Abstract
Human longevity is a multifactorial phenotype influenced by both genetic and environmental factors. Despite its heritability of 25–32 %, the genetic background of longevity is as yet largely unexplained. Apart from APOE status, variation in the FOXO3A gene is the only confirmed genetic contributor to survival into old age. On the other hand, FOXO3A activity is known to be downregulated in various cancers, and the gene was recently identified as a novel deletion hotspot in human lung adenocarcinoma. In view of the strong association between smoking and lung cancer, we set out to explore whether smoking modifies the known association between FOXO3A variation and longevity. To this end, we conducted a case–control study in two different populations, drawing upon extensive collections of old-aged individuals and younger controls available to us (1,613 German centenarians/nonagenarians and 1,104 controls; 1,088 Danish nonagenarians and 736 controls). In the German sample, 21 single nucleotide polymorphisms (SNPs) from the FOXO3A gene region were genotyped, whereas 15 FOXO3A SNPs were analyzed in the Danish sample. Eight SNPs were typed in both populations. Logistic regression analysis revealed that adjustment for smoking does not systematically alter the association between FOXO3A variation and longevity in neither population. Our analysis therefore suggests that the said association is not largely due to the confounding effects of lung cancer.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-013-9578-z) contains supplementary material, which is available to authorized users.
Keywords: Longevity, Aging, Smoking, Lung cancer, Confounder, Heritability
Introduction
Human longevity is a complex phenotype, and genetic factors have been estimated to account for ∼30 % of the overall variation in adult lifespan (Christensen et al. 2006; Finch and Tanzi 1997; Gogele et al. 2011; Herskind et al. 1996; Hjelmborg et al. 2006; Ljungquist et al. 1998; Skytthe et al. 2003). As yet, however, variation in only two genes has been confirmed to influence survival into old age, namely in those encoding apolipoprotein E (APOE), with ε4 being a mortality factor in the elderly (Blanché et al. 2001; Christensen et al. 2006; Deelen et al. 2011; Nebel et al. 2011; Schachter et al. 1994), and transcription factor forkhead box O3A (FOXO3A). A comparatively modest association between FOXO3A polymorphisms and longevity has been observed in different populations (Anselmi et al. 2009; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010; Willcox et al. 2008). However, the molecular mechanisms underlying this relationship still remain to be elucidated. Most of the longevity-associated FOXO3A single nucleotide polymorphisms (SNPs) analyzed so far are located in intronic regions, and the “functionally relevant” variants still have to be identified (Donlon et al. 2012; Flachsbart et al. 2012).
The FOXO3A protein is an evolutionarily conserved key regulator of the insulin-IGF1 signaling pathway (Hwangbo et al. 2004; Kenyon 2005; Kenyon et al. 1993; Lin et al. 1997; Ziv and Hu 2011). It also plays an important role in growth arrest, DNA repair, and apoptosis in response to DNA damage and oxidative stress (Furukawa-Hibi et al. 2002; Greer and Brunet 2005; Kops et al. 2002; Tran et al. 2002). Furthermore, FOXO3A has been implicated as a tumor suppressor (Fei et al. 2009; Hu et al. 2004; Mikse et al. 2010; Yang et al. 2008), and FOXO3A activity was consistently found to be downregulated in various cancers (Yang and Hung 2009; Greer and Brunet 2005). More recently, FOXO3A was also reported to stimulate a proapoptotic transcriptional program in response to a human lung carcinogen (Blake et al. 2010), and the FOXO3A gene region is a target of somatic deletion in lung adenocarcinoma (LAC) in both humans (Mikse et al. 2010) and mice (Herzog et al. 2009).
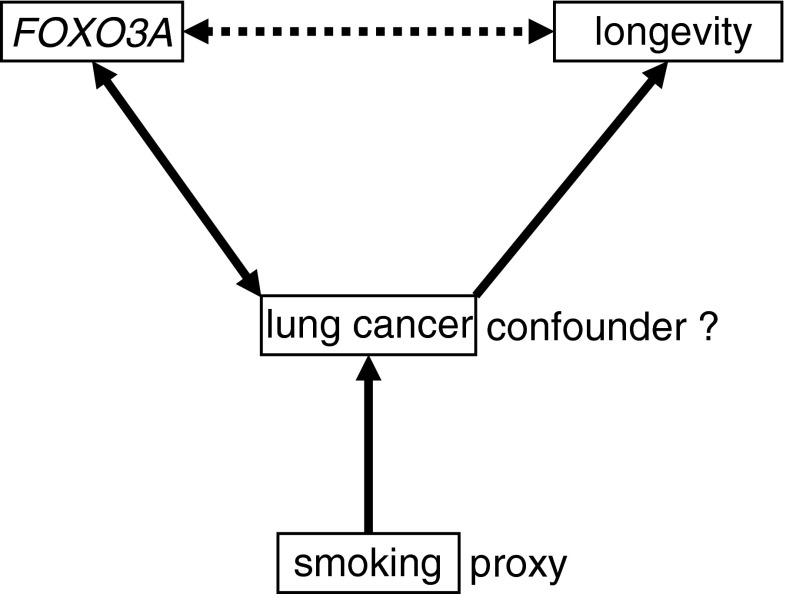
Since development of lung cancer clearly reduces an individual’s life expectancy, the above association implies that the known influence of FOXO3A on longevity may be due, at least partially, to confounding by lung cancer. Such provisos are not uncommon in epidemiology, so that the allowance for possible confounders in the respective analyses has become mandatory to avoid false positive results. However, since lung cancer is not directly observable in a case–control study of longevity, we had to resort to smoking as a proxy for the disease (Doll et al. 2004; Dayan 1986; Liu et al. 1998; Warner et al. 1989), knowing that many smokers never develop lung cancer, and that smoking has many effects upon longevity that are unrelated to lung cancer (Fig. 1). This incongruence implies that the effects of smoking adjustment on the FOXO3A–longevity relationship can only partly reflect the relevance of lung cancer. This notwithstanding, if the association between FOXO3A and longevity was indeed confounded by lung cancer, then the same association would be expected to be notably stronger among smokers than among nonsmokers.
