes that are morphologically similar to their vegetative progenitors. Gametes conjugate and form swimming diploid cells called planozygotes that remain in the plankton for about one week before transforming into diploid resting cysts (Anderson and Wall, 1978). A. fundyense exits the sexual cycle through meiotic division of a planomeiocyte, a swimming cell that emerges from the resting cyst stage (Pfiester and Anderson, 1987). The resulting haploid vegetative cells then resume mitosis.
Figure 1.
Life cycle diagram of Alexandrium fundyense (syn. A. tamarense Group I). The life cycle is comprised of the mitotic, reproductive cycle that has a period of 2-5 days and a slower, sexual cycle that has a minimum period of ~3 months. The mitotic cycle consists of an initial growth period (G1 phase), followed by DNA replication (S phase), a short growth phase (G2), then nuclear division (M phase) and cytokinesis (cell division). A. fundyense enters the sexual cycle through gametogenesis, a division that yields two gametes rather than new G1 phase vegetative cells. Gametes may be isogamous (similar in size) or anisogamous (one large and one small, pictured).
In this study, cells were classified according to their total DNA content which was quantified in increments ‘C’, where 1C is the DNA content of a haploid cell immediately after division (i.e. one complement of unreplicated chromosomes). Each stage in the mitotic cycle is haploid (1N) and contains either a single or double complement of chromosomes (1C or 2C DNA content). Sexual stages may contain 1C, 2C, or 4C DNA content. Gametes are haploid but the planozygote and resting cyst stages are diploid (2N). Premeiotic replication by A. fundyense, the process that would give rise to 4C DNA content cells, has not yet been observed but is presumed to occur during the planozygote or resting cyst stages.
Our primary objective in this study was to facilitate the identification of A. fundyense life cycle stages in natural populations through the use of a modified Imaging FlowCytobot (IFCB). IFCB is a custom-built flow cytometer that records particle images in addition to measuring fluorescence and laser scattering (Olson and Sosik, 2007). An associated image-classification pipeline is used to identify both rare and abundant species so that the fluorescence and image metrics of individual species can be compared between samples (Sosik and Olson, 2007). We configured IFCB to measure DNA content via fluorescence of the membrane permeable dye Hoechst 33342 and also to detect fluorescence from species-specific oligonucleotide probes. Data collected with the modified IFCB were then used to assess whether the combination of images and DNA measurements could be used to determine the timing and extent of shifts from vegetative division to the formation of planozygotes.
We also used the instrument to investigate the effects of infections by the parasitic dinoflagellate Amoebophrya on host A. fundyense since these infections might obscure increases in DNA content associated with A. fundyense sexual transitions. Amoebophrya are intracellular parasites that chronically infect many dinoflagellate species and may be a dominant factor in the termination of inshore blooms (Taylor, 1968; Coats, 1999; Chambouvet et al., 2008). Free-living Amoebophrya are small, 3 to 5 μm in diameter, and infect their hosts as single cells. Once inside, the parasite undergoes several rounds of replication to become a large, multi-nucleated trophont that distends its host, filling much of the host cytoplasm or nucleus as it matures. Ultimately, the host cell bursts and tens to hundreds of new infective Amoebophrya cells emerge. We examined the effects of Amoebophrya infection on the DNA content and size of A. fundyense cells in culture and by inspecting A. fundyense that were infected by Amoebophrya in the field.
2. Materials and methods
2.1 IFCB modification and sample analysis
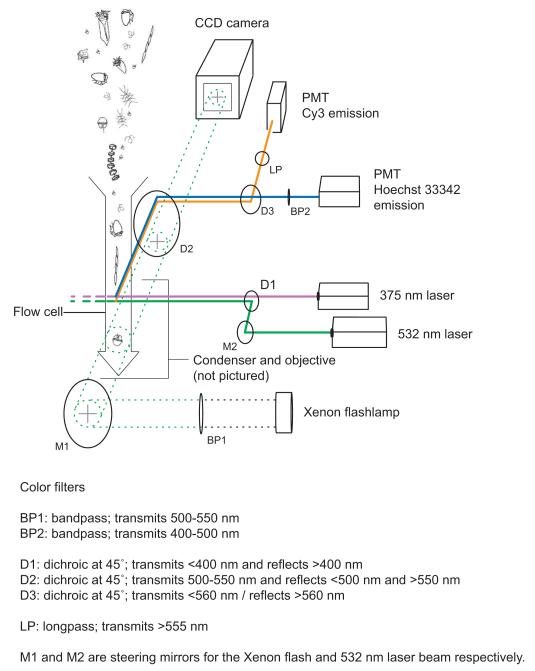
IFCB was developed to address the need for sustained, in situ observation of large nanoplankton and microplankton (cells 10 to >100 μm in length). In its original design, it was enclosed in a watertight aluminum housing and was configured to analyze raw seawater withdrawn directly from the environment (Olson and Sosik, 2007). For this study, IFCB was used as a laboratory instrument instead. This allowed samples to be concentrated and stained prior to analysis and also made it possible to install new lasers that do not fit within the standard underwater housing. IFCB’s red diode laser was replaced with a UV one (CUBE™375-16C, 375 nm, 16 mW by Coherent, Inc.) that excites the DNA-specific dye Hoechst 33342. A second, green laser (COMPASS™ 215M-10, DPSS-CW, 532 nm, 10 mW by Coherent, Inc.) was installed for the excitation of Cy3, a cyanine fluorophore that was attached to A. fundyense species-specific and Amoebophrya genus-specific oligonucleotide probes. Other changes to the optical system included the replacement of dichroic filters to allow the passage of blue light (Hoechst fluorescence, 400-500 nm wavelengths) to the trigger photomultiplier tube (PMT; Hamamatsu HC120-05) and yellow-orange light (Cy3 fluorescence, >555 nm wavelengths) to the second PMT. Both lasers were focused through a single pair of cylindrical lenses to produce horizontally elongated, elliptical beam spots across the sample channel. Because the lenses were not color corrected, the two beams had different focal points relative to the fluidic system’s sample core. This difference was diminished by adjusting the collimation of each beam independently with separate beam expanders. The beams were also separated in space so that cells first passed through the UV beam and then the green beam (Figs. 2 and 3).
Figure 2.
Optical layout of the modified Imaging FlowCytobot.
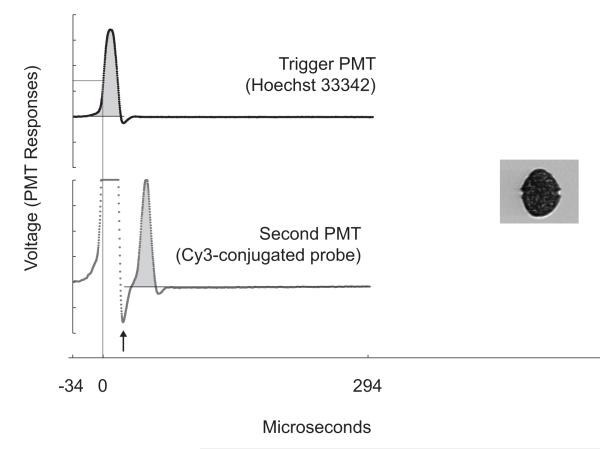
Figure 3.
PMT responses as recorded by the AD-2100-14 digitizer card (Chase Scientific Company) and image acquisition. Time zero for each particle ‘event’ was set by the trigger threshold for blue fluorescence. The minimum in orange-yellow fluorescence (arrow) was used to demarcate the boundary between PMT responses to the UV and green beams. The integrals used for assessing Hoechst fluorescence and identification of positive Cy3-conjugated probe staining are shaded gray.
