Abstract
Historically, the ribosome has been viewed as a complex ribozyme with constitutive rather than intrinsic regulatory capacity in mRNA translation. However, emerging studies reveal that ribosome activity may be highly regulated. Heterogeneity in ribosome composition resulting from differential expression and post-translational modifications of ribosomal proteins, ribosomal RNA (rRNA) diversity and the activity of ribosome-associated factors may generate ‘specialized ribosomes’ that have a substantial impact on how the genomic template is translated into functional proteins. Moreover, constitutive components of the ribosome may also exert more specialized activities by virtue of their interactions with specific mRNA regulatory elements such as internal ribosome entry sites (IRESs) or upstream open reading frames (uORFs). Here we discuss the hypothesis that intrinsic regulation by the ribosome acts to selectively translate subsets of mRNAs harbouring unique cis-regulatory elements, thereby introducing an additional level of regulation in gene expression and the life of an organism.
A major challenge in biology is to understand how protein expression is regulated with exquisite temporal and spatial precision to control unique cell behaviours and to give rise to the remarkable diversity of cell types that is characteristic of metazoan development. Decades of research have demonstrated numerous layers of regulation that orchestrate this process. Indeed, almost every step in the control of gene expression has evolved many levels of regulatory specificity: from exquisitely modified histone tails, to the countless small RNAs that fine-tune the abundance of transcripts, as well as the remarkable repertoire of post-translational modifications that steer protein behaviour. On the contrary, such regulatory control has not been fully realized at a crucial step in gene expression, that of protein production. In particular, although the ribosome is well known to be an immensely complex ‘molecular machine’, it is believed to function as somewhat of an automaton in translatin g the genetic code.
Recent findings are now, however, shifting the view of ribosome function, revealing unique and unexpected roles for the translation machinery itself in directing fundamental aspects of cell and organismal biology. Here we discuss the hypothesis that ‘specialized ribosomes’, which have a unique composition or specialized activity, confer regulatory control in gene expression. Examples of heterogeneity in ribosomes can include diversity in the composition and post-translational modifications of subsets of ribosomal proteins, variations in ribosomal RNA (rRNA) sequences or binding to distinct ribosome-associated factors, all of which may contribute to the occurrence of specialized ribosomes in different cell types. In addition, even core ribosome components that show little variation may exert more specialized activity by virtue of their interactions with specific cis-acting regulatory elements that are present within subsets of mRNAs. In a multicellular organism, greater regulation in ribosome activity may provide an important new layer for the spatiotemporal control of gene expression.
In this Review, we examine the evidence for hetero geneity in ribosomes, how regulatory elements in mRNAs may interface with ribosomes to confer transcrip t-specific regulation of gene expression, and how such mechanisms may bestow greater specificity in decoding the genomic template into unique cellular and molecular behaviours as well as morphology.
The ribosome as a ‘molecular machine’
The ribosome has been described as a “molecular machine par excellence”1,2. This metaphor can be traced back to an influential article written by Bruce Alberts, in which the analogy of the ‘molecular machine’ was first used to describe the highly coordinated behaviour of large complexes such as the DNA replication machinery or the ribosome3. The machine analogy influenced the manner in which the molecular comple x was conceptualized; that although it may be composed of a number of distinct elements, a high degree of coordination and precision of these parts serves a singular, unilateral function. The basic mechanisms of how the genetic code is translated into proteins fits well with this concept. For example, the ribosome has a universally conserved role in catalysing protein synthesis. Moreover, protein synthesis is remarkably fast. Elongation of the polypeptide chain by one amino acid occurs in approximately 60 ms and in a highly accurate fashion4. This requires a sophisticated level of precision and automation. However, the powerful metaphor of a machine has also perhaps stereotyped the view of the ribosome as a passive participant in translation. A crucial question is whether certain parts of the ribosome are changeable and/or activated by additional layers of regulation that depend on the context. To understand this principle, the molecular machine has to be examined at its very core, as ribosome biogenesis is a highly coordinated process (BOX 1). Indeed, ribosomes make up much of the cell’s mass, and ribosome biogenesis uses a significant proportion of the cell’s energy5. The eukaryotic ribosome contains 4 RNA species and 79 ribosomal proteins. Despite the constitutive nature of the ribosome, several components could also give rise to divergent regulation of the ‘molecular machine’. In understanding the construction of the ribosome (BOX 1), possible points of variation can also be envisioned. We will discuss the evidence, or the lack thereof, for individual components exerting unique and more regulatory control.
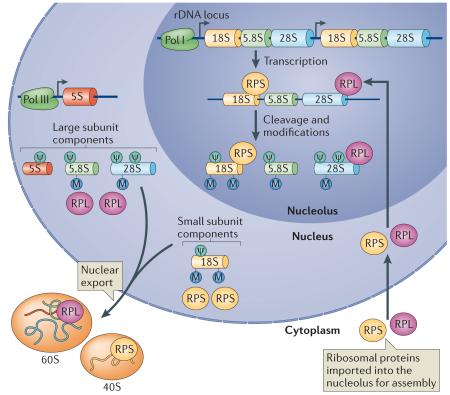
Box 1. Ribosome biogenesis.
Ribosome biogenesis is a highly orchestrated process involving all three RNA polymerases and over 150 non-ribosomal factors that take part in the maturation process129. Three of the four ribosomal RNA (rRNA) species, namely 18S, 5.8S and 28S, are transcribed as a single 47S transcript (pre-rRNA) by RNA polymerase I (Pol I) in the nucleolus (see the figure). The fourth rRNA, 5S, is transcribed by Pol III in the nucleus. Strikingly, pre-rRNAs are extensively modified by pseudouridylation (ψ) and methylation (M). This process is orchestrated by small nucleolar RNAs (snoRNAs) that guide site-specific modifications of ~200 nucleotides in the pre-rRNA. Although the role of rRNA modifications is not fully understood, they are important for ribosome function130. For example, recent findings demonstrate that loss of rRNA pseudouridylation decreases translational fidelity, transfer RNA (tRNA) binding and binding to structured RNA elements within mRNAs in an evolutionarily conserved manner from yeast to human cells87.
All ribosomal proteins are transcribed by Pol II and translated in the cytoplasm (not shown). They are then imported into the nucleus and are assembled on the pre-rRNA while it is being transcribed. Although less well studied, some ribosomal proteins have also been shown to be involved in rRNA processing, subunit assembly and export131-135. As the ribosomal proteins assemble, the pre-rRNA folds and undergoes a series of cleavages to generate the 18S, 5.8S and 28S rRNAs. The 18S rRNA along with 32 ribosomal proteins assemble into the small ribosomal 40S subunit. The 28S, 5.8S and 5S rRNA along with additional 47 ribosomal proteins assemble into the large 60S subunit. Both ribosomal subunits are exported into the cytoplasm in a coordinated process136,137. RPL, large subunit ribosomal protein; RPS, small subunit ribosomal protein.
Heterogeneity in ribosome function and activity
Francis Crick first proposed, in relation to the ‘RNA world’ hypothesis, that an ancestral proto-ribosome may have been comprised only of functional rRNAs, which shared homology with present-day rRNAs6. Biochemical and genetic experiments have revealed a direct involvement of conserved rRNA nucleotides, highlighting their central role in ribosome function7,8. Remarkably, early attempts in delineating the minimal components necessary for ribosome activity demonstrated that peptidyl transferase, the ribosomal activity responsible for the catalysis of peptide-bond formation, might only require rRNAs in the absence of the majority of ribo somal proteins9. This raises the question of the function of the ribosomal proteins that associate with the ribosome. Ribosomal proteins are likely to have co-evolved important roles in rRNA folding and function10,11 (BOX 1). However, we speculate that the emergence of ribosomal proteins might, in addition, imbue greater specificity to the RNA-based translation machinery to control protein synthesis. It is also important to consider whether ribosomal proteins may be actin g at multi ple times and/or locations to exert unique activities. For example, ribosomal proteins may have important roles in rRNA processing within the nucleolu s and also exert more specialized functions in translational control as part of the mature ribosome within the cytoplasm or even as components of extra ribosomal complexes (BOX 2). In this section, we examin e the possibility of further regulation in ribosome composition and activity with respect to the proteins and rRNAs that constitute the ribosome.
Box 2. Extraribosomal functions of ribosomal proteins in translational control.
Some ribosomal proteins have been shown to have additional functions besides their roles in ribosomes (for a review, see REF. 138). Most ribosomal proteins are RNA-binding proteins and some of these proteins can potentially bind to additional RNAs or protein complexes outside of the ribosome. For example, many ribosomal proteins regulate their own synthesis. In Saccharomyces cerevisiae, ribosomal protein Rpl30 inhibits splicing of its own transcript139. In human cells, RPS13 also inhibits splicing of its own transcript by binding to its intron140. Ribosomal proteins can also function in other protein complexes141.
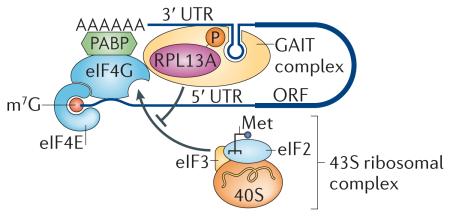
The most interesting extraribosomal protein that functions in translational control is RPL13A. RPL13A regulates the translation of specific mRNAs as part of a non-ribosomal complex (see the figure). Following interferon-γ (IFNγ) treatment, monocytes show a marked increase in transcription of ceruloplasmin mRNA, which is an acute phase inflammatory protein. Ceruloplasmin protein synthesis is terminated after 16 hours even though ceruloplasmin mRNA is still present142. The IFNγ-activated inhibitor of translation (GAIT) complex was found to bind to an RNA structure in the 3′ untranslated region (UTR) of ceruloplasmin mRNA to inhibit translation. RPL13A is one of the four proteins that constitute the GAIT complex. RPL13A is phosphorylated and released from the ribosome 24 hours after IFNγ stimulation143. Phosphorylated RPL13A binds strongly to cap-bound eukaryotic translation initiation factor eIF4G and blocks the recruitment of the 43S ribosomal complex144. It also has the same repressive effect on a number of other inflammatory response genes that have the same 3′ UTR RNA structure, such as many chemokines and chemokine receptors145. Following this paradigm, certain ribosomal proteins that are not essential for the catalytic activity of the ribosome may be co-opted for additional, specialized functions in translational control acting in complexes that function apart from the ribosome itself. m7G, 7-methylguanosine; Met, initiator tRNA; ORF, open reading frame; PABP, poly(A)-binding protein.