Fig. 1.
Study setting. Associations between variables are depicted by bold arrows. A dashed arrow is used to indicate that the association between FOXO3A and longevity may be due partially to confounding by lung cancer. Smoking serves as a proxy for lung cancer. Note that cases and controls differ by both their life expectancy and generation membership
To address the above questions, we performed a case–control study in two populations, namely a German sample comprising 1,613 long-lived individuals (95–110 years) and 1,104 younger controls, and a Danish sample of 1,088 cases aged 92–93 years and 736 younger controls (Table 1). Individuals were classified as either smokers (ever smokers) or nonsmokers (never smokers). One caveat has to be taken into account in the present study, however, namely that cases and controls belong to different generations. Smoking behavior is known to have changed during the twentieth century (Benowitz et al. 2005; Franceschi and Bidoli 1999) so that “smoking” may have meant different things in the two groups. Nevertheless, as regards possible confounding by lung cancer, we think that use of such a “noisy” proxy was still valid. In view of the important role of APOE allele ε4 in human longevity (Blanché et al. 2001; Christensen et al. 2006; Deelen et al. 2011; Nebel et al. 2011; Schachter et al. 1994), we also adjusted all analyses for the APOE ε4 status.
Table 1.
Study samples
| Median age* (in years) | Total number | Smokers# (%) | Number of FOXO3A SNPs analyzed | |
|---|---|---|---|---|
| German | ||||
| Old-aged individuals | 99 (95–110) | 1,613 Male: 436 (27.0 %) Female: 1,177 (73.0 %) |
27.1 | 21 |
| Controls | 67 (60–75) | 1104 Male: 283 (25.6 %) Female: 821 (74.4 %) |
50.7 | |
| Danish | ||||
| Old-aged individuals | 93 (92–93) | 1,088 Male: 313 (28.7 %) Female: 775 (71.2 %) |
45.9 | 15 |
| Controls | 50.5 (46–54) | 736 Male: 371 (50.4 %) Female: 365 (49.6 %) |
62.6 | |
| German and Danish | ||||
| Old-aged individuals | 96 (92–110) | 2,701 Male: 749 (27.7 %) Female:1,952 (72.3 %) |
35.4 | 8 |
| Controls | 63 (46–75) | 1,840 Male: 654 (35.5 %) Female:1,186 (64.5 %) |
55.5 | |
* Age range is given in parenthesis; # ever smokers
Material and methods
Study population
We analyzed two case–control samples from two different populations (Table 1). The German sample included 1,613 unrelated old-aged individuals, with an age range of 95 to 110 years (median age 99 years). Some 27 % of these were male. Old-aged individuals were recruited from different geographic regions of Germany and included a subset of 748 centenarians (median age 101 years). The 1,104 controls were 60 to 75 years old (median age 67 years), and were matched for ancestry, sex, and geographic origin within the country. The recruitment of the German sample has been described in detail elsewhere (Nebel et al. 2005).
The Danish cohort comprised of 1,088 old-aged individuals (29 % male) born in 1905 (Nybo et al. 2001). The 736 younger controls were randomly selected from the study of middle-aged Danish twins (Skytthe et al. 2002), but only one twin of each twin pair was included in the present study. In the controls, the sex ratio was approximately 1:1. At the time of interviewing, the old individuals were 92 to 93 years old (median age 93 years) and the younger controls were 46 to 54 years old (median age 50.5 years). The Danish study population also has been described in more detail elsewhere (Soerensen et al. 2010).
Genotyping
The German sample was genotyped on an automated platform (Hampe et al. 2001) using Taqman SNP genotyping assays (Life Technologies Corporation, Foster City, CA). A subset of the German sample and some SNPs have been analyzed before (Flachsbart et al. 2009), with six additional SNPs and 955 additional individuals investigated here. Genotyping of the Danish sample was performed on the Illumina GoldenGate platform (Illumina Inc) (Steemers and Gunderson 2005; Soerensen et al. 2010). All samples and SNPs have been investigated before (Soerensen et al. 2010). In total, 21 FOXO3A SNPs were analyzed in the German sample (Table 2) and 15 SNPs were analyzed in the Danish sample (Table 3). Eight of the FOXO3A SNPs and the two APOE SNPs rs7412 and rs429358 were analyzed in both populations (Table 4).
Table 2.