A fast digitizer/data acquisition card (model AD2100-14, Chase Scientific Company) was added to the IFCB computing stack. The digitizer card enabled the recording of complete traces of PMT responses at 6 MHz for each cell that passed through the laser beams. Integrals of the trace data were computed after acquisition to quantify the fluorescence from both stained nuclei (UV beam) and Cy3-conjugated oligonucleotide probes (green beam; Fig. 3). Trace data from the blue light detector also provided measurements of peak height, which were used to ensure that integrated signal values were not saturated. Coefficients of variation (CV) for the fluorescence distributions were estimated by measuring the full width at half maximum height and dividing by 2.36 (Shapiro, 2003).
All plankton and culture samples were concentrated by retention of cells on 20 μm Nitex. Plankton samples from Salt Pond were collected with 5-L Niskin bottles, then pre-screened with 100 μm mesh to remove large plankton and detritus that could clog the IFCB’s fluidics system. Plankton samples from the western Gulf of Maine red tide were collected with a bucket and measured into a 2-L bottle without a pre-sieving step. In both cases, fixation and pigment extraction were as described by Anderson et al. (2005). This procedure included the addition of unbuffered formalin (5% v/v), which is known to diminish Hoechst fluorescence over time (Cetta and Anderson, 1990). We attempted to minimize the effect of prolonged formalin fixation by keeping concentrated samples on ice in the field. After transport to the lab, samples were resuspended in 14 mL of methanol that had been chilled to −20 °C. The samples were stored up to 4 months at −20 °C before staining with oligonucleotide probes and Hoechst 33342 immediately prior to analysis.
Two Cy3-conjugated oligonucleotide probes were applied in separate experiments: the A. fundyense-specific NA1 ribosomal probe (5′-AGTGCAACACTCCCACCA-3′, Anderson et al., 1999) and a genus-specific Amoebophrya ribosomal probe (5′-CCTGCCGTGAACACTCTAAT-3′, provided by Mario Sengco). Hybridization was completed as described by Anderson et al. (1999), and samples were re-suspended in 5x SET/Hoechst solution (750 mM NaCl, 5 mM EDTA, 100 mM Tris-HCl, 5 μg mL−1 Hoechst 33342, pH 7.8). Stained samples were stored at 4 °C for no longer than six hours then stirred in an ice bath with a magnetic stir bar for up to 30 minutes prior to analysis.
The IFCB’s maximum acquisition rate is ~12 Hz and is limited by the time needed to acquire an image for each trigger event. As a consequence, if cells pass through the laser beams too frequently, many are not recorded. This may be problematic if the target species comprises only a small fraction of the microplankton assemblage or when sample volumes are limited. Two strategies were used to maximize the proportion of A. fundyense cells detected. First, a high trigger threshold was applied in order to maximize detection of particles with intense Hoechst fluorescence (dinoflagellates and ciliates with exceptionally high DNA content and also blue fluorescent detritus). Second, the analysis rate was kept below 5 Hz by dilution of the sample with 5X SET/Hoechst 33342 solution. The application of a high trigger threshold precluded the use of beads as internal standards of Hoechst fluorescence because their brightness was below the threshold necessary for efficient sample analysis. Instead, UV-excited AlignFlow Plus beads (A-7305, Invitrogen) were analyzed before and after each sample at a higher electronic gain setting. Data were not used if the mean fluorescence of the AlignFlow Plus beads differed by more than 10% before and after a sample was run. The overall mean of the AlignFlow Plus beads’ fluorescence was then used to normalize measurements of Hoechst fluorescence from the A. fundyense data. In a similar fashion, stability of the green laser was assessed once a day with phycoerythrin Calibrite beads (BD Biosciences). The CV values from both the AlignFlow Plus and Calibrite beads ranged from 5% to 10% throughout the two-week period when samples were analyzed.
Images were classified with a support vector machine (SVM) based image-processing pipeline that was developed for analysis of IFCB data (Sosik and Olson, 2007). Six species/image categories were defined: A. fundyense singlet, A. fundyense doublet, Dinophysis acuminata, Gonyaulax sp., Protoperidinium sp., and a tintinnid class. The performance of the SVM classifier varies substantially depending on the relative uniqueness of its categories. Errors were corrected by manually reviewing all SVM-based image classifications and reassigning them as necessary. Data from cell clumps were not categorized, nor were other ciliate and dinoflagellate cell types that were found in only one or two water samples. After image classification of field samples, A. fundyense classifications were confirmed by reviewing Cy3 fluorescence from the A. fundyense-specific oligonucleotide probe.
Image features assessed during classification included a wide range of particle metrics such as size, shading and orientation, but only diameter measurements were included in our post-classification analysis. Single A. fundyense cells were approximated as spheres to calculate their volume, which was of interest because it was expected to co-vary with DNA content during cell division and because previous studies had identified larger A. fundyense as planozygotes (Anderson et al., 1983; Lindquist and Anderson, 1985).
Data from IFCB were not used to estimate A. fundyense concentration. Our experience has been that flow-cytometer based A. fundyense counts are unreliable for fixed samples (probably due to sinking of the fixed cells). Therefore, A. fundyense abundance estimates were taken from data collected in conjunction with the same field studies but published elsewhere (NMS: Crespo et al., 2011; western GOM red tide: McGillicuddy et al., this issue).
2.2 Salt Pond sampling
Samples for IFCB analysis were taken in conjunction with weekly surveys of the NMS bloom from March to June 2009 (Fig. 4). Here, a brief overview of the NMS and the 2009 survey effort is provided (see Crespo et al., 2011 for a more detailed description).
Figure 4.
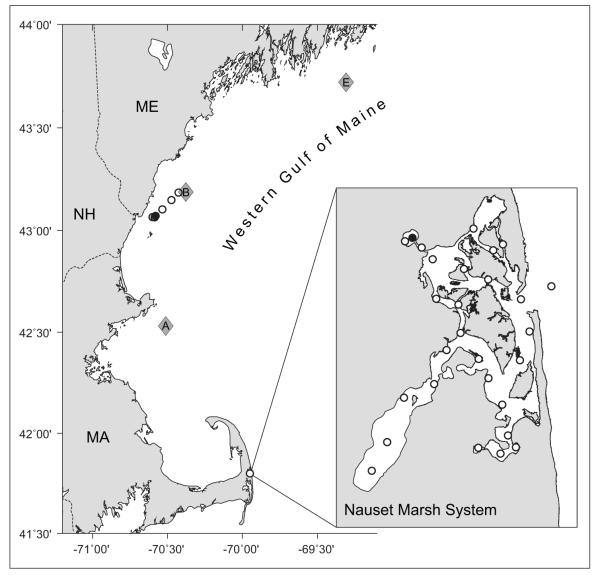
Western Gulf of Maine (WGOM) map. Stations sampled from the WGOM red tide are denoted by circles leading from Portsmouth Harbor to the Northeastern Regional Association of Coastal and Ocean Observing Systems (NERACOOS) Buoy B (diamond, also NERACOOS Buoys A and E). A subsample from station 1 (filled circle) was analyzed with the modified IFCB and with a FACSCalibur instrument. Inset. Nauset Marsh system (NMS) map. Thirty-one stations throughout the NMS were monitored weekly from late March to June 2009, but only samples from Salt Pond (filled circle) were analyzed for this study.