Ribosomal protein paralogues
In mammals, although there are ~2,000 sequences encoding ribosomal proteins, most of them are predicted to be pseudogenes12, and most functional ribosomal proteins are encoded by a single gene. By contrast, in yeast, plants and flies many ribosomal proteins are encoded by more than one gene. Saccharomyces cerevisiae, for example, arose from an ancient genome duplication, and 59 of the 78 ribosomal proteins retained two genomic copies13. Remarkably, despite having high sequence homology, the two ribo somal protein gene copies do not always seem to be functionally redundant. Deletions of individual paralogue s can give rise to completely unique phenotypes showing differences in cell size, bud size selection14, sporu lation15, susceptibility to virus16,17 and drug sensitivity18,19, and these phenotypes are as dissimilar as deletions in completely unrelated ribosomal proteins. For example, deletions of at least six ribosomal proteins, but not their paralogues, affect the localization of ASH1 mRNA to the bud tip of the emerging daughter cell, a process that requires tight translational regulation18. It is, however, important to note that distinct phenotypes observed among some ribosomal protein paralogues18 may be confounded by additional suppressor mutations that arise during the propagation of ribosomal protein-deletion strains20,21, and hence a more careful examination is needed to confirm these phenotypes. Despite these caveats, the many examples of unique functional roles for individual ribosomal protein paralogues suggest a differential requirement for individual paralogues in specific molecular and cellular events.
Recently, it has been shown that splicing of yeast ribosomal protein mRNAs can regulate the expression of both ribosomal protein paralogue genes at the transcript level, often in a non-reciprocal manner. For example, the removal of the intron in the gene encoding ribosomal protein S29A (Rps29a) reduces not only its own expression but also the expression of RPS29B19 (FIG. 1a). This is surprising, as a change in the expression of any single copy of the duplicated ribosomal protein gene pair would be expected to increase the expression of the other if it was simply required to maintain a stoichiometric level of expression. On the contrary, in wild-type cells ~70% of all duplicated ribosomal protein genes are asymmetrically expressed19. This suggests that in yeast cells the ratio of paralogues is regulated and that paralogues are not merely equivalent, functional substitutes. It is tempting to speculate that variation in ribosome composition, for example by incorporating different paralogues, may be a mechanism to provide unique regulatory activity to the ribosome. Although loss-of-function phenotypes in distinct ribosomal protein gene copies would support this possibility, little is known to date about the functional roles of distinct ribosomal protein paralogues in translational control.
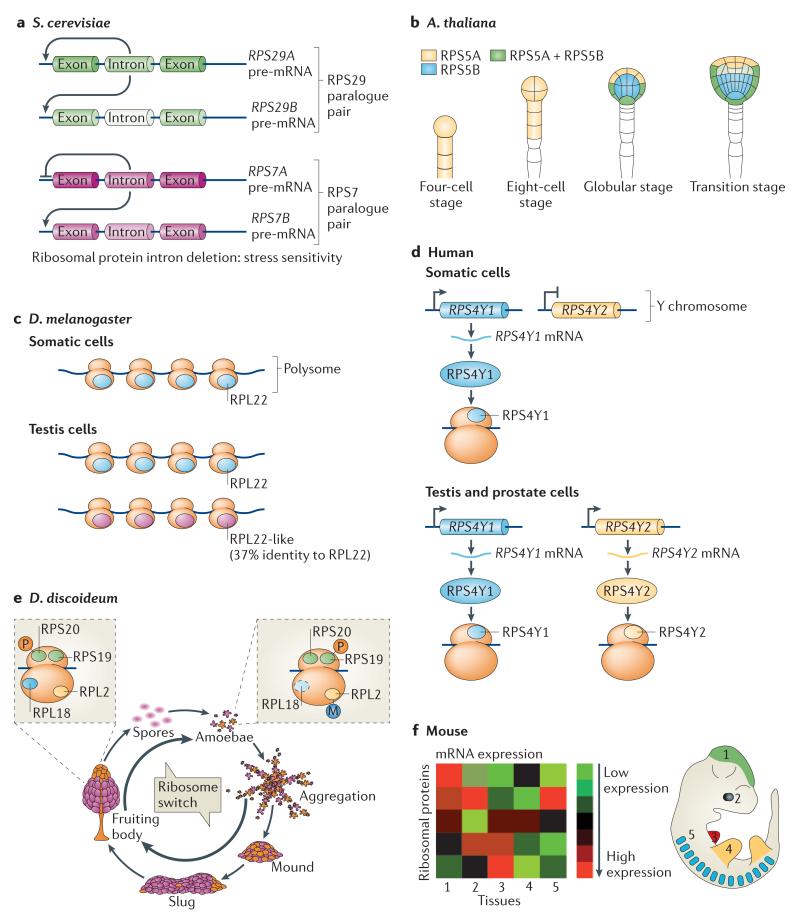
Figure 1. Heterogeneity in the ribosome at the level of ribosomal protein composition and post-translational modifications across different species.
a | Introns of genes encoding ribosomal proteins in Saccharomyces cerevisiae regulate the expression of both the intron-containing ribosomal protein genes and their paralogues, and this has important outcomes in cells: for example, increased fitness under stress. 70% of ribosomal protein paralogue pairs show differential expression. b | In plants, ribosomal protein paralogues have different functions and different expression patterns. For example, in Arabidopsis thaliana, RPS5A is expressed in rapidly dividing cells early in embryonic development, whereas RPS5B is expressed in cells undergoing differentiation. c | In Drosophila melanogaster, ribosomal protein paralogues show different expression patterns in the adult testes. For example, RPL22 is expressed ubiquitously, but RPL22-like protein levels are specifically increased in the testes. Both proteins are incorporated into translationally active ribosomes (called the polysomes). d | In humans, only some ribosomal protein paralogues have been identified; however, notable examples exist. RPS4Y1 is expressed ubiquitously, whereas RPS4Y2 is restricted to the testis and prostate. e | In the life cycle of the social amoebae Dictyostelium discoideum, ribosome switches are characterized by changes in ribosome composition at the level of ribosomal protein expression and post-translational modifications. For example, phosphorylation (P) on RPS19 and methylation (M) on RPL2 are lost as D. discoideum aggregates from a single cell amoebae to a multicellular fruiting body, while RPL20 gains phosphorylation marks. RPL18 is exclusively found in ribosomes of developing cells and not in those of the amoebae stage. f | In mice, the mRNA expression pattern of ribosomal proteins varies dramatically among tissues. This may potentially translate into unique ribosomal protein compositions of ribosomes in distinct cell types. Part a is modified, with permission, from REF. 19 © (2011) Elsevier. Part b is modified, with permission, from REF. 24 © (2001) The Company of Biologists. Part f is modified, with permission, from REF. 37 © (2011) Elsevier.
Plants have even more gene copies encoding a single ribosomal protein than yeast. Each ribosomal protein in Arabidopsis thaliana is encoded by two to seven genes22. Rapeseed, Brassica napus, has 966 genes encoding 79 ribosomal proteins23. Some of the ribosomal protein gene copies are identical in sequence, but many of the paralogues display sequence variations and are differentially expressed during development24,25. For example, in A. thaliana, RPS5A is strongly expressed in dividing cells, whereas RPS5B is expressed in cells undergoing differentiation24 (FIG. 1b). The gene expression of ribosomal protein L11B (RPL11B; previously known as RPL16B) is also high in all actively dividing cells, whereas the expression pattern of RPL11A is much more tissue-specific26. Moreover, the two A. thaliana RPL23A paralogues, which possess high sequence homology, are not of equivalent importance for normal plant development27,28. Deletion of RPL23AA but not of RPL23AB impeded growth and led to morphological abnormalities, revealing specialized functions for specific ribosomal protein paralogues. A genome-wide study of ribosomal proteins in B. napus showed that different paralogues of the same ribosomal protein are not regulated in the same pattern among tissues. Interestingly, more than two-thirds of ribosomal proteins are encoded by paralogues that have opposite expression patterns in embryos and seedlings, suggesting that a different set of ribosome paralogues might be required for early development23.
Heterogeneity in ribosomal protein paralogue expression was also found in Drosophila melanogaster. For example, some paralogues show differential expression levels in the adult testes29. RpL22 is transcribed ubiquitously, but RpL22-like is enriched in the testes (FIG. 1c). Both genes are essential in the fly, which suggests that they are not functionally redundant. Moreover, both proteins are incorporated into ribosomes as they are detected in polysomes (which are translationally active ribosomes) and 80S fractions but not in pre-ribosomal fractions30. In addition, RpS5b, RpS19a, RpL10Aa and RpL37b show enhanced expression in the testes compared to their paralogues29. Such heterogeneity in ribosomal protein expression in the gonads suggests that the development of germ cells may require tissue-specific variations in the translational machinery.
