Abstract
The small ubiquitin-related modifier (SUMO) system has been implicated in a number of biological functions, yet the individual components of the SUMO machinery involved in each of these activities were largely unknown. Here we report the first global SUMO system interactome. Using affinity purification coupled with mass spectrometry, we identify >450 protein–protein interactions surrounding the SUMO E2, Siz type E3s and SUMO-specific proteases in budding yeast. Exploiting this information-rich resource, we validate several Siz1- and Siz2-specific substrates, identify a nucleoporin required for proper Ulp1 localization, and uncover important new roles for Ubc9 and Ulp2 in the maintenance of ribosomal DNA.
Keywords: affinity purification-mass spectrometry, mass spectrometry, ribosomal DNA, S. cerevisiae , SUMO
Introduction
The small ubiquitin-related modifier (SUMO) system plays important roles in a number of diverse biological processes in all eukaryotes (Johnson, 2004; Kerscher et al, 2006). Like ubiquitin, SUMO modification is effected via covalent conjugation to an epsilon amine moiety of a lysine residue in a substrate protein, via the sequential action of SUMO-specific E1, E2 and E3 proteins. SUMO conjugation can be reversed by SUMO-specific proteases (Ulp1 and Ulp2; Johnson, 2004; Kerscher et al, 2006). Systematic proteomics screens have identified over 500 putative SUMO conjugates in budding yeast (Vertegaal et al, 2004; Wohlschlegel et al, 2004; Zhou et al, 2004; Denison et al, 2005; Hannich et al, 2005). However, the SUMO system components responsible for each of these specific conjugation and deconjugation events remain largely unknown. In addition, proteins that may be involved in specifying the localization of SUMO system components or the regulation of SUMO system activity have not been well described.
Results and discussion
Functional organization of the SUMO system
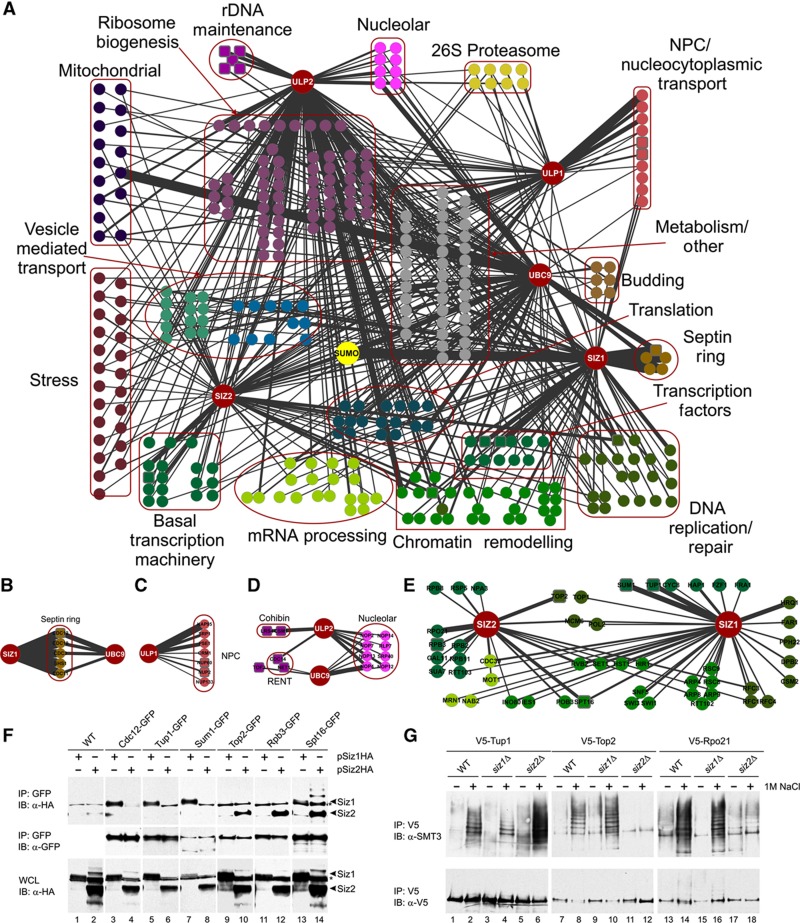
To begin to characterize the molecular organization of the budding yeast SUMO system, we identified interacting partners for the Saccharomyces cerevisiae SUMO E2 Ubc9, the E3 ligases Siz1 and Siz2, and the SUMO-specific proteases Ulp1 and Ulp2. Each of these ‘bait’ proteins was expressed with a C-terminal HA-ProtA epitope tag from a plasmid containing a galactose-inducible promoter (the mORF system; Gelperin et al, 2005) and subjected to affinity purification followed by mass spectrometry (AP-MS), essentially as in Breitkreutz et al (2010). At least four biological replicates were conducted for each bait (in two different parental yeast strains) and two technical replicates analyzed for each sample, for a total of 48 MS runs. As controls, an identical analysis of the HA-ProtA tag alone and three unrelated HA-ProtA tagged proteins expressed in the same yeast strains was conducted. Polypeptides identified with a ProteinProphet (Keller et al, 2002; Nesvizhskii et al, 2003) confidence value >0.80 (corresponding to a 1% false discovery rate in this analysis) and determined by the statistical analysis of interactomes (SAINT) algorithm (Liu et al, 2010; Choi et al, 2011) to be bona fide interactors with a confidence value >0.95 are presented in Figure 1A, Supplementary Figure 1A, and Supplementary Tables 1 and 2. A range of 4 to >300 peptides were identified for each of the interactors, with an average of 12. In total, 452 high-confidence interactions, encompassing 321 unique proteins, were identified. (n.b. This type of purification strategy is designed to preserve protein complexes, and thus identifies both direct and indirect protein–protein interactions.)
