Abstract
Extradiol catechol ring-cleaving dioxygenases function by binding both the organic substrate and O2 at a divalent metal center in the active site. They have proven to be a particularly versatile group of enzymes with which to study the O2 activation process. Here, recent studies of homoprotocatechuate 2,3-dioxygenase (HPCD) are summarized with the objective of showing how Nature can utilize the enzyme structure and the properties of the metal and the substrate to select among many possible chemical paths to achieve both specificity and efficiency. Possible intermediates in the mechanism have been trapped by swapping active site metals, introducing active site amino acid substituted variants, and using substrates with different electron donating capacities. While each of these intermediates could form part of a viable reaction pathway, kinetic measurements significantly limit the likely candidates. Structural, kinetic, spectroscopic and computational analysis of the various intermediates shed light on how catalytic efficiency can be achieved.
Introduction
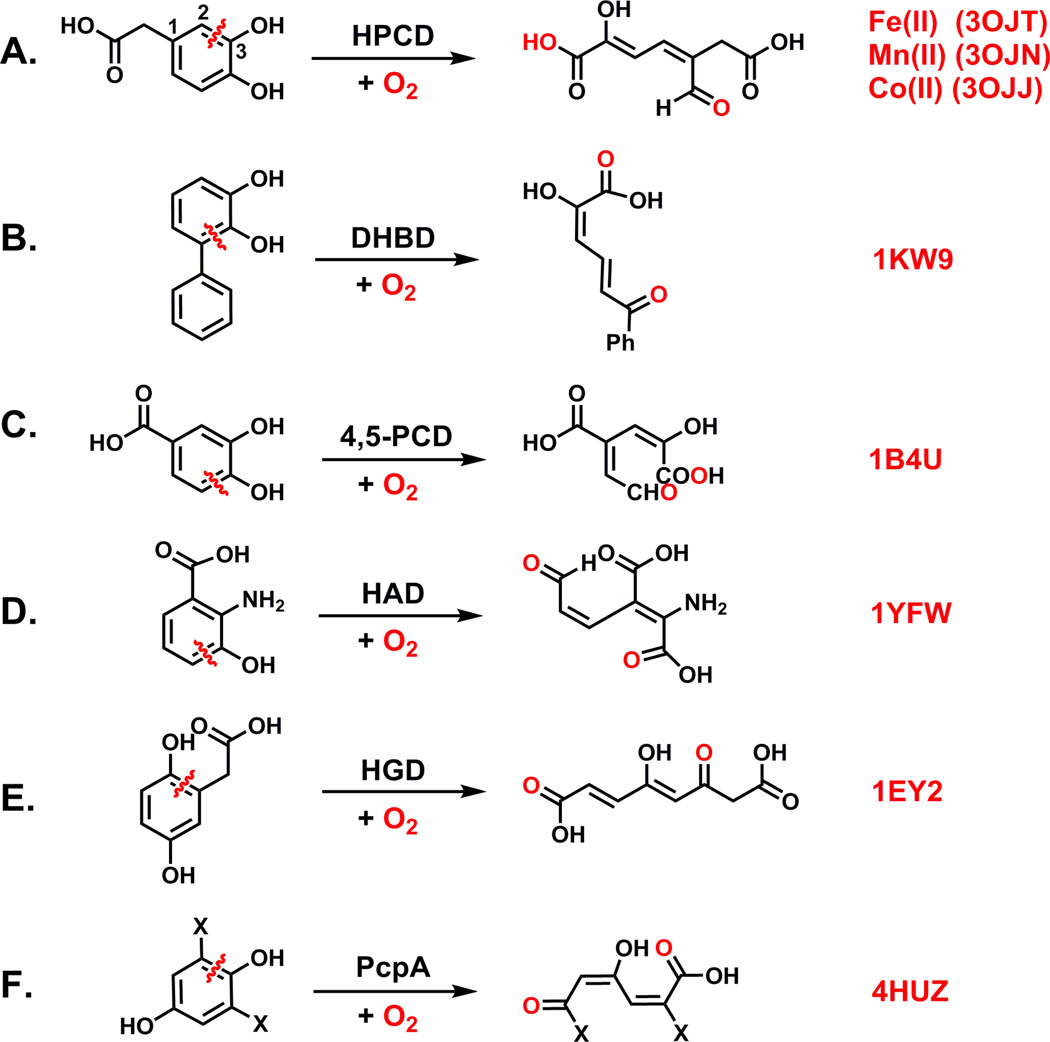
The extradiol cleaving catechol dioxygenases constitute a part of nature’s strategy for recycling the carbon atoms of aromatic molecules that naturally occur in the environment as a result of biodegradation. These enzymes are most often found in soil bacteria and serve to transform a range of substrates with 1,2-dihydroxybenzene moieties into ring-cleaved aliphatic products that eventually re-enter the Krebs cycle (Scheme 1A–C) [1]. Mechanistically similar enzymes that utilize substrates in which one of ortho-hydroxyl groups is replaced by an amine (Scheme 1D) [2] or where the hydroxyl groups are para to one another (Scheme 1E&F) have also been studied [3, 4]. Other extradiol cleaving catechol dioxygenases are involved in natural product biosynthesis pathways [5]. In humans, related dioxygenases are involved in the metabolism of aromatic amino acids, where mutations are associated with several severe diseases including the neurodegenerative disorder Huntington’s chorea and degenerative arthritis [6, 7].
Scheme 1.
Examples of reactions catalyzed by extradiol-cleaving catechol dioxygenases. (A) HPCD, homoprotocatechuate 2,3-dioxygenase [9]; (B) DHBD, 2,3-dihydroxybiphenyl 1,2-dioxygenase [10]; (C) 4,5-PCD, protocatechuate 4,5-dioxygenase [11]; (D) HAD, 3-hydroxyanthranilate-3,4-dioxygenase [2]; (E) HGD, homogentisate 1,2-dioxygenase[12]; (F) PcpA, 2,6-dichloro-p-hydroquinone 1,2-dioxygenase [4]. Four-letter codes adjacent to each reaction represent PDB files for respective enzymes.
The oxygenation of the dihydroxybenzene ring catalyzed by the extradiol cleaving catechol dioxygenases results in the cleavage of the C–C bond adjacent to a hydroxyl functionality to generate a carboxylate and an aldehyde on opposite ends of the ring-cleaved product (Scheme 1). This cleavage is distinct from that carried out by the intradiol cleaving catechol dioxygenases, which cleave the C–C bond of the enediol unit to form a dicarboxylate product [1]. The intradiol enzymes require an iron(III) center in the active site, while all of the extradiol enzymes have an active site metal(II) center [1]. Manganese(II) is bound instead of iron(II) in a small number of as-isolated extradiol cleaving enzymes [8]. The latter observation demonstrated for the first time that O2 activation can occur at a biological manganese center. Following these initial discoveries, methods were developed to replace the native metal in extradiol dioxygenases with a different metal [8, 9]. Some metal-substituted enzymes have also proven to be effective at carrying out oxidative cleavage. In this mini-review, we focus on the mechanistic insights gained through the study of wild-type and metal-substituted homoprotocatechuate 2,3-dioxygenases together with variants and alternative substrates.
Homoprotocatechuate 2,3-dioxygenase
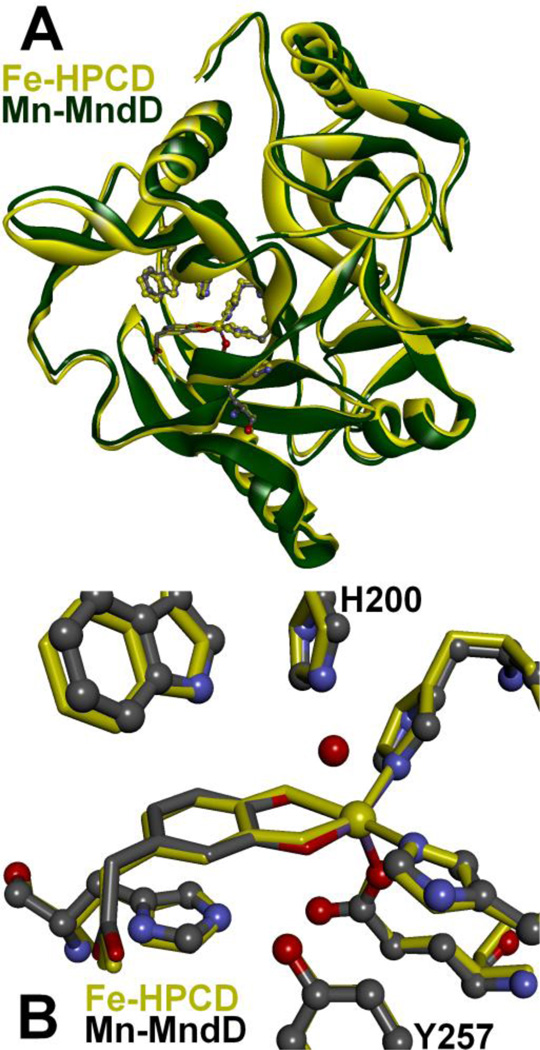
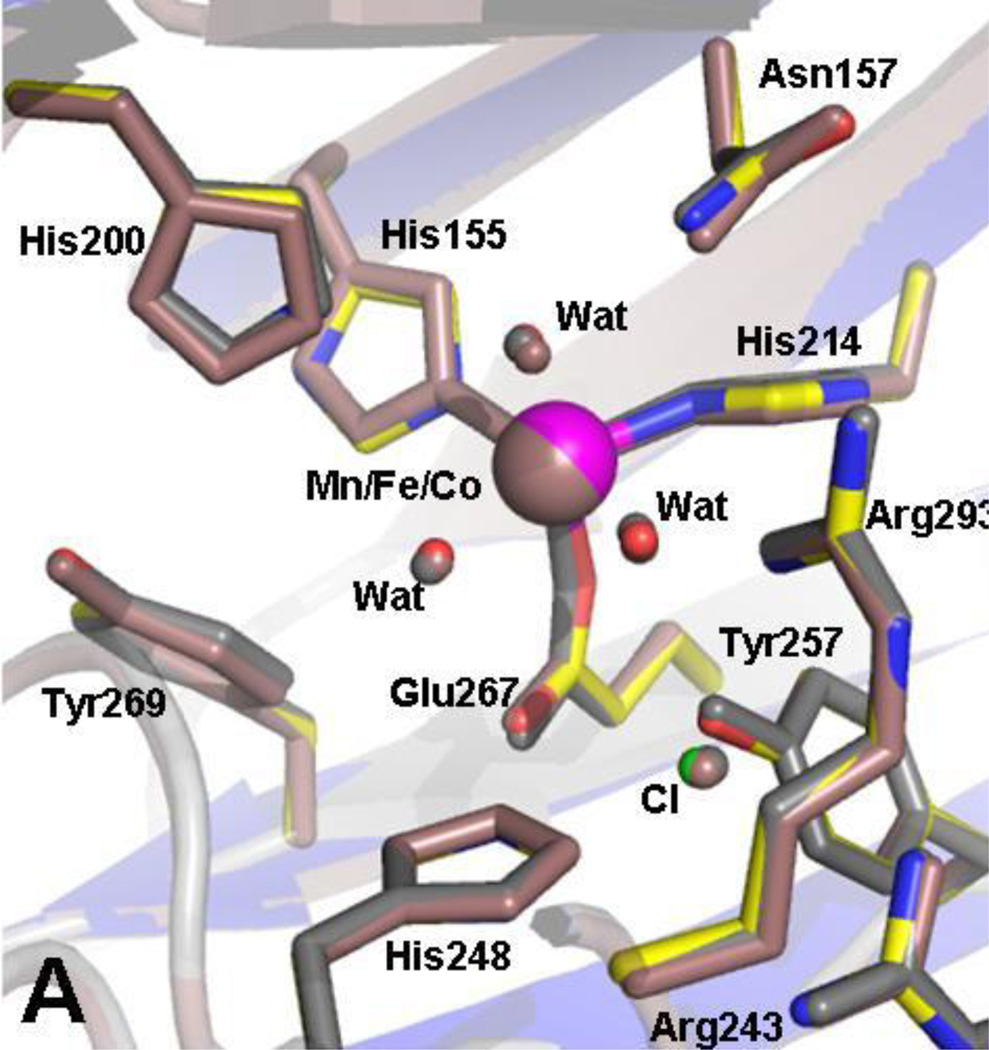
Homoprotocatechuate 2,3-dioxygenases are the best characterized of the extradiol cleaving catechol dioxygenases studied to date. They catalyze the cleavage of the C2-C3 bond (Scheme 1A; See Scheme 2 for numbering convention) of homoprotocatechuate (HPCA) with the incorporation of both oxygen atoms of O2 into the ring-cleaved product [13]. Crystal structures of the iron-containing enzyme from Brevibacterium fuscum (HPCD) and the manganese-containing enzyme from Arthrobacter globiformis (MndD) have been solved and reveal a remarkable structural similarity [8, 14, 15]. Both are homotetrameric enzymes that belong to the Type I family of extradiol dioxygenases (vicinal oxygen chelate superfamily [16]) with 83% sequence identity. Each monomer has four βαβββ modules forming structurally homologous N-terminal and C-terminal barrel-shaped domains (Fig. 1A). The metal center in each case is located in the C-terminal barrel and is bound in a 2-His-1-carboxylate facial triad motif [17–19] commonly found among O2 activating nonheme iron enzymes. The remaining coordination sites are occupied by two or three water molecules. The first and second coordination spheres of these enzymes are virtually identical (root mean square difference over all atoms, 0.19 Å). The substrate 2,3-dihydroxyphenylacetate (HPCA) binds to the metal in a bidentate fashion via the two hydroxyl functions at sites trans to the two His ligands. Spectroscopic and crystallographic studies show that only the C2 hydroxyl is likely to be deprotonated in the complex [15, 20, 21]. The remaining metal coordination site trans to the Glu residue in the facial triad is available for binding O2 pending release of any blocking solvent. A conserved second sphere His residue (His200) lies in close proximity to both the bound substrate and the O2 binding site in each enzyme. Similarly, Tyr257 is conserved in both enzymes and hydrogen bonds to the deprotonated C2-hydroxyl of the bound substrate.
