Abstract
Systems pharmacology models capable of accurately recapitulating sophisticated patient phenotypes have enabled the investigation of mechanisms responsible for therapeutic efficacy. Although omics data sets are capable of characterizing the operation of subcellular networks, their utility in mechanistically predicting quantitative, clinically accessible outcome measures has been limited. Developing insights into clinical outcomes from omics data sets will benefit from modeling approaches that can integrate molecular networks mechanistically with simulations of patient pathophysiology across compartments and scales.
Perhaps the most well-known model of pathophysiology with proven clinical utility for chronic disease was the minimal model of glucose homeostasis developed by Bergman et al.1 I recently watched Dr Bergman present his renowned work to an audience of young bioengineers at the University of California, San Diego. One student was not sure how to characterize the study and asked Dr Bergman whether his work was “systems biology.” The philosophical inclination of the current generation of biologically inclined engineers who are matriculating in a biological science environment that is data rich was palpable. At the very least, we have witnessed a paradigm shift in research driven by new technologies that has defined a mature field in some of our institutions of higher learning.
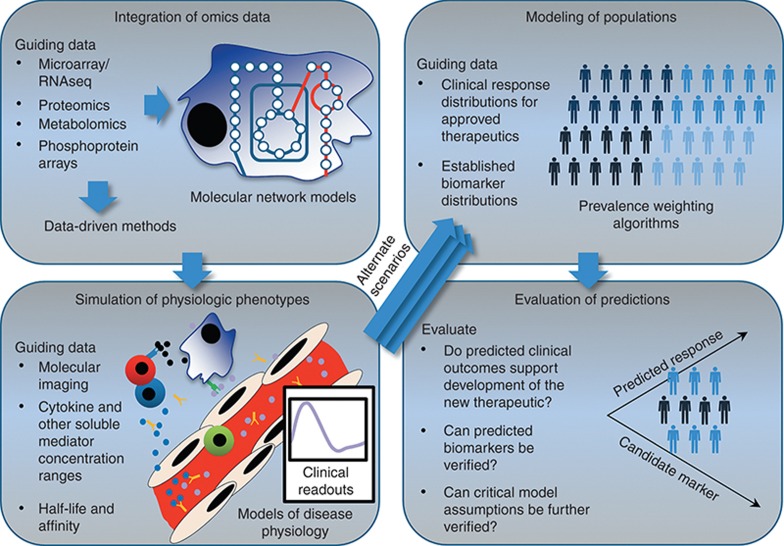
Before, and concurrently with, the aforementioned omics revolution, there has been a growth of systems pharmacology models, the development of which has been driven by the need to recapitulate dynamic and complex patient phenotypes.2 Many models with predictive value have been developed from medical research, with demonstrated tangible impacts on drug development from discovery through the clinical stage.3 Systems pharmacology models that mechanistically simulate clinical phenotypes have been presented in the peer-reviewed literature with validation.2 However, there is arguably substantial utility for modeling approaches that are both compatible with omics data sets and ultimately mechanistically insightful with respect to quantifiable clinical end points (clinical phenotypes) in medical research and drug development. Separate research studies have demonstrated individual, concrete steps to model pathophysiology mechanistically across scales, from detailed molecular mechanisms up to clinical outcomes for a simulated population, as will be discussed here and as shown in Figure 1.
Figure 1.
Large data sets can be used to guide mechanistic models and ultimately develop quantitative predictions of clinical phenotypes at the population level. Omics data may be used to inform network models directly, or instead, to inform data-driven methods to assess cellular states. The resulting predictions for cellular function (cellular “exhaustion,” dysfunctional or inactive pathways, etc.) can be used to inform simulations of physiologic phenotypes that can be assessed and quantified in a clinical setting. Calibration or validation of the simulated physiologic phenotypes has been performed with additional data sets such as molecular imaging data and mediator concentrations. Given alternate physiologic parameters or alterations in the underlying network, alternate clinical outcomes are predicted. Disease outcome data can then be used to calculate the prevalence of the modeled patient phenotypes. The results are a prediction for the frequency of occurrence of underlying mechanisms in the disease pathophysiology, a rich simulated data set to mine for markers, and a validated disease-modeling platform that can be mechanistically altered to hypothesize and prioritize opportunities for therapeutic intervention. Portions adapted from refs. 3 and 9.
