Abstract
We report a label-free real-time nanopore sensing method for the detection of anthrax lethal factor, a component of the anthrax toxin, by using a complementary single-stranded DNA as a molecular probe. The method is rapid and sensitive: sub-nanomolar concentrations of the target anthrax lethal factor DNA could be detected in ∼1 min. Further, our method is selective, which can differentiate the target DNA from other single-stranded DNA molecules at the single-base resolution. This sequence-specific detection approach should find useful application in the development of nanopore sensors for the detection of other pathogens.
Keywords: complementary DNA, anthrax lethal factor, label-free, real-time, nanopore sensor, pathogen
1. Introduction
Despite the significant advances in new diagnostic tools and therapeutic drugs, and better ways to prevent diseases, humans remain vulnerable to health threats posed by infectious diseases. According to the World Health Organization, infectious diseases cause 16.2% of global deaths each year, and are the second leading cause of death worldwide only after cardiovascular disease.1 Although proper use of personal protection, effective public policy, and advances in vaccine development are efficient means of controlling the spread of these diseases, early detection of related pathogens is crucial to enable efficient prevention and treatment of infectious diseases, and is vital to the development of an appropriate timely national response to an infectious disease outbreak or a bioterrorist attack, which can greatly reduce mortality.
Conventional and standard methods for pathogen detection include cell culture, enzyme immunoassay, and polymerase chain reaction.2−5 These methods are often laborious and time-consuming, which usually take hours or even days to provide results. To overcome these limitations, developing rapid, sensitive, and selective biosensors or nanobiotechnologies for pathogen detection is currently under intense investigation. Most of the bio- and nanomaterial-based pathogen sensors developed so far rely on antigen–antibody interaction or sequence-specific nucleic acid detection, and employ optical, electrochemical, piezoelectric, or mass-based transduction to achieve good detection limit and high specificity.6−15
In this work, we demonstrate a rapid nanopore sensing method for the sensitive and selective detection of pathogens. Nanopore technology is an emerging label-free and amplification-free technique for measuring single molecules. By monitoring the ionic current modulations produced by the passage of target analytes through a single nanopore bathed in high salt solutions at a fixed applied potential, the concentration of the analyte can be obtained by the frequency of occurrence of the blockage events, while its identity can be determined from the mean residence time of the analyte coupled with the extent of current blockage (amplitude). Under experimental conditions of constant electrolyte pH, temperature, and applied potential, the event blockage amplitude is related to the size (or diameter) of the analyte molecule, while the event residence time depends on the length of the analyte and the strength of the interaction between the analyte and the nanopore. Over the past 15 years, nanopore sensors have successfully been utilized for various applications, including biosensing,16−25 studying covalent and non-covalent bonding interactions,26,27 investigating biomolecular folding and unfolding,28,29 probing enzyme activity and kinetics,30−33and so on.
2. Experimental Section
2.1. Chemicals and Reagents
DNA samples with standard purification (desalting) were purchased from Intergrated DNA Technologies (Coralville, IA). All the other chemicals were ordered from Sigma-Aldrich (St. Louis, MO). All of the DNA samples and chemicals were dissolved in HPLC-grade water (ChromAR, Mallinckrodt Baker). All the stock solutions of DNA polymers were prepared at 5 mM each, and kept at −20 °C before and after use. Three electrolyte solutions were used in this work, which contained 0.15/1.0/3.0 M NaCl buffered with 10 mM Trizma base, with the pH adjusted to 7.5 using hydrochloride acid. Lipid 1, 2-diphytanoylphosphatidylcholine was obtained from Avanti Polar Lipids (Alabaster, AL). Teflon film (25 μm thick) was purchased from Goodfellow (Malvern, PA).
2.2. Preparation and Formation of Protein Pores
The mutant αHL M113F gene was constructed by site-directed mutagenesis (Mutagenex, Piscataway, NJ) with a wild-type αHL gene in a T7 vector (pT7-αHL).34 The mutant αHL monomers were first synthesized by coupled in vitro transcription and translation (IVTT) using the Escherichia coli T7 S30 Extract System for Circular DNA from Promega (Madison, WI). Subsequently, they were assembled into homoheptamers by adding rabbit red cell membranes and incubating for 1–2 h. The heptamers were then purified by SDS-polyacrylamide gel electrophoresis and stored in aliquots at −80°C.
2.3. Single-Channel Recording
A bilayer of 1,2-diphytanoylphosphatidylcholine was formed on an aperture (150 μm) in a Teflon septum that divided a planar bilayer chamber into cis and trans compartments. The formation of bilayer was achieved using Montal–Mueller method.35 Unless otherwise noted, all the experiments were performed under symmetrical buffer conditions with a 2.0 mL solution comprising 1 M NaCl, and 10 mM Tris·HCl (pH 7.5) at 26 ± 1 °C. Both the αHL protein and the DNA polymers were added to the cis compartment, which was connected to “ground”. The final concentration of the αHL proteins used for the single channel insertion was 0.2–2.0 ng·mL–1. The transmembrane potential, which was applied with Ag/AgCl electrodes with 3% agarose bridges containing 3 M KCl, was +180 mV, unless otherwise noted. A positive potential indicates a higher potential in the trans chamber of the apparatus. Currents were recorded with a patch clamp amplifier (Axopatch 200B, Molecular Devices; Sunnyvale, CA, USA). They were low-pass filtered with a built-in four-pole Bessel filter at 5 kHz and sampled at 50 kHz by a computer equipped with a Digidata 1322A/D converter (Molecular Devices).