The NMS is a network of shallow channels that link three brackish ponds to a central marsh area that, in turn, is connected to the Atlantic Ocean via Nauset inlet (Fig. 4 inset). The mean depth within the NMS is approximately 1.25 m but its three ponds (the northwestern, southwestern, and southeastern extremities of the system) are deeper (up to 10 m). Alexandrium fundyense cells are selectively retained in the NMS ponds in spite of strong tidal flushing. Their retention is caused by an interaction between the system’s bathymetry and the swimming behavior of the A. fundyense cells (Anderson and Stolzenbach, 1985). As a result, the population in Salt Pond (the northwestern extremity of the NMS) is isolated from the southern populations and experiences independent development and termination bloom phases (Crespo et al., 2011). We took advantage of this isolation to identify samples collected during the bloom’s development, peak and termination phases (May 11th, 18th, and 26th respectively; see Fig. 5). Samples for IFCB analysis were collected as previously described at a station over the deepest area of the pond (~9 m at high tide) and from three depths: the near-surface (0-1 m), mid-depth (approximately 5 m), and 1 m above the bottom.
Figure 5.
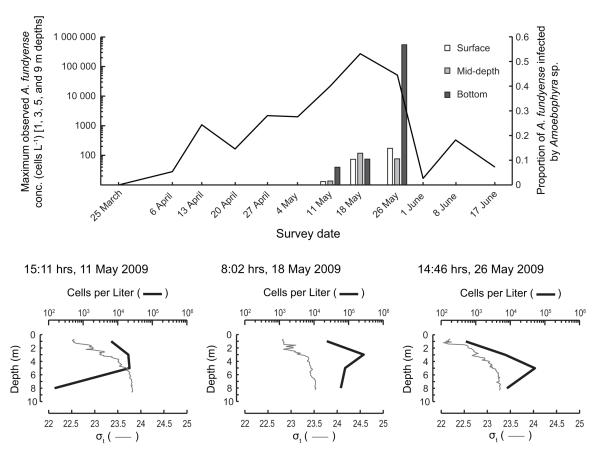
Salt Pond A. fundyense population history from weekly surveys, March-June 2009 and described by Crespo et al. (2011). Top panel. Line graph of maximum observed cell densities recorded from all depths sampled and bar graph of surface, mid-depth, and bottom sample rates of Amoebophrya infection. Bottom panel. Water density and cell abundance profiles taken during growth, peak, and termination phase of the Salt Pond bloom (11, 18, and 26 May 2009, respectively).
All of the NMS surveys were conducted at high tide and during daylight so that our boats could navigate through areas of the central marsh that were drained during low tide. The IFCB samples from the 11th and 26th of May (development and termination phases) were taken in the mid-afternoon. Samples from May 18th (the bloom’s peak) were taken approximately three hours after sunrise.
2.3 Western Gulf of Maine red tide sampling
An A. fundyense red tide (hereafter termed the “WGOM red tide”) was discovered serendipitously during a mooring deployment July 10, 2009 aboard the R/V Gulf Challenger (University of New Hampshire, Portsmouth, NH). Six surface samples were collected within discolored water along a transit line from the NERACOOS B buoy to the mouth of Portsmouth Harbor (Fig. 4). Cell concentrates were refrigerated on board after sieving and formalin fixation. Once in port, the samples were transferred to ice for transport to Woods Hole, MA where they were resuspended in methanol less than 12 hours after formalin fixation.
The monospecificity of A. fundyense in the GOM samples allowed us to analyze their DNA content with a commercial flow cytometer (FACSCalibur, BD Biosciences) in parallel with our IFCB analysis. A subsample from transect station 1 (where the A. fundyense cell concentration was highest) was stained with propidium iodide and treated with RNAse A as described by Taroncher-Oldenburg et al. (1997). The results from the two flow cytometers were compared through analysis of a clonal A. fundyense culture sample that had significant numbers of both G1 and G2+M phase cells (and therefore 1C and 2C modes in its DNA content distribution; see McGillicuddy et al., this issue).
2.4 Amoebophrya spp. studies
To study the effect of Amoebophrya infection on the DNA content and cell size of host A. fundyense cells, an Amoebophrya sp. isolate was propagated in cultures of the A. fundyense clone SP-E10-03 (cultures isolated in 2004 and 2001 respectively, both from Salt Pond). The SP-E10-03 culture was grown in f/2 media without silicate (Guillard and Ryther, 1962) and infected with suspensions of actively parasitized host cells. Prior to formalin fixation, infected host cells are readily identified by their green fluorescence under blue illumination. Amoebophrya trophont autofluorescence was used to monitor the progression of parasite infections. Transfers to new host cultures were completed when more than half of host cells were visibly infected (typically every 4-6 days), but no other effort was made to control the parasite-to-host ratio. An infected sample was fixed for IFCB analysis when large numbers of the host A. fundyense had begun to lyse in the culture.
The measurement of DNA content required extraction of all intracellular pigments with ice-cold methanol so that infected cells could be identified through staining with the Amoebophrya genus-specific ribosomal probe. Previously it was shown that the Amoebophrya genus-specific probe was effective for identifying infected A. fundyense cells except during the initial phase of infection (M. Sengco, pers. comm.). We ignored this limitation because mature infections could be identified unequivocally and it was these older infections that were expected to have the greatest effect on host DNA content and cell size. Culture samples were stained for analysis on the modified IFCB as described for field samples except that infected cells were identified by the fluorescence of the Amoebophrya-specific ribosomal probe.
Amoebophrya infection rates in the field were assessed by staining subsamples from methanol suspensions simultaneously with the Cy3 conjugated A. fundyense-specific probe and a fluorescein isothiocyanate (FITC) conjugated version of the Amoebophrya-specific probe. The dual labeled samples were then inspected by epifluorescence microscopy. Both infected and uninfected A. fundyense fluoresced yellow-orange under green illumination from the A. fundyense-specific Cy3 probe, but only infected cells fluoresced green under blue illumination due to staining by the Amoebophrya probe. The proportion infected was determined from observations of no fewer than 200 A. fundyense per field sample. Additional counts that distinguished small diameter and large diameter host A. fundyense (nominally ~30 and >45 μm in diameter, respectively) were performed for samples taken May 26th from mid-depth and near the bottom of the Salt Pond bloom.
3. Results and Discussion
3.1 Image classification and A. fundyense-specific probe staining
Particle detection and machine-based image classification for A. fundyense were completed essentially as described for the original IFCB (Olson and Sosik, 2007; Sosik and Olson, 2007). The most significant difference was the manner in which machine-based image classification errors were controlled. The performance of the SVM-based image classifier was mixed, with misclassification rates ranging from 1.8% to 19% for A. fundyense, but these errors were corrected during manual review of the classifier’s output. The modifications to the IFCB enabled us to refine our A. fundyense classifications by only using data from cells that were positively stained by the species-specific ribosomal probe (83 – 96% of post-review classifications). While these precautions were not necessary for all samples (e.g., the WGOM red tide and cultures in which the majority of particles were A. fundyense), several of the Salt Pond samples were dominated by unclassified particles (debris and cell clumps) and included several other species (e.g., D. acuminata and a Gonyaulax sp.) that were similar either in appearance or fluorescence to the probe-stained A. fundyense. Therefore, the modified IFCB was especially well suited for analyzing samples from Salt Pond.