Systems biology approaches have been applied to develop insights into alterations in molecular pathways and targets from omics data sets.2 For example, there are many illustrative studies that model complete cellular metabolism in both healthy and pathological states.4 Although it is recognized that there are still gaps in the knowledge of all of the metabolic conversions that occur in a human cell,4 metabolism is sufficiently well described to recreate functional cellular networks for many tissue types.3,4 Genome-scale models of metabolism (GEMs) have, therefore, served as frameworks for omics data integration and mechanistic interpretation that have been increasingly applied in medically relevant contexts.4 GEMs mechanistically model the conversion of thousands of metabolites through reactions and also contain mappings from transcripts to associated proteins, reactions, and metabolites for all of the known metabolic reactions in a cell. GEMs, therefore, facilitate the mechanistic integration of omics data and the impact of medically relevant interventions on the function of a cell's metabolic network.3,4
Notably, despite their successful application, the scope of GEMs is necessarily limited. For example, incorporating the capability to predict the mechanistic impact of additional cellular processes, such as signaling and transcriptional regulation, at the genome scale in human tissues presents additional, significant challenges. Furthermore, network functions that have been successfully modeled with GEMs of human cells may not be best suited for the measures that are clinically accessible or possess established clinical relevance. For example, GEMs directly model allowed rates of reaction or transport as flux. However, it is generally easier to measure the serum concentration for a panel of metabolites than to determine their rates of consumption and secretion from a given tissue. Recently, Krauss et al.5 developed an innovative genome-scale modeling approach to recapitulate phenotypes that have been quantitatively measured in the clinic. Krauss et al.5 integrated a GEM with a physiologically based pharmacokinetic model to simulate the effect of hepatic metabolic network function on plasma metabolite concentrations. A key development that facilitated the integrated approach was the availability of HepatoNet1,6 a GEM developed specifically to model liver metabolism. HepatoNet1 was coupled with the consumption and production of metabolites in the appropriate compartment of the physiologically based pharmacokinetic model. Krauss et al. demonstrated a number of applications of the integrated model, from the study of inborn errors of metabolism to drug-induced toxication. Notably, the integrated approach by Krauss et al. demonstrated a prediction of the quantitative concentration profile of metabolites, such as ammonia and uric acid, in the plasma that was consistent with clinical observations.
In analogy with population pharmacokinetic/pharmacodynamic modeling, the evaluation of systems models at the population scale presents an additional, important step in both validating and using models as a decision-making tool when developing new medical treatments. The characterization of diversity in a cohort of simulated patients has previously been reported in the context of selecting clinical trial end points and optimizing trial design.3 There have been a few published algorithms capable of linking the outcomes for simulated patients, often referred to as virtual patients, to clinical outcomes at the population scale.7,8,9 One primary element has been to manipulate the prevalence of the simulated patients so that each simulated clinical measure, including the response to medicines, rate of disease progression, and additional simulated biomarkers, exhibits a quantitatively similar average and diversity as the real clinical population. One method to simulate population responses has been developed by Klinke7 for a model of type 2 diabetes, and the method takes advantage of the data available from the National Health and Nutritional Examination Survey III. One strength of the approach, which used direct fitting of the multidimensional distribution data from the National Health and Nutritional Examination Survey III, was that it allowed for a comprehensive validation of the simulated population's multivariate diversity, including the correlation of clinical measures. A second approach to simulate population responses was used to develop simulated populations of drug-induced liver injury.8 Rather than adjusting weights after developing the simulated patients, this approach guides the creation of each individual simulated patient based on the comparison to available data for both the clinical outcomes and specified parameters. A third method for developing simulated populations was used to match the response of simulated patients from a model of rheumatoid arthritis to a number of large clinical trials of biologics.9 This approach enabled the assessment of how changes in the prevalence of individual patients in the population would influence the observed mean efficacy in a trial and the resulting impact on patient stratification biomarkers.
One important challenge that, if addressed, would enhance the utility of systems pharmacology models is an improvement in the ability to interface with the data sets of increasing comprehensiveness that are becoming available. There will be much utility in the development of models that simulate clinical end points and can also be readily informed with omics data sets to address the effects of interventions on clinically quantifiable disease outcome measures given the available knowledge of molecular factors. The model developed by Krauss et al.5 is an excellent example, because their model simulates plasma metabolite concentrations and also contains the mappings that define the dependencies of hepatic metabolic reactions on messenger RNA transcripts and enzymes. In other words, Krauss et al. used a complete metabolic network reconstruction as a bridge to calculate cellular metabolic pathway activity, which can in turn be used to interpret the implications of transcriptomics or proteomics data sets on plasma end points. Alternatively, systems models may be developed and refined to include critical pathways identified from data-driven approaches. There are clinical trials that have taken advantage of the results of mining patient data in their design. For example, the Biomarkers of Anti-TNF Treatment Efficacy in Rheumatoid Arthritis–Unresponsive Populations (BATTER-UP) trial is currently evaluating an eight-gene classifier to predict the patients who will not respond to anti–tumor necrosis factor-α agents (http://clinicaltrials.gov/, trial NCT01211678). Notably, the interpretation of omics data sets to refine model scope is an important step in the development of enhanced pharmacodynamic models.10 When effectively coupled to omics data sources, modeling patient physiology promises to lend insight into the mechanistic role for markers identified by systems biology methods in therapeutic responses and disease progression.