Scheme 2.
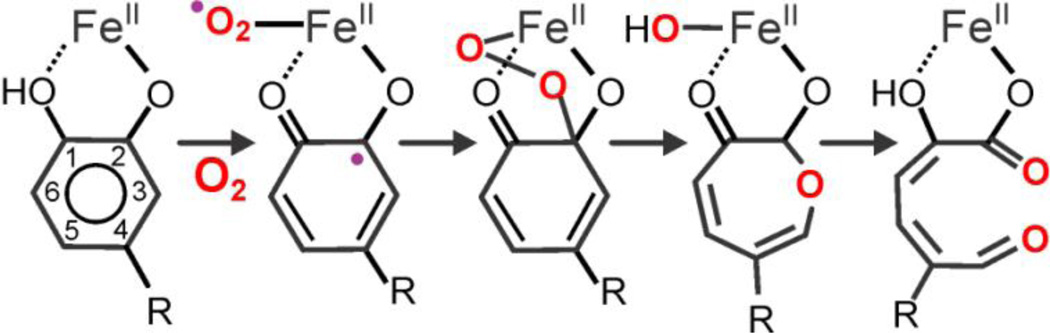
Original working mechanism for extradiol cleavage of catechols based on early observations of Lipscomb, Que, and Bugg [8, 30–35].
Fig. 1.
Overlays of the crystal structures of the enzyme-substrate complexes, [Fe-HPCD(HPCA)] (1.50 Å, PDB 4GHG)[22] and [Mn-MndD(HPCA)] (1.90 Å, PDB 1F1V) [15], showing (A) the protein fold and (B) the first and second coordination spheres.
Figure 1B illustrates the nearly superimposable Fe and Mn active sites in the two enzyme-substrate complexes [15, 22]. Steady-state kinetic measurements show that the two enzymes have similar kcat, KMO2, and KMHPCA values for HPCA turnover as shown in Table 1 [8, 13, 23]. The likelihood that the metal center in both enzymes can transfer electron density to O2 as it binds to the metal during the activation process presents an intriguing mechanistic conundrum. How can these two enzymes, which employ metal ions with standard aqueous M(III/II) redox potentials that differ by 0.79 V (Eo = 0.77 V vs SHE for the Fe(III/II) couple and 1.56 V for the Mn(III/II) couple [24]), carry out electron transfer to O2 with equal efficiency as measured by the equivalent kcat/KMO2 ratios? In most cases, the answer would be that variations in the protein structures tune the potentials of the metal ions to be similar, as seen for Fe/Mn superoxide dismutases [25–27]. However, the virtually identical structures of Fe- and Mn-HPCD suggest that Nature has found another solution [8].
Table 1.
Steady-state kinetic parameters of HPCDs and MndDs.
| Enzyme |
KMHPCA (μM) |
KMO2 (μM) |
kcat/[metal] (min−1) |
kcat/KMHPCA (μM−1 min−1) |
kcat/KMO2 (μM−1 min−1) |
|---|---|---|---|---|---|
| Fe-HPCD | 31 ± 6 | 60 | 470 ± 20 | 15 ± 2 | 7.8 ± 0.3 |
| Mn-HPCD | 35 ± 5 | 50 ± 4 | 370 ± 10 | 11 ± 2 | 7.4 ± 0.6 |
| Co-HPCD | 5 ± 1 | 1200 ± 100 | 215 ± 8a 590 ± 20b 1120 ± 70c |
43 ± 9 120 ± 20 220 ± 50 |
0.18 ± 0.02 0.49 ± 0.05 0.9 ± 0.1 |
| Mn-MndD | 14 ± 4 | 62 | 360 ± 20 | 25 ± 4 | 5.8 ± 0.3 |
| Fe-MndD | 15 ± 4 | 37 ± 6 | 420 ± 20 | 28 ± 4 | 11.4 ± 1.4 |
Metal ion substitution experiments
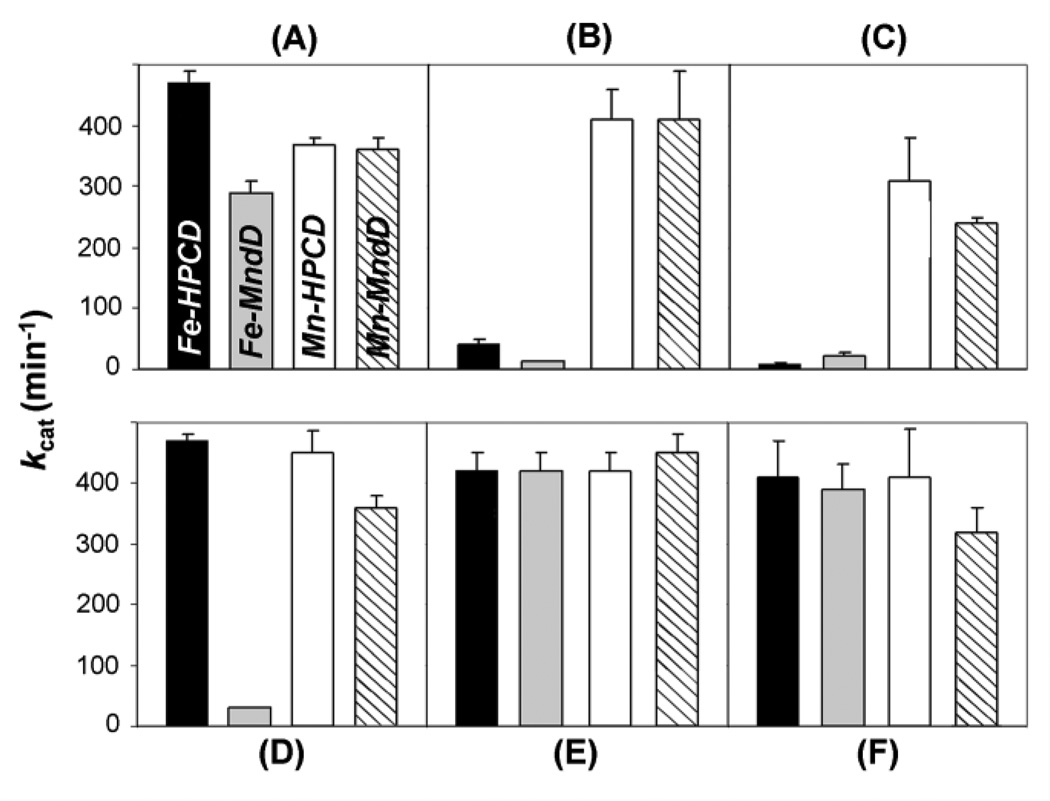
The crystallographic results reviewed above prompted metal ion substitution experiments in Fe-HPCD and Mn-MndD to make the corresponding Mn-HPCD and Fe-MndD derivatives [8]. Apoprotein reconstitution experiments with the native or alternative metal ion were unsuccessful. The successful strategy was to grow the cells in minimal media and then introduce the desired metal ion in large excess at an appropriate point in the growth cycle. This led to the formation of Mn-HPCD and Fe-MndD with very little cross contamination [8]. The four enzyme preparations could be distinguished by their respective metal contents and their responses to various oxidants and reductants (Fig. 2). In particular, note that only the Fe enzymes were sensitive to oxidation by H2O2 or K3Fe(CN)6 (Figs. 2B & 2C) and that only the Fe-MndD was easily inactivated by exposure to O2 (Fig. 2D). Presumably, this ease of oxidation is related to the low midpoint potential of the Fe(II) center. These observations emphasize that the inherently lower potential of the iron center relative to the manganese center has been retained when the metals are bound in the enzymes, as predicted by the nearly identical structures. All four forms exhibited comparable catalytic activity after treatment with ascorbate to ensure that all of the metal was reduced (Fig. 2E). This result shows that the dioxygenase active site accommodates either Fe or Mn and that either metal center can activate O2 with approximately equal efficiency.
Fig. 2.