In mammals, most ribosomal proteins are encoded by only a single gene copy. However, some notable exceptions exist. For example, RPS4 is encoded by three genes (namely RPS4X, RPS4Y1 and RPSY2) that are located on the X chromosome and the Y chromosome31,32. In human males, RPS4X and RPS4Y1 are expressed ubiquitously, but RPSY2 expression is restricted to the testis and prostate, suggesting a male-specific role for the ribosomal protein encoded by RPSY2 and the possibility of testis-specific ribosomes31 (FIG. 1d). Although the sequences of the three RPS4 proteins are very similar, distinct amino acids in the carboxyl terminus of RPS4Y2 possibly facilitate unique interactions with distinct, potentially testis-specific, ribosomal proteins or extraribosomal factors31. In addition, mice carry paralogues for RPL10, RPL22 and RPL39, which are also differentially expressed33. Ribosomes from rodent liver, mammary gland and testi s were isolated and analysed by two-dimensional (2D) gel electrophoresis and mass spectrometry. Surprisingly, RPL10-like and RPL39-like were only found in ribosomes from the testis but not in the liver or the mammary gland. Interestingly, RPL39-like localized to the nucleoli and was found in 80S and polysome fractions, which is consistent with RPL39-like being a ribosomal protein33. By contrast, RPL22-like was absent from the testes but is specifically expressed in the liver and mammary gland. Therefore, this suggests that ribosomes expressed in different tissues contain unique ribosomal protein paralogues, and similarly to the findings in D. melanogaster, variations in the expression patterns of ribosomal protein paralogues seem to be evolutionarily conserved and partic ularly eviden t in germ cells.
Ribosomal protein expression
In addition to the fact that some ribosomal proteins are encoded by paralogue genes, the expression of core ribosomal proteins may not be ubiquitous, at least under certain circumstances. It has previously been suggested that the equimolar production of ribosomal proteins is important for proper ribosome biogenesis and that ribosomal proteins are transcribed at the same rate and have similar lifespans34,35. However, most of these studies focused on only a handful of ribosomal proteins in a single cell type. Recent studies that examined ribosomal proteins more globally at an organismal level have shown that different ribosomal proteins are expressed at distinct levels in unique cell types36,37. In the social amoebae Dictyostelium discoideum, ribosomes are composed of different ribosomal proteins at two different states of its life cycle, the vegetative stage and the spore stage (FIG. 1e). Many ribosomal proteins are expressed at different levels in one state compared to the other state, suggesting that these proteins may be developmentally regulated during cell differentiation38,39.
In mice, the levels of Rpl38 transcripts exhibit a tissue-specific expression pattern37. For example, during embryonic development, Rpl38 expression is substantially increased in developing somites and in a specific subset of motor neurons within the spinal cord. Strikingly, this expression pattern mirrors, to a large extent, the tissue s that are affected by the loss of function of RPL38 in mutant embryos37. Moreover, Rpl38 is expressed only in some adult tissues40. RPL10 expression in mouse embryos is also highly tissue-specific as this ribosomal protein is enriched in the developing epidermis and limb buds41 and shows a very specific expression pattern in fetal bone42. Notably, RPL10 is one of the few mammalian ribosomal proteins that has a paralogue; however, this cannot solely account for its restricted expression pattern as the paralogue encoding RPL10-like is also differentially expressed33. RPL10-like has only been found to be expressed in the testes but not other tissues such as the liver.
More globally, a large-scale quantitative expression-profiling screen examined the expression of 72 ribo somal proteins in 14 tissues and cell types in the developing vertebrate embryo (FIG. 1f). This study revealed dramatic differences in the expression levels of ribosomal proteins among different tissues and cell types, and revealed coregulated expression patterns among specific groups of ribosomal proteins37. Additional studies also point to the possibility of greater heterogeneity in ribosomal protein expression in adult tissues36. Although most of these studies only examine the mRNA levels of ribo somal proteins, the huge range of variation in expression among tissues suggests that ribosomes may be heterogeneous in distinct cell and tissue types. These findings also raise the possibility that some ribosomes may contain more than one copy of certain, highly expressed ribosomal proteins. This is analogous to the dimerization of the acidic ribosomal proteins in the ribosomes of Escherichia coli, which contain two protein copies of RpL7 and RpL12 per ribosome43-45. Importantly, as ribosomal proteins may have extra-ribosomal functions, can be degraded at the protein level46 or are translationally controlled146, further proteomics studies will be key in confirming the contribution of heterogenous ribosomal protein expression in specialized ribosomal activity.
Post-translational modifications
In addition to the differences in ribosomal protein expression levels and patterns, post-translational modifications on ribosomal proteins can add another layer of specificity. In many organisms, ribosomal proteins are post-translationally modified. In yeast, at least 16 ribosomal proteins are either methylated or acetylated47,48. In humans, at least 11 large subunit ribosomal proteins and most of the small subunit proteins are also post-translationally modified49,50. In A. thaliana, 23 of the 80 ribosomal protein families contain at least one covalent modification48. Modifications of ribosomal proteins seem to be highly regulated. For example, ribosomes from different phases of the D. discoideum lifecycle show specific changes in ribosomal protein phosphorylation and methylation patterns that may facilitate unique translational needs of the cell51 (FIG. 1e).
Historically, the post-translational modifications on RPS6 are the best characterized. For example, in mammals, RPS6 is phosphorylated on five residues in response to mitogen and growth factor signalling52. It has long been believed that RPS6 phosphorylation has an important function in the translational control of a subclass of mRNAs that harbour a 5′ tract oligopyrimidine (5′ TOP) sequence, and that this level of regulation may imbue the ribosome with greater specificity. However, unexpectedly, a knock-in mouse, in which all five phosphorylated residues of RPS6 were mutated to Ala, did not show a decrease in general or 5′ TOP mRNA translation but paradoxically resulted in an increase in protein synthesis rates. Moreover, the in vivo physiological role of RPS6 phosphorylation seems to be rather limited as mice with defectiv e RPS6 phosphorylation only display decreased cell size and impaired glucose homeostasis53. Hence, the physio logical significance of ribosomal protein post-translationa l modifications needs to be carefully evaluate d in the future.
A recent study showed that in addition to acetylation, methylation and phosphorylation, many ribosomal proteins can be modified at Ser and Thr residues by the addition of O-linked β-d-N-acetylglucosamine (O-GlcNAc). Overexpression of O-GlcNAc transferase (OGT), the enzyme involved in O-GlcNAc modification, resulted in an increase in the 60S and 80S peaks in the polysome profile, suggesting that O-GlcNAc modificatio n might be important in ribosome biogenesis54.
The same ribosomal proteins exhibit different dynamics of specific post-translational modifications, suggesting an even more complex regulation of the ribosome activity54. In addition to being phosphorylated and O-GlcNAcylated, ribosomal proteins are regulated by ubiquitylation. For example, Rpl28 in S. cerevisiae is heavily ubiquitylated during S phase of the cell cycle, whereas ubiquitylation is reduced in G1 phase. Rpl28 is located at the peptidyl transferase centre, and its ubiquitylation has a stimulatory effect on translation. Ribosomes that carry polyubiquitylated Rpl28 are able to translate a reporter gene at a faster rate than ribosomes with monoubiquitylated Rpl28 (REF. 55). In addition, mass spectrometry studies have shown that ribosomal proteins can be methylated under different conditions50. Together, these studies identify unique post-translational modifications on specific ribosomal proteins that may affect the function of the ribosome.
Ribosome-associated factors
Proteomics studies of S. cerevisiae ribosomes identified 77 ribosome-associate d proteins, which suggests that apart from ribosomal protein s, many additional factors may also modulate ribosome activity. Cells lacking some of these proteins show defects in the rate and fidelity of protein synthesis as well as ribosome biogenesis56. Interestingly, ribosome-associated proteins may also regulate the translation of specific subsets of mRNAs. For example, in D. mela- nogaster, the pro-apoptotic protein Reaper binds directly and specifically to the 40S subunit and disrupts AUG recognition by the scanning 48S complex, thereby inhibiting cap-dependent translation (FIG. 2a). Interestingly, Reaper may facilitate the selective translation of specific pro-apoptotic mRNAs that harbour internal ribosome entry site (IRES) elements (see below), as these mRNAs can bypass scanning, thereby augmenting the apoptotic programme57. Another ribosome-associated protein is glycogen synthase 1 (GYS1). In HeLa cells, GYS1 associates with actively translating ribosomes. Knockdown of GYS1 causes a decrease in polysomes and a change in the subsets of mRNAs in polysomes58. It is therefore clear that the translation of mRNAs may be enhanced or inhibited by distinct ribosome-associated factors, adding a further level of complexity to the translation machinery.
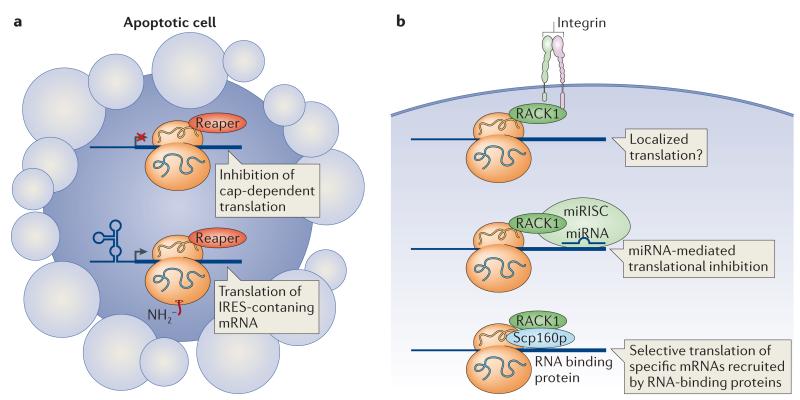
Figure 2. Ribosome-associated factors affect ribosome function.
a | In Drosophila melanogaster, Reaper protein associates with the 40S small ribosomal unit, disrupts the cap-scanning mechanism of translational initiation and promotes the translation of mRNAs containing internal ribosome entry site (IRES) elements that may be required for cell death. b | Ribosome-bound RACK1 can potentially localize ribosomes to the cell membrane by interacting with transmembrane receptors. RACK1 can recruit the microRNA (miRNA)-induced silencing complex (miRISC) to the ribosome to facilitate miRNA-mediated repression. RACK1 also binds to RNA-binding proteins such as yeast Scp160p, and this interaction may facilitate the recruitment of specific mRNAs to the ribosome.