Association between longevity and FOXO3A SNP genotype. German sample (1,613 old-aged individuals, 1,104 controls)
| No. | dbSNP ID | MAF controls (n = 1104) | MAF cases (n = 1613) | Risk allele | Unadjusted | Adjusted for smoking | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95 % CI | P | OR | 95 % CI | |||||
| 1 | rs2274776 | 0.490 | 0.486 | – | 0.899 | 1.009 | 0.881–1.154 | 0.928 | 1.007 | 0.871–1.164 |
| 2 | rs1571631 | 0.465 | 0.462 | – | 0.847 | 1.013 | 0.885–1.160 | 0.986 | 1.001 | 0.866–1.158 |
| 3 | rs6911407 | 0.378 | 0.415 | A | 0.032 | 1.163 | 1.013–1.336 | 0.010 | 1.215 | 1.048–1.409 |
| 4 | rs768023 | 0.382 | 0.415 | G | 0.049 | 1.149 | 1.001–1.319 | 0.016 | 1.201 | 1.035–1.392 |
| 5 | rs2802288 | 0.385 | 0.407 | – | 0.139 | 1.091 | 0.972–1.226 | 0.096 | 1.112 | 0.981–1.260 |
| 6 | rs2883881 | 0.093 | 0.087 | – | 0.659 | 1.047 | 0.855–1.281 | 0.463 | 1.086 | 0.872–1.353 |
| 7 | rs12200646 | 0.123 | 0.138 | – | 0.192 | 1.119 | 0.945–1.326 | 0.259 | 1.110 | 0.926–1.331 |
| 8 | rs2802290 | 0.387 | 0.417 | G | 0.083 | 1.130 | 0.984–1.298 | 0.025 | 1.185 | 1.022–1.375 |
| 9 | rs2802292 | 0.382 | 0.413 | G | 0.062 | 1.141 | 0.993–1.310 | 0.043 | 1.165 | 1.005–1.351 |
| 10 | rs13220810 | 0.259 | 0.251 | – | 0.561 | 1.039 | 0.913–1.184 | 0.607 | 1.037 | 0.902–1.193 |
| 11 | rs2764264 | 0.304 | 0.338 | C | 0.039 | 1.164 | 1.007–1.346 | 0.019 | 1.203 | 1.030–1.405 |
| 12 | rs7762395 | 0.153 | 0.175 | A | 0.092 | 1.174 | 0.974–1.416 | 0.023 | 1.266 | 1.034–1.551 |
| 13 | rs13217795 | 0.297 | 0.328 | C | 0.048 | 1.160 | 1.001–1.345 | 0.020 | 1.206 | 1.030–1.412 |
| 14 | rs9400239 | 0.295 | 0.331 | T | 0.028 | 1.180 | 1.018–1.369 | 0.007 | 1.245 | 1.062–1.460 |
| 15 | rs3800231 | 0.287 | 0.327 | A | 0.010 | 1.215 | 1.047–1.410 | 0.002 | 1.290 | 1.100–1.513 |
| 16 | rs4945816 | 0.289 | 0.316 | C | 0.062 | 1.126 | 0.994–1.276 | 0.031 | 1.160 | 1.014–1.326 |
| 17 | rs4946936 | 0.291 | 0.321 | G | 0.045 | 1.133 | 1.003–1.279 | 0.024 | 1.162 | 1.020–1.324 |
| 18 | rs1268170 | 0.347 | 0.377 | G | 0.060 | 1.147 | 0.994–1.323 | 0.017 | 1.204 | 1.033–1.403 |
| 19 | rs473268 | 0.337 | 0.367 | A | 0.077 | 1.139 | 0.986–1.316 | 0.023 | 1.197 | 1.025–1.398 |
| 20 | rs479744 | 0.199 | 0.229 | T | 0.020 | 1.225 | 1.032–1.454 | 0.005 | 1.304 | 1.085–1.567 |
| 21 | rs519007 | 0.186 | 0.183 | – | 0.798 | 1.023 | 0.860–1.217 | 0.865 | 1.016 | 0.844–1.223 |
MAF minor allele frequency, Risk allele “risk” allele for, attaining old age, P p value obtained from a risk allele-based case–control comparison using a Wald test, OR odds ratio for attaining old age, 95 % CI 95 % confidence interval. All analyses were adjusted for APOE status. In the combined sample, nationality was included as an additional influential variable. Nominally significant associations upon adjustment for smoking (p < 0.05) are printed in bold
Table 3.
Association between longevity and FOXO3A SNP genotype. Danish sample (1,088 old-aged individuals, 736 controls)
| No. | dbSNP ID | MAF controls (n = 736) | MAF cases (n = 1,088) | Risk allele | Unadjusted | Adjusted for smoking | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95 % CI | P | OR | 95 % CI | |||||
| 1 | rs9486902 | 0.178 | 0.203 | A | 0.057 | 1.192 | 0.881–1.154 | 0.048 | 1.205 | 1.002–1.450 |
| 2 | rs10499051 | 0.095 | 0.087 | – | 0.468 | 1.091 | 0.885–1.160 | 0.706 | 1.047 | 0.823–1.332 |
| 3 | rs12206094 | 0.258 | 0.282 | – | 0.181 | 1.111 | 1.013–1.336 | 0.132 | 1.129 | 0.964–1.321 |
| 4 | rs2802292 | 0.355 | 0.379 | – | 0.179 | 1.105 | 1.001–1.319 | 0.103 | 1.131 | 0.975–1.310 |
| 5 | rs13220810 | 0.286 | 0.258 | – | 0.116 | 1.129 | 0.972–1.226 | 0.076 | 1.149 | 0.985–1.339 |
| 6 | rs2764264 | 0.286 | 0.294 | – | 0.735 | 1.026 | 0.855–1.281 | 0.516 | 1.052 | 0.902–1.226 |
| 7 | rs7762395 | 0.143 | 0.158 | – | 0.260 | 1.115 | 0.945–1.326 | 0.209 | 1.132 | 0.933–1.374 |
| 8 | rs12207868 | 0.099 | 0.101 | – | 0.918 | 1.012 | 0.984–1.298 | 0.946 | 1.008 | 0.801–1.269 |
| 9 | rs13217795 | 0.275 | 0.289 | – | 0.506 | 1.053 | 0.993–1.310 | 0.363 | 1.075 | 0.920–1.255 |
| 10 | rs9400239 | 0.277 | 0.292 | – | 0.415 | 1.065 | 0.913–1.184 | 0.276 | 1.089 | 0.934–1.270 |
| 11 | rs12212067 | 0.101 | 0.102 | – | 0.984 | 1.002 | 1.007–1.346 | 0.962 | 1.005 | 0.803–1.259 |
| 12 | rs9398172 | 0.275 | 0.288 | – | 0.440 | 1.061 | 0.974–1.416 | 0.320 | 1.080 | 0.928–1.259 |
| 13 | rs3800231 | 0.276 | 0.293 | – | 0.343 | 1.076 | 1.001–1.345 | 0.222 | 1.101 | 0.943–1.284 |
| 14 | rs3800232 | 0.123 | 0.123 | – | 0.900 | 1.013 | 1.018–1.369 | 0.935 | 1.009 | 0.817–1.246 |
| 15 | rs479744 | 0.191 | 0.211 | – | 0.149 | 1.133 | 1.047–1.410 | 0.068 | 1.175 | 0.989–1.397 |
MAF minor allele frequency, Risk allele “risk” allele for attaining old age, P p value obtained from a risk allele-based case–control comparison using a Wald test, OR odds ratio for attaining old age, 95 % CI 95 % confidence interval. All analyses were adjusted for APOE status. In the combined sample, nationality was included as an additional influential variable. Nominally significant associations upon adjustment for smoking (p < 0.05) are printed in bold
Table 4.