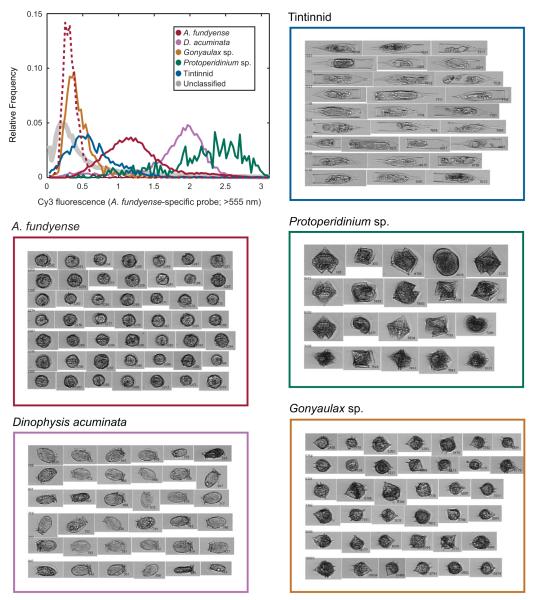
Staining with the Cy3-conjugated A. fundyense-specific rRNA probe effectively differentiated A. fundyense from most similarly sized, round cells in the Salt Pond samples, but the interpretation of the probe fluorescence measurements was not always simple. Besides A. fundyense, the most common round-celled dinoflagellate species in Salt Pond were Gonyaulax sp. and Protoperidinium sp. The Protoperidinium sp. often produced higher yellow-orange fluorescence signals than probe-stained A. fundyense (Fig. 6). Inspection of unstained Protoperidinium cells under several wavelengths of illumination confirmed that their fluorescence was broad spectrum. This autofluorescence did not affect IFCB analysis because (1) most of the Protoperidinium cells were distinguishable during manual inspection of the images and (2) the abundance of Protoperidinium was low relative to A. fundyense in all of the samples analyzed. However, in future studies it may be necessary to select more effective probe fluorophore and/or IFCB color filters, especially if Protoperidinium spp. or other broadly autofluorescent cells are abundant. Future improvements to the system’s image classifier may also reduce the time needed to manually correct A. fundyense and Protoperidinium assignments.
Figure 6.
Upper Left: Frequency distributions of >555 nm fluorescence (associated with the A. fundyense-specific probe) for all species classified in samples from Salt Pond (solid lines) and an unstained A. fundyense culture (dotted line). Staining with the A. fundyense-specific probe strongly differentiated A. fundyense cells from unstained A. fundyense and Gonyaulax sp. cells but autofluorescence from D. acuminata and Protoperidinium sp. cells was greater than the signal produced by cells stained with Cy3-conjugated probes. Unclassified particles (gray solid line) were the most abundant type overall and included debris, rare cell types and cell clumps. Image collages: Example images of the five most abundant classes in samples from Salt Pond.
In contrast to Protoperidinium, Gonyaulax sp. cells produced much lower yellow-orange fluorescence signals, similar to those of unstained A. tamarense cells (Fig. 6), so their discrimination was relatively easy via probe detection. Scrippsiella troichodea and A. ostenfeldii, dinoflagellates that resemble and frequently co-occur with A. fundyense, were not present in any of the field samples tested, but previous work has shown that the A. fundyense-specific NA1 probe has excellent specificity in samples that also contain these species (e.g., Anderson et al., 2005). Therefore, we expect taxonomic probe staining during IFCB analysis to be especially useful when cells morphologically similar to A. fundyense are abundant. For this approach to be used, however, a laboratory-based IFCB is needed, as it is not yet possible to apply ribosomal probes during in situ deployments.
While probe fluorescence was valuable for identifying A. fundyense cells, image classification was at least as important in this study. Both tintinnids and D. acuminata produced probe-like fluorescence that substantially overlapped that of A. fundyense cells, but in both cases cells were readily distinguished from A. fundyense using images (Fig. 6). In the case of D. acuminata, the overlap in yellow-orange fluorescence was due to poor methanol extraction of pigments from its kleptoplastids, and similar to Protoperidinium, the D. acuminata cells produced larger fluorescence signals than most of the probe-stained A. fundyense cells. Separation of D. acuminata and A. fundyense without images might have been achievable with a different conjugate to the A. fundyense probe. However, using only the Cy3 probe fluorescence (without images), the two dinoflagellates would have been difficult to distinguish from each other, or from similarly fluorescent debris that was sometimes more abundant than either species.
The combination of images and fluorescence measurements may also provide means for studying species interactions such as grazing. In the case of the tintinnids, a small proportion of the population had similar yellow-orange fluorescence to stained A. fundyense cells, and this fluorescence may actually have resulted from staining of A. fundyense that had been consumed as prey. The color of tintinnid fluorescence was similar to the Cy3 A. fundyense probe when observed under an epifluorescence microscope. The mean intensity of this fluorescence was also twice as high during the peak and termination phases of the bloom, suggesting a shift towards consumption of A. fundyense cells. Recent work by Haley et al. (2011) has shown that A. tamarense rDNA remains detectable in copepod concentrates several hours after ingestion. Similar characterization of the persistence of A. fundyense rRNA after ingestion by tintinnids may enable estimation of A. fundyense loss rates to these and other microzooplankton species using the modified IFCB.
3.2 Measurement of A. fundyense DNA content
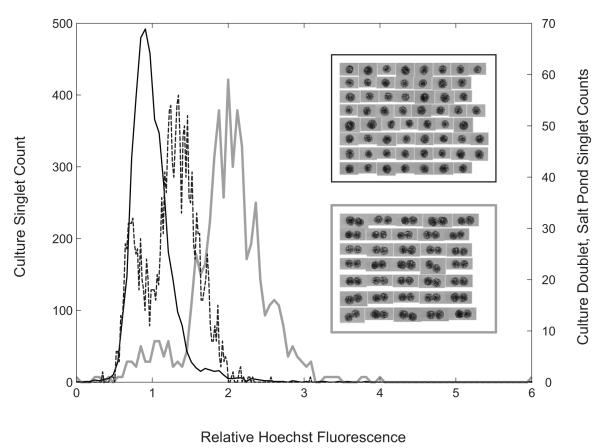
DNA measurements from A. fundyense cells were classified using the unit ‘C’ where 1C is the DNA content of an A. fundyense cell immediately after division, i.e. having one haploid complement of unreplicated chromosomes. Thus, gametes and G1 phase vegetative cells are 1C, and G2+M phase vegetative cells and planozygotes (at least newly formed ones) are 2C (Fig. 1). We confirmed the modified IFCB could distinguish 1C and 2C A. fundyense by comparing the Hoechst fluorescence of A. fundyense singlets and doublets from the same culture. The mode Hoechst fluorescence of doublets was twice that of singlets, as expected since doublets consist of two singlet cells attached anterior to posterior (Fig. 7). There was also little overlap between the singlet and doublet populations’ DNA distributions which had coefficients of variation of ~23% and ~16%, respectively.
Figure 7.
Frequency distributions of the Hoechst fluorescence from A. fundyense singlet and doublet populations in a single culture (black and gray lines respectively) and from singlet cells in the near-surface sample taken May 18th at Salt Pond (dashed line; see also Fig. 9, A and D). Inset collages are representative images of singlet and doublet cells from the culture.