RACK1, the receptor for activated protein kinase C (PKC), is a scaffold protein that associates with the 40S subunit in a 1:1 ratio (reviewed in REFS 59,60). Many signalling molecules interact with RACK1 on the ribosome and affect protein synthesis. For example, activated PKCβII binds to ribosome-bound RACK1 and phosphorylates the translation initiation factor eIF6, releasing it from the 60S subunit and allowing subunit joining61. Recent work has also suggested that RACK1 can recruit the microRNA (miRNA)-induced silencing complex (miRISC) to the ribosome, where this complex facilitates miRNA-mediated gene repression62. Other binding partners of RACK1 include integrin receptors60, which can potentially localize ribosomes to the cell membrane, and the yeast RNA-binding protein Scp160p63,64 (FIG. 2b). Scp160p associates with specific mRNAs, and loss of Scp160p reduces the association of those mRNAs with polysomes65. As Scp160p interacts with the ribosome through RACK1 (REFS 63,64), these results suggest that Scp160p brings specific mRNAs to the ribosome to enhance their translation. Yeast contain many ribosome-associated proteins56, and it is intriguing to hypothesize that additional intermediaries could associate with the ribosome and may contribute towards transcript-specific translational regulation.
Ribosome-associated factors can also function in processes that are unrelated to translation. For exampl e, mammalian target of rapamycin complex 2 (mTORC2) is a highly conserved protein kinase complex that integrates cell growth and metabolism in response to nutrients and growth factors66. Following insulin signalling mTORC2 binds to ribosomes, and this inter action activates mTORC2 independently of translation67. Moreover, it seems that ribosome-bound mTORC2 may phosphorylate its substrate AKT during or immediately after AKT mRNA translation68. Importantly, these findings suggest that ribosome-associated factors such as mTORC2 can also operate to control specific molecular events independently from a direct role in translational control.
Heterogeneity in rRNA
Heterogeneity in rRNA may also contribute to specialized ribosomal activity. The malaria-causing Plasmodium parasites carry two classes of ribosomal DNA genes, which allow the use of different ‘types’ of ribosomes during their life cycle. One form is predominantly found when the parasite is in the sporozoit e form (type S) in the mosquito, whereas the other form is found when it is in the gametocyte form (type A) in the bloodstream of the host after infection69 (FIG. 3). The two types of ribosomes seem to possess different functions, as substitution of the 25S rRNA of S. cerevisiae with the rRNA of type A P. falciparum produces no phenotype, whereas substitution with the rRNA of type S is lethal70. The differences between the two classes of rRNA in Plasmodium falciparu m are mainly found in the variable regions of the rRNA, and only few differences are evident in areas that are conserved among all eukaryotes71.
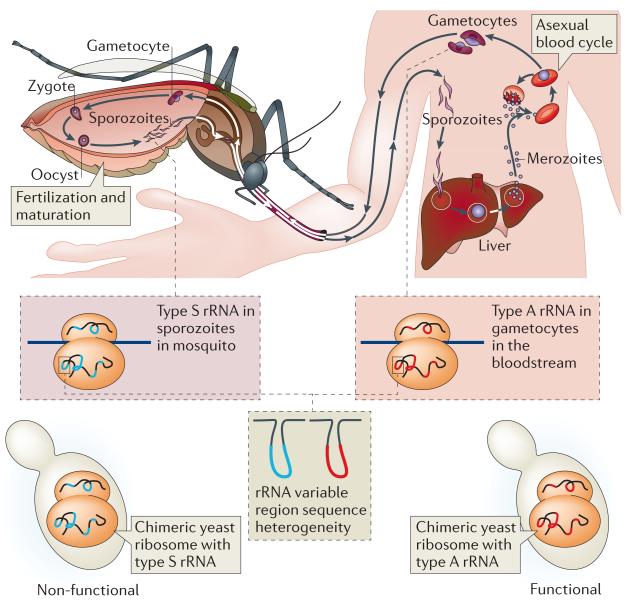
Figure 3. rRNA heterogeneity.
Plasmodium falciparum expresses different forms of ribosomal RNAs (rRNAs) when it is in sporozoite form (type S) in the mosquito and when it is in the asexual form (type A) in the bloodstream. The two forms of rRNA differ mainly in the variable regions . Chimeric yeast ribosomes in which part of the 25S rRNA was replaced with type S P. falciparum rRNA are non-functional, whereas replacement with type A P. falciparum rRNA does not produce any defects, suggesting the two types of rRNA are functionally distinct.
During evolution, from prokaryotes to eukaryotes, the ribosome has increased in size and the rRNA has changed substantially. For example, D. melanogaster has a 30 nucleotide 2S rRNA in addition to the 18S, 5.8S, 28S and 5S rRNAs72. The most drastic variations in the rRNA are found in the variable regions, which are also known as expansion segments. Expansion segments are generally located on the surface of the ribosome, are very flexible and may have more subtle roles in fine-tuning translational control. For example, the largest expansion segment, ES7L (also known as D2), has expanded from 53 nucleotides in E. coli to a dramatic 689 nucleotides in mice73. In yeast, ES7L (which consists of 216 nucleotides) was shown to interact with multiple eukaryotic specific ribosomal proteins such as P0 in vitro, and this inter action might result in the recruitment of G proteins to the ribosome74. It will therefore be important to delineate the functional roles of expansion segments and whether changes in these sequences influence ribosome function and translational control specificity.
Ribosomal RNA is heavily modified by methylation on the 2-hydroxyl group of ribose, conversion of uridine to pseudouridine and methylation of bases. Most of these modifications occur in important functional areas of the ribosome75. The 2-hydroxyl methylation and conversion of uridine to pseudouridine are facilitated by C/D box and H/ACA box small nucleolar RNAs (snoRNA s), respectively. There is evidence for the heterogeneity in the expression levels of both C/D box and H/ACA box snoRNAs in different tissues76. The expression of some C/D box snoRNAs has also been shown to oscillate in a circadian manner77. However, a careful examination of how tissue-specific differences in snoRNA expression directly affect rRNA modifications has not been carried out. Knockdown studies using morpholino oligomers showed that the deletion of different snoRNAs in zebrafish causes specific developmental phenotypes. For example, U26 snoRNA-deficient embryos have a decreased body size and specific defects in brain and eye development78. These results suggest that snoRNAs and snoRNA-mediated modifications on rRNA have very specific effects in an organism.
Transcript-specific control of ribosome function
Several key questions need to be addressed in order to determine how changes in ribosome composition, structure and/or activity affect translational control. A central unresolved question is the nature of specific cis-regulatory elements within mRNAs that may interface with specialized ribosomes to provide regulatory control and whether additional trans-acting factors such as RNA-binding proteins have a role in this process. In addition, even core components of the ribosome that remain constitutively associated with the translation machinery may exert regulative effects by interfacing with specific regulatory elements that are present on subsets of mRNAs. Therefore, a greater understanding of the cis-acting elements within mRNAs that promote translational control is an important avenue to explore. An intriguing hypothesis is that ribosomal proteins could themselves act as ‘specificity filters’, allowing the ribosome to interface with specific subsets of mRNAs to enhance translation79. However, the precise functions of only a handful of ribosomal proteins have been identified to date. Here we discuss the nature of regulatory elements in mRNAs that may imbue regulation specific to the ribosome in the recognition and the translation of specific subsets of mRNAs.
IRES elements
In eukaryotic translational initiation, the 40S ribosomal subunit is recruited to the mRNA by translation initiation factors2. The 40S ribosome complex, together with associated factors, scans along the 5′ UTR for the initiation codon. Translation initiation factors are released at the initiation codon, and the 60S ribosomal subunit joins to form a translationally active 80S ribosome. Most eukaryotic mRNAs are translated via this cap-dependent mechanism, but it is approximated that more than 10% of cellular mRNAs rely on cap-independent mechanisms of translational initiation involving IRES elements80. The IRES is an RNA structured element positioned at the 5′ UTR of specific mRNAs that can recruit the ribosome directly to the initiation region of mRNAs with a reduced requirement for canonical initiation factors2. IRES elements were first discovered in RNA viruses, which produce uncapped RNAs and require these elements for efficient translation of viral mRNAs81,82. Later, many additional cellular mRNAs were discovered to contain IRES elements in their 5′ UTR. This class of mRNAs is often thought to encompass stress response genes, such as hypoxia-inducible factor 1A (HIF1A), TP53 and X-linked inhibitor of apoptosis (XIAP), and the IRES elements within their 5′ UTRs are thought to be active during cellular stress conditions when cap-dependent translation is inhibited.
Modifications in rRNAs are important for IRES-mediated translation. In particular, alterations in rRNA pseudouridylation cause a specific defect in the translation of some IRES-containing mRNAs. In mice that are hypomorphic for dyskerin (which is the enzyme that converts rRNA uridine to pseudouridine) general cap-dependent translation is not impaired. However, translation of IRES-containing mRNAs including the tumour suppressors p27 and p53 is perturbed83-86. As a result, these mice have a higher propensity to develop cancer. Importantly, ribosomes that lack pseudouridine modifications show a direct impairment in binding to IRES elements87.