Association between longevity and FOXO3A SNP genotype. Combined sample (2,701 old-aged individuals, 1,840 controls)
| No. | dbSNP ID | MAF controls (n = 1840) | MAF cases (n = 2701) | Risk allele | Unadjusted | Adjusted for smoking | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P | OR | 95 % CI | P | OR | 95 % CI | |||||
| 1 | rs2802292 | 0.368 | 0.396 | G | 0.023 | 1.123 | 1.016–1.241 | 0.009 | 1.150 | 1.036–1.276 |
| 2 | rs13220810 | 0.270 | 0.254 | – | 0.143 | 1.076 | 0.975–1.188 | 0.099 | 1.091 | 0.984–1.209 |
| 3 | rs2764264 | 0.295 | 0.316 | C | 0.084 | 1.096 | 0.988–1.217 | 0.031 | 1.127 | 1.011–1.256 |
| 4 | rs7762395 | 0.148 | 0.166 | A | 0.046 | 1.145 | 1.002–1.308 | 0.012 | 1.197 | 1.041–1.377 |
| 5 | rs13217795 | 0.286 | 0.308 | C | 0.060 | 1.107 | 0.996–1.231 | 0.021 | 1.139 | 1.020–1.272 |
| 6 | rs9400239 | 0.286 | 0.311 | T | 0.033 | 1.122 | 1.010–1.248 | 0.007 | 1.164 | 1.042–1.300 |
| 7 | rs3800231 | 0.281 | 0.310 | A | 0.013 | 1.144 | 1.029–1.272 | 0.002 | 1.190 | 1.065–1.329 |
| 8 | rs479744 | 0.195 | 0.220 | T | 0.009 | 1.176 | 1.042–1.326 | 0.001 | 1.234 | 1.088–1.400 |
MAF minor allele frequency, Risk allele “risk” allele for attaining old age, P p value obtained from a risk allele-based case–control comparison using a Wald test, OR odds ratio for attaining old age, 95 % CI 95 % confidence interval. All analyses were adjusted for APOE status. In the combined sample, nationality was included as an additional influential variable. Nominally significant associations upon adjustment for smoking (p < 0.05) are printed in bold
Smoking information
For the German sample, information on smoking behavior was obtained via questionnaires. Individuals were asked whether they had ever smoked, whether they were recent smokers, for how many years they had smoked, and how many cigarettes they smoked per day. For the Danish sample, individuals were interviewed and classified as never, former, or current smokers. More detailed smoking information is provided in Supplementary Table S1.
Statistical analysis
Statistical analyses were performed using software R v.12.0 (Team RDC 2008). All tests were two-sided at the 5 % significance level. The genotyped SNPs were tested for Hardy–Weinberg equilibrium in the controls using an exact test as implemented in R-package genetics (Warnes et al. 2011). Associations between a SNP genotype and longevity were assessed for statistical significance by means of logistic regression analysis with and without interaction, followed by a Wald test (Wald 1943). SNP genotypes were coded by the dosage of the respective “risk allele” for attaining old age, thereby assuming multiplicativity of the odds ratios and ensuring that reported odds ratios were always larger than unity. All SNP association analyses were adjusted for APOE genotype coded by the number of APOE ε4 alleles (“dosage”). When German and Danish data were analyzed together, nationality was added as an additional influential variable.
Model selection involving all FOXO3A SNPs was impossible due to multicollinearity caused by linkage disequilibrium (LD). To avoid collinearity, we excluded SNPs in such a way that all remaining markers had pairwise r2 values <0.80 (see Supplementary Figures S1–S3). LD plots were prepared with Haploview v4.2 (Barrett et al. 2005). The following FOXO3A SNPs were eventually taken into account in the following model selection: rs2274776, rs1571631, rs2883881, rs1220646, rs2802290, rs2802292, rs13220810, rs2764264, rs7762395, rs13217795, rs473268, rs479744, and rs519007 for the German sample; rs9486902, rs10499051, rs12206094, rs2802292, rs13220810, rs7762395, rs12207868, rs3800232, and rs479744 for the Danish sample; and rs2802292, rs13220810, rs2764264, rs7762395, rs13217795, and rs479744 for the combined sample. Starting from the full model, we performed backward selection that kept only influential variables with a p value <0.05 in the final model.
Results
Association of APOE and smoking with longevity
As was to be expected, smoking (ever versus never) and APOE ε4 dosage both had a strong impact upon longevity (Table 5), with estimated odds ratios of 0.36 and 0.42, respectively, in the German sample and of 0.51 and 0.58 in the Danish sample. No statistically significant interaction was observed between the two factors.
Table 5.