2.4. Data Analysis
Data were analyzed with the following software: pClamp 10.3 (Molecular Devices), Origin 8.0 (Microcal, Northampton, MA), and SigmaPlot 12.0 (Systat Software Inc., San Jose, CA). Conductance values were obtained from the amplitude histograms after the peaks were fit to Gaussian functions. The values of τon (the mean interevent interval) and τoff (the mean residence time) for DNA polymers were obtained from the dwell time histograms by fitting the distributions to single exponential functions by the Levenberg–Marquardt procedure.36 Thermodynamics of hairpin folding and DNA hybridization was obtained from the DINAMelt web server.37
3. Results and Discussion
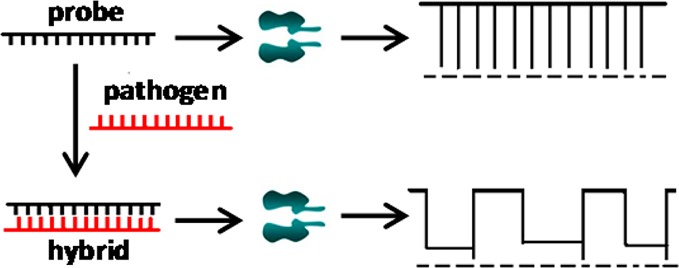
The principle for nanopore detection of pathogens is based on the hybridization between a characteristic single-stranded gene segment of the target pathogen and an unmodified complementary single-stranded DNA (cDNA) probe, which takes advantage of our finding that although their diameters are larger than the channel constriction, short double-stranded DNA (dsDNA) molecules could be rapidly unzipped through an appropriately engineered α-hemolysin (αHL) protein nanopore.38 As shown in Scheme 1, in the absence of the target pathogen gene segment, the translocation of the cDNA probe through the nanopore produces only one major type of current modulation events. In contrast, in the presence of the target DNA sequence, two complementary DNA monomers will be hybridized in the solution to form dsDNA molecules. Because of their larger molecular sizes than those of ssDNA molecules, the interaction between the dsDNA and the nanopore may result in a new type of current modulation events having different signatures from those of the cDNA probe and the single stranded pathogen gene segment, for example, with longer residence times or showing complicated substate current modulation features.39
Scheme 1. Detection of Pathogens in a Nanopore.
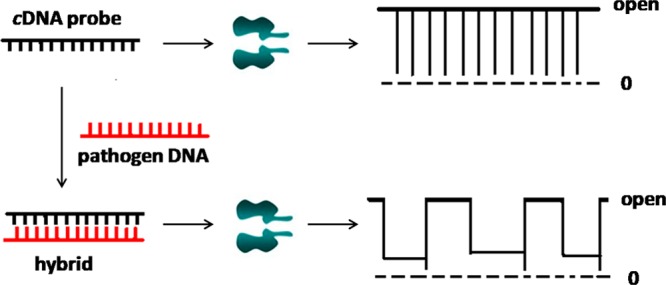
The hybridization of the target pathogen gene segment by an unlabeled complementary DNA probe produces current modulation events in the nanopore having significantly different signatures from those of the cDNA probe.
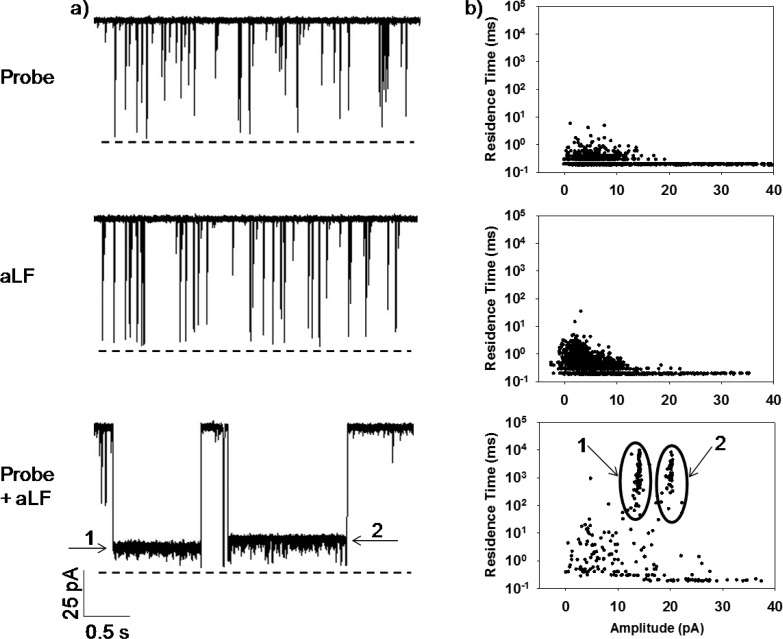
To demonstrate this concept, a characteristic 20-base gene segment (sequence 5′-GGATTATTGTTAAATATTGA-3′, Supporting Information, Table S1) of anthrax lethal factor (aLF), a component of the anthrax toxin, was employed as the target pathogen molecule, while an engineered version of the αHL protein, (M113F)7, was used as the nanopore sensing element. It has been reported that dsDNA could be unzipped much more rapidly in the (M113F)7 protein than in the wild-type αHL pore.38 Our initial experiments were carried out at an applied potential bias of +120 mV and using a 20-base ssDNA (sequence 5′-TCAATATTTAACAATAATCC-3′) as the molecular probe, which can form blunt-ended double-stranded DNA with the target analyte. The experimental results (Figure 1) showed that, without the target aLF DNA sample, the single-stranded cDNA probe produced only one major type of current modulation events in the nanopore, with a mean residence time of 0.20 ± 0.01 ms. However, upon addition of the target aLF DNA molecules to the cDNA