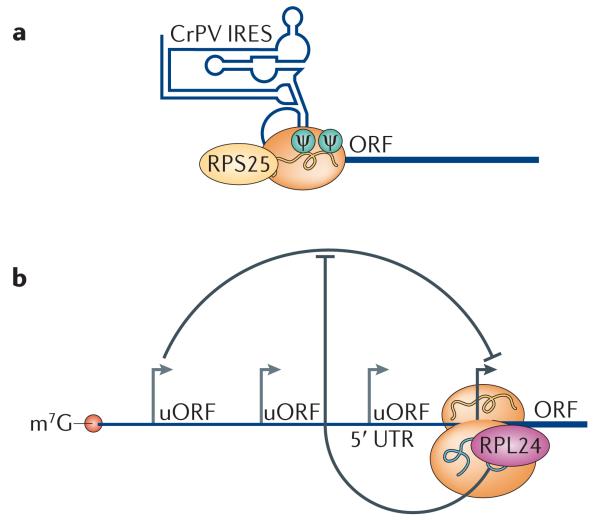
Emerging evidence suggests that ribosomal proteins themselves may function to recognize specific IRES elements. For example, RPS25 is specifically required for IRES-dependent translation of viral mRNAs88. In yeast harbouring a deletion in RPS25, cap-dependent translational initiation and ribosome fidelity are not affected. However, the translation of two distinct viral IRES elements from the hepatitis C virus (HCV) and cricket paralysis virus (CrPV) is defective (FIG. 4a). Moreover, Rps25-deficient 40S subunits are unable to bind to the CrPV IRES element. Structural studies have placed Rps25 in the head of the 40S subunit, where it can interact with two stem-loops in the CrPV IRES89. The function of RPS25 in IRES-mediated translation of viral mRNAs is conserved in mammalian cells88. However, it remains unknown whether RPS25 can regulate the translation of mRNAs that contain specific cellular IRES elements. Consistent with this idea, RPS19 and RPL11, two ribosomal proteins that are mutated in patients with Diamond-Blackfan anaemia (DBA), regulate IRES-dependent translation of at least two mRNAs that are important for erythroid differentiation, namely BCL-2-associated athanogene (BAG1) and cold shock domain containing protein E1 (CSDE1)90. In addition, mass spectrometry studies have shown that ribosomes bound to the HCV IRES have a different methylation pattern from ribosomes bound to host mRNAs, indicating a role of methylated ribosomal proteins in IRES-mediated translational control50. These studies may reflect an emerging theme: that ribosomal proteins may confer more specialized regulation of translation by interfacing with specific IRES elements in subsets of mRNAs.
Figure 4. Cis-regulatory elements within mRNAs that interface with ribosomes or ribosomal proteins to confer transcript-specific regulation of gene expression.
a | Internal ribosome entry site (IRES) elements: the ribosomal protein RPS25 is necessary for the recruitment of the small ribosomal subunit 40S to the cricket paralysis virus (CrPV) intergenic region IRES to initiate translational initiation by a cap-independent mechanism in yeast and mammals. Ribosomal RNA (rRNA) modifications (such as pseudouridylation (Ψ)) are important for ribosome binding to IRES elements. b | Upstream open reading frames (uORFs): RPL24 promotes the reinitiation of translation following an uORF, thereby allowing the translation of the downstream main ORF. m7G, 7-methylguanosine, UTR, untranslated region.
Upstream open reading frames
Almost half of all human and mouse transcripts contain at least one upstream open reading frame (uORF)91. Moreover, ribosomal profiling has recently uncovered many near-cognate uORFs that do not start with an AUG codon and, therefore, have been difficult to predict computationally92,93. uORFs can regulate the translation of the downstream main ORF. In cap-dependent translation, the ribosome scans along the 5′ UTR to start translation at the first start codon. As it is difficult to reinitiate translation after translating an uORF, most uORFs repress to the translation of the downstream main ORF. However, ribosome occupancy on some uORFs with near-cognate, non-AUG start codons correlates positively with the translation of the main ORF. For example, many mRNAs needed for sporulation in yeast contain near-cognate uORFs. Ribosome occupancy at such uORFs is higher in meiotic cells than in vegetative cells, suggesting that there is a shift in the translation machinery during meiosis to translate uORFs94. This transition can change the types of mRNAs that are translated, promoting the translation of mRNAs with non-AUG uORFs over those with cognate ones.
In plants, RPL24 is implicated in translational reinitiation of polycistronic mRNAs and mRNAs containing uORFs (FIG. 4b). A. thaliana carrying a mutation in RPL24B have developmental defects in vascular patterning and gynoecium structure owing to a reduction in translation of auxin response genes. Interestingly, many genes of the auxin response factor E class contain uORFs that inhibit the translation of the main ORF. The removal of the uORFs in these genes results in partial suppression the RPL24B phenotype95. RPL24 is located at the interface between 40S and 60S subunits and may help in subunit joining during translational re initiation74. The cauliflower mosaic virus (CaMV) takes advantage of this RPL24 function to promote infection. RPL24 binds to the CaMV transactivator (TAV), and this binding is essential for the translation of the second ORF from the polycistronic CaMV RNA. TAV also binds eIF3, which is the initiation factor that stimulates the binding of the ternary complex to the 40S. TAV can remain associated with the 40S subunit after termination at the first ORF and potentially help in recruiting eIF3 and RPL24-containing 60S subunits for reinitiation96. It will be interesting to see whether RPL24 also has a role in overcoming uORF-mediated repression in mammalian cells as a mechanism to promote translational control of specific subsets of uORF-containing mRNAs. Moreover, it will be important to determine whether additional ribosomal proteins modulate uORF-mediated regulatio n of translational control.
Ribosome specificity in cell biology
As translation rates are a much better predictor of protein levels than mRNA levels, translational control is an important mechanism to regulate the expression of many genes97. The possibility of greater specificity in translation through a more regulatory function of the ribosome opens an important avenue of exploration into the cellular and developmental processes that are regulated by this new level of control. This is a very rapidly emerging field, and although much remains to be addressed, clear examples of specialized ribosome functions are becoming evident across all species.
Ribosome specificity in bacteria
In bacteria, a new paradigm is emerging for heterogeneity in ribosomes as a mechanism for stress adaptation. Remarkably, the formation of ‘stress-ribosomes’ that contain rRNAs with sequence-specific differences allows for the selective translation of specific mRNAs (FIG. 5a). In E. coli, the sequence-specific endonuclease MazF, which preferentially cleaves single-stranded mRNAs at ACA sequences, is activated following stress. Although MazF induction blocks the synthesis of most proteins, the translation of ~10% of all transcripts remains active, and some of these mRNAs encode proteins that promote cell death. Strikingly, MazF is able to selectively promote translation of these transcripts by cleaving ACA sites at, or closely upstream of, the AUG start codon to gen erate leaderless mRNAs. MazF also changes the type of ribosomes that are formed to translate these leaderless mRNAs by cleaving 16S rRNAs near the 3′ end, thereby releasing the anti-Shine-Dalgarno sequence. In bacteri a, translational initiation depends heavily on base pairing between the Shine-Dalgarno sequence in the mRNA and the anti-Shine-Dalgarno sequence in the 16S rRNA98. The removal of the anti-Shine-Dalgarno sequence by MazF renders a subpopulation of specialized ribosomes that are less efficient in translating most mRNAs but improves translation of leaderless mRNAs that no longer possess the Shine-Dalgarno sequence99.
Figure 5. Ribosome specificity in cell and developmental biology.
a | In Escherichia coli, MazF cleaves the 3′ end of the 16S ribosomal RNA (rRNA) and releases the anti-Shine-Dalgarno sequence, thereby generating specialized ribosomes that translate leaderless mRNAs that are involved in the stress response. b | Mitochondrial ribosomes (green) in the germ plasm (yellow area) of Drosophila melanogaster embryos are found outside of mitochondria and may translate certain cytoplasmic germ cell-specific mRNAs. c | Gene expression of Rpl38 (pink) is highly increased in somites and the neural tube of developing mouse embryos and is responsible for translating specific homeobox (Hox) mRNAs that are necessary for axial skeletal patterning and the specification of PEA3-expressing motor neurons. d | Localized translation takes place in dendrites, far away from the cell body. mRNAs for ribosomal proteins are found in dendrites and can potentially form dendrite-specific ribosomes.
The role of ribosome heterogeneity in plants
In plants, mutations in ribosomal proteins result in various phenotypes. Loss-of-function mutations in a number of ribosomal proteins are embryonic lethal, whereas others have more varied patterning and developmental defects100,101. For example, mutations in RPL10A, RPL9 and RPL5 give a piggyback phenotype, in which mutants have pointed leaves and pronounced marginal serrations. These genes are crucial in dorsal-ventral patterning in leaves, possibly through translational regulation of dorsal genes of the class III homeodomain-Leu zipper (HD-ZIPIII) family and ventral genes of the KANADI family102. A. thaliana with mutantions in RPL24B have developmental defects in vascular patterning and gynoecium structure owing to a reduction in translation of auxin response genes95. As discussed, the effect of RPL24 may be specifically at the level of translation reinitiation of mRNAs that harbour uORFs (see above). Although a clear mechanism for translational regulation by these ribosomal proteins is not fully understood, it is clear that ribosomal proteins have an unexpected and important role in regulating specific developmental processes and pattern formation100.
Ribosome specificity in invertebrate embryonic development
In D. melanogaster, the loss of a copy of several ribosomal proteins leads to a Minute phenotype. Minute flies are smaller than their wild-type counterparts, have delayed larval development and short, thin bristles29. This phenotype has been attributed to a general decrease in protein synthesis. However, some ribosomal protein mutants show additional patterning defects. For example, some Minute flies have a reduction in wing size, whereas at least two additional ribosomal protein mutants display an increase in wing size and wing vein patterning defects103,104. This suggests that some ribosomal proteins are specifically required for embryonic development and patterning.