Association between longevity and either APOE status or smoking
| Simple logistic regression analysis | Multiple logistic regression analysis | |||
|---|---|---|---|---|
| P | OR | P | OR | |
| German | ||||
| APOE | <2 × 10−16 | 0.427 | <2 × 10−16 | 0.424 |
| Smoking status | <2 × 10−16 | 0.361 | <2 × 10−16 | 0.379 |
| Danish | ||||
| APOE | 3.2 × 10−8 | 0.580 | 2.9 × 10−8 | 0.495 |
| Smoking status | 2.9 × 10−12 | 0.505 | 2.0 × 10−8 | 0.572 |
P p values were obtained using a Wald test, OR odds ratio for attaining old age; simple logistic regression analysis, both influential variables were analyzed separately; multiple logistic regression analysis, both influential variables were analyzed jointly. No statistically significant interaction was observed in any of the analyses; APOE number of APOE ε4 alleles; smoking status, ever versus never smokers with never smokers as reference category
Association between FOXO3A and longevity, with and without adjustment for smoking
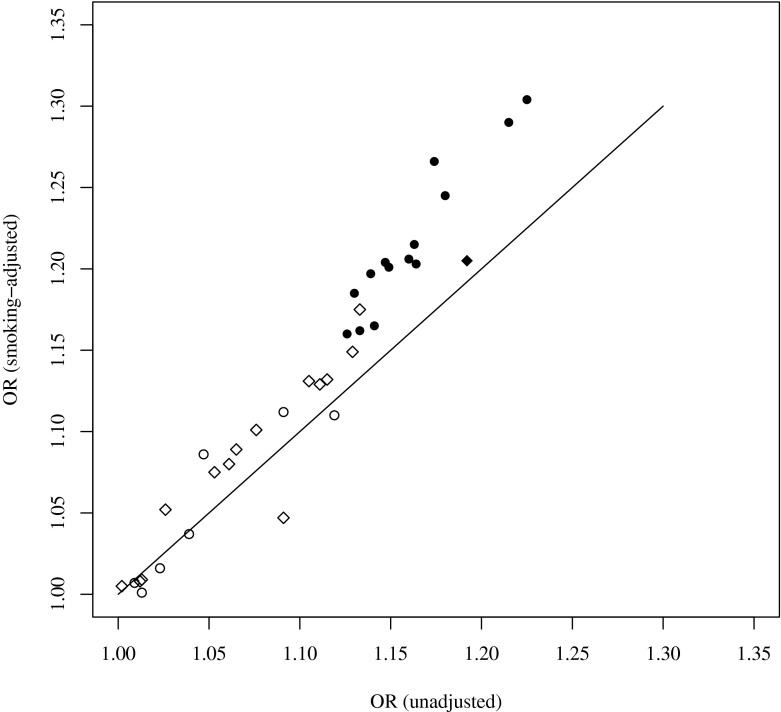
We analyzed the association between longevity and genetic variation in the FOXO3A gene region for 21 SNPs in the German sample and 15 SNPs in the Danish sample. Eight of the SNPs that had been genotyped in both groups were analyzed in both samples combined. All SNPs were in Hardy–Weinberg equilibrium in the control samples, and all analyses were adjusted for APOE ε4 dosage. In the German sample, eight SNPs (rs6911407, rs768023, rs2764264, rs13217795, rs9400239, rs3800231, rs4946936, and rs479744) were significantly associated with longevity (Table 2). In the Danish sample, no significant association between a SNP and longevity was detected (Table 3). In the combined sample, five SNPs (rs2802292, rs7762395, rs9400239, rs3800231, and rs479744) were found to be significantly associated with longevity (Table 4). After adjustment for smoking, all significant genotype–phenotype relationships remained significant (Fig. 2). In addition, some of the SNPs acking an association in the unadjusted analysis became nominally significant after adjustment for smoking as follows: rs2802290, rs2802292, rs7762395, rs4945816, rs1268170, and rs473268 in the German sample, rs2764264 and rs13217795 in the combined sample set, and rs9486902 in the Danish sample (Tables 2, 3, and 4).
Fig. 2.
FOXO3A odds ratios for attaining old age, with and without adjustment for smoking. Odds ratios were obtained by logistic regression analyses of longevity and FOXO3A SNP genotype, with (vertical axis) and without smoking adjustment (horizontal axis). Circles German sample; diamonds Danish sample. Significant associations are marked by filled symbols. All analyses were adjusted for APOE genotype
Model selection with and without adjustment for smoking
In order to assess the joint effects upon longevity of FOXO3A variation, APOE genotype, nationality, and smoking, allowing for a possible correlation between these variables (e.g., due to linkage disequilibrium), we performed two types of logistic regression analysis with backward model selection. Initial model M1 included only the FOXO3A SNPs and APOE ε4 dosage (plus nationality, when appropriate), whereas model M2 also included smoking. In the German sample, the same FOXO3A SNPs occurred in final models M1 and M2 (Table 6), namely rs1200646 and rs479744. Moreover, APOE and smoking (in the case of model M2) were also included in the models. Final model M1 for the Danish sample did not include any FOXO3A SNP, but only APOE ε4 dosage. Final model M2 included APOE, FOXO3A SNP rs13220810, and smoking. For the combined sample, both final models included FOXO3A SNP rs479744 and APOE ε4 dosage. Apart from smoking, nationality was also found to be a significantly influential variable in model M2 (but not in M1). To test the robustness of the model selection, we added smoking to final model M1. All previously selected influential variables remained statistically significant in all three samples (German, Danish, and combined). No statistically significant pairwise interaction between the different influential variables, particularly not between smoking and any FOXO3A genotype, was observed.
Table 6.