probe-containing solution, a new type of events with a mean residence time of 620 ± 63 ms was observed. Since the aLF sample alone also only produced short-lived events with a mean residence time of 0.31 ± 0.01 ms, the three-order increase in the event residence time suggests that the cDNA probe and the target aLF indeed formed DNA duplexes. According to the theoretical prediction (using the DINAMelt web server),34 both the probe and target DNA molecules could form thermodynamically stable hairpin loop structures (the Tm values were 30.9 °C and 34.9 °C, respectively). Since the event residence times of these DNA hairpins were not significantly different from those of the well-studied ssDNA molecules with the same length,40 our experimental results suggest that the closed states of these hairpin loop structures could be rapidly unfolded and driven through the nano-channel under our experimental conditions. Since the target aLF DNA has a slightly larger folding free energy (ΔG = −0.72 vs −0.42 kcal/mol) and hence more stable than the cDNA probe, it is not unreasonable that the aLF sample is more difficult to be unfolded, thus producing events with a longer mean residence time than the cDNA probe (0.31 vs 0.20 ms). This phenomenon has been reported in our previous dsDNA unzipping study.38 In contrast, when both the cDNA probe and the target aLF monomers are present in the solution, they would be able to form a very stable fully-matched DNA duplex with ΔG of −12.2 kcal/mol. Hence, significantly longer residence time events were observed.
Figure 1.
Monitoring the hybridization of the target aLF strand by the cDNA probe. (a) Representative single-channel current recording trace segments, and (b) the corresponding scatter plots of event residence time vs. amplitude. The experiments were performed at +120 mV with the (M113F)7 αHL pore in a 1 M NaCl solution buffered with 10 mM Tris·HCl (pH 7.5).
Similar to the previous observations made by our group and other researchers,41−44 these long-lived dsDNA events also exhibited sub-state current modulations, a clear indication that they might be attributed to the unzipping and translocation of the DNA duplex through the αHL channel. This interpretation was further supported by the voltage dependence study, where the mean residence time of the long-lived events decreased as the applied potential bias increased (Supporting Information, Figure S1). More interestingly, we noticed that these sub-state current modulation events showed two different intermediate levels, level 1 and level 2 as shown in Figure 1. One likely reason is that these events were attributed to the two different orientations in which the dsDNA entered the nanopore. Note that observation of two types of events with different blockage amplitudes and/or residence times has been previously reported in the experiments with the translocation of single-stranded polynucleotides through the αHL pores.39,45 According to DINAMelt, in a mixture of equal amounts of the cDNA probe and the target aLF molecules, the cDNA-aLF hybrid duplexes (with one end containing a GC base pair and the other having a AT base pair) are the dominant species (>99.9%), while other DNA species or structures such as homodimers and hairpins could be neglected.