Activation and inhibition of HPCD and MndD activity in 50 mM MOPS pH 7.8. The average activity for each enzyme is displayed as one bar within each panel. (A) Activities of as-isolated enzymes. (B and C) Data collected after the enzymes were treated with 1 mM H2O2 or K3Fe(CN)6, respectively. (D) Activities of the as-isolated enzymes after exposure to air for 1 hr. (E and F) Activity after treatment of the air-exposed enzymes in D with 1 mM sodium ascorbate and or K4Fe(CN)6, respectively. Reproduced from ref. [8] with permission.
Further metal-substitution experiments on HPCD showed that the native Fe(II) metal cofactor of HPCD could also be substituted with Co(II) to yield catalytically active Co-HPCD, despite the 1.15 V higher M(III/II) standard redox potential of Co(II) compared with Fe(II) [9]. As observed for Mn-HPCD, Co-HPCD is not inactivated by treatment with H2O2. Crystallographic studies of Co-HPCD showed that its active site is superimposable with those of Fe-HPCD and Mn-HPCD. (Fig. 3). There are no discernible perturbations around the metal center to suggest differential tuning by the protein environment to compensate for the large difference in the redox potentials of the metal centers [9].
Fig. 3.
Structure superposition of the metal centers in the resting state of Fe-HPCD (PDB 3OJT, 1.70 Å resolution; bronze), Mn-HPCD (PDB, 1.65 Å resolution; 3OJN; gray) and Co-HPCD (PDB 3OJJ, 1.72 Å resolution; color-coded). Reproduced from ref. [9] with permission.
The steady-state kinetic parameters for the three HPCDs and the two MndDs are compared in Table 1. With respect to substrate, Co-HPCD exhibits the highest apparent affinity during turnover (KMHPCA = 5 µM), decreasing slightly for the two MndD enzymes (KMHPCA = 15 µM), and slightly more for the Fe- and Mn-HPCD (KMHPCA = 33 µM). More interesting is the observation that [Co-HPCD(HPCA)] has a very low apparent affinity during turnover for O2 with KMO2 = 1.2 ± 0.1 mM O2 compared to 40–60 µM for the other enzyme-substrate complexes [9]. Under ambient conditions, the kcat value of 215 min−1 for Co-HPCD is about half of that for the other enzymes, but this value triples under O2-saturating conditions. Extrapolation of the kcat vs. [O2] dependence plot gives a maximum kcat value of 1100 min−1, so that it has by far the highest turnover value of the enzyme forms examined. In contrast, the kcat/KMO2 value for Co-HPCD is an order of magnitude smaller than those for Fe- and Mn-HPCD. The kcat/KMO2 value is essentially the second order rate constant for the overall process of O2 binding, and it is comprised of all the rate constants from O2 binding through the first irreversible step (presumably O-O bond cleavage) [28]. Thus, the lower kcat/KMO2 value of Co-HPCD relative to those of Fe- and Mn-HPCD reflects its lower efficiency at capturing and activating O2. This difference likely arises from the expected higher M(III/II) redox potential of the Co-HPCD active site, based on the 1.15 V gap in the standard potentials of aqueous Co(II) vs. Fe(II). A gap of this magnitude would cause a very large decrease in the rate of O2 activation if the energy of activation results purely from the difference in redox potential. Consequently, the relatively small changes in kcat/KMO2 value for the various forms of HPCD examined suggest that the activation energy for the key step is not directly related to the redox potential of the metal. The hypothesis we advanced to rationalize this observation was that the redox state of the metal in the activated metal-O2 complex is the same as that of the metal before O2 binding. Thus, the bound substrate must ultimately provide the initial electron required to activate O2.
Single-turnover stopped-flow experiments on the reaction of the various anaerobic enzyme-substrate complexes with O2 shed light on what gives rise to the differences in the steady-state kinetic parameters. The formation rate constant of the yellow extradiol ring-cleaved product by Fe-HPCD showed no O2-concentration dependence and was, in fact, equal to the kcat value measured under steady-state conditions. This indicates that the rate-limiting step is separated from the O2 binding step by one or more irreversible steps and is likely to be either the penultimate product-formation step or the final product-release step of the reaction cycle [29]. In contrast, the corresponding rate for Co-HPCD exhibited a significant O2-concentration dependence, suggesting that O2 binding or a step reversibly connected with O2 binding becomes rate determining for Co-HPCD [9, 29]. The larger kcat value achievable by Co-HPCD in comparison to Fe-HPCD indicates that product formation or release steps are faster for Co-HPCD than for Fe-HPCD. Thus, under low O2 concentrations, a step in the process of O2 binding to [Co-HPCD(HPCA)] is rate limiting, but a subsequent step becomes rate determining under saturating conditions. This step is faster than the rate-determining step for turnover of HPCA by Fe-HPCD.
Proposed mechanism for O2 activation and substrate oxidation
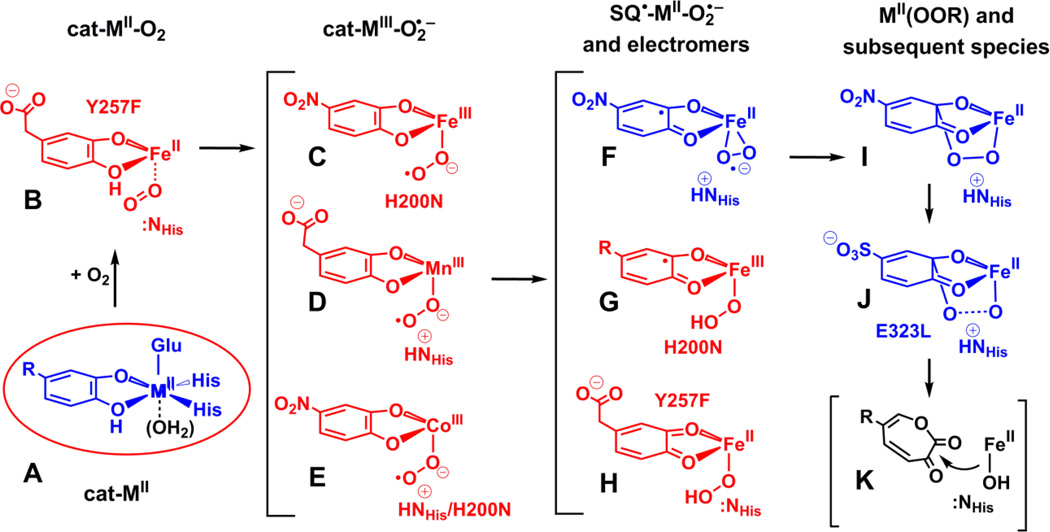
Scheme 2 represents the working mechanistic hypothesis for the extradiol-cleaving catechol dioxygenases prior to the discovery of the various intermediates summarized in this review. In its simplest form, the activated metal(II)(semiquinone)superoxo diradical species would be formed by concerted one-electron transfers from metal to O2 and substrate to metal so that there is literally no change in oxidation state of the metal. The possibility of additional complexity in this simple scheme was introduced by the discovery of oxygenated intermediates with metal(III) oxidation states in enzyme variants of a second-sphere residue (see below). While no experimental evidence has yet been obtained for any Fe(III) intermediates in the native Fe-HPCD enzyme, the metal(III) intermediates observed in other enzyme forms suggest two additional potential mechanistic scenarios: (1) stepwise electron transfer between the metal and O2 and then the substrate and the metal, or (2) reaction of the metal(III)-superoxo species with unactivated substrate at a rate that is too fast to detect experimentally. The first scenario would suggest that both transfers are fast and occur before attack of the activated O2 species so that no net change in metal oxidation state occurs during O2 activation. For metals with higher standard M(III/II) redox potentials than Fe, the rates of the two steps would be self-compensating so that slower transfer from the metal to O2 for a higher potential metal would be compensated by a faster transfer from substrate to the metal. This might allow the reactions to proceed with little change in kcat/KMO2 and also permit detection of a metal(III) intermediate under conditions that slowed the second electron-transfer step. The second scenario would rely on the reaction being pulled rapidly forward by the subsequent irreversible formation of the alkylperoxo intermediate in order to account for the lack of observable metal redox potential dependence.
Several subsequent observations supported the original mechanistic hypothesis and provided the means to test it. For the [Fe-HPCD(HPCA)] enzyme-substrate complex, structural studies showed that binding of the catecholic substrate to the active-site metal center displaced the two water molecules bound trans to the two His ligands, as shown by the crystal structure of the [Fe-HPCD(HPCA)] complex (Fig. 1). The two Fe–OHPCA bonds differ in length by 0.2 Å, leading to the suggestion that the longer Fe–O bond arises from the presence of a proton on the O1 atom [15]. Structural studies show that this proton is close enough to be hydrogen bonded to the nearby H200 residue, while the ionized O atom on C2 is stabilized by hydrogen bonding to the nearby Y257 residue [14, 15]. As presented below, the H200 and Y257 residues play important roles in modulating the reaction of the enzyme-substrate complex with O2. The monoanionic state of the bound HPCA is also supported by the UV resonance Raman studies of Eltis that directly probe the protonation state of the bound substrate in solution [20, 21].
The crystal structure of the [Fe-HPCD(HPCA)] complex shows that the water molecule trans to the Glu ligand is weakened or displaced upon substrate binding to the iron center [15], thereby poising the metal center to bind O2 in the next step (Scheme 3A). It is noteworthy, that O2 does not bind to the metal center before the aromatic substrate. Since the substrate does not compete directly with the solvent blocking the O2 binding site, its effects must be indirect, perhaps by either decreasing the M(III/II) redox potential of the metal or supplying electron density via the metal to destabilize the solvent and stabilize the metal-O2 bond. Upon formation of a ternary enzyme-substrate O2-adduct (ESO2), electron reorganization among the actors in this mechanism leads to C–O bond forming and O–O/C–C bond breaking steps that eventually produce the ring-cleaved product, as outlined in Scheme 2. By using substrates with different oxidation potentials, replacing either of the key second-sphere residues H200 and Y257, as well as substituting the metal center, a range of intermediates has been characterized that differ in the extent of electron transfer among the trio of redox-active species in this active site, namely the substrate, the metal center, and O2. These species, listed in Table 2, shed light on the fine tuning required for the oxidative cleavage reaction to work efficiently in the HPCD active site.
Scheme 3.
Intermediates that have been observed spectroscopically (in red) or crystallographically (in blue) in the reactions of HPCDs with substrate and O2. Enzyme variants are specifically noted for particular intermediates; otherwise wild-type HPCD was used. Note that intermediate K has not been observed and in no case has any Fe(III) species been found with the wild-type HPCD. When used, R = -CH2COO− or –NO2 for that particular intermediate.
Table 2.
