Abstract
Long noncoding RNAs (lncRNAs) are increasingly implicated in cancer biology, contributing to essential cancer cell functions such as proliferation, invasion, and metastasis. In prostate cancer, several lncRNAs have been nominated as critical actors in disease pathogenesis. Among these, expression of PCGEM1 and PRNCR1 has been identified as a possible component in disease progression through the coordination of androgen receptor (AR) signaling (Yang et al., Nature 2013, see ref. [1]). However, concerns regarding the robustness of these findings have been suggested. Here, we sought to evaluate whether PCGEM1 and PRNCR1 are associated with prostate cancer. Through a comprehensive analysis of RNA-sequencing data (RNA-seq), we find evidence that PCGEM1 but not PRNCR1 is associated with prostate cancer. We employ a large cohort of >230 high-risk prostate cancer patients with long-term outcomes data to show that, in contrast to prior reports, neither gene is associated with poor patient outcomes. We further observe no evidence that PCGEM1 nor PRNCR1 interact with AR, and neither gene is a component of AR signaling. Thus, we conclusively demonstrate that PCGEM1 and PRNCR1 are not prognostic lncRNAs in prostate cancer and we refute suggestions that these lncRNAs interact in AR signaling.
Keywords: prostate cancer, long noncoding RNA, androgen receptor
INTRODUCTION
Long noncoding RNAs (lncRNAs) have emerged as a critical element in cell biology, contributing to a wide variety of cellular behaviors and functions [2]. In cancer, lncRNAs have been the subject of much research during the past five years. Notably, lncRNAs are known to coordinate aggressive phenotypes of several common tumors, including breast cancer and prostate cancer [3, 4]. Large profiling studies have suggested that upwards of 10,000 lncRNAs may exist in the human genome [5]; yet only a fraction of these entities have been characterized. Thus, the identity and function of lncRNAs in cancer remains largely unknown.
In prostate cancer, several lncRNAs, including PCA3 and PCAT-1, have been shown to be upregulated in patients with cancer [6-9]. Recently, two lncRNAs, PCGEM1 and PRNCR1, have been suggested in prostate cancer to act as mediators of castration-resistance disease by binding, in a direct and sequential fashion, to the androgen receptor (AR), causing ligand-independent activation of its gene expression programs [1]. While PCGEM1 has been observed in prostate cancer previously [6, 10], PRNCR1 is a poorly characterized transcript, and we were concerned that PRNCR1 had been nominated by previous global profiling studies of prostate cancers [7, 11-14].
We therefore sought to investigate PRNCR1 and PCGEM1 in prostate cancer. In specific, we sought to reproduce three core observations suggested by Yang et al published in Nature (see [1]) and include: 1) that PRNCR1 and PCGEM1 are highly overexpressed in aggressive forms of prostate cancer, 2) that these two lncRNAs bind to AR under ligand-stimulated conditions, and 3) that the coordination of PRNCR1 and PCGEM1 interact with AR via specific post-translational modifications of the AR protein. Here, we report that none of these three findings is fully reproducible.