Figure 1.
(A) Functional organization of the budding yeast SUMO system. AP-MS was conducted to identify SUMO system component interactors. Large red nodes indicate proteins used as ‘baits’. Smaller nodes indicate interactors (‘prey’). Edge width is proportional to the average number of peptides identified for each prey protein. Square nodes indicate interactions confirmed using a second method. (B–E) Close-up of selected sub-networks. (B) Siz1 and Ubc9 localize to the septin ring during mitosis, and interact with septin proteins in our AP-MS. (C) Ulp1 localizes to the nuclear pore complex via interactions with several different karyopherins. (D) Ulp2 and Ubc9 interact with a number of nucleolar proteins, including components of the RENT and cohibin complexes. (E) The Siz1 and Siz2 interactomes are enriched for proteins involved in transcriptional control and chromatin remodeling. (F) Verification of Siz1 and Siz2 interactions via co-immunoprecipitation. GFP strains were transformed with Siz1- or Siz2-HA-ProtA mORF plasmids. GFP affinity purification was conducted, followed by immunoblotting using an antibody directed against HA (upper panel). An anti-GFP antibody was used to track the efficiency of each pulldown (middle panel), and whole-cell lysates (WCL) were evaluated with anti-HA to track the efficiency of protein induction (lower panel). A strain lacking GFP was used as negative control (WT). Siz1 and Siz2 migration are indicated by arrowheads. The asterisk indicates a non-specific band. (G) V5-tagged proteins identified as putative substrates in the AP-MS study were expressed in wt, siz1Δ and siz2Δ cells. Proteins of interest were subjected to V5 IP and western analysis, using antibodies directed against SUMO (top) or the V5 epitope (bottom).
Consistent with our earlier synthetic genetic array dataset (Makhnevych et al, 2009) and a more recent study of SUMO chain function in budding yeast (Srikumar et al, 2013), gene ontology analysis highlighted significant enrichment for proteins involved in a number of biological functions, including ribosome biogenesis, chromatin remodeling, nuclear-cytoplasmic trafficking, transcriptional regulation and bud site selection (Supplementary Table 3). SUMO system interactors were significantly enriched in proteins previously reported to be SUMO targets (77/321 proteins, P<0.0001; Supplementary Figure 1B). In total, our mapping effort increased the number of SUMO system interacting partners >10-fold (Supplementary Figure 1C) and for the first time, linked each putative target/regulatory protein with specific components of the SUMO machinery.
The SUMO E2 and E3s
Ninety-seven high-confidence Ubc9 interacting proteins were identified in our analysis. Four of these proteins were previously reported to be Ubc9 interactors, and 37 previously identified as SUMO conjugates (Supplementary Table S1). Similar to the SUMO system as a whole, GO enrichment for Ubc9 interactor functions included ribosome biogenesis, bud site selection, nuclear export, translation initiation and chromatin remodeling.
The Siz1 (77 interactors) and Siz2 (76 interactors) interaction maps displayed ∼25% overlap (Supplementary Table 4), consistent with previous reports suggesting that the Siz-type E3s target both shared and unique substrates (Reindle et al, 2006). Both Siz1 and Siz2 interacted with proteins involved in transcriptional control, DNA replication and chromatin remodeling (e.g., Spt16, Set1, Hst1, Rfc1 and Rvb2; Figure 1E). As expected, Siz1 (but not Siz2) specifically interacted with the septin complex in our analysis (Siz1 and Ubc9 localize to the septin ring in mitotic cells; Figure 1B; Johnson and Gupta, 2001; Takahashi et al, 2001). Siz1 also interacted with histone chaperones, chromatin remodeling proteins and transcriptional co-repressors; e.g., Tup1, Sum1, Hap1 and components of the Swi/Snf and Ino80 protein complexes (Figures 1A and E). Siz2 did not interact with Tup1, Sum1 or Hap1, but specifically interacted with components of the RNA Pol I, II and III core transcription complexes (Figure 1E).
Very few Siz1- or Siz2-specific targets have been characterized to date. We thus validated several of the Siz1/2 binding partners identified in our AP-MS analysis via co-immunoprecipitation (IP)-western analysis (Figure 1F). Strains expressing GFP-tagged fusion proteins (chromosomally tagged at the C-terminus of each ORF, under the control of their own promoters; Huh et al, 2003) were transfected with Siz1- or Siz2-HA-ProtA mORF plasmids, as above. GFP pulldowns were conducted, and proteins eluted with Laemmli buffer. Western blotting with an anti-GFP antibody (Invitrogen HRP-conjugated anti-GFP) was used to track the efficiency of each pulldown, and an anti-HA antibody (Covance HA.11) was used to assay for the presence of Siz1 and Siz2. As expected, Siz1 interacted specifically with the septin component Cdc12 (Figure 1F, lanes 3 and 4). Consistent with our AP-MS data, Siz1 also specifically interacted with the transcriptional co-regulators Sum1 and Tup1 (lanes 5–8). No Siz2 protein was detected in these immunoprecipitates. Conversely, Siz2 interacted specifically with the DNA topoisomerase Top2 (lanes 9 and 10), and the RNA Pol II core component Rpb3 (lanes 11 and 12). Finally, also consistent with our AP-MS data, both Siz1 and Siz2 interacted with the FACT component Spt16 (lanes 13 and 14).