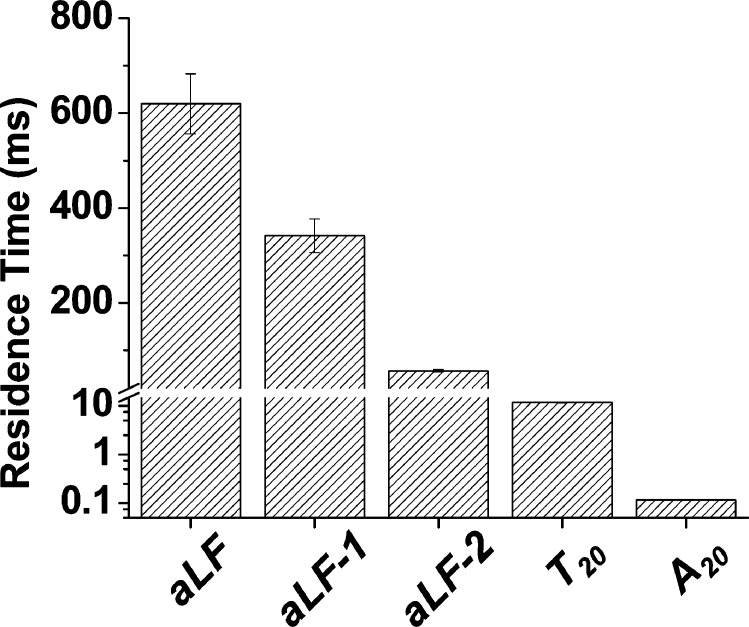
To study the aLF sensor selectivity, two other DNA samples, aLF-1 and aLF-2 (Supporting Information, Table S1), were examined at +120 mV. These two samples had sequences of 5′-GGATTATTGTGAAATATTGA-3′ and 5′-GGATTATGGTGAAATATTGA-3′, respectively, which were different from the target analyte aLF (sequence 5′-GGATTATTGTTAAATATTGA-3′) only by a single base and two bases (note that the mismatch portion are highlighted). Our experimental results showed that the event mean residence times of the aLF-1 and aLF-2 samples were 342 ± 35 ms and 57.4 ± 2.5 ms, respectively. These values were about two-folds and ten-folds smaller than the residence time of the target aLF sample. The results are not unreasonable considering that the aLF-1/aLF-2 and the cDNA probe are able to form double-stranded DNA having single base-pair/two base-pair mismatches, which are less stable than the fully-matched aLF-cDNA duplexes, thus needing less time to be unzipped by the nanopore. This interpretation is supported by the predicted theoretical hybridization free energy between these DNA samples and the cDNA probe (using the DINAMelt web server), which were −12.2, −10.9 kcal/mol, and −9.6 kcal/mol for aLF, aLF-1, and aLF-2, respectively. As added multiple-base mismatch controls, two additional DNA polymers A20 (sequence AAAAAAAAAAAAAAAAAAAA) and T20 (sequence TTTTTTTTTTTTTTTTTTTT) were also examined. Their event residence times were 11.7 ± 0.2 and 0.12 ± 0.01 ms, respectively, which was in agreement with our observation that with an increase in the number of the base-pair mismatches of the dsDNA sample, the DNA duplex became less stable, leading to a decrease in the event mean residence time (Figure 2). Taken together, our experimental results (Supporting Information, Figure S3) suggest that the hybridization free energy between the cDNA probe and an analyte DNA could potentially be inferred from the mean residence time of the DNA duplex events.
Figure 2.
Selectivity of the aLF nanopore sensor. The experiments were performed with the (M113F)7 αHL pore in a buffer solution comprising 1.0 M NaCl and 10 mM Tris·HCl (pH 7.5) at +120 mV in the presence of a 20-base ssDNA (sequence 5′-TCAATATTTAACAATAATCC-3′) as the molecular probe.
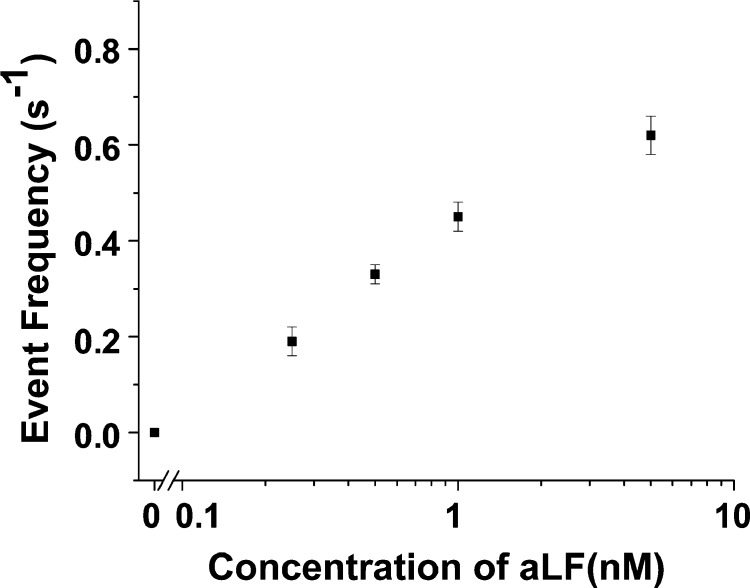
Since the (M113F)7 protein pore/cDNA probe system can selectively detect the target aLF sequence at the single-base resolution, the sensitivity of this nanopore biosensor was further investigated. Unlike most of the other nucleic acid hybridization-based sensors, which rely on labeled nucleic acid probes or require time-consuming incubation of the probe and the target DNA/RNA molecules to achieve sensitive detection,46−48 in our nanopore experiments, detection of aLF molecules were monitored real time in the presence of an unlabeled cDNA probe. Under a symmetric electrolyte condition with 1 M NaCl in both the cis and trans compartments, aLF could be detected with a detection limit (defined as the concentration corresponding to three times the standard deviation of a blank signal) as low as 15 nM (Supporting Information, Figure S2). To further improve the sensitivity of this nanopore sensor, hybridization between the target aLF and the cDNA probe was monitored at +180 mV in a salt gradient of 3 M NaCl (trans)/0.15 M NaCl (cis) (Supporting Information, Figure S4). It has been well established that, use of an asymmetric electrolyte gradient instead of the conventional symmetric electrolyte solution can significantly increase the event frequency for the translocation of DNA/RNA molecules through a nanopore, thus improving the sensor sensitivity.49,50 Our experimental results showed that such a physical condition change didn’t significantly affect the open channel current (160 ± 1.0 vs 160 ± 1.5 pA) and the blockage residual current (7.9 ± 0.3 vs 8.2 ± 0.4 pA) of the DNA duplex events, but would lead