Germline formation in D. melanogaster may require specific ribosomes that are only produced in the mitochondria (FIG. 5b). Mitochondrial ribosomes are typically smaller than cytoplasmic ribosomes, resemble prokaryotic ribosomes and are required to translate the mitochondrial genome. Strikingly, however, these ribosomes can be localized outside of mitochondria in polar granules, which are large ribonucleoprotein complexes found at the posterior end of D. melanogaster embryos and are required for germ cell formation105,106. Polysomes on polar granules contain both mitochondrial and cytoplasmic ribosomes. Blocking mitochondrial ribosomes with the prokaryotic translation inhibitors kasugamycin and chloramphenicol, both of which do not affect cytoplasmic ribosomes, causes a decrease in the expression levels of the germ cell-specific protein GCL in polar granules and a defect in germ cell formation. The translation of another germ cell mRNA, namely Nanos (Nos), is not affected by these drugs, suggesting that mitochondrial ribosomes may only be responsible for the proper translation of a subset of mRNAs107. Importantly, further studies on the codon usage of the GCL mRNA are urgently required to determine whether mitochondrial ribosomes are indeed participating in this level of translational control. Although it is not entirely understood why mitochondrial ribosomes that are located in the cytoplasm may possess unique properties that endow them with the ability to translate specific cytoplasmic mRNAs, these studies suggest a specificity in translational regulation governed at the level of heterogeneous ribosomes.
Ribosome specificity in vertebrate embryonic development
Ribosomal proteins are important regulators of vertebrate embryonic development. For example, morpholino knockdown of ribosomal proteins in zebrafish produces phenotypes that can be both unique and specific, resulting in abnormalities in the development of the brain, body trunk, eye and ear108. How can mutations in ribosomal proteins cause tissue-specific effects? One important paradigm that provided insight into the role of ribosomal proteins in controlling specialized ribosome activity stems from the analysis of RPL38 function, as the results linked this component of the ribosome to the formation of the mammalian body plan (FIG. 5c). In a forward genetic screen, Rpl38 was identified as the gene that is deleted in tail short (Ts) mice. These mice have highly specific defects in the formation of the axial skeleton37. These defects include homeotic transformations and a change in the identity of one skeletal element from another, which suggests an unexpected regulatory role for ribosomes during tissue patterning. Homeobox (Hox) genes have been shown to be key regulators of vertebrae segment identity, and the loss of function of specific Hox genes causes homeotic transformations109,110. Ts/+ mouse embryos also show a defect in the specification of PEA3-positive motor neurons, which also requires HOX-directed patterning. Strikingly, Ts/+ embryos show no defects in general protein synthesis, however, the translation of 8 of the 39 Hox mRNAs is specifically reduced. At the molecular level, RPL38 regulates the formation of the 80S complex during translation initiation on these mRNAs, while being part of the ribosome. The effects of RPL38 are highly specific as other ribosomal protein deficiencies in mice do not give rise to phenotypes similar to the phenotypes observed in RPL38-deficient mice, even when the general rate of protein synthesis is decreased. Interestingly, Ts/+ phenotypes are inherited in a dominant form, as haploinsufficiency in RPL38 expression is sufficient for patterning defects to manifest. This suggests that RPL38 is rate-limiting for the translation of selective transcripts such as HOX mRNAs. Moreover, this is consistent with the fact that HOX mutants also display phenotypes in a heterozygous condition, thereby revealing that precise levels of HOX proteins are necessary for establishing the mammalian body plan. It will be important to determine whether RPL38 recognizes unique sequences or structures within subsets of mRNAs either directly or through interactions with other RNA-binding proteins.
The tissue specific phenotypes associated with RPL38 loss of function are also, at least in part, explained by the observation that Rpl38 expression is highly regulated during embryonic development. A striking enrichment of Rpl38 transcripts is evident in specific regions of the embryo such as developing somites and within the neural tube. These regions mirror where the tissue specific phenotypes associated with loss of RPL38 expression are observed37. These findings suggest that the increased expression of specific ribo somal proteins may produce heterogeneous ribosomes in distinct cell and tissue types with unique specificities in translating specific classes of mRNAs.
In addition to Ts/+ mice, the few mouse models with mutated ribosomal proteins that have been generated or characterized to date display specific phenotypes. For example, mice carrying a dominant mutation in Rpl24 display a white ventral midline spot, white hind feet and a kinked tail. These mice also have reduced numbers of retinal ganglion cells and extra digits in the limbs111. On the contrary, Rpl29−/− mice are smaller in size and possess specific defects in osteogenesis associated with increased bone fragility112. These phenotypes suggest that additional ribosomal proteins may have more specialized roles during embryonic development.
Linking ribosome specificity to disease
Loss of some ribosomal proteins has been associated with certain morphological abnormalities, and these phenotypes may rely on the activation of a p53-dependent stress response. A subset of ribosomal proteins are able to interact with MDM2, an inhibitor of p53, and thereby affect p53 activity113. Morpholino knockdown of rpl11, rps7 and rps19 in zebrafish causes an upregulation of p53, and certain phenotypes can be partially rescued by knockdown of tp53 (REFS 114-116). Importantly, increased p53 protein levels cannot fully explain the phenotypes observed in ribosomal protein mutants. For example, the tissue-specific defects associated with Rpl38 loss of function were not reversed following the reduction of p53 protein levels37. Moreover, the mouse embryos carrying Rps6+/− and p53−/− mutations have tissue-specific phenotypes, including defects in placenta and liver development117.
In humans, accumulating evidence links mutations in the genes of several ribosomal proteins, including RPS19, RPS24, RPS17, RPL5, RPL11 and RPL35A, to Diamond-Blackfan anaemia (DBA)118. Notably, patients with DBA carrying ribosomal protein mutations also exhibit tissue-specific defects including limb defects, cleft palate, abnormalities in heart development, growth failure and a predisposition for cancer. Mutations in different ribosomal proteins also give rise to different types of birth defects. For instance, most patients with RPL5 mutations have a cleft palate, whereas most individual s with RPL11 mutations do not have any craniofacial defects119. In addition, mutations in RPL21 have been identified in hereditary hypotrichosis simplex (HHS), a disorder that is characterized by progressive hair loss early in childhood120. Together, patients with genetic disorders that are linked to mutations in ribosomal proteins show remarkably specific phenotypes, which suggests that ribosomal proteins have unique functions in different tissues.
Intracellular localization of specialized ribosomes
A clue for the possible roles of specialized ribosomal protein function in specific cell types comes from research in neurons. Neurons are extremely polarized cells in which axons and dendrites perform very different functions from the cell body. Local protein synthesis in neuro nal dendrites is important in remodelling synaptic connections and has a role in learning and memory121 (FIG. 5d). Long-term potentiation (LTP), a cellular measure of memory formation, can be induced by localized translation in isolated hippocampal dendrites, which have been removed from the cell bodies122. Polysomes are found in dendrites, and the number of polysomes in dendritic spines increases after the initiation of LTP123. Thereby, mRNAs are translated in an extremely localized manner, although the mechanism for this regulation is poorly understood.
Strikingly, transcripts for specific subsets of ribosomal proteins are enriched up to tenfold at these sites compared to the cell body124. As most ribosomal proteins are assembled onto the ribosome in the nucleus, it is surprising to find pools of specific ribosomal protein transcripts in neuronal processes. Additional components of the translational machinery have also been found in neuronal processes and are transported as part of large ribonucleoprotein particles that contain mRNA, ribosomes and other RNA binding proteins125,126. It has been suggested that ribosomal subunits are stored as inactive forms in RNA granules and are activated by the addition of locally synthesized ribosomal proteins126. Thus, ribosomes at these neuronal processes may be distinct from ribosomes in the cell body owing to the presence of newly, locally synthesized ribosomal proteins that may influence the translation of mRNAs that are localized in the dendrites. In support of this hypothesis, recent studie s have shown that the transmembrane receptor DCC, which participates in axon guidance, associates with specific ribosomal proteins and spatially regulates translation in neuronal axons and dendrites127. Interestingly, some mRNAs present in dendrites are known to contain IRES elements128. By extension, simila r localized mechanisms for translational control may provide a unique regulation in multiple cellular contexts, including translational regulation in polarized cells, at the leading edge of migrating cells or during specific steps in cell differentiation.
Conclusions and future perspectives
Several parallels can be drawn between what has been termed a DNA code and a possible ‘ribosome code’. Like DNA, rRNA is extensively modified. Histones, once considered as boring housekeeping proteins, are now clearly recognized as active participants in chromatin remodelling and transcriptional control through exquisite post-translational modifications identified within histone tails. Likewise, the view of ribosomal proteins as only carrying out rote-like functions is undergoing a paradigm shift. Post-translational modifications and differences in ribosomal protein composition may add greater regulatory specificity to the RNA-based translation machinery, as do histones to DNA. However, several outstanding questions need to be addressed before a ribosome code for translational control could be accepted.
First, as discussed in this Review, many examples now exist across all species suggesting that great heterogeneity characterizes the ribosome, both at the level of rRNA and ribosomal proteins. However, despite these examples, this hypothesis has not been explored in a systematic manner. Most ribosomes are purified from tissues that contain an amalgamation of multiple cell types, and any differences in ribosome composition in a particular cell may be lost in such an ensemble. Studies in cell lines have the advantage of being able to identify heterogeneity in a single cell type; however, this approach lacks the dynamics of an in vivo system in which more variations may be present. A systems approach is necessary to examine the extent of variation in ribosomes more globally across different cell types, perhaps during early stages of organogenesis or during specific steps in cell differentiation. It is also tempting to speculate that a heterogeneous pool of ribosomes may already exist in a single cell, for example in germ cells and neurons. If so, how is the stoichiometry and the formation of such heterogeneous pools of ribosomes regulated? What regulates the intracellular localization of heterologous ribosomes in a single cell?
Second, although examples of heterogeneity are evident, little is known on how heterogeneity affects translational control. For example, to date classes of mRNAs that may respond to specialized ribosomes remain elusive. The discovery of a specific requirement for RPL38-containing ribosomes in facilitating translational regulation of subsets of HOX mRNAs hints at exciting possible future work into this problem.