Logistic regression model selection
| Sample | Final model without smoking (M1) | M1 + Smoking | Final model with smoking (M2) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Influential variables | P | OR | Influential variables | P | OR | Influential variables | P | OR | |
| German | rs1200646 rs479744 APOE |
0.021 0.019 8.5 × 10−12 |
1.289 1.242 2.278 |
rs1200646 rs479744 APOE Smoking |
0.031 0.006 7.1 × 10−11 1.3 × 10−14 |
1.288 1.314 2.351 2.402 |
rs1200646 rs479744 APOE Smoking |
0.031 0.006 7.1 × 10−11 1.3 × 10−14 |
1.288 1.314 2.351 2.402 |
| Danish | APOE | 1.2 × 10−8 | 1.801 |
APOE
Smoking |
1.1 × 10−8
1.1 × 10−12 |
1.827 2.110 |
rs13220810 APOE Smoking |
0.036 1.3 × 10−8 2.7 × 10−13 |
1.041 1.821 2.136 |
| Combined | rs479744 APOE |
0.014 < 2 × 10−16 |
1.170 1.937 |
rs47944 APOE Smoking |
0.003 < 2 × 10−16 < 2 × 10−16 |
1.219 1.932 2.115 |
rs479744 APOE Smoking Nationality |
0.002 < 2 × 10−16 <2 × 10−16 1.8 × 10−4 |
1.225 1.982 2.220 1.331 |
P p values were obtained using a Wald test, OR odds ratio for attaining old age, nationality was encoded as 1 for German and 0 for Danish, APOE number of APOE ε4 alleles, smoking status ever versus never smokers with never smokers as reference category
Robustness of results
The above results were based upon the use of two smoking categories, namely “ever” and “never”. We also adopted alternative classifications of smoking status, particularly for the German sample, where more information on smoking behavior was available. For example, we considered “never and former” versus “current” and “smoking for more than 5 years” versus “never”. The results of these analyses turned out to be very similar to those obtained with the original classification (data not shown). We also performed a sex-stratified analysis, with results both for males and females that were very similar to the results of the non-stratified analysis (see Supplementary Tables S2 and S3). In the Danish males, however, one SNP (rs13220810) ceased to show a significant association with longevity upon adjustment for smoking.
Discussion
Variation in the FOXO3A gene is not only associated with longevity (Anselmi et al. 2009; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010; Willcox et al. 2008) but also plays a role in the etiology of various neoplasias, including lung cancer (Donlon et al. 2012; Greer and Brunet 2005; Herzog et al. 2009; Mikse et al. 2010; Willcox et al. 2008; Yang and Hung 2009). In the present study, we therefore set out to investigate whether the association observed between FOXO3A and longevity may have been due, at least partially, to the confounding effects of lung cancer. Unfortunately, lung cancer mortality is difficult, if not impossible, to address in retrospective case–control studies. However, since smoking is strongly associated with lung cancer (Dayan 1986; Doll et al. 2004; Liu et al. 1998; Warner et al. 1989), we thought that a combined analysis of longevity and FOXO3A variation using smoking as a proxy for lung cancer still appeared well warranted. If confounding by the disease played an important role, such an analysis should have yielded a smaller or even absent residual effect of FOXO3A variation on aging, or a significant interaction between FOXO3A genotype and smoking. Contrary to this expectation, our study revealed an unaltered genotype–phenotype association after adjustment for smoking in both single and multiple SNP analyses. The latter ultimately invoked only smoking and one or two FOXO3A SNPs as significantly influential variables, and no interaction with smoking became apparent.
Interestingly, both the number and the magnitude of significant FOXO3A SNP associations differed considerably between the German and the Danish samples analyzed here, with the German sample showing the stronger associations. This discrepancy is potentially explicable by the fact that the association between FOXO3A and longevity is much stronger in centenarians than in nonagenarians (Flachsbart et al. 2009). In the Danish sample, the age range was only 92 to 93 years, compared to 95 to 110 years in the Germans, who even included a subset of 748 centenarians. Moreover, in the first report of an association between FOXO3A and longevity in the Danish population, the association was observed only for males and only using specific modes of inheritance (Soerensen et al. 2010). Such differences notwithstanding, smoking adjustment did not alter the said genotype–phenotype relationship in neither population samples.
Our study was not intended to investigate the effect of smoking on longevity, but rather addressed the possible confounding effects of lung cancer. Smoking prevalence varied considerably in the past and therefore differed between our cases and controls as well simply because the two groups belonged to different generations (Deutsches Krebsforschungszentrum 2008; Peto et al. 2000). Notably, the quality of smoking has also changed, for example, by a trend towards so-called “light” cigarettes that stimulate smokers to inhale more deeply (Benowitz et al. 2005; Franceschi and Bidoli 1999). This means that our proxy for lung cancer may have been somewhat imprecise. However, several aspects seem to support the general validity of our conclusion that the FOXO3A-longevity association is not strongly confounded by lung cancer. First, we obtained similar results in two different populations, and with several SNPs. Second, although smoking prevalence and smoking habits are known to differ considerably between males and females, we obtained similar results in a sex-stratified as in the non-stratified analyses. Finally, a lack of an effect of smoking adjustment was consistently seen with all smoking classifications employed here.
In our study, the number of smokers was considerably higher in the Danish sample than in the German sample (Table 1). This was true for the controls, but even more so for the old-aged individuals. Not surprisingly, a stronger association between smoking and longevity was, therefore, seen in the German sample. There are several possible explanations for this population difference in smoking prevalence. Danish individuals were recruited in a cohort study and were observed over a long period of time. Therefore, their smoking behavior could be recorded more precisely than in the German sample, which followed a case–control design with the usual drawback of recall bias, especially at exceptionally old age. However, the difference in smoking prevalence did not qualitatively change the effect of smoking adjustment on the association between FOXO3A variation and longevity.
Our study followed a case–control design, which is known to be inferior to a cohort design in many respects. Most importantly, case–control studies are liable to recall bias so that environmental exposure data may lack the accuracy of analogous information from direct follow-up. Moreover, as was mentioned in the introduction, the relationship between an exposure and a phenotype may become confounded by age if the typology of the exposure, in this case smoking, has changed over time. However, since the association between FOXO3A variation and longevity only becomes apparent for extremely old age (≥95 years of age), prospective studies are difficult because of the extremely low prevalence of the phenotype (0.01–0.02 %, Perls 2006) and the long follow-up period required. Therefore, case–control studies almost inevitably have become the most popular design for the investigation of longevity (Deelen et al. 2013). Moreover, as has been pointed out above, our study was not intended to investigate the effect of smoking on longevity per se, which would be difficult to do in a case–control design anyway. Instead, we were interested in the possible confounding effect of lung cancer (with smoking used as a proxy), which should be detectable in case–control studies as well. Finally, it must be emphasized that the association between FOXO3A variation and longevity was originally detected (Willcox et al. 2008) and subsequently confirmed in case–control studies (Anselmi et al. 2009; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010). Only one study was prospective in nature (Soerensen et al. 2010). Bearing in mind the methodological limitations of case–control studies and their possible impact on statistical power, the question whether an association was confounded by a given candidate can, thus, only sensibly be clarified using the same study design.