to a ∼2-fold increase in the event mean residence time (7.7 ± 0.3 vs 16.7 ± 0.6 ms). Similar to the observation made in the case of symmetric electrolyte solution, the mean residence time of the DNA duplex events under an asymmetric electrolyte gradient was significantly (∼80 folds) larger than those of the cDNA probe alone or aLF alone, which allows the DNA duplex events to be readily differentiated from those of the free ssDNA molecules (Supporting Information, Figure S5). With this approach, the detection limit for aLF can be lowered to 100 pM (Figure 3). At present, polymerase chain reaction (PCR) is the dominant method to detect nucleic acid sequences. Although PCR is highly accurate and sensitive, it is also laborious and time-consuming, and challenging to be used outside a laboratory. Therefore, rapid, sensitive, and accurate detection of specific DNA or RNA sequences that could eliminate the requirement for a PCR step is highly desired in a variety of applications including identification of a disease or pathogen. Although the sensitivity of our nanopore aLF sensor has the potential to be significantly improved by utilizing a PNA probe51,52 instead of the cDNA probe and/or replacing the (M113F)7 protein pore with another engineered αHL nanopore (e.g., K131D7/K147D7),53 which could catalyze the translocation of biomolecules, the detection limit of our present unoptimized sensor is still comparable with those (ranging from 1 pM to 10 nM) of most of the other cDNA-based approaches to detect gene sequences.7,11,47,54
Figure 3.
Dose–response curve for aLF detection. The experiments were performed with the (M113F)7 αHL pore at +180 mV in the presence of a 20-base ssDNA (sequence 5′-TCAATATTTAACAATAATCC-3′) as the molecular probe. An asymmetric buffer condition (with 3 M NaCl and 10 mM Tris·HCl (pH 7.5) in the trans compartment, while 0.15 M NaCl and 10 mM Tris·HCl (pH 7.5) in the cis compartment) was used. The event frequency was calculated by dividing the number of long-lived DNA duplex events by the recording time. The concentration of the cDNA probe used was 10 nM.
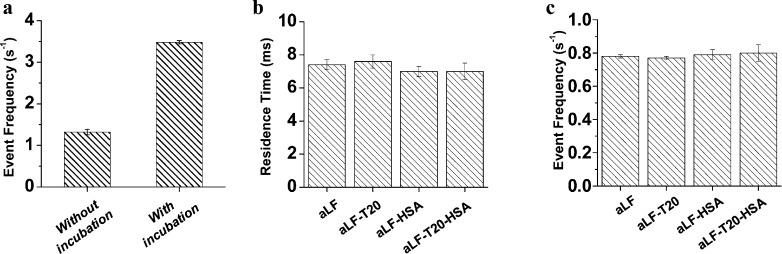
It should be noted that, Figure 3 shows the DNA blockage frequency increases linearly with the logarithm of the analyte concentration. One interpretation for the nonlinear relationship calibration curve is that under our experimental condition (i.e., detection of DNA hybridization real time without pre-incubation of the cDNA probe and the target analyte DNA), not 100 percent of the aLF molecules were able to hybridize with the cDNA probe to form DNA duplexes. We speculate that if the cDNA probe and the target aLF molecules have been incubated for a period of time before single-channel recording, more DNA duplexes could be formed, thus improving the sensitivity and the detection limit for nanopore detection of aLF. For this purpose, the probe and aLF mixture solution was heated at 90°C for 5 min, and then gradually cooled to room temperature. Then, this solution was examined using the (M113F)7 protein pore. Our experimental results showed that the frequency of the long-lived events of the incubated DNA sample was ∼2.5 fold larger than that of the unincubated DNA sample (Figure 4a), thus confirming our hypothesis.
Figure 4.
(a) Effect of incubation on the frequency of the long-lived DNA duplex events and effect of matrix components on the (b) mean residence time and (c) frequency of the aLF-cDNA duplex events. The experiments were performed at +180 mV using the (M113F)7 αHL pore in a solution comprising 1.0 M NaCl and 10 mM Tris·HCl (pH 7.5). In the experiment with the unincubated aLF sample, the electrolyte solution contained an additional 20-base ssDNA (sequence 5′-TCAATATTTAACAATAATCC-3′) as the molecular probe. The concentrations of HSA, aLF, T20, and cDNA probe used in experiments shown in panels b and c were 10 μg/mL, 100 nM, 100 nM, and 1 μM, respectively.