Third, virtually nothing is known about the upstream regulatory signals that might produce ribosome heterogeneity in the first place. The realization that components of the ribosome may be more highly regulated opens a new avenue of exploration in identifying upstream signalling pathways and trans-acting factors that control their expression. Lastly, it will be crucial to identify cis-acting ‘translational regulons’ within mRNAs that interface with specialized ribosomes to confer translational specificity. As discussed, examples of these important elements may include IRES elements and uORFs. Moreover, it is equally importan t to determine whether even constitutive components of the ribosome that show little variation may also confer more specialized activity by virtue of their ability to interact with such specific ‘translation regulons’. Emerging evidence suggests that there is a great disconnect between the expression of the genome at the level of the transcriptome versus the proteome, thereby leaving ample room for regulation at the level of translational control97. Thus, variations in gene expression at the level of translational control have tremendous importance to cell biology. Therefore, we hypothesize that it will be important to conceptualize translational control in the same light as transcriptional control — as a process in which enhancers or attenuators may fine-tune protein abundance that culminates in unique readout s with important biological significance.
Ultimately, a key question is how ribosome-mediate d gene regulation may shape our view of organismal biolog y. It has long been known that a small handful of signalling molecules (such as sonic hedgehog (SHH), fibroblast growth factors (FGFs), WNT ligands and bone morphogenetic proteins (BMPs)) act many times during embryonic development and in distinct locations to produce entirely different tissues or structures (for example, limbs versus motor neurons). Transcriptional profiling has not provided a complete explanation for these different mechanisms of action. The hypothesis that developmentally regulated signals directly converge on ribosome specificity opens an entirely new avenue of exploration into whether rapid changes in the production of proteins from pre-existing pools of mRNAs may instead influence fundamental aspects of cell and embryonic development.
Acknowledgements
The authors would like to thank D. Ruggero, C. Bellodi, M. McMahon, C. Stumpf and members of the Barna laboratory for discussion and critical reading of the manuscript. S.X. is supported by the Agency of Science, Technology and Research of Singapore. This work was supported by the Program for Breakthrough Biomedical Research, UCSF (to M.B.) and the National Institutes of Health (NIH) Director’s New Innovator Award,1DP2OD008509 (to M.B.).
Glossary
- Nucleolus
A conserved organelle that assembles around ribosomal DNA genes. The nucleolus is the site of ribosomal RNA transcription and the site of ribosome subunit assembly.
- Paralogues
Homologous genes that are separated by a duplication event and that have evolved new functions
- ASH1
(Asymmetric synthesis of homothallic switching endonuclease (HO)). A gene encoding a repressor that inhibits the transcription of HO — an endonuclease that causes mating-type switching in Saccharomyces cerevisiae. ASH1 mRNA is transported to the bud before translation. In the bud, Ash1 prevents the daughter cell from switching its mating type following cell division.
- Polysomes
Or polyribosomes; clusters of two or more ribosomes attached at different sites on the same strand of mRNA. mRNAs bound to polysomes are being actively translated.
- Somites
Blocks of mesoderm on either side of the neural tube of a developing vertebrate embryo. Somites will develop into structures including the vertebrae.
- Mammalian target of rapamycin complex 2
(mTORC2). A protein kinase complex that includes mTOR, RICTOR and other proteins. mTORC2 regulates cell growth, metabolism and survival in response to environmental cues such as nutrients and growth factors.
- Sporozoite
The cellular form of Plasmodium parasites when they infect a new host.
- Expansion segments
A region of ribosomal RNA (rRNA) that has dramatically increased in length from prokaryotes to eukaryotes during evolution.
- Small nucleolar RNAs
(snoRNAs). Small RNA molecules that function in ribosome biogenesis in the nucleolus by guiding the assembly of macromolecular complexes on the target RNA to allow site-specific modifications or processing reactions to occur.
- Upstream open reading frame
(uORF). An uORF is defined by a start codon and an in-frame stop codon in the 5′ untranslated region of an mRNA.
- Shine–Dalgarno sequence
A ribosomal binding site of approximately eight nucleotides in the mRNA of bacteria, located upstream of the initiation codon. Helps to recruit the small ribosomal subunit to the mRNA to initiate protein synthesis.
- Cleft palate
A craniofacial abnormality that results from a failure to fuse the left and right palatal shelves at the midline during embryogenesis. It can be caused by several environmental and genetic factors, including defects in sonic hedgehog signalling.
- Long-term potentiation
(LTP). A long-lasting increase in the size of the postsynaptic response to synaptic transmissions. LTP is thought to be a key mechanism for learning and long-term memory formation in the brain.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Maria Barna’s homepage: http://biochemistry.ucsf.edu/labs/barna/Site/Welcome.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Frank J. The ribosome — a macromolecular machine par excellence. Chem. Biol. 2000;7:R133–R141. doi: 10.1016/s1074-5521(00)00127-7. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. Reviews the mechanism of translation initiation and its regulation..
- 3.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 4.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 6.Crick FHC. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 7.Nissen P. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 8.Noller HF, Hoang L, Fredrick K. The 30S ribosomal P site: a function of 16S rRNA. FEBS Lett. 2005;579:855–858. doi: 10.1016/j.febslet.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 9.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl extraction transferase to protein procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 10.Held WA, Mizushima S, Nomura M. Reconstitution of Escherichia coli 30S ribosomal subunits from purified molecular components. J. Biol. Chem. 1973;245:5720–5730. [PubMed] [Google Scholar]
- 11.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balasubramanian S, et al. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 2009;10:R2. doi: 10.1186/gb-2009-10-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 14.Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;54:47, 54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohtake Y, Wickner RB. Yeast virus propagation depends critically on free 60S ribsomal subunit concentration. Mol. Cell. Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll K, Wickner RB. Translation and M1 double-stranded RNA propagation: MAK18 = RPL41B and cycloheximide curing. J. Bacteriol. 1995;177:2887–2891. doi: 10.1128/jb.177.10.2887-2891.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parenteau J, et al. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–331. doi: 10.1016/j.cell.2011.08.044. Shows that introns in ribosomal protein genes regulate the expression of both the intron-containing genes and their paralogues.
- 20.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nature Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 21.Steffen K, et al. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barakat A, et al. The organization of cytoplasmic ribosomal protein genes in the arabidopsis genome. Plant Physiol. 2001;127:398–415. [PMC free article] [PubMed] [Google Scholar]
- 23.Whittle CA, Krochko JE. Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell. 2009;21:2203–2219. doi: 10.1105/tpc.109.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weijers D, et al. An. Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a ribosomal protein S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. Shows differential expression of RPS5 paralogues in A. thaliana development.
- 25.Falcone Ferreyra ML, Pezza A, Biarc J, Burlingame AL, Casati P. Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 2010;153:1878–1894. doi: 10.1104/pp.110.157057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams ME, Sussex IM. Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana. Plant J. 1995;8:65–76. doi: 10.1046/j.1365-313x.1995.08010065.x. [DOI] [PubMed] [Google Scholar]
- 27.Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008;147:128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degenhardt RF, Bonham-Smith PC. Transcript profiling demonstrates absence of dosage compensation in Arabidopsis following loss of a single RPL23a paralog. Planta. 2008;228:627–640. doi: 10.1007/s00425-008-0765-6. [DOI] [PubMed] [Google Scholar]
- 29.Marygold SJ, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearse MG, Chen AS, Ware VC. Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res. 2011;39:2701–2716. doi: 10.1093/nar/gkq1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes AM, et al. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol. Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. Shows the differential expression of human RPS4 paralogues and predicts the differences in protein structures between the paralogues.
- 32.Fisher EM, et al. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 33.Sugihara Y, et al. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 34.Hariharan N, Kelley DE, Perry RP. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989;3:1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- 35.Kim CH, Warner JR. Messenger RNA for ribosomal proteins in yeast. J. Mol. Biol. 1983;165:79–89. doi: 10.1016/s0022-2836(83)80243-5. [DOI] [PubMed] [Google Scholar]
- 36.Bortoluzzi S, D'Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 37.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. First paper that shows heterogeneity in ribosomal protein expression in different tissues of a developing vertebrate embryo. It also shows that loss of a single ribosomal protein can have profound but specific effects on translational regulation in vivo.
- 38.Ramagopal S, Ennis HL. Regulation of synthesis of cell-specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proc. Natl Acad. Sci. USA. 1981;78:3083–3087. doi: 10.1073/pnas.78.5.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramagopal S. Induction of cell-specific ribosomal proteins in aggregation-competent nonmorphogenetic Dictyostelium discoideum. Biochem. Cell Biol. 1990;68:1281–1287. doi: 10.1139/o90-190. Demonstrates dramatic changes in ribosome composition as D. discoideum shifts from vegetatively growing to aggregation-competent.