In the future, it would be interesting to explore the potential relationship between genetic variation in FOXO3A and various cancers from the viewpoint that the former is a potential tumor suppressor (Fei et al. 2009; Hu et al. 2004; Mikse et al. 2010; Yang et al. 2008). So far, little is known about the link between germ line FOXO3A variation and a predisposition to cancer (Campa et al. 2011). For example, deletions in the FOXO3A gene region in LAC were explored in somatic cancer cells only (Mikse et al. 2010). Moreover, an investigation of the role of FOXO3A variation in cancers other than LAC might provide further independent insight into the importance of FOXO3A for human aging and longevity (Donlon et al. 2012).
Electronic supplementary material
(DOC 407 kb)
Acknowledgments
We thank all the study participants for their cooperation. This study was supported by the Deutsche Forschungsgemeinschaft (NE1191/1-1), the RESOLVE project (FP7-HEALTH-F4-2008-202047), the Excellence Cluster “Inflammation at Interfaces,” and the INTERREG 4 A program Syddanmark-Schleswig-K.E.R.N. (with funds from the European Regional Development Fund). Data for the Danish samples were made available from the Danish Aging Research Center.
Footnotes
Carolin Däumer and Friederike Flachsbart contributed equally to this work.
References
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12(2):95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Jacob P, 3rd, Bernert JT, Wilson M, Wang L, Allen F, Dempsey D. Carcinogen exposure during short-term switching from regular to “light” cigarettes. Cancer Epidemiol Biomarkers Prev. 2005;14(6):1376–1383. doi: 10.1158/1055-9965.EPI-04-0667. [DOI] [PubMed] [Google Scholar]
- Blake DC, Jr, Mikse OR, Freeman WM, Herzog CR. FOXO3a elicits a proapoptotic transcription program and cellular response to human lung carcinogen nicotine-derived nitrosaminoketone (NNK) Lung Cancer. 2010;67(1):37–47. doi: 10.1016/j.lungcan.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Blanché H, Cabanne L, Sahbatou M, Thomas G. A study of French centenarians: are ACE and APOE associated with longevity? C R Acad Sci III. 2001;324(2):129–135. doi: 10.1016/S0764-4469(00)01274-9. [DOI] [PubMed] [Google Scholar]
- Campa D, Husing A, Dostal L, Stein A, Drogan D, Boeing H, Tjonneland A, Roswall N, Ostergaard JN, Overvad K, Rodriguez L, Bonet C, Sanchez MJ, Larranaga N, Huerta JM, Ardanaz E, Khaw KT, Wareham N, Travis RC, Allen NE, Trichopoulou A, Zylis D, Karapetyan T, Palli D, Sieri S, Tumino R, Vineis P, Bueno-de-Mesquita HB, Lenner P, Johansson M, Jenab M, Cox D, Siddiq A, Kaaks R, Canzian F. Genetic variability of the forkhead box O3 and prostate cancer risk in the European Prospective Investigation on Cancer. Oncol Rep. 2011;26(4):979–986. doi: 10.3892/or.2011.1359. [DOI] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7(6):436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan R (1986) Tobacco: a major international health hazard. Proceedings of an international meeting. Moscow, 4–6 June 1985. IARC Sci Publ, vol 74, 1986/01/01 edn [PubMed]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van de Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Capri M, Franceschi C, Slagboom PE. Identifying the genomic determinants of aging and longevity in human population studies: progress and challenges. Bioessays. 2013;35(4):386–396. doi: 10.1002/bies.201200148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsches Krebsforschungszentrum (Eds) (2008) Frauen und Rauchen in Deutschland, Heidelberg
- Doll R, Peto R, Boreham J, Sutherland I. Mortality in relation to smoking: 50 years’ observations on male British doctors. BMJ. 2004;328(7455):1519. doi: 10.1136/bmj.38142.554479.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon TA, Curb JD, He Q, Grove JS, Masaki KH, Rodriguez B, Elliott A, Willcox DC, Willcox BJ. FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci. 2012 doi: 10.1093/gerona/gls067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei M, Zhao Y, Wang Y, Lu M, Cheng C, Huang X, Zhang D, Lu J, He S, Shen A. Low expression of Foxo3a is associated with poor prognosis in ovarian cancer patients. Cancer Invest. 2009;27(1):52–59. doi: 10.1080/07357900802146204. [DOI] [PubMed] [Google Scholar]
- Finch CE, Tanzi RE. Genetics of aging. Science. 1997;278(5337):407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachsbart F, Moller M, Daeumer C, Gentschew L, Kleindorp R, Krawczak M, Caliebe A, Schreiber S, Nebel A. Genetic investigation of FOXO3A requires special attention due to sequence homology with FOXO3B. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi S, Bidoli E. The epidemiology of lung cancer. Ann Oncol. 1999;10(Suppl 5):S3–S6. doi: 10.1093/annonc/10.suppl_5.S3. [DOI] [PubMed] [Google Scholar]
- Furukawa-Hibi Y, Yoshida-Araki K, Ohta T, Ikeda K, Motoyama N. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J Biol Chem. 2002;277(30):26729–26732. doi: 10.1074/jbc.C200256200. [DOI] [PubMed] [Google Scholar]
- Gogele M, Pattaro C, Fuchsberger C, Minelli C, Pramstaller PP, Wjst M. Heritability analysis of life span in a semi-isolated population followed across four centuries reveals the presence of pleiotropy between life span and reproduction. J Gerontol A Biol Sci Med Sci. 2011;66(1):26–37. doi: 10.1093/gerona/glq163. [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Hampe J, Wollstein A, Lu T, Frevel HJ, Will M, Manaster C, Schreiber S. An integrated system for high-throughput TaqMan based SNP genotyping. Bioinformatics. 2001;17(7):654–655. doi: 10.1093/bioinformatics/17.7.654. [DOI] [PubMed] [Google Scholar]