To validate applicability of our nanopore sensor to samples resembling those relevant for clinical analysis, two samples were initially examined with the (M113F)7 protein pore in the presence of the cDNA probe and under the symmetric electrolyte condition. One sample contained human serum album (HSA), while the other contained a mixture of aLF and HSA (note that HSA is the dominant protein in human blood). As shown in Figure S6 (Supporting Information), without aLF, the HSA sample only produced short-lived events with a mean residence time of 0.04 ms. Since in the absence of the cDNA probe, HSA alone didn’t produce any current modulations (Supporting Information, Figure S6), it is apparent that these rapid events were attributed to the cDNA probe. In sharp contrast, the mixture sample produced significantly longer duration events with a mean residence time of 7.5 ms, which was similar to that of the single aLF standard. Furthermore, the frequency of the long-lived events of the aLF-cDNA duplexes didn’t change significantly in the absence/presence of HSA. To demonstrate the feasibility of our developed nanopore sensor in the analysis of aLF in the presence of other DNA molecules or in more complicated mixtures, two additional samples were examined: one contained a mixture of aLF and T20, while the other was consisted of aLF, T20, and HSA. Our experimental results (Figure 4) showed that both the residence time and the frequency of the long-lived events for the mixture samples were similar to those of aLF alone. Taken together, the combined results suggest that our developed nanopore sensor can effectively detect aLF in the presence of other matrix components.
4. Conclusions
In summary, a rapid and sensitive nanopore sensing method for the label-free real-time detection of anthrax lethal factor was developed. Unlike other reported nucleic acid hybridization-based nanopore sensors, which rely on time-consuming incubation of the cDNA probe and the target DNA/RNA molecules to achieve sensitive detection,46−48 in our nanopore sensor design, detection of a target single-stranded aLF gene segment was achieved by real-time monitoring of the hybridization interaction between the target DNA and the cDNA probe. Sub-nanomolar concentrations of aLF DNA could be detected in ∼1 min. Further, our method is selective, and other ssDNA molecules including those differing by only a single base will not interfere with the detection of the target aLF DNA. This sequence-specific detection approach should find useful application in the development of nanopore sensors for the detection of other pathogens.
Acknowledgments
This work was financially supported by the National Institutes of Health (1R15GM110632), and Department of Homeland Security (HSHQDC-09-C-00091).
Supporting Information Available
Additional table listing the nucleic acid sequences of the cDNA probe and DNA samples and figures showing voltage bias, dose–response curve for aLF detection, effect of DNA hybridization free energy, 150 s single-channel current recording trace segment, nanpore detection of aLF, and trace segments showing matrix components. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- World Health Organization. The Global Burden of Disease: 2004 Update; World Health Organization: Genève, 2008. [Google Scholar]
- Foudeh A. M.; Fatanat Didar T.; Veres T.; Tabrizian M. Microfluidic Designs and Techniques Using Lab-on-a-Chip Devices for Pathogen Detection for Point-of-Care Diagnostics. Lab Chip 2012, 12, 3249–3266. [DOI] [PubMed] [Google Scholar]
- França R. F.; da Silva C. C.; De Paula S. O. Recent Advances in Molecular Medicine Techniques for the Diagnosis, Prevention, and Control of Infectious Diseases. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Poshtiban S.; Evoy S. Recent Advances in Bacteriophage Based Biosensors for Food–Borne Pathogen Detection. Sensors 2013, 13, 1763–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. Y.; Kim B. Lab-on-a-Chip Pathogen Sensors for Food Safety. Sensors 2012, 12, 10713–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt O.; Hoheisel J. D. Peptide Nucleic Acids on Microarrays and Other Biosensors. Trends Biotechnol. 2004, 22, 617–622. [DOI] [PubMed] [Google Scholar]
- Du H.; Disney M. D.; Miller B. L.; Krauss T. D. Hybridization-Based Unquenching of DNA Hairpins on Au Surfaces: Prototypical “Molecular Beacon” Biosensors. J. Am. Chem. Soc. 2003, 125, 4012–4013. [DOI] [PubMed] [Google Scholar]
- Gilmartin N.; O’Kennedy R. Nanobiotechnologies for the Detection and Reduction of Pathogens. Enzyme Microb. Technol. 2012, 50, 87–95. [DOI] [PubMed] [Google Scholar]