Intermediates characterized in the reactions of O2 with M-HPCD enzyme-substrate complexes.a
| Intermediateb | Metal Oxidation State (System Spin State) |
λmax (nm) (ε, M−1cm−1) |
Proposed Assignment |
Ref |
|---|---|---|---|---|
| Fe-HPCAInt1 (I) | hs-Fe(II) (S = 2) | – | [Fe(II)(OOR)] | [29, 36] |
| H200N-Fe-HPCAInt1 (G) | hs-Fe(III)/radical (S = 2) | 395 (3,200), 610(1,100) |
[Fe(III)(SQ•)(OO(H))] | [33, 36] |
| H200N-Fe-HPCAInt2 (I) | hs-Fe(II) (S = 2) | – | [FeII(OOR)] | [33, 36] |
| H200N-Fe-4NCInt1 (C) | hs-Fe(III)/radical (S = 2) | 506 (~10,000), 630 (~1,200) |
[Fe(III)(4NC)(O2•−)] | [33, 37] |
| H200N-Fe-4NCInt2 (G) | hs-Fe(III)/radical (S = 2) | 405 (~15,000), 675 (~1,000) |
[Fe(III)(SQ•)(OO(H)] | [33, 37] |
| Y257F-Fe-HPCAInt1 (B) | hs-Fe(II) (S = 2) | – | [Fe(II)(HPCA)(O2)] | [38] |
| Y257F-Fe-HPCAInt2 (H) | hs-Fe(II) (S = 2) | 425 (~10,500) | [Fe(II)(Q)(OO(H))] | [38] |
| Mn-HPCAInt1 (D or G) | hs-Mn(III)/radical (S = 5/2) | – | [Mn(III)(HPCA)(O2•−)] or [Mn(III)(SQ•)(OO(H))] |
[9] |
| Mn-HPCAInt2 (I) | hs-Mn(II) (S = 5/2) | – | [Mn(II)(OOR)] | [9] |
| wt/H200N-Co-4NCInt (E) | ls-Co(III) (S = 1/2) | 390 (~7,000), 526 (~13,000) |
[Co(III)(4NC)(O2•−)] | [39] |
Abbreviations used: HPCA, homoprotocatechuate; 4NC, 4-nitrocatechol; SQ• semiquinone; Q, quinone; O2•− superoxide radical anion; hs, high-spin; ls, low-spin.
Labels in this column refer to structures in Scheme 3.
Intermediates observed in reactions carried out in solution
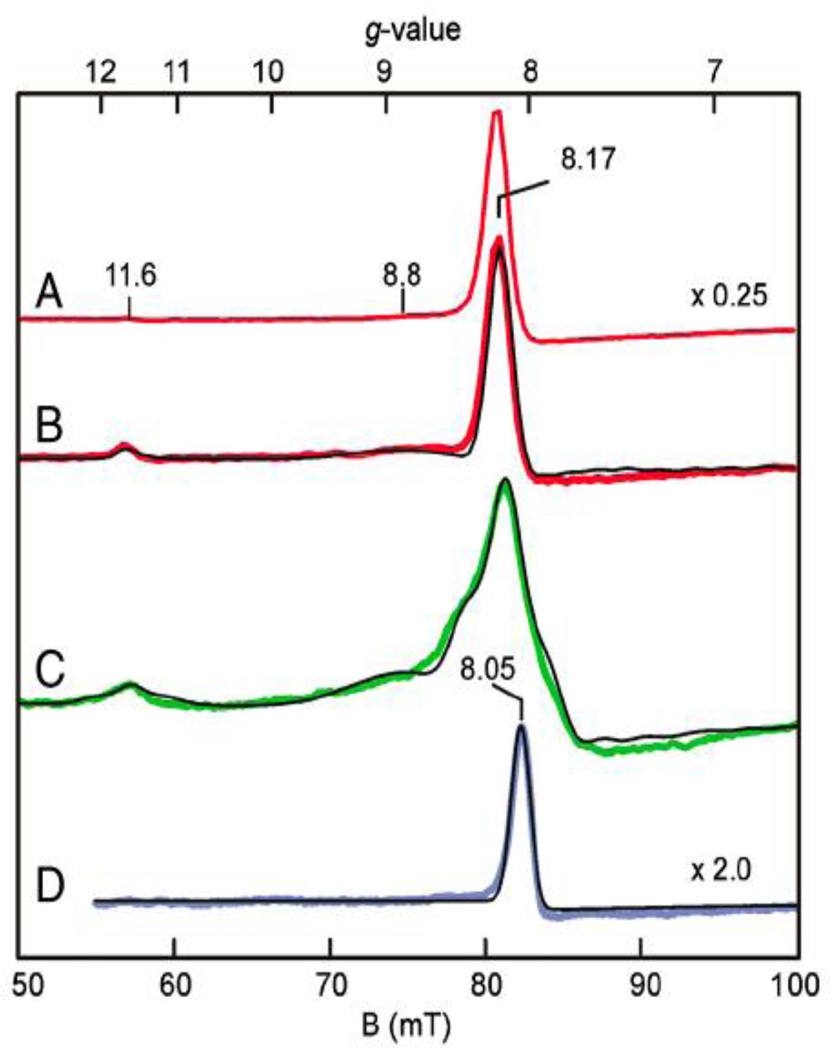
The use of the H200N Fe-HPCD variant and the replacement of HPCA with the less easily oxidized 4-nitrocatechol (4-NC) afforded an intermediate (H200N-Fe-4NCInt1) that is formulated as an Fe(III)-superoxide species (Scheme 3C), the first example of such an intermediate to be found in the mononuclear nonheme iron oxygenase family. H200N-Fe-4NCInt1 has been characterized spectroscopically to have a high-spin (S1 = 5/2) Fe(III) center that is antiferromagnetically (AF) coupled to an S2 = 1/2 radical. The iron center had Mössbauer parameters (δ = 0.55 mm/s; ΔEQ 0.33 mm/s) that indicated a high-spin ferric state, but it was nearly EPR-silent in perpendicular mode (the mode typically used to detect species with half-integer spin). Instead, relatively intense EPR signals were observed in parallel mode, indicating the intermediate to be an integer-spin species. Therefore, the high-spin ferric center must be coupled to a radical moiety. At 2 K, a dominant EPR signal was found at g = 8.17 that arose from an S = 2 ground state (Fig. 4A); at higher temperatures signals at g = 8.8 and 11.6 increased in intensity and were associated with an S = 3 excited state (Fig. 4B). The temperature dependences of the EPR signals could be fit with an S1 = 5/2/S2 = ½ model with weak AF coupling (J ≈ 6 cm−1). Importantly, all three signals were broadened significantly with the use of 17O2 (I = 5/2) to generate the intermediate (A17O ≈ 180 MHz, Fig. 4C), indicating the presence of substantial unpaired spin density on the bound dioxygen unit and fully consistent with the AF-coupled Fe(III)-O2−• formulation. This first intermediate decayed over the course of 10 minutes at 4 °C to form a second intermediate, H200N-Fe-4NCInt2, with Mössbauer spectra that also show an AF-coupled S = 2 Fe(III)-radical complex. H200N-Fe-4NCInt2 also exhibited a parallel-mode EPR signal at g = 8.05 (Fig. 4D), but this signal arose from an excited state of a species with an inverted zero-field splitting. It was only slightly broadened by 17O2 (A17O ≈ 5 MHz with <3% spin delocalization) indicating little spin delocalization onto the bound O2. The optical spectrum of H200N-Fe-4NCInt2 was similar to that of 4NC quinone or semiquinone. Based on the accumulated data, H200N-Fe-4NCInt2 was proposed to be a 4NSQ•-Fe(III)-peroxo species (Scheme 3G), which decayed over the course of hours to release hydrogen peroxide and the observed 4-nitroquinone product and restore the Fe(II) active site.
Fig. 4.
Parallel-mode EPR spectra (colored lines) and simulations (thin black lines). (A) 2 K spectrum 10 s after mixing H200N-4NC complex with O2-saturated buffer (200 mM MOPS pH 7.5) at 4 °C. (B) Sample A at 9 K. (C) Spectrum at 10 K for a sample prepared as in A but with 70% enriched 17O2. (D) Sample from A at 9 K after 10 min incubation at 4 °C. Reprinted from ref. [37] with permission.
A similar peroxo-Fe(III)-semiquinone radical species was also observed with the H200N-Fe-HPCD variant and the native substrate HPCA, H200N-Fe-HPCAInt1 (Scheme 3G) [36]. In addition to an absorption band from the substrate semiquinone, this intermediate exhibited a blue chromophore at 610 nm, which may arise from the Fe(III)-peroxo unit by comparison with model complexes, but attempted resonance Raman experiments have been unsuccessful in characterizing these chromophores. H200N-Fe-HPCAInt1 was formed in nearly stoichiometric yield, but it rapidly (400 ms at 4 °C) converted to another intermediate H200N-Fe-HPCAInt2 with a maximum of 10% yield before decaying to form the ring-cleaved product. The latter intermediate contained only Fe(II) and did not exhibit an unique visible chromophore, so H200N-Fe-HPCAInt2, may represent the alkylperoxo intermediate of the reaction cycle (similar to Scheme 3I). Reaction of wt Fe-HPCD with HPCA resulted in stoichiometric formation of an intermediate within the dead time of the stopped-flow instrument (1 ms), which contained only Fe(II) (wt-Fe-HPCAInt1). The Mössbauer spectrum of this species resembled that of H200N-Fe-HPCAInt2, suggesting that it may also be an alkylperoxo intermediate similar to that shown in Scheme 3I. This intermediate decayed rapidly to yield the yellow ring-cleaved product and resting enzyme.