- Herskind AM, McGue M, Holm NV, Sorensen TI, Harvald B, Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97(3):319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- Herzog CR, Blake DC, Jr, Mikse OR, Grigoryeva LS, Gundermann EL. FoxO3a gene is a target of deletion in mouse lung adenocarcinoma. Oncol Rep. 2009;22(4):837–843. doi: 10.3892/or_00000507. [DOI] [PubMed] [Google Scholar]
- Hjelmborg JB, Iachine I, Skytthe A, Vaupel JW, McGue M, Koskenvuo M, Kaprio J, Pedersen NL, Christensen K. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119(3):312–321. doi: 10.1007/s00439-006-0144-y. [DOI] [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117(2):225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signaling in brain and fat body. Nature. 2004;429(6991):562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120(4):449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366(6454):461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419(6904):316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18(24):4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Liu BQ, Peto R, Chen ZM, Boreham J, Wu YP, Li JY, Campbell TC, Chen JS. Emerging tobacco hazards in China: 1. Retrospective proportional mortality study of one million deaths. BMJ. 1998;317(7170):1411–1422. doi: 10.1136/bmj.317.7170.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungquist B, Berg S, Lanke J, McClearn GE, Pedersen NL. The effect of genetic factors for longevity: a comparison of identical and fraternal twins in the Swedish Twin Registry. J Gerontol A Biol Sci Med Sci. 1998;53(6):M441–M446. doi: 10.1093/gerona/53A.6.M441. [DOI] [PubMed] [Google Scholar]
- Mikse OR, Blake DC, Jr, Jones NR, Sun YW, Amin S, Gallagher CJ, Lazarus P, Weisz J, Herzog CR. FOXO3 encodes a carcinogen-activated transcription factor frequently deleted in early-stage lung adenocarcinoma. Cancer Res. 2010;70(15):6205–6215. doi: 10.1158/0008-5472.CAN-09-4008. [DOI] [PubMed] [Google Scholar]
- Nebel A, Croucher PJ, Stiegeler R, Nikolaus S, Krawczak M, Schreiber S. No association between microsomal triglyceride transfer protein (MTP) haplotype and longevity in humans. Proc Natl Acad Sci U S A. 2005;102(22):7906–7909. doi: 10.1073/pnas.0408670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Aging Dev. 2011;132(6–7):324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Nybo H, Gaist D, Jeune B, Bathum L, McGue M, Vaupel JW, Christensen K. The Danish 1905 cohort: a genetic-epidemiological nationwide survey. J Aging Health. 2001;13(1):32–46. doi: 10.1177/089826430101300102. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8(4):460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perls TT. The different paths to 100. Am J Clin Nutr. 2006;83(2):484S–487S. doi: 10.1093/ajcn/83.2.484S. [DOI] [PubMed] [Google Scholar]
- Peto R, Darby S, Deo H, Silcocks P, Whitley E, Doll R. Smoking, smoking cessation, and lung cancer in the UK since 1950: combination of national statistics with two case–control studies. Br Med J. 2000;321(7257):323–329. doi: 10.1136/bmj.321.7257.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6(1):29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Kyvik K, Holm NV, Vaupel JW, Christensen K. The Danish Twin Registry: 127 birth cohorts of twins. Twin Res. 2002;5(5):352–357. doi: 10.1375/136905202320906084. [DOI] [PubMed] [Google Scholar]
- Skytthe A, Pedersen NL, Kaprio J, Stazi MA, Hjelmborg JV, Iachine I, Vaupel JW, Christensen K. Longevity studies in GenomEUtwin. Twin Res. 2003;6(5):448–454. doi: 10.1375/136905203770326457. [DOI] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an association of variation in the FOXO3A gene with human longevity using both case–control and longitudinal data. Aging Cell. 2010;9(6):1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemers FJ, Gunderson KL. Illumina, Inc. Pharmacogenomics. 2005;6(7):777–782. doi: 10.2217/14622416.6.7.777. [DOI] [PubMed] [Google Scholar]
- Team RDC (2008) A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296(5567):530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- Wald A. On a statistical generalization of metric spaces. Proc Natl Acad Sci U S A. 1943;29(6):196–197. doi: 10.1073/pnas.29.6.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner KE, Davis RM, Holbrook JH, Novotny TE, Ockene JK, Rigotti NA (1989) The Surgeon General’s 1989 Report on Reducing the Health Consequences of Smoking: 25 Years of Progress, vol 38 Suppl 2. MMWR Morb Mortal Wkly Rep, 1989/03/24 edn [PubMed]
- Warnes G, Gorjanc G, Leisch F, Man M (2011) genetics: Population Genetics
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37):13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15(3):752–757. doi: 10.1158/1078-0432.CCR-08-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, Huang H, Kuo HP, Lee DF, Li LY, Lien HC, Cheng X, Chang KJ, Hsiao CD, Tsai FJ, Tsai CH, Sahin AA, Muller WJ, Mills GB, Yu D, Hortobagyi GN, Hung MC. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10(2):138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, Hu D. Genetic variation in insulin/IGF-1 signaling pathways and longevity. Aging Res Rev. 2011;10(2):201–204. doi: 10.1016/j.arr.2010.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 407 kb)