- Rai V.; Hapuarachchi H. C.; Ng L. C.; Sohn S. H.; Leo Y. S.; Toh C. S. Ultrasensitive cDNA Detection of Dengue Virus RNA Using Electrochemical Nanoporous Membrane-Based Biosensor. PLoS One 2012, 7, e42346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velusamy V.; Arshak K.; Korostynska O.; Oliwa K.; Adley C. An Overview of Foodborne Pathogen Detection: In the Perspective of Biosensors. Biotechnol. Adv. 2010, 28, 232–254. [DOI] [PubMed] [Google Scholar]
- Zhang B.; Dallo S.; Peterson R.; Hussain S.; Tao W.; Ye J. Detection of Anthrax Lef with DNA-Based Photonic Crystal Sensors. J. Biomed. Opt. 2011, 16, 127006. [DOI] [PubMed] [Google Scholar]
- Ghosh N.; Gupta N.; Gupta G.; Boopathi M.; Pal V.; Goel A. Detection of Protective Antigen, An Anthrax Specific Toxin in Human Serum by Using Surface Plasmon Resonance. Diagn. Microbiol. Infect. Dis. 2013, 77, 14–19. [DOI] [PubMed] [Google Scholar]
- Mechaly A.; Cohen N.; Weiss S.; Zahavy E. A Novel Homogeneous Immunoassay for Anthrax Detection Based on the AlphaLISA Method: Detection of B. Anthracis Spores and Protective Antigen (PA) in Complex Samples. Anal. Bioanal. Chem. 2013, 405, 3965–3972. [DOI] [PubMed] [Google Scholar]
- Kim D. J.; Park H. C.; Sohn I. Y.; Jung J. H.; Yoon O. J.; Park J. S.; Yoon M. Y.; Lee N. E. Electrical Graphene Aptasensor for Ultra-Sensitive Detection of Anthrax Toxin with Amplified Signal Transduction. Small 2013, 9, 3352–3360. [DOI] [PubMed] [Google Scholar]
- Farrow B.; Hong S.; Romero E.; Lai B.; Coppock M.; Deyle K.; Finch A.; Stratis-Cullum D.; Agnew H.; Yang S.; Heath J. A Chemically Synthesized Capture Agent Enables the Selective, Sensitive, and Robust Electrochemical Detection of Anthrax Protective Antigen. ACS Nano 2013, 7, 9452–9460. [DOI] [PubMed] [Google Scholar]
- Gu L.; Braha O.; Conlan S.; Cheley S.; Bayley H. Stochastic Sensing of Organic Analytes by a Pore-Forming Protein Containing a Molecular Adapter. Nature 1999, 398, 686–690. [DOI] [PubMed] [Google Scholar]
- Kang X.; Cheley S.; Guan X.; Bayley H. Stochastic Detection of Enantiomers. J. Am. Chem. Soc. 2006, 128, 10684–10685. [DOI] [PubMed] [Google Scholar]
- Liu A.; Zhao Q.; Guan X. Stochastic Nanopore Sensors for the Detection of Terrorist Agents: Current Status and Challenges. Anal. Chim. Acta 2010, 675, 106–115. [DOI] [PubMed] [Google Scholar]
- Siwy Z.; Trofin L.; Kohli P.; Baker L. A.; Trautmann C.; Martin C. R. Protein Biosensors Based on Biofunctionalized Conical Gold Nanotubes. J. Am. Chem. Soc. 2005, 127, 5000–5001. [DOI] [PubMed] [Google Scholar]
- Stefureac R. I.; Madampage C. A.; Andrievskaia O.; Lee J. S. Nanopore Analysis of the Interaction of Metal Ions with Prion Proteins and Peptides. Biochem. Cell Biol. 2010, 88, 347–358. [DOI] [PubMed] [Google Scholar]
- Wang G.; Wang L.; Han Y.; Zhou S.; Guan X. Nanopore Stochastic Detection: Diversity, Sensitivity, and Beyond. Acc. Chem. Res. 2013, 46, 2867–2877. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Jayawardhana D. A.; Wang D.; Guan X. Study of Peptide Transport through Engineered Protein Channels. J. Phys. Chem. B 2009, 113, 3572–3578. [DOI] [PubMed] [Google Scholar]
- Baran C.; Smith G. S. T.; Bamm V. V.; Harauz G.; Lee J. S. Divalent Cations Induce a Compaction of Intrinsically Disordered Myelin Basic Protein. Biochem. Biophys. Res. Commun. 2010, 391, 224–229. [DOI] [PubMed] [Google Scholar]
- Mereuta L.; Schiopu I.; Asandei A.; Park Y.; Hahm K. S.; Luchian T. Protein Nanopore-Based, Single-Molecule Exploration of Copper Binding to an Antimicrobial-Derived, Histidine-Containing Chimera Peptide. Langmuir 2012, 28, 17079–17091. [DOI] [PubMed] [Google Scholar]
- Asandei A.; Schiopu I.; Iftemi S.; Mereuta L.; Luchian T. Investigation of Cu2+ Binding to Human and Rat Amyloid Fragments Aβ (1–16) with a Protein Nanopore. Langmuir 2013, 29, 15634–15642. [DOI] [PubMed] [Google Scholar]
- Luchian T.; Shin S. H.; Bayley H. Single-Molecule Covalent Chemistry with Spatially Separated Reactants. Angew. Chem., Int. Ed. 2003, 42, 3766–3771. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Jayawardhana D. A.; Guan X. Stochastic Study of the Effect of Ionic Strength on Noncovalent Interactions in Protein Pores. Biophys. J. 2008, 94, 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J. W.; Tan Q.; Gu L. Single-Molecule Detection of Folding and Unfolding of the G-Quadruplex Aptamer in a Nanopore Nanocavity. Nucleic Acids Res. 2009, 37, 972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaga D. S.; Li J. Single-Molecule Protein Unfolding in Solid State Nanopores. J. Am. Chem. Soc. 2009, 131, 9287–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G.; Talukdar P.; Matile S. Fluorometric Detection of Enzyme Activity with Synthetic Supramolecular Pores. Science 2002, 298, 1600–1602. [DOI] [PubMed] [Google Scholar]