We next wished to validate the specific role of each Siz-type E3 ligase in the sumoylation of a set of predicted substrates. Since the levels of SUMO-modified proteins under standard laboratory growth conditions can be quite low (for most SUMO substrates, only a few percent of the protein is modified at any given time; Johnson, 2004; Hay, 2005), we analyzed three Siz1/2 interactors identified in our AP-MS screen that were previously reported to be sumoylated in response to environmental stress (Zhou et al, 2004; Takahashi et al, 2008). As expected, when wt cells expressing V5-tagged Tup1, Top2 or Rpo21 were subjected to hyperosmotic shock (1 M NaCl for 15 min), a significant increase in sumoylation was observed for all three proteins (Figure 1G). Consistent with our interaction mapping results, Tup1 was sumoylated in NaCl-treated siz2 cells, but much less efficiently modified in NaCl-treated cells lacking Siz1 (Figure 1G). Conversely, Top2 and Rpo21 were robustly sumoylated in NaCl-treated wt and siz1 cells, but not in cells lacking Siz2 (Figure 1G). As predicted by our AP-MS study, Top2 and Rpo21 sumoylation is thus dependent on Siz2, whereas Tup1 SUMO modification is largely dependent on Siz1.
Together, these results highlight the quality of our interactome data, and suggest that while both Siz-type SUMO E3s are likely to be important for transcriptional control, they appear to regulate different components of the transcription machinery. Further study will be required to understand the specific contributions of each SUMO E3 ligase to transcriptional control.
The SUMO-specific proteases
The Ulp1 and Ulp2 interactomes were almost completely non-overlapping (<10% shared interactions; Supplementary Table 4). These results agree with earlier data indicating that the two budding yeast SUMO-specific proteases display very different intracellular localization patterns (Li and Hochstrasser, 2000; Makhnevych et al, 2007) and appear to target different substrates (Panse et al, 2003).
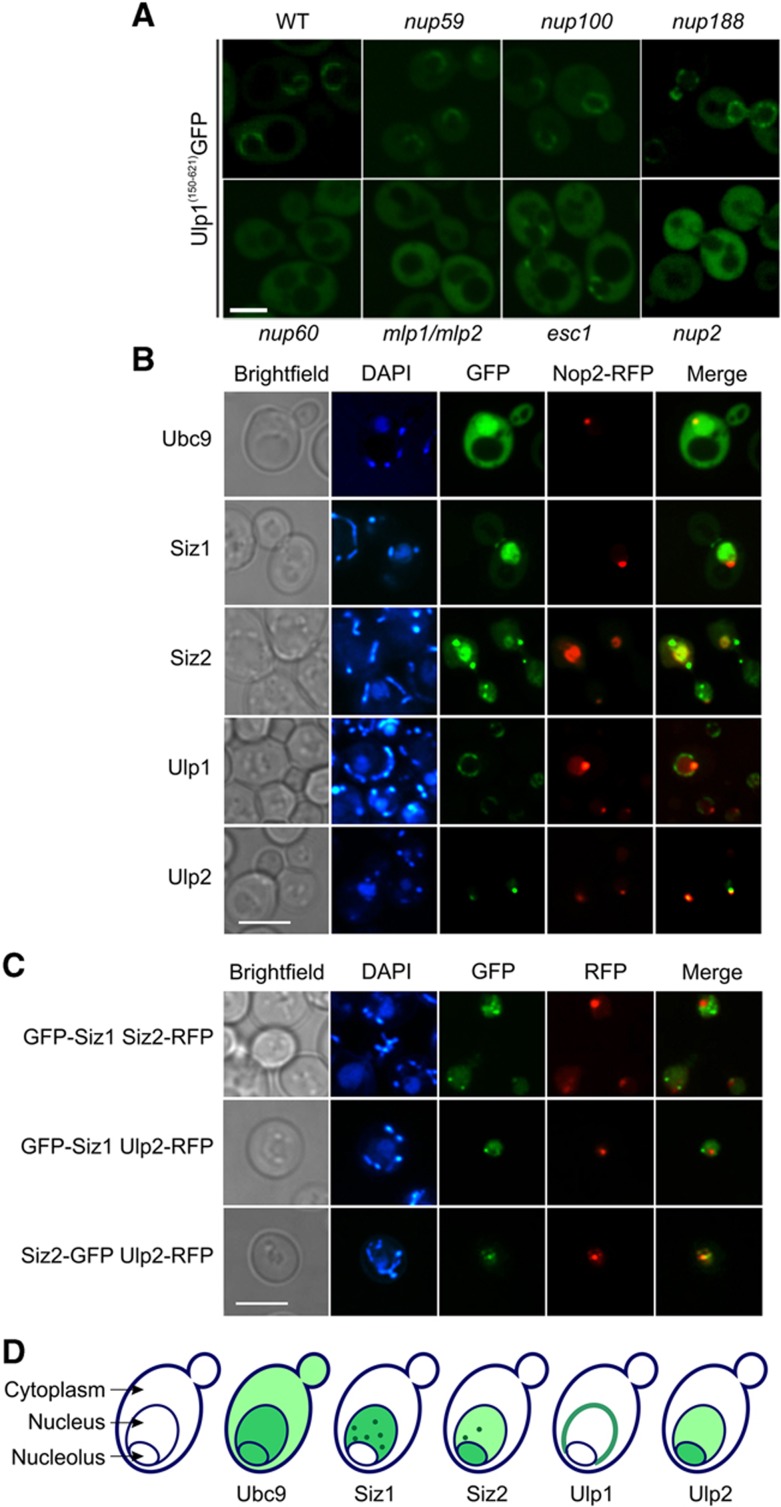
Ulp1 is tethered to the nuclear face of the nuclear pore complex (NPC) via unconventional interactions with the karyopherins Kap121 and Kap95/Kap60 (Panse et al, 2003; Makhnevych et al, 2007). Our AP-MS data agree with these earlier reports (Figure 1C). Interestingly, however, we also detected a previously unreported NPC-associated Ulp1 interacting partner, Nup2 (Figure 1C). While several different NPC components have been demonstrated to be required for proper Ulp1 localization (Panse et al, 2003; Makhnevych et al, 2007), Nup2 was not previously implicated in Ulp1 function. To explore the role of this interaction in Ulp1 localization, we expressed a Ulp1–GFP protein fragment (Ulp1150–621GFP) that lacks the Kap121 binding site and localizes to the NPC in a Kap95/Kap60- and Nup60-dependent manner (Makhnevych et al, 2007). Consistent with previous data, the Ulp1150–621GFP protein is mislocalized in nup60, mlp1/2 and esc1 deletion strains (Figure 2A). Ulp1150–621GFP localization is unaffected in strains lacking other nuclear pore components, but is completely mislocalized in nup2 deletants (Figure 2A). These data for the first time implicate Nup2 in Ulp1 localization.