Mixtures of 1C and 2C cells were present in several field samples based on the bimodal distributions of A. fundyense Hoechst fluorescence (Figs. 7, 8 and 9). However, the intensity of fluorescence from field samples was diminished relative to cultures (Fig. 7). This result is similar to earlier work that showed Hoechst fluorescence to be inversely related to formalin concentration and fixation time (Cetta and Anderson, 1990). Though the fixation time was always less than 10 min for our cultures, for field samples it was sometimes several hours and was not well controlled between days. Therefore, we attribute the reduction in Hoechst fluorescence to formalin fixation, which was used because it is the standard protocol in our field program. We could not apply a correction for the fixation effect because we lack reliable estimates of fixation times in the field. Instead, DNA measurements from field samples were interpreted by assuming that (1) bimodal populations represented 1C and 2C cells, (2) the fluorescence of samples taken from the same depth profile was diminished to a similar extent, and (3) the Hoechst fluorescence of field samples was always diminished relative to cultures. DNA modes in field samples were classified by applying these assumptions in a hierarchical fashion.
Figure 8.
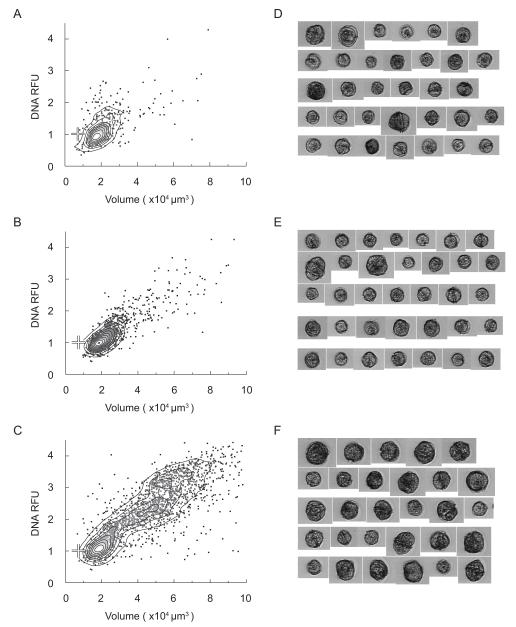
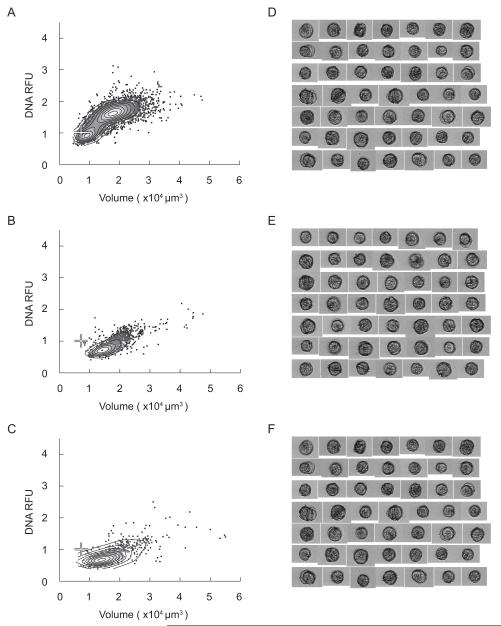
Density scatter plots (A-C) and collages of representative images (D-F) of A. fundyense collected during the development phase of the Salt Pond bloom (May 11th) at the near-surface (A and D), mid-depth (B and E), and near-bottom (C and F). White crosses on the density scatter plots represent the mode DNA content and cell volume of a reference culture of G1 phase vegetative cells (see Section 3.2).
Figure 9.
Density scatter plots (A-C) and collages of representative images (D-F) of A. fundyense collected near the peak of the Salt Pond bloom (May 18th) at the near-surface (A and D), mid-depth (B and E), and near-bottom (C and F). White crosses on the density scatter plots represent the mode DNA content and cell volume of a reference culture of G1 phase vegetative cells (see Section 3.2).
3.3 Changes in A. fundyense DNA content over the course of the Salt Pond bloom
The progression of A. fundyense blooms is associated with a transition from vegetative reproduction (mitotic cell division) during the early development phase toward the production of gametes, planozygotes, and cysts (sexual recombination) as the bloom matures and declines. Both of these processes — cell division and sexual recombination — are associated with life cycle stages that have 2C DNA content (dividing vegetative [G2+M] cells and planozygotes respectively; Fig. 1). The 2C DNA content ‘division stage’ cells are only abundant during early morning hours because A. fundyense divisions are controlled by the diel light-dark cycle (Rubin, 1981; Taroncher-Oldenburg et al., 1999). In contrast, planozygotes — the 2C DNA content ‘sexual stage’ — mature over the course of about a week (Anderson and Lindquist, 1985) and are present at all times of day. Therefore, we expected to observe a sexual transition during the course of the Salt Pond bloom through an increase in the relative proportion of 2C cells over time, especially at times of day when mitotic divisions would not typically occur.
Another potential marker of sex for dinoflagellates is the presence of 4C cells, zygotes that have undergone premeiotic replication (PR). Such cells provide an unambiguous marker of sexual recombination in the dinoflagellate Prorocentrum micans because only its planozygote stage has 4C DNA content (Bhaud et al., 1988). The same may be true of A. fundyense but the timing of PR has not yet been established — A. fundyense may complete PR as planozygotes or during one of its later zygotic stages (resting cyst or planomeiocyte). Modification of the IFCB has provided a means to characterize the DNA content distribution of A. fundyense’s zygotic stages, but interpretation of the data can be complex, especially when examining natural populations.
In assessing the ability of IFCB to characterize the sexual transition of the Salt Pond A. fundyense bloom, we first established the bloom’s phase — development or decline — during each of our weekly NMS surveys. As a well-isolated population, the Salt Pond bloom was an ideal subject because its phases could be assessed directly from cell abundance estimates and without consideration of advective fluxes to and from the pond. A. fundyense cells were detected in Salt Pond during our first survey conducted March 24th. From that time, the bloom grew in a sustained logarithmic phase through its peak May 18th, then declined precipitously after May 26th (Crespo et al., 2011; Fig. 5). Samples collected May 11th , 18th, and 26th were chosen for IFCB analysis because A. fundyense abundance was relatively high at all depths and because these dates captured the bloom’s transition from development to decline.
Samples taken May 11th were expected to be dominated by vegetative cells because A. fundyense abundance grew with an approximate doubling time of 2 days in the weeks immediately before and after that date (Fig. 5). Since samples were collected during mid-afternoon (when division does not typically occur), the DNA distribution of A. fundyense was expected to be heavily skewed toward 1C (G1 phase vegetative cells). Indeed, most cells were 1C except at the near-surface, where 2C were dominant but where cell concentrations were also several fold lower than at mid-depth (Figs. 5 and 8). This near-surface 2C subpopulation must have been comprised of either planozygotes or G2 phase vegetative cells (which can divide to produce either gametes or new G1 phase vegetative cells; Fig. 1). In either case, the near-surface localization of 2C cells indicates a difference in their behavior that is likely important for sexual transitions.
The air-sea interface is a reliable barrier where either gametes or G2 phase vegetative cells could aggregate. The case for G2 phase cells is especially intriguing because this cell type lies at the intersection of A. fundyense’s division and sexual cycles. The likelihood of a G2 cell’s forming gametes (as opposed to new vegetative cells) may be enhanced by interactions with other cells or surface associated physical cues (Brosnahan et al., 2010; Figueroa et al., 2011), so that aggregation of G2 cells at the surface could be linked to gametogenesis. Alternatively, if the 2C cells near the surface were planozygotes, they may have been newly formed by the fusion of 1C gametes. In either case, the near-surface 2C subpopulation represented only a small fraction of the bloom as a whole. The bulk of the population (integrated through the full depth of the pond) appeared to be vegetative and in the G1 phase of the cell cycle, consistent with the rapid accumulation of cells that continued through May 11th and up to the next sampling date (May 18th).