- Macrae M. X.; Blake S.; Jiang X.; Capone R.; Estes D. J.; Mayer M.; Yang J. A Semi-Synthetic Ion Channel Platform for Detection of Phosphatase and Protease Activity. ACS Nano 2009, 11, 3567–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd S.; Yusko E. C.; MacBriar A. D.; Yang J.; Mayer M. Gramicidin Pores Report the Activity of Membrane-Active Enzymes. J. Am. Chem. Soc. 2009, 131, 16119–16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q.; Wang D.; Jayawardhana D. A.; de Zoysa R. S.; Guan X. Real-Time Monitoring of Peptide Cleavage Using a Nanopore Probe. J. Am. Chem. Soc. 2009, 131, 6324–6325. [DOI] [PubMed] [Google Scholar]
- Song L.; Hobaugh M. R.; Shustak C.; Cheley S.; Bayley H.; Gouaux J. E. Structure of Staphylococcal Alpha-Hemolysin, A Heptameric Transmembrane Pore. Science 1996, 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- Montal M.; Mueller P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. U. S. A. 1972, 69, 3561–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Zhao Q.; Kang X.; Guan X. Probing Mercury(II)–DNA Interactions by Nanopore Stochastic Sensing. J. Phys. Chem. B 2013, 117, 4763–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N. R.; Zuker M. DINAMelt Web Server for Nucleic Acid Melting Prediction. Nucleic Acids Res. 2005, 33, W577–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.; Zhao Q.; Krishantha D. M.; Guan X. Unzipping of Double-Stranded DNA in Engineered α-Hemolysin Pores. J. Phys. Chem. Lett. 2011, 2, 1372–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzard J.; Martinho M.; Mathe J.; Bockelmann U.; Viasnoff V. DNA Translocation and Unzipping through a Nanopore: Some Geometrical Effects. Biophys. J. 2010, 98, 2170–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathe J.; Visram H.; Viasnoff V.; Rabin Y.; Meller A. Nanopore Unzipping of Individual DNA Hairpin Molecules. Biophys. J. 2004, 87, 3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman V. S.; Lee C. C.; Deamer D. W.; Vercoutere W. A. Sequence-Dependent Gating of an Ion Channel by DNA Hairpin Molecules. Nucleic Acids Res. 2006, 34, 6425–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S.; Bayley H. Probing Distance and Electrical Potential within a Protein Pore with Tethered DNA. Biophys. J. 2002, 83, 3202–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K.; He Z.; Wang Y.; Chen S.; Gu L. Designing a Polycationic Probe for Simultaneous Enrichment and Detection of MicroRNAs in a Nanopore. ACS Nano 2013, 7, 3962–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoutere W.; Winters-Hilt S.; Olsen H.; Deamer D.; Haussler D.; Akeson M. Rapid Discrimination among Individual DNA Hairpin Molecules at Single-Nucleotide Resolution Using an Ion Channel. Nat. Biotechnol. 2001, 19, 248–252. [DOI] [PubMed] [Google Scholar]
- Mathe J.; Aksimentiev A.; Nelson D. R.; Schulten K.; Meller A. Orientation Discrimination of Single-Stranded DNA inside the Alpha-Hemolysin Membrane Channel. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 12377–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.; Neumann R.; Ensinger W. Sequence-Specific Recognition of DNA Oligomer Using Peptide Nucleic Acid (PNA)-Modified Synthetic Ion Channels: PNA/DNA Hybridization in Nanoconfined Environment. ACS Nano 2010, 4, 7267–7274. [DOI] [PubMed] [Google Scholar]
- Esfandiari L.; Monbouquette H. G.; Schmidt J. J. Sequence-Specific Nucleic Acid Detection from Binary Pore Conductance Measurement. J. Am. Chem. Soc. 2012, 134, 15880–15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zheng D.; Tan Q.; Wang M. X.; Gu L. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6, 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M.; Morrison W.; Grosberg A. Y.; Meller A. Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient. Nat. Nanotechnol. 2010, 5, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S.; Zeng T.; Liu L.; Zhao K.; Zhao Y.; Liu X.; Wu H. C. Highly Sensitive and Selective DNA-Based Detection of Mercury(II) with α-Hemolysin Nanopore. J. Am. Chem. Soc. 2011, 133, 18312–18317. [DOI] [PubMed] [Google Scholar]
- Micklitsch C. M.; Oquare B. Y.; Zhao C.; Appella D. H. Cyclopentane-Peptide Nucleic Acids For Qualitative, Quantitative, and Repetitive Detection of Nucleic Acids. Anal. Chem. 2013, 85, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.; Appella D. H. Colorimetric Detection of Anthrax DNA with a Peptide Nucleic Acid Sandwich-Hybridization Assay. J. Am. Chem. Soc. 2007, 129, 8424–8425. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J.; Mohammad M. M.; Cheley S.; Bayley H.; Movileanu L. Catalyzing the Translocation of Polypeptides Through Attractive Interactions. J. Am. Chem. Soc. 2007, 129, 14034–14041. [DOI] [PubMed] [Google Scholar]
- Zhu L.; Luo L.; Wang Z. DNA Electrochemical Biosensor Based on Thionine-Graphene Nanocomposite. Biosens Bioelectron. 2012, 35, 507–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.