Figure 2.
Physical organization of the SUMO system. (A) Nup2 is required for proper Ulp1 localization. Ulp1150–621GFP is mislocalized in nup60, mlp1/mlp2, esc1 and nup2 deletants, but not in nup59, nup100 or nup188 deletion strains. Scale bar=5 μm. (B) Localization of GFP-tagged SUMO system components relative to the RFP-tagged nucleolar marker Nop2. (C) Cells were co-transfected with the indicated plasmids and imaged, as above. (D) Summary of localization for each SUMO system component. DAPI, 4′,6-diamidino-2-phenylindole; scale bar=5 μm.
The Ulp2 interactome was significantly enriched in proteins involved in ribosome biogenesis, nucleolar maintenance and ribosomal DNA (rDNA) transcription, suggesting that this SUMO-specific protease may have an important role in nucleolar function (see below).
A role for the SUMO system in rDNA organization and maintenance
Live cell microscopy
Several earlier reports have linked the SUMO system to nucleolar function (Heun, 2007; Takahashi et al, 2008), but the molecular details of this relationship were not well understood. As our AP-MS data highlighted interactions between SUMO system components and a number of proteins that are important for ribosome biogenesis (Figure 1A and Supplementary Table 3), we scrutinized this sub-network of proteins more closely.
The intracellular localization of SUMO system components is thought to have an important role in their specificity (Gong et al, 2000; Nishida et al, 2000; Zhang et al, 2002; Johnson, 2004; Di Bacco et al, 2006; Kerscher et al, 2006; Kroetz et al, 2009). Immunofluorescence microscopy was previously used to map the intracellular localization of the SUMO system components (Strunnikov et al, 2001; Takahashi et al, 2001; Kroetz et al, 2009). However, very little information was available regarding nucleolar localization. To better characterize nucleolar localization of the yeast SUMO system components, we used live cell confocal microscopy and analyzed co-localization with the nucleolar marker Nop2. Consistent with previous reports (Huh et al, 2003), GFP-Ubc9 displayed a diffuse signal throughout the cytoplasm and nucleus, including the nucleolus (Figure 2B). GFP-Siz2 was also present throughout the nucleus, and was enriched in a small number of nuclear foci and the nucleolus (Figure 2B). GFP-Ulp2 displayed a nuclear signal (Strunnikov et al, 2001; Kroetz et al, 2009) and partially co-localized with Nop2, indicating that it is also present (and actually enriched in some cells) in the nucleolus (Figure 2B). Finally, GFP-Siz1 also exhibited a diffuse nuclear signal (as reported in Johnson and Gupta, 2001; Takahashi et al, 2001) along with multiple nuclear foci (the function and composition of which remain unknown). Notably, however, GFP-Siz1 displayed no overlap with Nop2-RFP (Figure 2B). This was not previously reported, and suggested that this SUMO E3 ligase is excluded from the nucleolus.
This observation was confirmed using multiple tagging configurations (GFP versus RFP, fused to both the N- and C-termini; Supplementary Figure 2), and co-expression of pairwise combinations of exogenous and endogenous GFP/RFP-tagged Ubc9, Ulp2, Siz1 and Siz2 (Figures 2C and D and Supplementary Figure 2). Indeed, this analysis confirmed for the first time that while Siz2, Ubc9 and Ulp2 are partially localized to the nucleolus, Siz1 is specifically excluded from this organelle.
A new model for SUMO-mediated regulation of rDNA organization
The Fob1 protein binds to replication fork barrier sites in the non-transcribed spacer region 1 of rDNA, which separate the ∼150 rDNA tandem repeats on chromosome XII in budding yeast (Johzuka and Horiuchi, 2009). Fob1 also interacts with the Tof2 protein and the RENT complex (Net1, Cdc14 and Sir2). Via interactions with Tof2 and RENT, the cohibin complex (Lrs4 and Csm1) acts as a bridge to link the rDNA to Src1/Heh1 and Nur1, polypeptides that are anchored at the inner nuclear membrane (INM; Chan et al, 2011). These interactions are required for the preservation of nucleolar structure, rDNA silencing, and the maintenance of rDNA copy number (Chan et al, 2011).