Intermediates from metal-replaced HPCDs
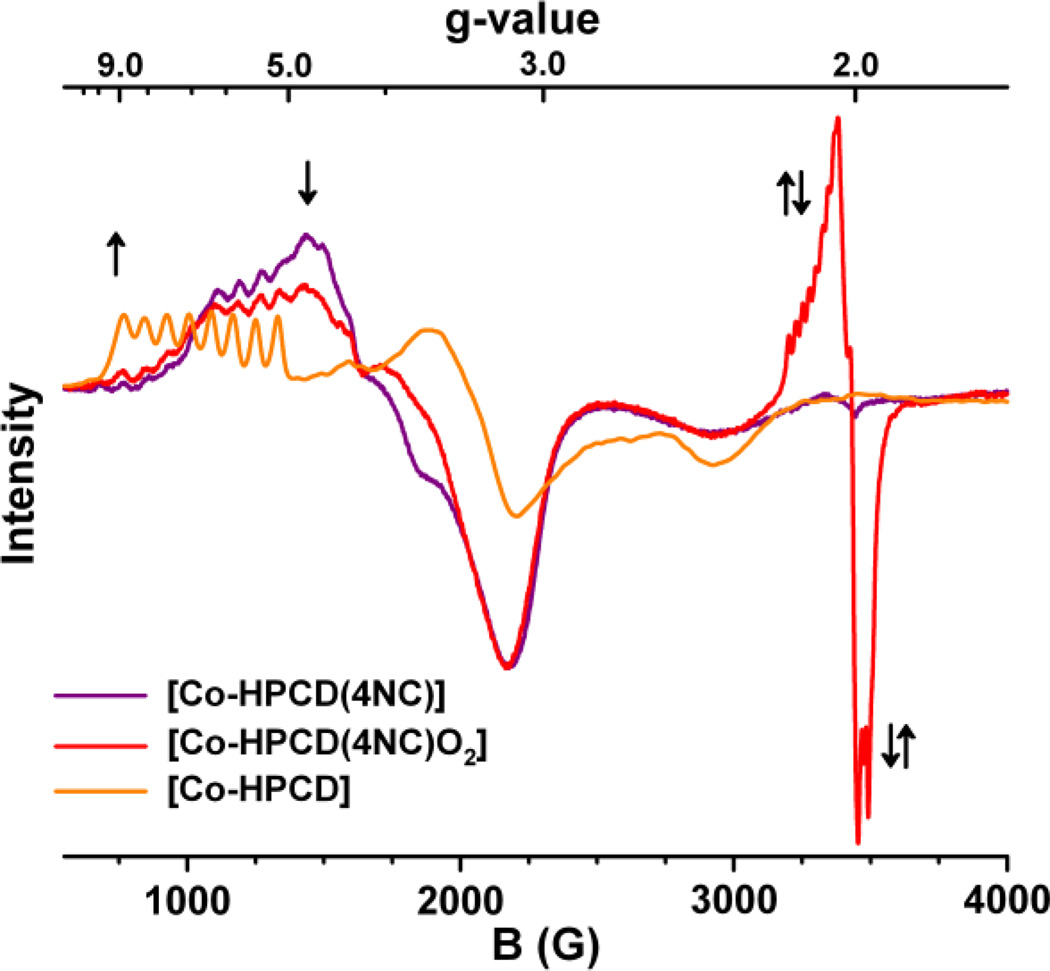
Intermediates were also observed when the iron center was replaced by cobalt or manganese. These metal centers differ from iron by one electron, so conventional perpendicular-mode EPR methods can be used to probe them in their divalent oxidation states. Co-HPCD is able to catalyze extradiol cleavage of the electron-poor substrate 4-nitrocatechol, but at a rate one thousand-fold slower than for the native HPCA substrate. The [Co-HPCD(4NC)] complex exhibits an EPR spectrum typical of a high-spin Co(II) S = 3/2 center with 59Co hyperfine splitting (A = 80 G) observable at the g = 6.7 resonance. Upon exposure to O2, this signal was replaced by an S = 1/2 EPR signal exhibiting 59Co hyperfine splitting (A = 24 G), which represented 10% of the enzyme in the sample. Upon decay of this intermediate, the EPR spectrum reverted to that of Co-HPCD. The EPR signal observed for the intermediate is typical of a low-spin Co(III)-superoxide complex (Scheme 3E). Because the rates of formation and decay of this intermediate were very slow in comparison to the analogous steps for turnover of 4NC by Fe-HPCD, the intermediate could be observed without resorting to rapid-freeze-quench techniques. Consistent with the low-spin Co(III)-O2−• formulation, the use of 17O2 resulted in significant broadening of the S = 1/2 signal. Interestingly, the H200N derivative afforded the same results, but the analogous O2 adduct could be accumulated to 50% of the enzyme in the sample. However, H200N-Co-HPCD does not effect the cleavage of 4NC, and this O2 adduct is a dead-end complex. This comparison demonstrates the important role of H200 in facilitating the step beyond O2 binding.
In contrast to the S = 2 high-spin Fe(III)-superoxide species observed for H200N-Fe-4NCInt1, both wt- and H200N-Co-4NCInt can be described as S = 1/2 species with a superoxo moiety bound to a low-spin Co(III) center, which is diamagnetic due to its d6 electronic configuration [39]. One might expect large kinetic barriers for the apparent spin-state change from high- to low-spin upon O2 binding [40, 41] and for the reverse spin transition in the subsequent electron transfer step upon oxidation of the electron-poor 4NC substrate, making the Co-O2 adduct relatively stable. These apparent spin-state changes may provide the rationale for the shift in rate-determining step to an earlier phase of the catalytic cycle for Co-HPCD [42].
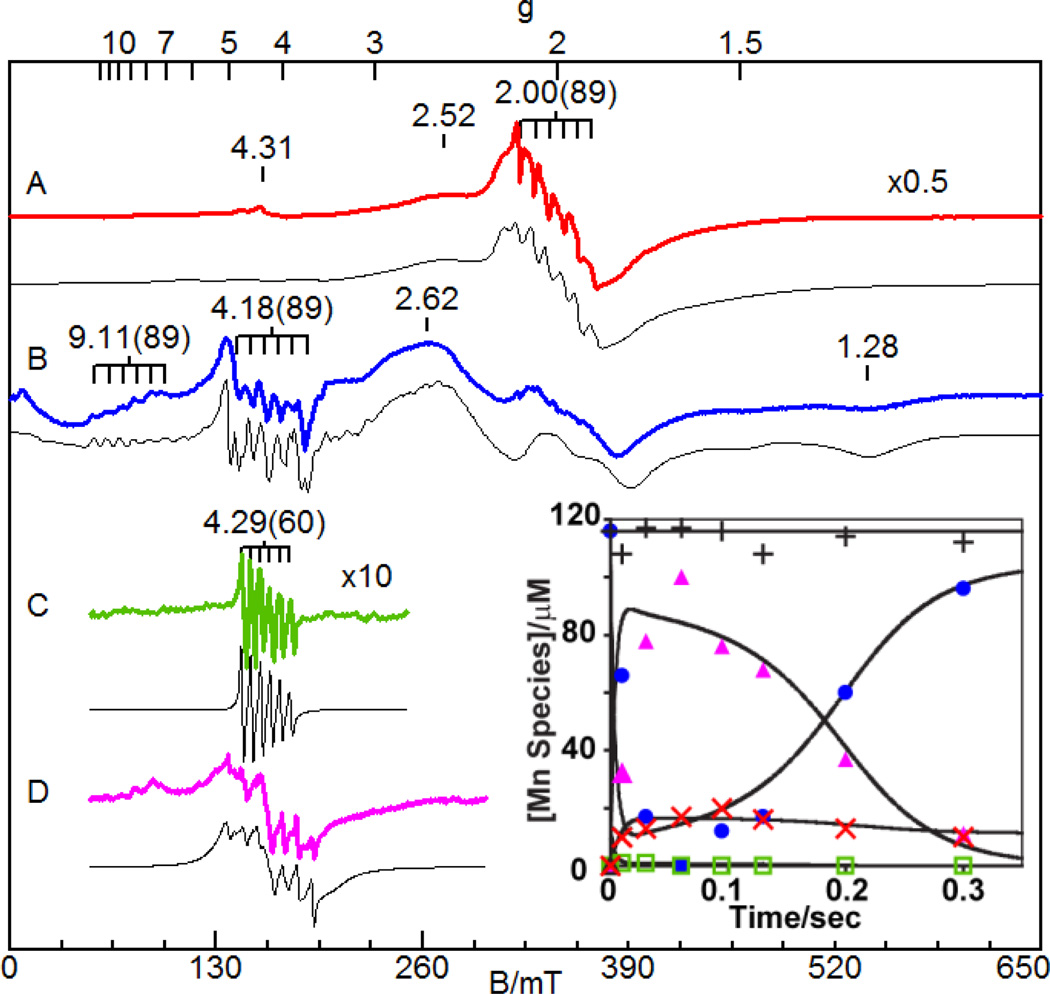
Transient species were also found in corresponding studies on Mn-HPCD. Its EPR spectrum showed a main feature at g = 2 with 55Mn hyperfine splitting (A = 89 G), typical of a six-coordinate Mn(II) center with a small zero-field splitting (D = 0.055 cm−1) (Fig. 6A). Upon binding of HPCA, this signal was replaced by signals that span a much broader field range due to an increase in the zero-field splitting (D = 0.090 cm−1) (Fig. 6B); simulation demonstrated that the spectrum arises from a single Mn(II) center. There were two six-line hyperfine splitting patterns at g = 4.18 and 9.11, both with A = 89 G, consistent with the expected retention of the Mn(II) oxidation state upon substrate binding. Rapid-freeze-quench EPR samples obtained upon reacting the [Mn-HPCD(HPCA)] complex with O2 for 15 ms revealed an intermediate in approximately 5% yield at g = 4.29. This species exhibited 55Mn hyperfine splitting (A = 60 G, Fig. 6C) and an axial zero-field splitting value of (D = 2.5 cm−1). The decrease in the A value and the large jump in zero-field splitting strongly suggest the Mn(II) center had been oxidized by one electron. However, the fact that its EPR signal is still observable in perpendicular mode requires the presence of a radical moiety that is ferromagnetically coupled to this Mn(III) center to generate a half-integer spin system. As with the other intermediates discussed above, this radical may reside on the bound O2 or on the substrate. Thus, wt-Mn-HPCAInt1 could be analogous in formulation to H200N-Fe-4NCInt1 (Scheme 3C) or (wt or H200N)-Co-4NCInt1 (Scheme 3D), or alternatively, to H200N-Fe-4NCInt2 (Scheme 3G). Further evolution of this intermediate gave rise to a second species, wt-HPCD-Mn-HPCAInt2, which was almost fully formed at 34 ms. This species exhibited an EPR signal with 55Mn hyperfine splitting of A = 89 MHz (Fig. 6D), suggesting a return to the Mn(II) oxidation state at this stage. Finally, this intermediate decayed to form product and eventually restore the resting form of the enzyme. These results provided the first evidence that a transient oxidation of the metal can occur during the process of O2 activation in a non-enzyme variant with the electron-rich substrate HPCA.
Fig. 6.
EPR spectra (colored lines) and simulations (black lines) of (A) Mn-HPCD, (B) [Mn-HPCD(HPCA)] complex, (C) Int1, and (D) Int2. See ref. [43] for experimental and simulation parameters. Inset: Concentration of Mn species vs. time for RFQ samples with [O2]/[ET] = 2.9: E( ), ES(
), ES( ), I1(
), I1( ), I2(
), I2( ), Total Mn(+). Reprinted from ref. [43] with permission.
), Total Mn(+). Reprinted from ref. [43] with permission.