- 40.Sahin F, et al. RPL38, FOSL1, and UPP2 are predominantly expressed in the pancreatic ductal epithelium. Pancreas. 2005;30:158–167. doi: 10.1097/01.mpa.0000151581.45156.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills AA, Mills MJ, Gardiner DM, Bryant SV, Stanbridge EJ. Analysis of the pattern of QM expression during mouse development. Differentiation. 1999;64:161–171. doi: 10.1046/j.1432-0436.1999.6430161.x. [DOI] [PubMed] [Google Scholar]
- 42.Green H, et al. The ribosomal protein QM is expressed differentially during vertebrate endochondral bone development. J. Bone Miner. Res. 2000;15:1066–1075. doi: 10.1359/jbmr.2000.15.6.1066. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian AR. Copies of proteins L7 and L12 and heterogeneity of the large subunit of Escherichia coli ribosome. J. Mol. Biol. 1975;95:1–8. doi: 10.1016/0022-2836(75)90330-7. [DOI] [PubMed] [Google Scholar]
- 44.Hardy SJS. The stoichiometry of the ribosomal proteins of Escherichia coli. Mol. General Genet. 1975;140:253–274. doi: 10.1007/BF00334270. [DOI] [PubMed] [Google Scholar]
- 45.Oleinikov AV, Jokhadze GG, Traut RR. A single-headed dimer of Escherichia coli ribosomal protein L7/L12 supports protein synthesis. Proc. Natl Acad. Sci. USA. 1998;95:4215–4218. doi: 10.1073/pnas.95.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SW, et al. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl Acad. Sci. USA. 2002;99:5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll AJ, Heazlewood JL, Ito J, Millar AH. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics. 2008;7:347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Odintsova TI, et al. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J. Protein Chem. 2003;22:249–258. doi: 10.1023/a:1025068419698. [DOI] [PubMed] [Google Scholar]
- 50.Yu Y, Ji H, Doudna JA, Leary JA. Mass spectrometric analysis of the human 40S ribosomal subunit: native and HCV IRES-bound complexes. Protein Sci. 2005;14:1438–1446. doi: 10.1110/ps.041293005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramagopal S. Covalent modifications of ribosomal proteins in growing and aggregation-competent Dictyostelium discoideum: phosphorylation and methylation. Biochem. Cell Biol. 1991;69:263–268. doi: 10.1139/o91-040. [DOI] [PubMed] [Google Scholar]
- 52.Krieg J, Hofsteenge J, Thomas G. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J. Biol. Chem. 1988;263:11473–11477. [PubMed] [Google Scholar]
- 53.Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeidan Q, Wang Z, De Maio A, Hart GW. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell. 2010;21:1922–1936. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spence J, et al. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 56.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colon-Ramos DA, et al. Direct ribosomal binding by a cellular inhibitor of translation. Nature Struct. Mol. Biol. 2006;13:103–111. doi: 10.1038/nsmb1052. Demonstrates that the translation regulator Reaper can bind directly to the 40S ribosome subunit to inhibit cap-dependent translational initiation.
- 58.Fuchs G, Diges C, Kohlstaedt LA, Wehner KA, Sarnow P. Proteomic analysis of ribosomes: translational control of mRNA populations by glycogen synthase GYS1. J. Mol. Biol. 2011;410:118–130. doi: 10.1016/j.jmb.2011.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams DR, Ron D, Kiely PA. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nilsson J, Sengupta J, Frank J, Nissen P. Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 2004;5:1137–1141. doi: 10.1038/sj.embor.7400291. Reviews, together with reference 59, the multiple functions of RACK1 as a protein that associates with the ribosome.
- 61.Ceci M, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:574–579. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- 62.Jannot G, et al. The ribosomal protein RACK1 is required for microRNA function in both C. elegans and humans. EMBO Rep. 2011;12:581–586. doi: 10.1038/embor.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baum S, Bittins M, Frey S, Seedorf M. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem. J. 2004;380:823–830. doi: 10.1042/BJ20031962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Coyle SM, Gilbert WV, Doudna JA. Direct link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li A-M, Watson A, Fridovich-Keil JL. Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res. 2003;31:1830–1837. doi: 10.1093/nar/gkg284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature Rev. Mol. Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. Shows that mTORC2 interacts with the ribosome and this interaction activates mTORC2 independently of translation.
- 68.Oh WJ, et al. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gunderson J, et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. Shows that different forms of rRNAs are expressed in different stages in the lifecycle of Plasmodium berghei.
- 70.Velichutina IV, Rogers MJ, McCutchan TF, Liebman SW. Chimeric rRNAs containing the GTPase centers of the developmentally regulated ribosomal rRNAs of Plasmodium falciparum are functionally distinct. RNA. 1998;4:594–602. doi: 10.1017/s1355838298980049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogers MJ, et al. Structural features of the large subunit rRNA expressed in Plasmodium falciparum sprozoites that distinguish it from the asexually expressed large subunit rRNA. RNA. 1996;2:134–145. [PMC free article] [PubMed] [Google Scholar]
- 72.Pavlakis GN, Jordan BR, Wurst RM, Vournakis JN. Sequence and secondary structure of Drosophila melanogaster 5.8 S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979;7:2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hassouna N, Michot B, Bachellerie JP. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 75.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 76.Castle JC, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012 Apr 3; doi: 10.1101/gr.128876.111. doi:10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higa-Nakamine S, et al. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012;40:391, 398. doi: 10.1093/nar/gkr700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc. Natl Acad. Sci. USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 81.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 82.Jang SK, et al. A segment of the 5ʹ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoon A, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. First paper that shows the importance of rRNA pseudouridylation in viral and cellular IRES-mediated translation.
- 84.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bellodi C, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montanaro L, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 87.Jack K, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell. 2011;44:660–666. doi: 10.1016/j.molcel.2011.09.017. Shows that rRNA pseudouridylation affects IRES-dependent translational initiation and translational fidelity.
- 88.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. Shows that a ribosomal protein interacts directly with an IRES element and regulates translation mediated by the IRES.
- 89.Muhs M, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horos R, et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 91.Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218, 223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishimura T, Wada T, Yamamoto KT, Okada K. The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell. 2005;17:2940–2953. doi: 10.1105/tpc.105.036533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park HS, et al. A plant viral "reinitiation" factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. Shows, together with reference 95, that RPL24 is important for the translational reinitiation of uORF containing mRNAs in plants.
- 97.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 98.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 99.Vesper O, et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–157. doi: 10.1016/j.cell.2011.07.047. Shows that MazF in E.coli cleaves both rRNA and specific mRNAs, generating specialized ribosomes without the anti-Shine-Dalgarno sequence. These specialized ribosomes are competent to translate the leaderless mRNAs generated by MazF..
- 100.Byrne ME. A role for the ribosome in development. Trends Plant Sci. 2009;14:512–519. doi: 10.1016/j.tplants.2009.06.009. Reviews the myriad of ribosomal protein mutants in plants.
- 101.Szakonyi D, Byrne ME. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 2011;65:269–281. doi: 10.1111/j.1365-313X.2010.04422.x. [DOI] [PubMed] [Google Scholar]
- 102.Pinon V, et al. Three piggyback genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135:1315–1324. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- 103.Brehme KS. Development of the Minute phenotype in Drosophila melanogaster. A comparative sudy of the growth of three Minute mutants. J. Exp. Zool. 1941;88:135–160. [Google Scholar]
- 104.Marygold SJ, Coelho CM, Leevers SJ. Genetic analysis of RpL38 and RpL5, two Minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics. 2005;169:683–695. doi: 10.1534/genetics.104.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi S, Amikura R, Okada M. Presence of mitochondrial large ribosomal RNA outside mitochondria in germ plasm of Drosophila melanogaster. Science. 1993;260:1521–1524. doi: 10.1126/science.7684857. [DOI] [PubMed] [Google Scholar]
- 106.Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc. Natl Acad. Sci. USA. 2001;98:9133–9138. doi: 10.1073/pnas.171286998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amikura R, Sato K, Kobayashi S. Role of mitochondrial ribosome-dependent translation in germline formation in Drosophila embryos. Mech. Dev. 2005;122:1087–1093. doi: 10.1016/j.mod.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 108.Uechi T, et al. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS ONE. 2006;1:e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 110.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 111.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oristian DS, et al. Ribosomal protein L29/HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J. Orthop. Res. 2009;27:28–35. doi: 10.1002/jor.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS ONE. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Duan J, et al. Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. Int. J. Biochem. Cell Biol. 2011;43:1218–1227. doi: 10.1016/j.biocel.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 116.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 117.Panic L, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Boria I, et al. The ribosomal basis of Diamond-Blackfan anemia: mutation and database update. Hum. Mutat. 2010;31:1269–1279. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gazda HT, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou C, et al. Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum. Mutat. 2011;32:710–714. doi: 10.1002/humu.21503. [DOI] [PubMed] [Google Scholar]
- 121.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nature Rev. Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 122.Vickers CA, Dickson KS, Wyllie DJ. Induction and maintenance of late-phase long-term potentiation in isolated dendrites of rat hippocampal CA1 pyramidal neurones. J. Physiol. 2005;568:803–813. doi: 10.1113/jphysiol.2005.092924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ostroff LE, Fiala JC, Allwardt B, Harris KM. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron. 2002;35:535–545. doi: 10.1016/s0896-6273(02)00785-7. [DOI] [PubMed] [Google Scholar]
- 124.Moroz LL, et al. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. Characterizes the mRNAs found in neuronal processes and finds an enrichment of ribosomal protein mRNAs in processes as compared to the soma.
- 125.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7:1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 126.Moccia R, et al. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J. Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl Acad. Sci. USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 130.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schafer T, et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 132.Martin-Marcos P, Hinnebusch AG, Tamame M. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 2007;27:5968–5985. doi: 10.1128/MCB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ferreira-Cerca S, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 134.Peltz SW, et al. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol. Cell. Biol. 1999;19:384–391. doi: 10.1128/mcb.19.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rhodin MH, Rakauskaite R, Dinman JD. The central core region of yeast ribosomal protein L11 is important for subunit joining and translational fidelity. Mol. Genet. Genomics. 2011;285:505–516. doi: 10.1007/s00438-011-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnson AW, Lund E, Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002;27:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- 138.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. Examines the extraribosomal function of many ribosomal proteins.
- 139.Dabeva MD, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 140.Malygin AA, Parakhnevitch NM, Ivanov AV, Eperon IC, Karpova GG. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007;35:6414–6423. doi: 10.1093/nar/gkm701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wan F, et al. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 142.Mazumder B, Fox PL. Delayed translational silencing of ceruloplasmin transcript in γ-interferon-activated U937 monocytic cells: role of the 3ʹ untranslated region. Mol. Cell. Biol. 1999;19:6898–6905. doi: 10.1128/mcb.19.10.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mazumder B, et al. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 144.Kapasi P, et al. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol. Cell. 2007;25:113–126. doi: 10.1016/j.molcel.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vyas K, et al. Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in γ-interferon-activated monocytes. Mol. Cell. Biol. 2009;29:458–470. doi: 10.1128/MCB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]