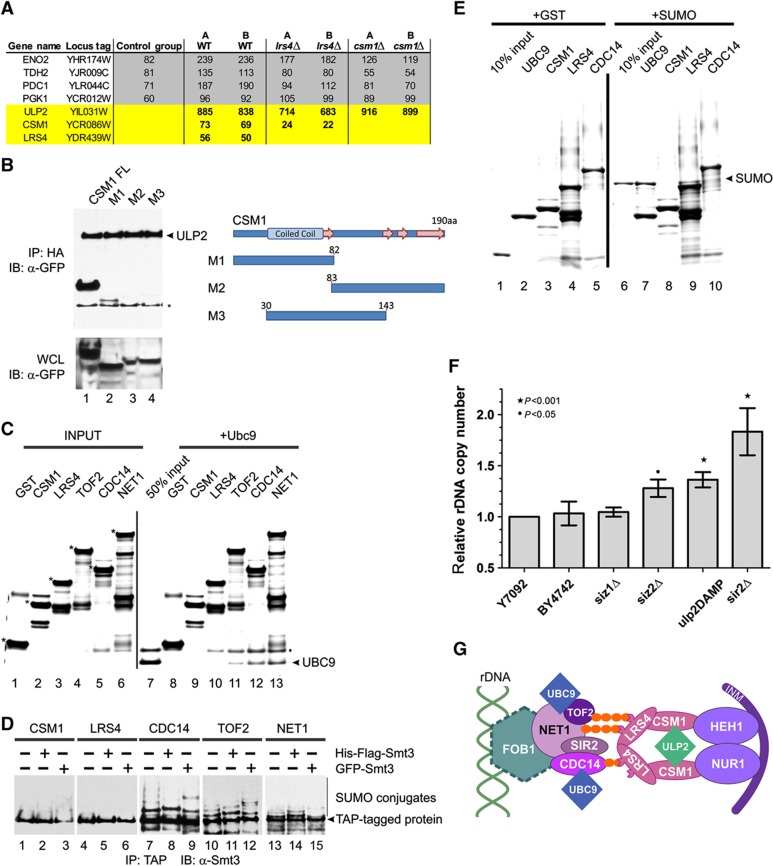
Our AP-MS analysis revealed an interaction between Ulp2 and cohibin (Figure 1D). To begin to characterize the molecular architecture of the Ulp2-cohibin complex, we expressed a Ulp2-HA-ProtA protein in wt yeast, and lrs4 and csm1 deletion strains. While the Csm1 protein was isolated in Ulp2-HA AP-MS from lrs4 deletants, neither Lrs4 nor Csm1 were detected in Ulp2-HA pulldowns from the csm1 knockout strain (Figure 3A). These data suggest that Ulp2 interacts directly with Csm1 in the context of the cohibin complex, and are consistent with a previously published yeast two-hybrid analysis identifying Ulp2 as a Csm1 binding partner (Wysocka et al, 2004). We validated these data by co-IP western analysis (Figure 3B). By expressing several different deletion mutants in csm1 cells, we were also able to map the Ulp2 binding region of Csm1 to its N-terminus (aa 1–30; Figure 3B).
Figure 3.
A role for the SUMO system in rDNA organization. (A) Ulp2 binds to the cohibin complex via Csm1. Ulp2-HA was purified from wt, lrs4Δ or csm1Δ strains and subjected to LC-MS/MS. Shown are spectral counts for several non-specific background proteins (control group; full data set in Supplementary Table S1), as well as Ulp2, Csm1 and Lrs4. (B) Ulp2 binding assay. Ulp2-HA-ProtA and Csm1–GFP (either full-length or truncation mutants M1–M3, as indicated) were co-expressed in wt yeast. Ulp2-HA was immunopurified, and western blotting conducted using an antibody directed against GFP (n.b. this antibody recognizes both GFP and the protein A moiety in the HA tag). (C) Ubc9 binding assay. Purified recombinant proteins (full-length protein indicated by asterisk, left panel) were incubated with recombinant Ubc9 protein (+Ubc9, right panel), and washed several times with binding buffer. Bound proteins were eluted with Laemmli buffer and subjected to 4–12% SDS–PAGE. The gel was stained with Coomassie blue. The location of Ubc9 is indicated with an arrowhead. (D) In vivo sumoylation assay. HisFlag- or GFP-tagged SUMO protein was expressed in the indicated TAP strains. TAP proteins were immunopurified, and subjected to western blotting using an antibody directed against yeast SUMO (this antibody recognizes both SUMO and the protein A moiety in the TAP tag). Position of TAP-tagged proteins is indicated with an arrowhead. (E) In vitro SUMO binding assay. As in (C), recombinant GST-tagged proteins (as indicated) were purified and incubated with GST or the GST–SUMO protein, washed, eluted with Laemmli buffer and visualized on a coomassie-stained polyacrylamide gel. (F) rDNA copy number (relative to the wild-type strain Y7092) was measured by qPCR using the ΔΔCt method. Reactions were performed in triplicate; error bars indicate standard deviation. (G) Model: SUMO conjugation/deconjugation has a role in the interaction between rDNA repeats and the INM. Note: the stoichiometry of this complex, SUMO conjugation sites and SUMO binding sites remain to be identified.