Intermediates from reactions carried out in enzyme crystals
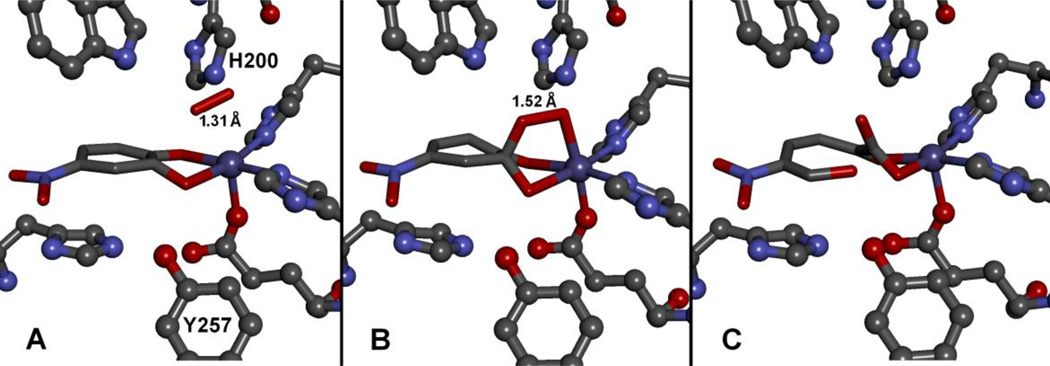
The most definitive evidence for the proposed intermediates of Fe-HPCD illustrated in Scheme 2 derives from a crystallographic study in which three different species along the reaction pathway were trapped in different active sites within the asymmetric unit of a single crystal of the [Fe-HPCD(4NC)] complex [14]. This allowed the structures of the intermediates to be determined. The catalytic reaction was initiated in crystallo by exposing the crystal to a very low concentration of O2 before being frozen. Presumably, different intermediates were trapped because the nominally identical subunits of the homotetrameric enzyme were exposed to different packing constraints from the crystal lattice. In one subunit, electron density for a diatomic molecule, presumably O2 was found bound side-on to the iron in a position adjacent to the chelated 4NC substrate analog (Fig. 7A, Scheme 3F). The side-on orientation would allow reaction with the C2 position of the substrate with only minor shifts in the position of the bound O2, a common feature of enzyme-catalyzed reactions. Interestingly, the aromatic ring of the 4NC is nonplanar in this intermediate with the maximum distortion occurring at C2. It was proposed that the ring is buckled because an electron had been transferred from the 4NC to the bound O2. The bond lengths from the iron to the O2 in this complex were relatively long, suggesting that the iron is ferrous, and thus the iron acted as “wire” to transfer one electron between the substrates. This would give both substrates radical character and promote recombination of the radicals to form an alkylperoxo intermediate. Accordingly, the next intermediate observed in two other active sites in the asymmetric unit is an alkylperoxo species in which the putative Fe(II)-superoxo species has attacked the substrate radical at C2 to form a tetrahedral carbon at this position (Fig. 7B, Scheme 3I). The O–O bond is intact in this intermediate, showing that the mechanism involves attack by a superoxo (or potentially a peroxy) species rather than a high valent iron-oxo species derived from O–O bond cleavage prior to attack on the aromatic substrate. The latter strategy is utilized by several other members of the broad 2-His-1-carboxylate facial triad family [44]. In the final subunit within the asymmetric unit, the ringopened product complex was observed (Fig. 7C). This intermediate shows that the enzyme can carry out the full catalytic cycle in the crystal, helping to validate the other observed intermediates. It had not previously been observed because attempted diffusion of the product back into a crystal from solution results in loss of crystal diffraction.
Fig. 7.
Intermediates and the enzyme-product complex observed in different subunits of the homotetrameric [Fe-HPCD(4NC)] enzyme-substrate complex contained within the asymmetric unit of the enzyme crystal after reacting with O2 (PDB 2IGA, 1.95 Å) [14]. Structures of (A) [Fe-HPCD(4NSQ•)superoxo] exhibiting substrate ring puckering at C2, suggesting a localized semiquinone substrate radical, (B) the [Fe-HPCD-alkylperoxo] intermediate, and (C) the enzyme-product complex.
The relationship of crystal packing forces to the ability of an enzyme crystal to stabilize intermediates was tested by making a surface mutation in Fe-HPCD. The E323L mutation caused a dramatic change in the organization of enzyme molecules within the unit cell with a corresponding change in crystal packing forces. This allowed a new intermediate to be trapped when 4-sulfonyl catechol was used as the substrate [45]. For the wild-type enzyme, the [Fe-HPCD-4-SO3-catechol] reaction with O2 in the crystal resulted in no single high-occupancy intermediate, but the [Fe-HPCD(gem-diol)] species was trapped in the E323L variant (Scheme 3J). This stable intermediate may be construed as related by one-electron reduction to the gem-diol radical intermediate predicted computationally by Siegbahn [46].
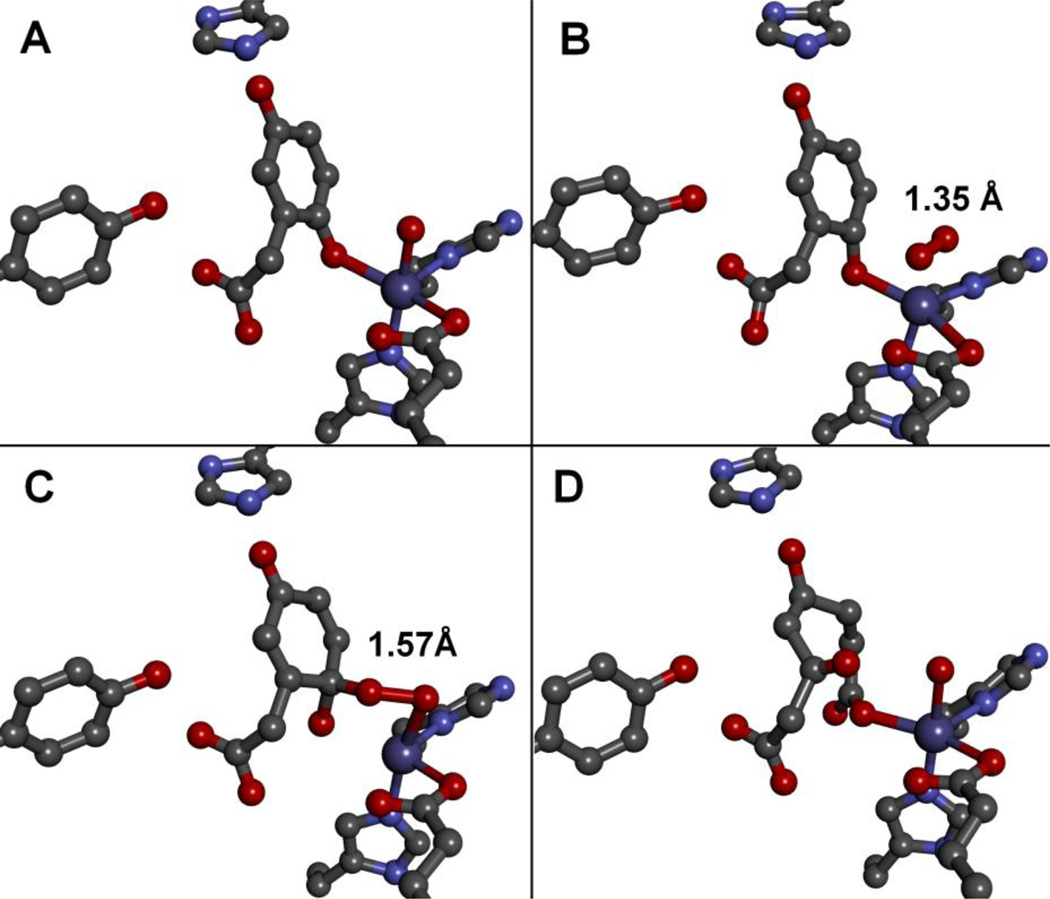
Very recently, three reaction intermediates were also found in an in crystallo study of homogentisate dioxygenase (HGD) [47]. Homogentisate is an isomer of HPCA, differing in having the two hydroxyl groups para to each other rather than ortho. HGD catalyzes the oxidative cleavage of the C–C bond to which the carboxylate and one hydroxyl group are connected, a transformation that can be related to alkaptonuria in humans [12, 48, 49]. As a consequence of the para positioning of the two hydroxyl groups, the substrate binds to the HGD iron center only with the hydroxyl group adjacent to the acetate substituent. As with HPCD, the other hydroxyl group is not ionized and is hydrogen bonded to a His residue, suggesting that the modulation of its protonation state is important for the O2 activation mechanism. Interestingly, the same three reaction intermediates as those of HPCD were found in an oxygenated crystal of the HGD-homogentisate complex, namely the superoxo/semiquinone (Fig. 8B and Scheme 3F), the alkylperoxo (Fig. 8C and Scheme 3I), and the product-bound intermediates (Fig. 8D), despite the distinct protein folds adopted by HGD (cupin) and HPCD (VOC) and the different relative positioning of the two hydroxyl groups. Common features include a) the side-on bound dioxygen moiety and the nonplanar substrate in Fig. 8B, and b) the increase in the O–O bond length upon going from 1.35 Å in Fig. 8B to 1.57 Å in Fig. 8C. These observations support the assignment of these species as the superoxo/semiquinone (Scheme 3F) and the alkylperoxo (Scheme 3I) intermediates. However there are some notable differences. In the HGD structure, dioxygen binds trans to a His residue,, rather than trans to the glutamate residue observed in the HPCD structure. Furthermore, residues corresponding to the critical H-bonding H200 and Y257 residues in HPCD are absent in the HGD structures and appear to be replaced by water molecules in the HGD active site pocket. This is an amazing example of convergent evolution where two nonhomologous enzymes HGD and HPCD share the same reaction mechanism for cleavage of dihydroxybenzene rings with O2 that is catalyzed by a common iron(II) center with a 2-His-1-carboxylate facial triad motif.
Fig. 8.
Enzyme-substrate, intermediates and the enzyme-product complex observed in different subunits of the homogentisate dioxygenase (HGD) enzyme-substrate complex contained within the asymmetric unit of the enzyme crystal after reacting with O2 (PDB 3ZDS, 1.70 Å) [47]. Structures of the (A) [HGD(substrate)], (B) [HGD(semiquinone)(superoxo)], (C) [HGD(alkylperoxo)], and (D) the enzyme-product complexes.
Substrate aromatic ring distortion stabilized by Tyr 257
The manner in which the enzyme promotes and stabilizes substrate ring distortion and electron transfer to the metal and O2 was investigated by using the Y257F variant of Fe-HPCD. Structural studies revealed two different types of interactions between Y257 and Fe-bound HPCA or 4NC substrates in the wild-type enzyme complex [22]. First, van der Waals interaction between the Y257-OH and the aromatic ring forces a ring distortion, because the substrate is otherwise rigidly bound between the iron and an anion binding pocket at the rear of the active site. Second, a specific hydrogen bond between the Y257-OH and the deprotonated C2-O− of the substrate stabilizes the hydroxyl oxygen out of the plane of the ring. Localized distortion of the ring favors the semiquinone state over the normally planar fully aromatic dihydroxybenzene or quinone states. This would promote one electron transfer out of the ring to O2 as it binds. When Y257 was replaced by phenylalanine, both types of interactions with the substrate were lost, and structural studies showed that the ring is planar for both HPCA and 4NC [22]. Although the rate of catalytic turnover was decreased only 75%, rate constants for normally fast individual steps within the catalytic cycle were decreased over 100-fold [38].
An intermediate (Y257F-Fe-HPCAInt1) was trapped by reacting the Y257F variant [Fe-HPCD(HPCA)] complex with O2 [38]. At pH 5.5, this intermediate formed in a second order reaction during the first 25 ms of the reaction, and thus it appears to be an oxy complex. Surprisingly, it exhibited no visible chromophore that would arise from a semiquinone or quinone form of the substrate. Mössbauer spectra of the intermediate showed that the high-spin Fe(II) center of the resting enzyme was retained but with slightly different parameters. It was proposed that this intermediate contains O2 weakly bound to the Fe(II) in the substrate complex, and thus represents the extreme case of no electron transfer within the substrate-Fe-O2 system (Scheme 3B). In the absence of Y257, the planar aromatic substrate does not promote single electron transfer from Fe(II) to O2, accounting for the initial weak O2 binding. The Y257F-Fe-HPCAInt1 intermediate evolves over the next second to form another intermediate (Y257F-Fe-HPCAInt2) which exhibits the characteristic optical spectrum of the HPCA quinone (Scheme 3H), Mössbauer spectroscopy again shows only Fe(II) in this intermediate, suggesting that the two electrons produced by substrate oxidation have been transferred to O2 to reduce it to the peroxide state (Scheme 3H). In the absence of distortions caused by Y257 the planar quinone state is apparently favored.