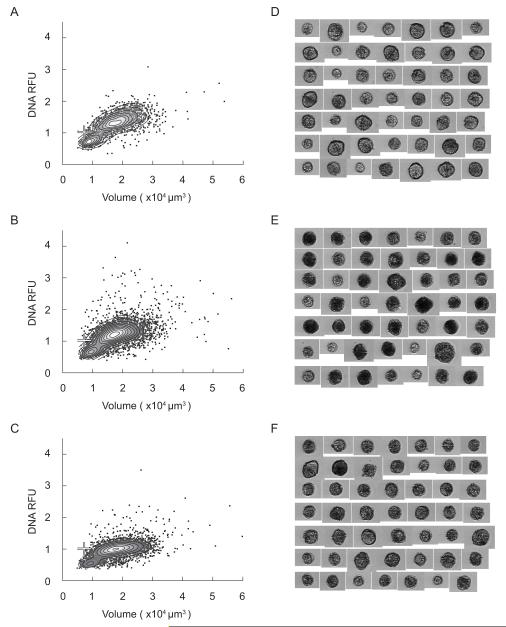
The highest cell concentrations observed during the 2009 bloom in Salt Pond occurred May 18th. In this case the survey was completed shortly after daybreak when large numbers of vegetative cells were likely to be in their G2+M cell cycle phase (2C DNA content, Fig. 1). IFCB analysis showed that the May 18th profile contained mixtures of 1C and 2C A. fundyense at all depths but the ratio of 2C to 1C was always greater than one and increased slightly from the near-surface (~3:1) to the near-bottom (~5:1; Fig. 9). Because A. fundyense divisions are phased to occur near the time-of-day of the May 18th survey, the ratio of 2C to 1C could be used to estimate A. fundyense’s specific growth rate, μ, by the fmax method (Chisholm, 1981). The calculation relies on the assumptions (1) that all dividing A. fundyense were 2C when sampled (i.e., that phasing of division was complete), and (2) that planozygote abundance was not significant. This approach provides estimated doubling times of 1.14 to 1.24 days, substantially higher than the net accumulation rate leading up to the sampling date.
Though the doubling time estimate should predict faster growth than the net accumulation rate (since the former does not account for grazing or other losses), A. fundyense cultures only achieve doubling times less than 2 days under optimal conditions, typically at temperatures several degrees higher than what was experienced by the Salt Pond bloom (see Watras et al., 1982; Stock et al., 2005 and references therein). Therefore the high estimate of μ — near the upper limit of the A. fundyense growth envelope — constitutes one line of evidence indicating a relatively high proportion of planozygotes on May 18th since subtraction of planozygotes from the 2C pool would reduce the μ estimate. A second line of evidence is the high proportion of large, high DNA content A. fundyense found in the samples collected one week later (May 26th; Fig. 10) since both traits – large cell size and high DNA content – are markers of mature planozygotes.
Figure 10.
Density scatter plots (A-C) and collages of representative images (D-F) of A. fundyense collected during the termination phase of the Salt Pond bloom (May 26th) at the near-surface (A and D), mid-depth (B and E), and near-bottom (C and F). White crosses on the density scatter plots represent the mode DNA content and cell volume of a reference culture of G1 phase vegetative cells (see Section 3.2).
From May 18th to May 26th, A. fundyense abundance in Salt Pond fell several-fold and cells were nearly undetectable by June 1st (Fig. 5). DNA content modes in the May 26th samples were poorly resolved, particularly in the near-bottom sample where maximum Hoechst fluorescence and cell volumes far exceeded what was observed in samples from earlier surveys (compare Fig. 10 and Figs. 8 and 9). Because mode populations were not well defined, cell DNA content was classified through comparison to a reference culture of G1 phase vegetative cells (1C DNA content); cells were classified as 2C and greater if their Hoechst fluorescence was higher than the reference mode. Large, 2C and greater cells were a significant proportion of the A. fundyense population at mid-depth and near the bottom (approximate proportions of 30% and 70%, respectively), but near the surface (where the cell concentration was ~20-fold lower) most cells were 1C and had ‘normal’ volumes, comparable to vegetative cells collected during the development and peak phases of the bloom (Figs. 5, 8, 9 and 10).
The DNA distribution of the large, near-bottom cells was diffuse with two weakly defined modes that were roughly two and three times the 1C culture reference, but their presence also coincided with an exceptionally high rate of infection by Amoebophrya (Figs. 5 and 10C). On the basis of their Hoechst fluorescence alone, large cells clustered around the highest mode in DNA content and volume might be classified as 4C cells, i.e. planozygotes that had completed PR. Upon examination by microscopy, however, we found that most of the large Salt Pond cells’ nuclei had been consumed by parasites, complicating the interpretation of DNA fluorescence measurements. As discussed in section 3.4, an infected A. fundyense culture of vegetative cells exhibited neither large size nor high DNA-associated fluorescence, so we attribute the increased size of the large Salt Pond cells to planozygote maturation rather than Amoebophrya infection.
3.4 Distinguishing the effects of sex and parasitism in A. fundyense
The co-occurrence of high rates of parasitism and the large A. fundyense cell size during the decline of the Salt Pond bloom challenged an assumption made in past studies that only planozygote maturation causes large cell size. The linkage between large size and planozygote maturation was also challenged by the absence of large cells in the WGOM red tide where most cells are presumed to have been planozygotes and where no evidence of parasitism was observed.
The conclusion that most cells in the WGOM red tide sample were planozygotes is informed by our DNA staining analysis. Samples from the WGOM red tide were collected in the late afternoon, a time when most vegetative cells would have been in the G1 phase of the cell cycle, but 1C cells were not observed. Instead, the vast majority of A. fundyense were 2C (Fig. 11C; McGillicuddy et al., this issue), indicating that the cells were most likely planozygotes. Two subsequent surveys showed that the high A. fundyense concentrations dissipated within 10 days of their discovery (McGillicuddy et al., this issue), and that a large deposition of cysts occurred beneath and downstream from the area where red water was observed (Anderson et al., this issue). There was also no evidence of Amoebophrya infection in the WGOM red tide. These observations and the absence of abundant grazers in the red tide samples led us to conclude that the termination of the GOM bloom was primarily due to sexual encystment, rather than to a combination of factors (e.g., encystment combined with grazing and parasite infection) as was observed in Salt Pond.
Figure 11.
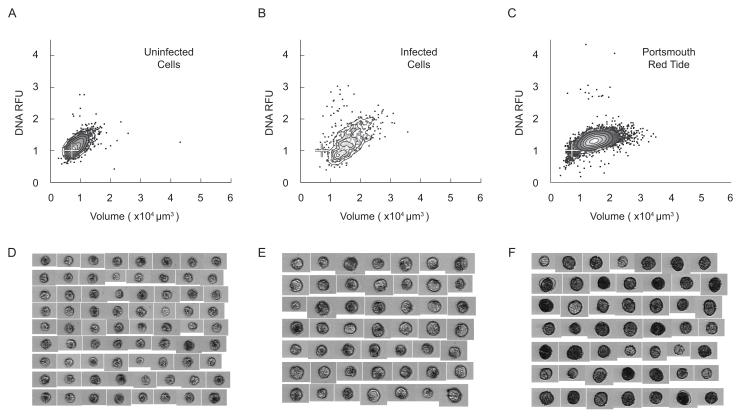
(A-C) Density scatter plots of an uninfected A. fundyense culture of the clone SPE10-03 (A), vegetative SPE10-03 infected by an Amoebophrya sp. isolate (B), and cells collected at station 1 during the Portsmouth red tide (C). White crosses mark the mode DNA content and cell volume of G1 phase vegetative cells that were minimally affected by formalin fixation and were used as a reference for classifying DNA modes in field samples. The measurement of Hoechst fluorescence in all field samples (including the Portsmouth red tide [C]) are considered underestimates because of prolonged fixation times (and therefore diminished Hoechst staining; see Section 3.2). (D-F) Associated image collages from the reference SPE10-03 culture (D), the infected SPE10-03 cells (E), and the Portsmouth red tide (F).