Our AP-MS analysis also indicated that Ubc9 interacts with Tof2 and Net1 (Figure 1D). Consistent with these data, we found that a recombinant Ubc9 polypeptide can interact directly with both of these proteins in vitro, but does not interact with Lrs4 or Csm1 under the same conditions (Figure 3C). To determine the sumoylation state of Tof2, RENT and cohibin in vivo, yeast SUMO proteins with two different N-terminal epitope tags (6xHis-Flag and GFP) were expressed in strains containing chromosomally tagged TAP (tandem AP) proteins (Ghaemmaghami et al, 2003). TAP was conducted (as in Sydorskyy et al, 2010) followed by western analysis using an antibody directed against the yeast SUMO protein (Figure 3D; n.b. this antibody detects both SUMO and the protein A moiety in the TAP tag). Consistent with our binding data (and with earlier proteomics screens for yeast SUMO targets; Wohlschlegel et al, 2004; Denison et al, 2005), we observed that Net1, Cdc14 and Tof2 are all sumoylated (Figure 3D). Consistent with the presence of a SUMO protease (Ulp2) in the complex and a lack of Ubc9 binding in vitro, no SUMO conjugation was detected on Lrs4 or Csm1 (Figure 3D).
We also conducted in vitro SUMO binding assays with this group of proteins (as in Makhnevych et al, 2009). As expected, while Ubc9 displayed no interaction with GST alone, it strongly interacted with a GST–SUMO protein (Figure 3E, lane 7). Csm1 and Cdc14 did not interact with GST or GST–SUMO (lanes 8 and 10). Notably, however, while Lrs4 did not interact with GST alone (lane 4), it displayed a highly reproducible interaction (albeit weaker than Ubc9) with the GST–SUMO protein (lane 9).
Alterations in nucleolar structure can affect the maintenance of rDNA repeats: e.g., lrs4 and csm1 deletion mutants display altered rDNA repeat copy number (Mekhail et al, 2008). To determine whether SUMO system components are also required for rDNA repeat maintenance, we analyzed rDNA repeat numbers in wt, siz1, siz2 and ulp2-damp strains (DAmP; decreased abundance by mRNA perturbation; Yan et al, 2008) using quantitative PCR, as in (Johzuka and Horiuchi, 2009). Consistent with our localization data, the siz1 deletion strain displayed no change in rDNA copy number as compared to two standard parental laboratory strains (Figure 3F). Notably, however, the siz2 deletant displayed a 28±8% increase (P<0.05) in rDNA copy number, and the ulp2-damp strain displayed a 36±8% increase in rDNA copies (P<0.001; Figure 3F).
Together, our data indicate that: (i) Tof2 and the RENT complex components Net1 and Cdc14 can interact with Ubc9 in vitro and are sumoylated in vivo; (ii) Lrs4 is a SUMO binding protein; (iii) Csm1 interacts with the SUMO protease Ulp2; (iv) the Siz2, Ubc9 and Ulp2 proteins are partially localized to the nucleolus, whereas Siz1 is specifically excluded from this organelle, and (v) while deletion of the siz1 gene has no effect, deletion of the siz2 gene or decreased expression of Ulp2 significantly affects rDNA repeat copy number. Together, these findings suggest that SUMO conjugation and deconjugation (as mediated by Ubc9, Siz2 and Ulp2) have a role in the interaction between rDNA repeats and the INM (Figure 3G). Further study will be required to refine this model.
In summary, our S. cerevisiae protein–protein interaction map represents the first description of the functional organization of the SUMO system in any organism. As the SUMO machinery is conserved throughout the plant and animal kingdoms, many of the functional and physical interactions identified here are likely to be relevant for all eukaryotes.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strains used in this study were derivatives of BY4741/2 haploid cells unless otherwise specified, and are listed in Supplementary Table 5. Yeast genetic manipulations were performed according to established procedures. Unless otherwise noted, yeast strains were grown at 30°C to mid-logarithmic phase in YPD or selective minimal medium supplemented with appropriate nutrients and 2% glucose. Transformations were performed as described previously (Delorme, 1989).
Cloning
The coding sequences of Ubc9, Siz1, Siz2, Ulp1, Ulp2, Nup60 and Nop2 were PCR amplified from genomic DNA and incorporated into the pDONR201 vector using BP clonase II (Invitrogen), according to the manufacturer’s instructions. Sequence-verified entry vectors were used in LR reactions with BG1805 (Gelperin et al, 2005), pAG416/5-GPD-ccdB-GFP/DsRED or pAG416/5-GPD-GFP-ccdB (Alberti et al, 2007) destination vectors.