Insights from DFT calculations
Computational studies performed on the Fe- and Mn-HPCD enzymes support the notion of a finely tuned reaction surface on which the two electrons involved in the first phase of substrate oxidation can be distributed over the three redox-active components [50–55]. They also provide additional insights not obtainable from X-ray crystallography or spectroscopy. In general, the various intermediates shown in Scheme 3 have reasonable energies, and the activation barriers for their formation are not too high.
In early DFT work using a relatively small active site model, Siegbahn and coworkers [50, 53] found support for many of the steps proposed in Scheme 2. They found that O2 binding to the enzyme-substrate complex resulted in facile electron transfer from the catechol substrate to O2 via the metal center to form a Fe(II)(semiquinone)-superoxo diradical pair. This computational result corroborated the idea of the active-site metal acting as a conduit for electron transfer, which was proposed as a means to rationalize how three distinct metal centers in the HPCD active site with very different standard M(III/II) redox potentials can catalyze the oxidative cleavage of HPCA at comparable rates [9]. The calculations further showed a specific role for the H200 residue in proton transfer from the monoanionic substrate to the bound superoxide that occurred concomitant with the electron transfer from substrate to O2. Once formed, the radical components of the (SQ•)FeII(O2•−) species are combined in a barrier-less attack of the superoxide at C2 of the HPCA semiquinone to form the alkylperoxo intermediate that subsequently rearranges to give rise to the ring-cleaved product. Hydrogen bonding interactions from His200H+ and Tyr257 residues to the bound O2 and substrate were found to be important for creating an active site that directed the attack of the superoxide on the semiquinone ring at the correct position and in the subsequent rearrangement of the bridging alkylperoxo species to form the correct proximal extradiol ring-cleaved product.
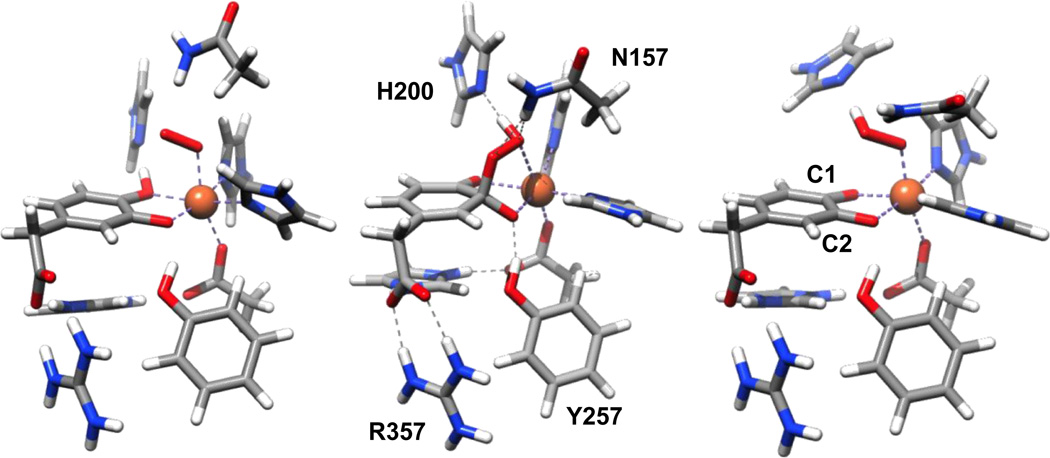
In more recent work, Neese and coworkers used a larger cluster model consisting of residues from the primary coordination sphere (H155, H214, and E267) and those in the second sphere (H200, N157, Y257, R243, and H248) that were found by crystallography to have interactions with the substrate, O2 or the E267 ligand [54]. With this model, an earlier intermediate involving only the iron(II) center and O2 could be found. This initially formed Fe-HPCD-HPCA-O2 adduct was described as a (monoanionic substrate)-(hs-Fe(III))-(end-on bound superoxo) moiety, with very little unpaired density found on the aromatic ring of the catecholate substrate (Fig. 9 left). Mössbauer parameters calculated for this model matched well with the experimental values found for the FeIII-superoxo moiety characterized for H200N-Fe-4NCInt1 (Scheme 3C) [37]. Although the ferromagnetically coupled septet represented the ground state for the FeIII-superoxo moiety, the antiferromagnetically coupled quintet that is consistent with the spectroscopic data for H200N-Fe-4NCInt1 was found to be only 3 kJ/mol higher in energy.
Fig. 9.
Calculated structures for the initially formed (catH)Fe(III)-superoxo adduct (left), the Fe(II)-alkylperoxoH intermediate that affords ring cleavage (middle), and the dead-end [Fe(III)(semiquinone)OOH] byproduct (right) [54].
The electronic nature of the Fe(III)-superoxo species was not significantly perturbed in three subsequent species, where the proton originally bound to O1 of the monoanionic substrate was first transferred to the H200 residue and, upon reorientation of the H200 residue, became H-bonded to the superoxide via the distal oxygen atom first and then the proximal oxygen atom. The last change brought the distal oxygen atom to a position above C2 of the substrate, thereby setting the stage for C–O bond formation leading to the Fe(II)-alkylperoxoH intermediate (Fig. 9 right). The key role of the H200 residue in mediating the proton transfer needed to advance the reactants to the next stage was corroborated by the trapping of the Fe(III)-superoxo moiety in H200N-Fe-4NCInt1 where the residue at position 200 was replaced by one incapable of accepting a proton [33, 37].
Their calculations also predicted the formation of an antiferromagnetically coupled high-spin [(SQ•)Fe(III)-OOH] species (Fig. 9 right) that was too stable to be on the catalytic pathway [54]. As an indication of the progression of the initially formed Fe(III)-superoxo moiety to an Fe(III)-hydroperoxide, the Fe–Oproximal distance of 2.10 Å found in the initial O2 adduct shortened to 1.92 Å in the dead-end species. This models the nature of H200N-Fe-4NCInt2, which was found to decay without ring cleavage to form the corresponding quinone and H2O2 [33, 37]. However, product was found to form in the case of H200N-Fe-HPCAInt2, albeit rather slowly [33, 36].
QM/MM calculations on the HPCD mechanism were reported very recently by Lai and coworkers. With this approach, they found evidence for the formation of both the (catH)Fe(III)-superoxo adduct and the (SQ•)Fe(II)(O2•−) diradical pair upon O2 binding to the ES complex. Moreover, they also located a species with hybrid (catH)Fe(III)(O2•−)/(SQ•)Fe(II)(O2•−) character with a lower activation barrier for the formation of the critical C–O bond of the alkylperoxo intermediate, making this hybrid species the reactive oxygen species in their mechanism.
Notably, none of the DFT studies thus far have modeled the side-on binding mode of the dioxygen moiety and the nonplanarity of the semiquinone moiety observed crystallographically for the initial ESO2 adducts of both the HPCD [14] and HGD[47]. Both of these structural features are presumably of significance mechanistically. Reproducing these features will be important in enhancing our understanding of the extradiol cleavage mechanism.
Conclusions and challenges for the future
The crystallographic, spectroscopic, kinetic, mutagenic, metal replacement, and computational studies of HPCD reviewed here highlight the delicate electronic balance that is required for specific and efficient ring-cleavage chemistry. We have shown that intermediates with very different distributions of electrons within the substrate-metal-O2 bonding network can be trapped by mutagenesis of active site residues, metal swapping, or selection of alternative substrates. This “shell game” for electrons exhibits remarkably facile transfer of electron density within the system, but three aspects of the catalytic process appear to be particularly important. First, the Y257 residue enforces the nonplanar distortion of the aromatic ring in HPCD as a means of localizing radical character at C2 and facilitating coupling with the superoxo radical to form the C–O bond; replacing Y257 significantly slows catalysis [22, 38]. Second, substrates that do not have a proton on C1-hydroxyl (e.g. 4NC) when bound in the active site are much less active and in some cases are converted to an alternative product, so the availability of a substrate-derived proton promotes the correct catalysis [29, 33, 37, 39]. As shown in the computational studies, this proton is likely to be used to facilitate the O–O bond cleaving reaction [8, 14, 23, 30, 50]. Finally, all residues other than His substituted at position 200 slow catalysis and in some cases lead to the incorrect product [33, 36, 37, 39]. Potential roles unique to His include: acid-base catalysis, steric effects that stabilize side-on O2 binding, and the development of a localized positive charge when it acts in the base-catalyst mode. The latter may be important in stabilizing the one-electron transfer to bound O2 from the iron or substrates. The specific metal that is present has some effect on catalysis based on properties; for instance, the change in case of Co-HPCD from high-spin to low-spin and back as the reaction proceeds may slow the reaction and reveal steps that would normally be hidden [39–41]. However, in no case is the magnitude of the effect on catalytic rate in line with the change in standard M(III/II) midpoint potential of the metal. It is likely that the most important properties that define a functional metal are the ability to bind in an octahedral coordination using the 2-His-1-carboxylate ligands, and the ability to form an initial weak interaction with O2 that can be strengthened by electron transfer from the substrate. We believe that the latter has been demonstrated crystallographically for two enzymes and is an essential aspect of the extradiol dioxygenase mechanism, but no DFT calculation has been able to reproduce this feature thus far.
The observation of intermediates in modified enzyme systems is useful in that it reveals possible reactions at the metal center. However, they must be interpreted with caution in the context of the actual mechanism used by a given enzyme. The actual mechanism is always a blend of the chemistry that is possible and the rate at which this chemistry occurs. It is often the case that the chemistry can take more than one course, but nature can bias the course actually taken by controlling the kinetics. For example, the computational studies indicate that end-on O2 binding is preferred and a reactive metal(III)-superoxo species is formed. However, only side-on O2 binding has been observed in crystallographic studies when His200 is present. Variants that do allow end-on O2 binding exhibit very slow turnover. Nature is apparently able to bias the binding orientation and substrate alignment to optimize specificity and turnover rate [14]. Similarly, computational studies show that the planar aromatic substrate is reactive with the activated metal-O2 species, but a planar substrate is not observed in structural studies so long as Y257 is in its normal position. This observation includes both a variety of enzyme-substrate complexes and the Fe(II)(semiquinone)superoxide intermediates [22]. It seems likely that when the substrate distortion is eliminated in the Y257F variant, the attack of the activated O2 species is slowed so that there is time to proceed to the structurally characterized substrate quinone-Fe(II)-peroxo intermediate (Scheme 3H) [22]. This species can also undergo ring cleavage in some cases, but the rate is slow. Although the peroxo intermediate is active and thus could represent a key part of the mechanism of extradiol dioxygenases [33, 36–38], nature has apparently selected against it in favor of the more rapid and specific radical recombination [36]. The metal(III)-superoxo species can be formed in each of the metal variants of HPCD, but the reactions of these species with substrates are very slow in comparison to the reactions that occur in the unmodified metal(II)-enzyme with the natural substrate [39, 43]. It is possible that the metal(III)-superoxo state is a species on the reaction path, but if so, its lifetime becomes vanishing short in the most efficient HPCD systems [36]. To the extent that electron transfer between metal and O2 and between substrate and metal become simultaneous, the reaction becomes independent of the metal potential as observed to a good approximation for extradiol dioxygenases functioning with their native substrate.