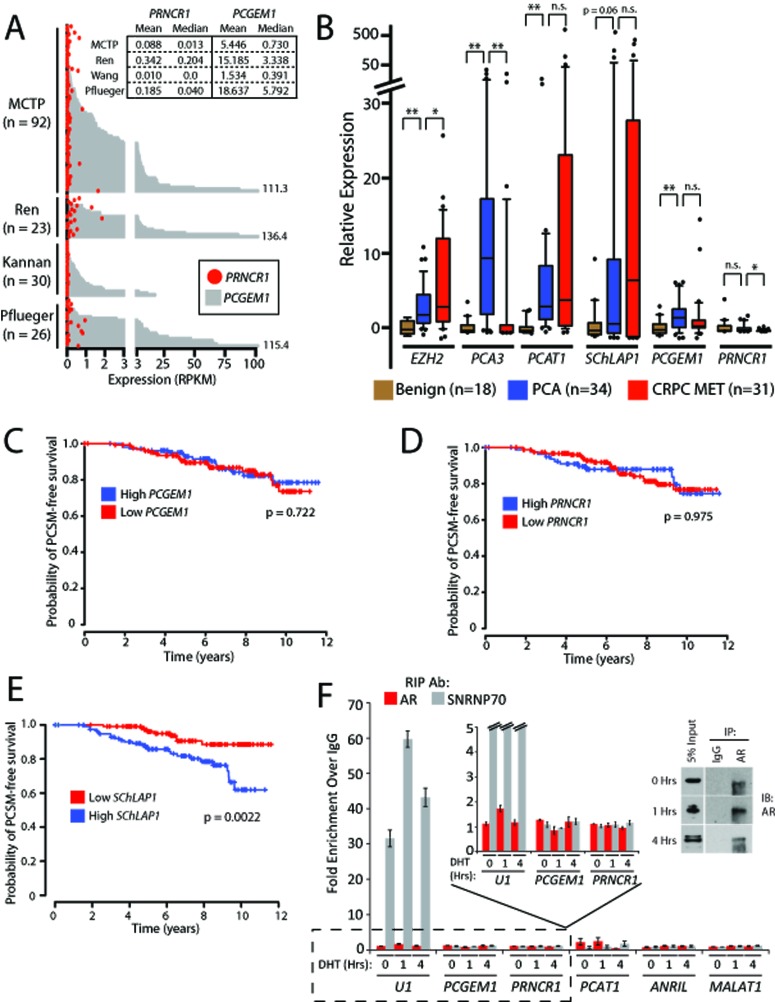
First, we asked whether PCGEM1 and PRNCR1 are highly overexpressed in aggressive prostate cancer, as suggested by others (see [1, 15]). Indeed, while some have argued that these lncRNAs are critical in castration-resistant prostate cancer [1], there has been no study that evaluated the expression of these lncRNAs in tissue samples from human castrate-resistant prostate cancers (CRPC). To evaluate these lncRNAs in more detail, we first assessed their expression levels in 171 human prostatic tissues using RNA sequencing data aggregated from four independent studies of prostate cancer, including our own internal datasets [1, 12-14] (Fig. 1A). Whereas we found robust expression of PCGEM1 in a subset of prostate tissues (RPKM >1 in 82 samples; RPKM >10 in 27 samples), we observed scant levels of PRNCR1 in all samples (RPKM >1 in only 3 samples; RPKM >10 in 0 samples) (Supplementary Table 1). This does not lend confidence to PRNCR1 as a significant entity in this disease. For comparison, we used the prostate cancer lncRNA SChLAP1 as a positive control. We found extreme overexpression of SChLAP1 in samples from all datasets (RPKM >1 in 69 samples; RPKM >10 in 26 samples) (Supplementary Fig. 1). To rule out the possibility that PRNCR1 was a non-poly-adenylated RNA, we verified experimentally that PRNCR1 was observed in the poly-A fraction of RNA that was used to generate the RNA-seq data (Supplementary Fig. 2).
Figure 1. PCGEM1 and PRNCR1 are not associated with prostate cancer progression and do not bind the androgen receptor.
(A) Plot showing PCGEM1 (grey bars) and PRNCR1 (red circles) expression levels (Reads per Kilobase per Million Reads, or RPKM) across 171 samples from four RNA-Seq studies of prostate cancer: Michigan Center for Translational Pathology (MCTP, internal data and dbGAP, phs000443.v1.p1), Ren et al. [13] (EGA, ERP00550), Kannan et al. [14] (GEO, GSE22260), and Pflueger et al. [12] (dbGAP, phs000310.v1.p1). Inset box shows descriptive statistics for each study. (B) Quantitative PCR for PCGEM1 and PRNCR1 in a cohort of prostate cancer tissues, benign (n = 18), localized cancer (n =34), metastatic cancer (n = 31). An asterisk (*) indicates p < 0.05. Two asterisks (**) indicate p < 0.01. n.s. = non-significant. P values were determined by a two-tailed Student's t-test. Data for SChLAP1 is obtained and re-analyzed from a prior publication (ref. [4]). (C) PCGEM1 expression does not predict for prostate cancer-specific mortality (PCSM). (D) PRNCR1 expression does not predict for PCSM. (E) High SChLAP1 expression is a powerful predictor of PCSM (p = 0.0022). Data in (E) is reproduced from a prior publication (ref. [4]). P values in (C-E) are determined using a log-rank test. (F) RNA- immunoprecipitation (RIP) for AR following stimulation of LNCaP cells with 100nM DHT does not show binding of PRNCR1 or PCGEM1 to AR. U1 binding to SNRNP70 is used as a positive control. PCAT-1, ANRIL, and MALAT1 serve as negative controls. Inset: Western blot confirmation of AR protein pull-down by the immunoprecipitation assays. Error bars represent S.E.M.