Affinity purification
Coding sequences for Ubc9, Siz1, Siz2, Ulp1 and Ulp2 in the vector BG1805 were transformed into BY4742 and Y7092 cells (Gelperin et al, 2005). Colonies were isolated and expression of epitope-tagged proteins confirmed by western blotting analysis of the whole-cell lysate from 5 ml overnight (O/N) cultures grown in CSM Ura with 2% galactose. For AP-MS experiments, O/N cultures in CSM Ura- (with 2% raffinose) were inoculated two nights before the experiment. Cultures were diluted the next morning and maintained in log phase throughout the day. Two hundred fify milliliters of CSM Ura- with 2% raffinose and 0.05% galactose were inoculated with logarithmically growing cultures, to obtain an OD600 ∼0.6 culture density in ∼16 h. Cultures were collected by centrifugation, transferred to 2 ml flat top tubes with 1 ml ice-cold water and pelleted. Pellets were snap frozen in an ethanol-dry ice bath. Frozen pellets were resuspended in 2 volumes (∼500 μl) of lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, 5 mM DTT, 10 mM NEM and 1 × Sigma fungal protease inhibitor) and snap frozen again. Five hundred microliters of glass beads and 250 units of benzonase nuclease were added to each tube, and cells were mechanically lysed with a beadbeater (BioSpec Products) using 4 × 5 min pulses (with 5 min breaks on ice). Tubes were centrifuged at 16k RCF for 10 min and the supernatant transferred to a chilled eppendorf tube. A 6 μl slurry of Dynal ProteinG beads (Invitrogen), pre-incubated with 2.5 μg of HA.11 (Covance Clone 16B12) and equilibrated with lysis buffer, was added to each cell lysate and incubated end-over-end for 1 h. Beads were gently washed once with 500 μl of lysis buffer and all residual lysis buffer carefully removed to avoid introduction of detergent into the mass spectrometer. Proteins were eluted by sequential elution (4 × ) with 25 μl of freshly prepared 125 mM ammonium hydroxide (pH 11). Pooled eluates were centrifuged, placed on a magnetic holder and the supernatant carefully transferred to a new eppendorf tube to avoid bead carryover. The eluate was then lyophilized and digested with 1 μg of trypsin (Promega, MS grade) in 200 μl of 50 mM ammonium bicarbonate (pH 8.3) for 16 h at 37°C. Trypsin (0.5 μg) was added and incubated for an additional 4 h. Samples were lyophilized, and peptides resuspended in 35 μl of 0.1% formic acid. Five microliters of the sample was analyzed per MS run.
Lysates for GFP APs were performed as above, except that 150 ml cultures were used. Clarified lysates were incubated with 20 μl slurry of GFP-Trap-M (ChromoTek) beads, pre-equilibrated with lysis buffer, for 1 h at 4°C. Beads were gently washed once, and proteins eluted by incubation with 20 μl of Laemmli buffer at 95°C for 10 min. Samples were then placed on a magnetic holder, and the supernatant loaded on to a 4–12% Criterion XT Bis-Tris gel (Bio-Rad). Proteins were transferred to a nitrocellulose membrane (Pall) and probed with HA.11 (Covance) antibody or anti-GFP HRP-conjugated antibody (Invitrogen). Antibody signal was visualized using ECL (Immuno-Star HRP, Bio-Rad). Goat anti-mouse HRP-conjugated secondary antibody was used to detect HA.11 binding.
Mass spectrometry
Analytical columns (75 μm inner diameter) and pre-columns (100 μm) for LC-MS analysis were prepared in-house from silica capillary tubing (InnovaQuartz, Phoenix, AZ), and packed with 3 μm 100 Å C18-coated silica particles (Burker-Michrom). Analytical columns were fitted with metal emitters (Thermo Proxeon) using zero dead volume connections. Peptides were subjected to nanoflow liquid chromatography–electrospray ionization–tandem mass spectrometry (nLC-ESI-MS/MS), using a 90-min reversed phase (10–40% acetonitrile, 0.1% formic acid) buffer gradient running at 250 nl/min on a Proxeon EASY-nLC pump in line with a hybrid linear quadrupole ion trap (Velos LTQ) Orbitrap mass spectrometer (Thermo Fisher Scientific). A parent ion scan was performed in the Orbitrap, using a resolving power of 60 000. Simultaneously, up to the 40 most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using standard CID fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a 10-p.p.m. window, exclusion list size 500) detected three times within 45 s were excluded from analysis for 30 s.
Thermo.raw files were uploaded to the ProHits (Liu et al, 2010) analytical suite and converted to .mzXML format using ReAdW software. Data were searched using X!Tandem (Craig and Beavis, 2004) against the yeast ORF (and reversed database; Saccharomyces Genome Database, December 2005). Search parameters specified a parent MS tolerance of ±15 p.p.m., and an MS/MS tolerance of 0.4 Da, with up to two missed cleavages for trypsin. Oxidation of methionine was allowed as a variable modification. Statistical validation of the results was performed using PeptideProphet and ProteinProphet (Keller et al, 2002; Nesvizhskii et al, 2003) as part of the trans-proteomic pipeline. For each search, the ProteinProphet probability at 1% error rate was used as a cutoff value to generate SAINT-compatible input files. Data for each bait protein were collapsed at the BAIT level using average spectral counts. SAINT parameters were as follows: 5000 iterations, low mode On (1), minFold 1 and no normalization (0) (Choi et al, 2011).
Significance of enrichment was calculated using Z-score test for proportions (77 known sumoylated proteins out of 321 identified by AP-MS versus 613 known sumoylated proteins out of 5045 verified yeast ORFs).
All raw files (and .mzXML files) have been uploaded to the data repository at TRANCHE and can be accessed using the following hash key: zKpG/K0LDH+UL6UbldWm0HQ2DqVfpoGPsHG6RUXn4PaB009ej1SidjWUGNcpm5GCaxfx00U7h4N/q6VnkXF5bCpaPN0AAAAAAABsjA==
Protein–protein interaction data have been submitted to BioGrid.
qPCR
Strains were grown O/N in YPD (+cloNAT 200 μg/ml or +G418 100 μg/ml for mutants), diluted in the morning to OD600 of 0.2. Cultures were grown to OD600 0.8 and 10 ml aliquots snap frozen. MasterPureYeast DNA Purification Kit (Epicenter) was used to isolate genomic DNA according to the manufacturer’s protocol. Samples were incubated with DNase-free RNase for 2 h in TE before storing at −20°C. DNA was quantified on a Nanodrop 1000 (Thermo). Power Sybr Green PCR kit was used in 20 μl reactions containing 1 ng of DNA and 50 nM of each primer as per manufacturer’s protocols, on a Stratagene Mx3000P. Primers used are: rDNA-F: 5′-TACTGCGAAAGCATTTGCCAAGGACG-3′; rDNA-R: 5′-TCCCCCCAGAACCCAAAGACTTTGAT-3′; act1-F: 5′-CTTTCAACGTTCCAGCCTTC-3′; and act1-R: 5′-CCAGCGTAAATTGGAACGAC-3′.