Many challenges remain on both the experimental and computational fronts of non-heme Fe(II) oxygenase O2 activation chemistry. The application of intermediate trapping and structural characterization in crystallo is already showing that very similar species are found for HPCD [13] and homogentisate dioxygenase [47]. To date, transient kinetic studies of only a few active site variants of HPCD and a limited range of substrates have been studied with the goal of trapping intermediates in sufficient yield for spectroscopic studies. Additional studies along these lines would clearly be beneficial. Moreover, the studies described here show that it is likely that similar intermediates will be detected and trapped for many members of the Fe(II) oxygenase family. This, in turn, will allow computational studies to be extended to larger active site models and to more enzymes as intermediates are discovered and structurally characterized. Finally, expanded small molecule model studies may allow the results from computational and experimental studies to be examined and tested in a common, accessible framework.
Fig. 5.
EPR spectra of freeze-quench samples of anaerobic enzyme-substrate complex [Co-HPCD(4NC)] (purple) rapidly mixed with O2-saturated buffer under 2 atm O2 at 22 °C in 50 mM MES buffer pH 6.0, showing formation of the [Co-HPCD(4NC)O2] intermediate (red) at 2 min and subsequent decay to Co-HPCD (orange) and the extradiol ring-cleaved product after 60 min. Initial concentration of reactants were 0.5 mM [Co-HPCD(4NC)] and 2.75 mM O2. Reprinted from reference [39] with permission.
References
- 1.Vaillancourt FH. Crit Rev Biochem Mol Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Colabroy KL, Begley TP, Ealick SE. Biochemistry. 2005;44:7632–7643. doi: 10.1021/bi047353l. [DOI] [PubMed] [Google Scholar]
- 3.Harpel MR, Lipscomb JD. J Biol Chem. 1990;265:22187–22196. [PubMed] [Google Scholar]
- 4.Hayes RP, Green AR, Nissen MS, Lewis KM, Xun L, Kang C. Mol, Microbiol. 2013;88:523–536. doi: 10.1111/mmi.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colabroy KL, Smith IR, Vlahos AH, Markham AJ, Jakubik ME. Biochim. Biophys Acta. 2014 doi: 10.1016/j.bbapap.2013.12.005. In press: http://dx.doi.org/10.1016/j.bbapap.2013.1012.1005. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Cañón J, Granadino B, de Bernabé DB-V, Renedo M, Fernández-Ruiz E, Peñalva M, de Córdoba SR. Nat Genet. 1996;14:19–24. doi: 10.1038/ng0996-19. [DOI] [PubMed] [Google Scholar]
- 7.Schwarcz R, Okuno E, White RJ, Bird ED, Whetsell WO., Jr Proc Natl Acad. Sci USA. 1988;85:4079–4081. doi: 10.1073/pnas.85.11.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerson JP, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr Proc Natl. Acad Sci USA. 2008;105:7347–7352. doi: 10.1073/pnas.0711179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr J Biol Inorg. Chem. 2011;16:341–355. doi: 10.1007/s00775-010-0732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato N, Uragami Y, Nishizaki T, Takahashi Y, Sazaki G, Sugimoto K, Nonaka T, Masai E, Fukuda M, Senda T. J Mol Biol. 2002;321:621–636. doi: 10.1016/s0022-2836(02)00673-3. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto K, Senda T, Aoshima H, Masai E, Fukuda M, Mitsui Y. Structure. 1999;7:953–965. doi: 10.1016/s0969-2126(99)80122-1. [DOI] [PubMed] [Google Scholar]
- 12.Titus GP, Mueller HA, Burgner J, Rodríguez dCS, Peñalva MA, Timm DE. Nat Struct Biol. 2000;7:542–546. doi: 10.1038/76756. [DOI] [PubMed] [Google Scholar]
- 13.Miller MA, Lipscomb JD. J Biol Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 14.Kovaleva EG, Lipscomb JD. Science. 2007;316:453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vetting MW, Wackett LP, Que L, Jr, Lipscomb JD, Ohlendorf DH. J Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Babbitt PC, Gerlt JA. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 17.Hegg EL, Que L., Jr Eur J Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 18.Bruijnincx PCA, van Koten G, Klein Gebbink RJM. Chem Soc Rev. 2008;37:2716–2744. doi: 10.1039/b707179p. [DOI] [PubMed] [Google Scholar]
- 19.Koehntop KD, Emerson JP, Que L., Jr J Biol Inorg Chem. 2005;10:83–97. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 20.Vaillancourt FH, Barbosa CJ, Spiro TG, Bolin JT, Blades MW, Turner RFB, Eltis LD. J Am Chem Soc. 2002;124:2485–2496. doi: 10.1021/ja0174682. [DOI] [PubMed] [Google Scholar]
- 21.Yam KC, Addison CJ, Farquhar ER, Mbughuni MM, Fielding AJ, Lipscomb JD, Blades MW, Que LJ, Turner RFB. unpublished manuscript [Google Scholar]
- 22.Kovaleva EG, Lipscomb JD. Biochemistry. 2012;51:8755–8763. doi: 10.1021/bi301115c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emerson JP, Wagner ML, Reynolds MF, Que L, Jr, Sadowsky MJ, Wackett LP. J Biol Inorg Chem. 2005;10:751–760. doi: 10.1007/s00775-005-0017-1. [DOI] [PubMed] [Google Scholar]
- 24.Bratsch SG. J Phys Chem Ref Data. 1989;18:1–21. [Google Scholar]
- 25.Jackson TA, Brunold TC. Acc Chem Res. 2004;37:461–470. doi: 10.1021/ar030272h. [DOI] [PubMed] [Google Scholar]
- 26.Vance CK, Miller A-F. J Am Chem Soc. 1998;120:461–467. [Google Scholar]
- 27.Yikilmaz E, Porta J, Grove LE, Vahedi-Faridi A, Bronshteyn Y, Brunold TC, Borgstahl GEO, Miller A-F. J Am Chem Soc. 2007;129:9927–9940. doi: 10.1021/ja069224t. [DOI] [PubMed] [Google Scholar]
- 28.Northrop DB. J Chem Ed. 1998;75:1153–1157. [Google Scholar]
- 29.Groce SL, Miller-Rodeberg MA, Lipscomb JD. Biochemistry. 2004;43:15141–15153. doi: 10.1021/bi048690x. [DOI] [PubMed] [Google Scholar]
- 30.Arciero DM, Lipscomb JD. J Biol Chem. 1986;261:2170–2178. [PubMed] [Google Scholar]
- 31.Shu L, Chiou YM, Orville AM, Miller MA, Lipscomb JD, Que L., Jr Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 32.Bugg TDH. Tetrahedron. 2003;59:7075–7101. [Google Scholar]
- 33.Groce SL, Lipscomb JD. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 34.Lipscomb JD. Curr Opin Chem Biol. 2008;18:644–649. doi: 10.1016/j.sbi.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spence EL, Langley GJ, Bugg TDH. J Am Chem Soc. 1996;118:8336–8343. [Google Scholar]
- 36.Mbughuni M, Chakrabarti M, Hayden JA, Meier KK, Dalluge JJ, Hendrich MP, Münck E, Lipscomb JD. Biochemistry. 2011;50:10262–10274. doi: 10.1021/bi201436n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mbughuni MM, Chakrabarti M, Hayden JA, Bominaar EL, Hendrich MP, Münck E, Lipscomb JD. Proc Natl Acad Sci USA. 2010;107:16788–16793. doi: 10.1073/pnas.1010015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mbughuni MM, Meier KK, Münck E, Lipscomb JD. Biochemistry. 2012;51:8743–8754. doi: 10.1021/bi301114x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fielding AJ, Lipscomb JD, Que L., Jr J Am Chem Soc. 2012;134:796–799. doi: 10.1021/ja2095365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RD, Summerville DA, Basolo F. Chem Rev. 1979;79:139–179. [Google Scholar]
- 41.Smith TD, Pilbrow JR. Coord Chem Rev. 1981;39:295–383. [Google Scholar]
- 42.Fielding AJF, Kovaleva EG, Farquhar ER, Lipscomb JD, Que LJ. J Biol Inorg. Chem. 2010;16:341–355. doi: 10.1007/s00775-010-0732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunderson WA, Zatsman AI, Emerson JP, Farquhar ER, Que L, Jr, Lipscomb JD, Hendrich MP. J Am Chem Soc. 2008;130:14465–14467. doi: 10.1021/ja8052255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs C, Galonić Fujimoori D, Barr EW, Walsh CT, Bollinger JM., Jr Acc. Chem Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovaleva EG, Lipscomb JD. Biochemistry. 2008;47:11168–11170. doi: 10.1021/bi801459q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borowski T, Georgiev V, Siegbahn Per EM. J Mol Model. 2010;16:1673–1677. doi: 10.1007/s00894-010-0652-5. [DOI] [PubMed] [Google Scholar]
- 47.Jeoung J-H, Bommer M, Lin T-Y, Dobbek H. Proc Natl Acad Sci USA. 2013;110:12625–12630. doi: 10.1073/pnas.1302144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zatkova A. J Inherit Metab Dis. 2011;34:1127–1136. doi: 10.1007/s10545-011-9363-z. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez JM, Timm DE, Titus GP, Beltrán-Valero de Bernabé D, Criado O, Mueller HA, Rodríguez dCS, Peñalva MA. Hum Mol Genet. 2000;9:2341–2350. doi: 10.1093/oxfordjournals.hmg.a018927. [DOI] [PubMed] [Google Scholar]
- 50.Siegbahn PEM, Haeffner F. J Am Chem Soc. 2004;126:8919–8932. doi: 10.1021/ja0493805. [DOI] [PubMed] [Google Scholar]
- 51.Bassan A, Borowski T, Siegbahn Per EM. Dalton T. 2004:3153–3162. doi: 10.1039/B408340G. [DOI] [PubMed] [Google Scholar]
- 52.Georgiev V, Borowski T, Siegbahn Per EM. J Biol Inorg Chem. 2006;11:571–585. doi: 10.1007/s00775-006-0106-9. [DOI] [PubMed] [Google Scholar]
- 53.Georgiev V, Borowski T, Blomberg MRA, Siegbahn Per EM. J Biol Inorg. Chem. 2008;13:929–940. doi: 10.1007/s00775-008-0380-9. [DOI] [PubMed] [Google Scholar]
- 54.Christian GJ, Ye S, Neese F. Chem Sci. 2012;3:1600–1611. [Google Scholar]
- 55.Dong G, Shaik S, Lai W. Chem Sci. 2013;4:3624–3635. [Google Scholar]