Given the low support for PRNCR1 in the RNA-seq data, we next confirmed these findings using quantitative PCR (qPCR) in a large set of prostate cancer tissues including 34 PCAs and 31 CRPC tumors as well as 18 benign adjacent tissues. As shown in Fig. 1B, PCGEM1 is upregulated in clinically localized cancer, confirming the known literature [6, 10]; however PRNCR1 expression does not demonstrate a convincing association with prostate cancer. We found a borderline decrease in PRNCR1 expression in metastatic castrate-resistant cancer (p = 0.047, Student's t-test). We used PCAT1, EZH2, and SChLAP1 as control genes, all of which have elevated expression in prostate cancer metastases. Conversely, we used PCA3 as a control gene that is known to be upregulated in localized prostate cancer but not metastatic prostate cancer. Finally, while PCGEM1 is upregulated in cancer patients from matched tumor/benign samples, PRNCR1 does not convincingly exhibit this pattern of upregulation (Supplementary Fig. 3).
Next, an independent analysis of 235 high-risk prostate cancer tissues demonstrated that neither PCGEM1 nor PRNCR1 is associated with aggressive prostate cancer, and neither lncRNA stratifies prostate cancer-specific mortality (Fig. 1C,D and Supplementary Tables 2,3). An analysis of intermediate endpoints such as biochemical recurrence and progression to metastatic disease demonstrated a trend for PCGEM1 and PRNCR1 to be associated with less aggressive disease and favorable outcomes (Supplementary Fig. 4), which contradicts previous claims that these lncRNAs are involved in an aggressive patient clinical course [1, 15]. Using an independent validation cohort of tissues we verified that neither PCGEM1 nor PRNCR1 is associated with aggressive prostate cancer (Supplementary Table 2). By contrast, we have used these datasets to confirm the prognostic utility of the lncRNA SChLAP1 in prostate cancer, and high expression of SChLAP1 is a powerful predictor for poor patient survival (Fig. 1E) [4].
Next, we examined whether PCGEM1 and PRNCR1 interacted with AR. We performed RNA-IP (RIP) assays using two independent AR antibodies, including the same antibody that was previously used to show an interaction between these lncRNAs and AR [1]. In accordance with the published literature, we performed a time-series of RIP experiments following AR stimulation, because prior data suggests that these lncRNAs bind AR from 1-2 hours after AR stimulation but not at 4 hours post-stimulation [1]. In our RIP experiments, we could not confirm that AR binds to PCGEM1 or PRNCR1 at either 1 hour or 4 hours post-stimulation with DHT (Fig. 1F and Supplementary Fig. 5). Similarly, in cells grown at steady-state, we used a second AR antibody and did not observe binding between AR and PCGEM1 or PRNCR1 (Supplementary Fig. 6). DHT-stimulated cells also demonstrated no induction in PCGEM1 or PRNCR1 expression (Supplementary Fig. 7). These results imply that PCGEM1 and PRNCR1 are not AR-interacting lncRNAs.
Finally, earlier data propose that PCGEM1 and PRNCR1 interact with AR via specific post-translational modifications (PTMs), specifically K349 methylation (K349Me) for PCGEM1 and K631/K634 acetylation (K631Ac/K634Ac) for PRNCR1 [1]. To search for these PTMs, we independently performed mass spectrometry for AR in the LNCaP cell line, achieving 95% coverage of all possible tryptic peptides. We were unable to confirm that these PTMs (K349Me, K631Ac, or K634Ac) are present on AR (Supplementary Fig. 8 and Supplementary Table 4). To examine this discrepancy further, we re-analyzed prior AR MS data (found in [1]). Although this MS dataset was obtained with a trypsin digestion to prepare samples for MS, we found no fully tryptic peptides supporting the nomination of K349Me, K631Ac, or K634Ac (Supplementary Fig. 8). In fact, in the MS data for these PTMs in ref. [1], almost all peptides harboring these PTMs are non-tryptic, which are generally considered to be analysis artifacts since true non-tryptic peptides are exceedingly rare following a trypsin digestion [16-18] (Supplementary Discussion). Non-tryptic peptides are also associated with a high false-discovery rate [19]. All peptides nominating the K349Me, K631Ac, or K634Ac PTMs in ref. [1] also had multiple additional PTMs that were nominated, indicating non-specificity. These included extraordinarily rare and unusual PTMs such as oxidated lysine and deamidated asparagine, which suggest technical artifacts given the negligible likelihood of multiple rare and unusual PTMs occurring on true non-trypic peptides. The statistical confidence for these non-tryptic peptides is <5%, whereas the corresponding fully tryptic peptides for these amino acid residues had statistical confidences >90%.