In vitro binding assays
Ubc9, Cdc14, Csm1 and Lrs4 were expressed as GST-fusion proteins in E. coli and captured on glutathione sepharose resin. After extensive washing in lysis buffer (see AP-MS protocol above), the sepharose-bound proteins were incubated (3 h at 4°C) with purified 15 mM GST or a 15 mM GST-SUMO fusion protein. Following several washes with lysis buffer, bound proteins were eluted with Laemmli buffer. The eluate was subjected to 4–12% SDS–PAGE and the resulting gel stained with coomassie brilliant blue.
Confocal microscopy
Mid-log phase cells were collected from 1 ml cultures, washed in water containing 2% glucose and mounted on a glass slide. Cells were imaged using a 100 × /1.40 NA PlanApo lens on an Olympus IX80 inverted microscope (Olympus Canada) fitted with a Yokogawa CSU10 spinning disk confocal scanner unit (Quorum Technologies ) and a 512 × 512 EM-CCD camera (Hamamatsu, Japan). The system was controlled with Volocity 5.5 software (Perkin–Elmer). The CCD camera was operated at maximum resolution. Exposure times, gain and sensitivity varied by protein. Settings were maintained for all subsequent images of the same strain. Further processing of images was performed using Volocity and Adobe Photoshop CS4 (Adobe Systems).
Supplementary Material
Acknowledgments
We thank Drs C Boone and B Andrews for yeast strains, and Drs A Nesvizhskii, H Choi and A–C Gingras for their expert assistance with MS data analysis, and critical reading of the manuscript. TS was funded by a Canadian Institutes of Health Research student fellowship. BR holds the Canada Research Chair in Proteomics and Molecular Medicine. Funding for this work was provided by CIHR grant MOP81268.
Author contributions: TS performed the AP-MS experiments and follow-up experiments. MCL performed the confocal microscopy experiments. BR conceived the project. BR and TS wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alberti S, Gitler AD, Lindquist S (2007) A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JN, Poon BP, Salvi J, Olsen JB, Emili A, Mekhail K (2011) Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell 20: 867–879 [DOI] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Meth 8: 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Delorme E (1989) Transformation of Saccharomyces cerevisiae by electroporation. Appl Environ Microbiol 55: 2242–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4: 246–254 [DOI] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G (2006) The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol 26: 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET (2000) Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem 275: 3355–3359 [DOI] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M (2005) Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Heun P (2007) SUMOrganization of the nucleus. Curr Opin Cell Biol 19: 350–355 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T (2009) The cis element and factors required for condensin recruitment to chromosomes. Mol Cell 34: 26–35 [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kroetz MB, Su D, Hochstrasser M (2009) Essential role of nuclear localization for yeast Ulp2 SUMO protease function. Mol Biol Cell 20: 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang J, Larsen B, Stark C, Breitkreutz A, Lin ZY, Breitkreutz BJ, Ding Y, Colwill K, Pasculescu A, Pawson T, Wrana JL, Nesvizhskii AI, Raught B, Tyers M, Gingras AC (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol 28: 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Ptak C, Lusk CP, Aitchison JD, Wozniak RW (2007) The role of karyopherins in the regulated sumoylation of septins. J Cell Biol 177: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, Raught B (2009) Global map of SUMO function revealed by protein–protein interaction and genetic networks. Mol Cell 33: 124–135 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D (2008) Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Nishida T, Tanaka H, Yasuda H (2000) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem 267: 6423–6427 [DOI] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, Hurt E (2003) Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol 5: 21–27 [DOI] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119: 4749–4757 [DOI] [PubMed] [Google Scholar]
- Srikumar T, Lewicki MC, Costanzo M, Tkach JM, van Bakel H, Tsui K, Johnson E, Brown GW, Andrews B, Boone C, Giaever G, Nislow C, Raught B (2013) Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J Cell Biol 201: 145–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Aravind L, Koonin EV (2001) Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydorskyy Y, Srikumar T, Jeram SM, Wheaton S, Vizeacoumar FJ, Makhnevych T, Chong YT, Gingras AC, Raught B (2010) A novel mechanism for SUMO system control: regulated Ulp1 nucleolar sequestration. Mol Cell Biol 30: 4452–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, Strunnikov A (2008) Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet 4: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Toh-e A, Kikuchi Y (2001) A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275: 223–231 [DOI] [PubMed] [Google Scholar]
- Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI (2004) A proteomic study of SUMO-2 target proteins. J Biol Chem 279: 33791–33798 [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR 3rd (2004) Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279: 45662–45668 [DOI] [PubMed] [Google Scholar]
- Wysocka M, Rytka J, Kurlandzka A (2004) Saccharomyces cerevisiae CSM1 gene encoding a protein influencing chromosome segregation in meiosis I interacts with elements of the DNA replication complex. Exp Cell Res 294: 592–602 [DOI] [PubMed] [Google Scholar]
- Yan Z, Costanzo M, Heisler LE, Paw J, Kaper F, Andrews BJ, Boone C, Giaever G, Nislow C (2008) Yeast Barcoders: a chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat Meth 5: 719–725 [DOI] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ (2002) Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mole Cell Biol 22: 6498–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H (2004) Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279: 32262–32268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.