The size of the WGOM red tide 2C cells was similar to G2 phase vegetative cells observed in culture and during the development phase of the Salt Pond bloom (~2 × 104 μm3; compare Figs. 8, 9 and 11C), much smaller than the planozygotes observed during the Salt Pond bloom’s decline (Fig. 10). The larger A. fundyense from the decline of the Salt Pond bloom were also unique among the samples we analyzed in that they had extraordinarily high DNA-associated fluorescence and a high rate of parasite infection (Figs. 5 and 10C). Together, these observations suggest that large size and/or high DNA-associated fluorescence could have been caused by the parasites rather than planozygote development.
We addressed the question of whether Amoebophrya infection could have caused large size and high Hoechst fluorescence by examining an infected A. fundyense culture with the modified IFCB. Samples from the culture were stained with an Amoebophrya specific probe rather than the A. fundyense probe used for our analysis of field samples, and only the most heavily infected cells were identified by probe fluorescence. The mean Hoechst fluorescence of these heavily infected A. fundyense was only slightly higher than that of uninfected cells, and their mean cell volume was more similar to 1C vegetative cells than to the large, heavily infected cells from Salt Pond (Figs 10 and 11). Thus, we conclude that infection by Amoebophrya is not sufficient by itself to cause the large cell size or high DNA content observed during the Salt Pond bloom’s decline, and that the largest Salt Pond cells were most likely matured planozygotes.
With regard to high DNA content, the possibility remains that the combination of Amoebophrya infection and planozygote development caused the high DNA-associated fluorescence; our culture infection test did not include planozygotes. It may also be that the higher infection rate of these large planozygotes occurred because infection stimulates the formation of planozygotes. Such an effect is suggested from recent work by Chambouvet et al. (2011) who showed that S. troichodea increases its rate of encystment when exposed to Amoebophrya.
Our finding that Amoebophrya infection did not cause large cell size in culture does not reconcile the substantial difference in planozygote size between the May 26th Salt Pond samples and the WGOM red tide. The differences between these populations may instead reflect differences in their histories leading up to our sampling times or genetic differences — A. fundyense in the GOM may simply form smaller planozygotes than those within the NMS. Several studies have shown significant differences among strains isolated from the coastal GOM bloom and the Salt Pond and NMS populations, including differences in their germination behavior (Anderson and Keafer, 1987), toxin profiles (Anderson et al., 1994), and microsatellite loci (Richlen et al., in prep). It is also possible that planozygotes in the red tide samples grew larger as they matured after our initial (and only) observation.
4. Conclusions
Here we present results from an IFCB that was modified to improve its ability to make quantitative fluorescence measurements. In an initial application of the modified IFCB, the instrument was used to investigate the population dynamics of A. fundyense through DNA staining and a species-specific ribosomal probe. The combination of imaging, single-cell DNA measurements and detection of a species-specific ribosomal probe enabled the characterization of two naturally occurring A. fundyense blooms. Data from the IFCB have revealed unexpected patterns in the occurrence of 2C cells and the large-scale conversion of A. fundyense to its diploid form during bloom termination in both inshore and offshore environments. The latter finding emphasizes the importance of sexual encystment for this organism’s life cycle.
This type of analysis does raise some outstanding challenges. Further method development, especially of sample fixation procedures, is likely to improve the reliability of comparisons between samples collected at different times and locations. Our observations were also complicated by the large proportion of A. fundyense that were infected by the parasite Amoebophrya during the termination of the Salt Pond bloom. More observations are needed to resolve the impacts of parasites on A. fundyense DNA content and planozygote maturation and to explore other interactions between species in natural assemblages. Finally, our inability to resample the WGOM red tide in spite of a prodigious effort underscores the value of studying A. fundyense in the context of isolated inshore blooms like those in Salt Pond. The retention of cells within the NMS ponds facilitated A. fundyense monitoring which, in turn, aided interpretation of changes to the distribution of the cells’ DNA content. Future studies implementing imaging flow cytometry to characterize the DNA contents of protist species will benefit from repeated sampling of target populations. Knowledge of mitotic rhythmicity, metabolic lifestyle, and the co-occurrence of parasites was also crucial for understanding changes in the frequency of A. fundyense DNA types and is likely to be needed for studies of other target species as well.
Acknowledgements
We are grateful to B. Crespo, K. Norton, and many other members of the Anderson lab for their contributions to the NMS surveys, to D. Kulis and M. Sengco (US EPA) for their guidance in the rearing of Amoebophrya, to the crews of the R/V Gulf Challenger and the R/V Tioga for their work during the WGOM red tide and event response, to D. McGillicuddy and D. Ralston for helpful discussions, and to two anonymous reviewers for valuable input. This work was supported by NSF grants OCE-0430724 and OCE-0911031 to DMA through the Woods Hole Center for Oceans and Human Health, National Park Service Cooperative Agreement H238015504 with DMA, by an EPA STAR graduate fellowship (FP-91688601) to MLB, and by grants from NSF, ONR, NOAA, NASA, and the Gordon and Betty Moore Foundation to RJO and HMS. This is ECOHAB contribution 733.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DM, Chisholm SW, Watras CJ. Importance of life cycle events in the population dynamics of Gonyaulax tamarensis. Mar. Biol. 1983;76(2):179–189. [Google Scholar]
- Anderson DM, Keafer BA. An endogenous annual clock in the toxic marine dinoflagellate Gonyaulax tamarensis. Nature. 1987;325(6105):616–617. doi: 10.1038/325616a0. [DOI] [PubMed] [Google Scholar]
- Anderson DM, Keafer BA, Kleindinst JL, McGillicuddy DJ, Martin JL, Norton K, Pilskaln CH, Smith JL. Alexandrium fundyense cysts in the Gulf of Maine: time series of abundance and distribution, and linkages to past and future blooms. Deep Sea Res. Part II Top. Stud. Oceanogr. 2013;(this issue) doi: 10.1016/j.dsr2.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Kulis DM, Doucette GJ, Gallagher JC, Balech E. Biogeography of toxic dinoflagellates in the genus Alexandrium from the northeastern United States and Canada. Mar. Biol. 1994;120(3):467–478. [Google Scholar]
- Anderson DM, Kulis DM, Keafer BA. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antidbody probes: variability in labeling intensity with physiological condition. J. Phycol. 1999;35:870–883. [Google Scholar]