Published studies have, therefore, begun to illustrate the link from molecular systems biology models to dynamic, quantifiable clinical outcomes at the population level. The establishment of new, integrative disease-modeling platforms that facilitate developing quantitative predictions for simulated patient outcomes from omics data sets will both fundamentally advance systems biology and improve medical research and drug development. However, the challenge of optimally deploying systems modeling to impact the development of new medicines is twofold. As has been discussed, mechanistic systems models can be developed and intelligently deployed to serve as a useful framework to interpret the content of omics data sets in the context of their functional implications. In addition, for maximum utility and confidence in model predictions, meaningful efforts to refine model parameters, pathways, and scope must also be performed. This requires prospectively developing quantitative hypotheses, challenging them with new data sets and experiments, and subsequent model refinement. Using systems modeling approaches well, therefore, also requires the development of new experiments that have not been previously performed, which requires time and planning.10 Mechanistic simulation and omics scientists are limited to available parameter and pathway information from databases and the literature, which may be inconsistent or incomplete. Life scientists must, therefore, also provide input into model development and refinement. Model development and revision are, therefore, best viewed as an active, nontrivial undertaking,3,10 requiring concerted planning and efforts by preclinical and clinical scientists with varied expertise.
Conflict of Interest
The author declared no conflict of interest.
References
- Bergman R.N., Ider Y.Z., Bowden C.R., Cobelli C. Quantitative estimation of insulin sensitivity. Am. J. Physiol. 1979;236:E667–E677. doi: 10.1152/ajpendo.1979.236.6.E667. [DOI] [PubMed] [Google Scholar]
- Vicini P., van der Graaf P.H. Systems pharmacology for drug discovery and development: paradigm shift or flash in the pan. Clin. Pharmacol. Ther. 2013;93:379–381. doi: 10.1038/clpt.2013.40. [DOI] [PubMed] [Google Scholar]
- Schmidt B.J., Papin J.A., Musante C.J. Mechanistic systems modeling to guide drug discovery and development. Drug Discov. Today. 2013;18:116–127. doi: 10.1016/j.drudis.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A., Gatto F., Nielsen J. Genome-scale modeling of human metabolism - a systems biology approach. Biotechnol. J. 2013;8:985–996. doi: 10.1002/biot.201200275. [DOI] [PubMed] [Google Scholar]
- Krauss M., Schaller S., Borchers S., Findeisen R., Lippert J., Kuepfer L. Integrating cellular metabolism into a multiscale whole-body model. PLoS Comput. Biol. 2012;8:e1002750. doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille C., et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol. Syst. Biol. 2010;6:411. doi: 10.1038/msb.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke D.J., 2nd Integrating epidemiological data into a mechanistic model of type 2 diabetes: validating the prevalence of virtual patients. Ann. Biomed. Eng. 2008;36:321–334. doi: 10.1007/s10439-007-9410-y. [DOI] [PubMed] [Google Scholar]
- Howell B.A., et al. In vitro to in vivo extrapolation and species response comparisons for drug-induced liver injury (DILI) using DILIsym™: a mechanistic, mathematical model of DILI. J. Pharmacokinet. Pharmacodyn. 2012;39:527–541. doi: 10.1007/s10928-012-9266-0. [DOI] [PubMed] [Google Scholar]
- Schmidt B.J., Casey F.P., Paterson T., Chan J.R. Alternate virtual populations elucidate the type I interferon signature predictive of the response to rituximab in rheumatoid arthritis. BMC Bioinformatics. 2013;14:221. doi: 10.1186/1471-2105-14-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar R., Zhao S., Chung S.W., Mager D.E., Gallo J.M. Merging systems biology with pharmacodynamics. Sci. Transl. Med. 2012;4:126ps7. doi: 10.1126/scitranslmed.3003563. [DOI] [PMC free article] [PubMed] [Google Scholar]