In summary, we have been unable to show a convincing role for PCGEM1 or PRNCR1 in aggressive prostate cancer or AR signaling. First, our data analysis of numerous human prostate cancer tissues from multiple independent laboratories indicates that neither PCGEM1 nor PRNCR1 are associated with castration-resistant prostate cancer. Second, we were unable to verify that PCGEM1 and PRNCR1 bind to the androgen receptor. Lastly, we are unconvinced that the K349Me, K631Ac, or K634Ac AR PTMs represent a plausible mechanism for interaction between AR and PCGEM1 and PRNCR1. While our results challenge the notion that PCGEM1 and PRNCR1 play a causal role in prostate cancer, we regard lncRNAs as an emerging field of study in cancer [3, 6, 20, 21] and we are encouraged by the interest in lncRNAs in prostate cancer.
METHODS
Prostate tissues were obtained from the radical prostatectomy series and Rapid Autopsy Program at the University of Michigan tissue core. All tissue samples were collected with informed consent under an Institutional Review Board (IRB) approved protocol at the University of Michigan. Outcomes analyses were performed on a cohort of Mayo Clinic prostate cancer radical prostatectomy samples obtained under an IRB-approved protocol as described previously. Cell lines were maintained according to standard conditions. For RIP experiments, cells were deprived of androgen for 48 hours prior to stimulation with 100nM DHT. RIP experiments were performed as previously described [1, 4]. Bioinformatics analyses utilized publicly available RNA-Seq data. Please see Supplementary Methods for details.
SUPPLEMENTARY FIGURES AND TABLES
Acknowledgments
We thank Xia Jiang and Shruthi Subramaniam for technical assistance. This work was supported in part by the NIH Prostate Specialized Program of Research Excellence grant P50CA69568, the Early Detection Research Network grant UO1 CA111275, the US National Institutes of Health R01CA132874-01A1, and the Department of Defense grant PC100171 (A.M.C.). A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, the Prostate Cancer Foundation, and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. F.Y.F. was supported by the Prostate Cancer Foundation, the Department of Defense grant PC094231. J.R.P. was supported by a Prostate Cancer Foundation Young Investigator Award. A.S. was supported by the NIH Predoctoral Fellowship 1F30CA180376-01. M.K.I. was supported by the Department of Defense Predoctoral Fellowship BC100238. R.M. was supported by the Department of Defense Post-doctoral Fellowship W81XWH-13-1-0284. J.R.P., M.K.I., and A.S. are Fellows of the University of Michigan Medical Scientist Training Program.
The University of Michigan has filed a patent on lncRNAs in prostate cancer, including SChLAP1, in which A.M.C.,J.R.P. and M.K.I. are named as co-inventors. Wafergen, Inc. has a non-exclusive license for creating commercial research assays for lncRNAs in prostate cancer. GenomeDx Biosciences Inc has an exclusive license for creating tissue assays for lncRNAs in prostate cancer. A.M.C. is a co-founder and advisor to Compendia Biosciences, which supports the Oncomine database. He also serves on the Scientific Advisory Board of Wafergen; neither Life Technologies or Wafergen had any role in the design or experimentation of this study, nor have they participated in the writing of the manuscript. I.A.V. and E.D. are employees of GenomeDx Biosciences Inc.
J.R.P., R.M., M.K.I., A.S. and A.M.C. designed the project and directed experimental studies. J.R.P, R.M, A.S. and B.C. performed in vitro studies. M.K.I. performed bioinformatics analysis. I.A.A. and A.P. performed AR mass spectrometry. I.A.V., R.B.J., E.D., and M.A. acquired human tissue samples and performed statistical outcomes analyses for PCGEM1 and PRNCR1 expression. J.R.P., M.K.I., A.S., R.M., F.Y.F. and A.M.C. designed experiments, interpreted data, and wrote the manuscript.
REFERENCES
- 1.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62(9):2695–2698. [PubMed] [Google Scholar]
- 9.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6(5):255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 10.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K, Buzard GS, Mostofi FK, McLeod DG, Moul JW, Srivastava S. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97(22):12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, Cao X, Barrette T, Tewari AK, Chee MS, Chinnaiyan AM, Rickman DS, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21(1):56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012;22(5):806–821. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci U S A. 2011;108(22):9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW, Srivastava S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23(2):605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 16.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6(9):1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Kim JS, Monroe ME, Camp DG, 2, Smith RD, Qian WJ. In-source fragmentation and the sources of partially tryptic peptides in shotgun proteomics. J Proteome Res. 2013;12(2):910–916. doi: 10.1021/pr300955f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics. 2007;6(9):1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3(6):608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493(7431):231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.