- Anderson DM, Kulis DM, Keafer BA, Gribble KE, Marin R, Scholin CA. Identification and enumeration of Alexandrium spp. from the Gulf of Maine using molecular probes. Dee Sea Res. Part II Top. Stud. Oceanogr. 2005;52(19-21):2467–2490. [Google Scholar]
- Anderson DM, Lindquist NL. Time-course measurements of phosphorus depletion and cyst formation in the dinoflagellate Gonyaulax tamarensis Lebour. J. Exp. Mar. Biol. Ecol. 1985;86:1–13. [Google Scholar]
- Anderson DM, Stolzenbach KD. Selective retention of two dinoflagellates in a well-mixed estuarine embayment: the importance of diel vertical migration and surface avoidance. Mar. Ecol. Prog. Ser. 1985;25:39–50. [Google Scholar]
- Anderson DM, Wall D. Potential importance of benthic cysts of Gonyaulax tamarensis and G. excavata in initiating toxin dinoflagellate blooms. J. Phycol. 1978;14(2):224–234. [Google Scholar]
- Bhaud Y, Soyer-Gobillard M-O, Salmon JM. Transmission of gametic nuclei through a fertilization tube during mating in a primitive dinoflagellate, Prorocentrum micans. Ehr. J. Cell Sci. 1988;89(2):197–206. [Google Scholar]
- Brosnahan ML, Kulis DM, Solow AR, Erdner DL, Percy L, Lewis J, Anderson DM. Outbreeding lethality between toxic Group I and nontoxic Group III Alexandrium tamarense spp. isolates: predominance of heterotypic encystment and implications for mating interactions and biogeography. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010;57(3-4):175–189. doi: 10.1016/j.dsr2.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetta CM, Anderson DM. Cell cycle studies of the dinoflagellates Gonyaulax polyedra Stein and Gyrodinium uncatenum Hulburt during asexual and sexual reproduction. J. Exp. Mar. Biol. Ecol. 1990;135(1):69–84. [Google Scholar]
- Chambouvet A, Alves-de-Souza C, Cueff V, Marie D, Karpov S, Guillou L. Interplay between the parasite Amoebophrya sp. (Alveolata) and the cyst formation of the red tide dinoflagellate Scrippsiella trochoidea. Protist. 2011;162(4):637–649. doi: 10.1016/j.protis.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Chambouvet A, Morin A, Marie D, Guillou L. Control of toxic marine dinoflagellate blooms by serial parasitic killers. Science. 2008;322(5905):1254–1257. doi: 10.1126/science.1164387. [DOI] [PubMed] [Google Scholar]
- Chisholm SW. Temporal patterns of cell division in unicellular algae. Can. Bull. Fish. Aquatic. Sci. 1981;210:150–181. [Google Scholar]
- Coats DW. Parasitic life styles of marine dinoflagellates. J. Eukaryot. Microbiol. 1999;46(4):402–409. [Google Scholar]
- Crespo BG, Keafer BA, Ralston DK, Lind H, Farber D, Anderson DM. Dynamics of Alexandrium fundyense blooms and shellfish toxicity in the Nauset Marsh System of Cape Cod (Massachusetts, USA) Harmful Algae. 2011;12:26–38. doi: 10.1016/j.hal.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa RI, Vázquez JA, Massenet A, Murado MA, Bravo I. Interactive effects of salinity and temperature on planozygote and cyst formation of Alexandrium minutum (Dinophyceae) in culture. J. Phycol. 2011;47(1):13–24. doi: 10.1111/j.1529-8817.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine plankton diatoms I. Cyclotella nana Hustedt and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Haley ST, Juhl AR, Keafer BA, Anderson DM, Dyhrman ST. Detecting copepod grazing on low-concentration populations of Alexandrium fundyense using PCR identification of ingested prey. J. Plankton Res. 2011;33(6):927–936. [Google Scholar]
- Hartwell AD. Hydrographic factors affecting the distribution and movement of toxic dinoflagellates in the western Gulf of Maine. In: Lo Cicero VR, editor. Toxic Dinoflagellate Blooms. Massachusetts Science and Technology Foundation; Massachusetts: 1975. pp. 47–68. [Google Scholar]
- Hasle GR. Phototactic vertical migration in marine dinoflagellates. Oikos. 1950;2(2):162–175. [Google Scholar]
- Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS ONE. 2009;4(9):e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U, Fensome RA, Medlin LK. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense “species complex” (Dinophyceae) Mol. Biol. Evol. 2003;20(7):1015–1027. doi: 10.1093/molbev/msg105. [DOI] [PubMed] [Google Scholar]
- Lilly EL, Halanych KM, Anderson DM. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae) J. Phycol. 2007;43(6):1329–1338. [Google Scholar]
- McGillicuddy DJ, Brosnahan ML, Couture DA, He R, Keafer BA, Manning JP, Martin JL, Pilskaln CH, Townsend DW, Anderson DM. A red tide of Alexandrium fundyense in the Gulf of Maine. Deep Sea Res. Part II Top. Stud. Oceanogr. 2013;(this issue) doi: 10.1016/j.dsr2.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan HF. Oceanographic factors associated with New England red-tide blooms. In: Lo Cicero VR, editor. Toxic Dinoflagellate Blooms. Massachusetts Science and Technology Foundation; Massachusetts: 1975. pp. 23–40. [Google Scholar]
- Olson RJ, Sosik HM. A submersible imaging-in-flow instrument to analyze nano- and microplankton: Imaging FlowCytobot. Limnol. Oceanogr. Methods. 2007;5:195–203. [Google Scholar]
- Pfiester LA, Anderson DM. Dinoflagellate reproduction. In: Taylor FJR, editor. The Biology of Dinoflagellates. Blackwell Scientific Publications; UK: 1987. pp. 611–648. [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol. Rev. 1994;58(2):233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin CG. Measurements of in situ growth rates of Gonyaulax tamarensis: the New England red tide organism. Massachusetts Institute of Technology; Cambridge, Massachusetts: 1981. [Google Scholar]
- Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 1994;30(4):744–754. [Google Scholar]
- Scholin CA, Hallegraeff GM, Anderson DM. Molecular evolution of the Alexandrium tamarense ‘species complex’ (Dinophyceae): dispersal in the North American and West Pacific regions. Phycologia. 1995;34(6):472–485. [Google Scholar]
- Shapiro HM. Practical Flow Cytometry. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2003. [Google Scholar]
- Sosik HM, Olson RJ. Automated taxonomic classification of phytoplankton sampled with imaging in-flow cytometry. Limnol. Oceanogr. Methods. 2007;5:204–216. [Google Scholar]
- Stock CA, McGillicuddy DJ, Solow AR, Anderson DM. Evaluating hypotheses for the initiation and development of Alexandrium fundyense blooms in the western Gulf of Maine using a coupled physical-biological model. Deep Sea Res. Part II Top. Stud. Oceanogr. 2005;52(19-21):2715–2744. [Google Scholar]
- Taroncher-Oldenburg G, Kulis DM, Anderson DM. Toxin variability during the cell cycle of the dinoflagellate Alexandrium fundyense. Limnol. Oceanogr. 1997;42(5):1178–1188. [Google Scholar]
- Taroncher-Oldenburg G, Kulis DM, Anderson DM. Coupling of saxitoxin biosynthesis to the G1 phase of the cell cycle in the dinoflagellate Alexandrium fundyense: temperature and nutrient effects. Nat. Toxins. 1999;7:207–219. doi: 10.1002/1522-7189(200009/10)7:5<207::aid-nt61>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Taylor FJR. Parasitism of the toxin-producing dinoflagellate Gonyaulax catenella by the endoparasitic dinoflagellate Amoebophrya ceratii. J. Fish. Res. Bd. Can. 1968;25[g883](10):2241–2245. [Google Scholar]
- Watras CJ, Chisholm SW, Anderson DM. Regulation of growth in an estuarine clone of Gonyaulax tamarensis Lebour: salinity-dependent temperature responses. J. Exp. Mar. Biol. Ecol. 1982;62:25–37. [Google Scholar]