Abstract
The water flea Daphnia magna has been used as an animal model in ecology, evolution, and environmental sciences. Thanks to the recent progress in Daphnia genomics, genetic information such as the draft genome sequence and expressed sequence tags (ESTs) is now available. To investigate the relationship between phenotypes and the available genetic information about Daphnia, some gene manipulation methods have been developed. However, a technique to induce targeted mutagenesis into Daphnia genome remains elusive. To overcome this problem, we focused on an emerging genome editing technique mediated by the clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system to introduce genomic mutations. In this study, we targeted a functionally conserved regulator of eye development, the eyeless gene in D. magna. When we injected Cas9 mRNAs and eyeless-targeting guide RNAs into eggs, 18–47% of the survived juveniles exhibited abnormal eye morphology. After maturation, up to 8.2% of the adults produced progenies with deformed eyes, which carried mutations in the eyeless loci. These results showed that CRISPR/Cas system could introduce heritable mutations into the endogenous eyeless gene in D. magna. This is the first report of a targeted gene knockout technique in Daphnia and will be useful in uncovering Daphnia gene functions.
Introduction
The water flea Daphnia magna is a planktonic crustacean ubiquitously found in the fresh water environment. It has been used as a model organism in ecology and toxicology because it is sensitive to artificial chemicals and environmental changes [1]. Moreover, researchers find it interesting that Daphnia can switch their reproduction mode from asexual to sexual in response to environmental stimuli [2]. Recent progress in genomics involved analyses of expressed sequence tags (ESTs) [3] and the draft genome sequence of D. magna. In addition, the genome sequence of a related organism, D. pulex, has recently been completed [4]. Therefore, a vast amount of genetic information on Daphnia is now available. To investigate the relationship between available genetic information and phenotypes, gene manipulation tools such as RNA interference (RNAi) and non-homologous integration with plasmid DNA have been developed in D. magna [5], [6]. However, there is still no technique to induce inheritable targeted gene disruptions.
The clustered regularly interspaced palindromic repeats/CRISPR-associated (CRISPR/Cas) system is a recently developed tool to induce targeted mutagenesis. CRISPR/Cas was initially identified as the bacterial immune system to bacteriophages [7]. In this system, a CRISPR RNA (crRNA) interacts with a transactivating CRISPR RNA (tracrRNA) and forms the tracrRNA:crRNA duplex, which acts as a guide RNA (gRNA) that directs the endonuclease Cas9 to its cognate target DNA and induces double-strand breaks (DSBs) [8]. Importantly, the cleavage site is often imperfectly repaired by the error-prone non-homologous end-joining (NHEJ) mechanism, resulting in gene disruptions through the introduction of small insertions or deletions (in-dels). Recent studies reported that a single chimeric RNA, a crRNA fused with a tracrRNA, could also function as a gRNA [8]. The approximately 20 bp targetable sites are limited by the requirement for the protospacer adjacent motif (PAM; 5′-NGG-3′) at their 3′ end [8]. Further constraint that target sites start with a GG dinucleotide is often required when gRNAs are synthesized in vitro by T7 polymerase. However, recent studies suggested some alternatives to circumvent the latter limitation without significant reduction of cleavage efficiency [9]–[11].Therefore, to induce targeted mutagenesis, co-expression of the customized gRNA with the Cas9 nuclease has been used in various organisms [9]–[25] (Figure 1A). Compared with the other targeted mutagenesis tools such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), the experimental design of CRISPR/Cas is remarkably simple and rapid [26]. However, nobody has applied these emerging techniques to Daphnia.
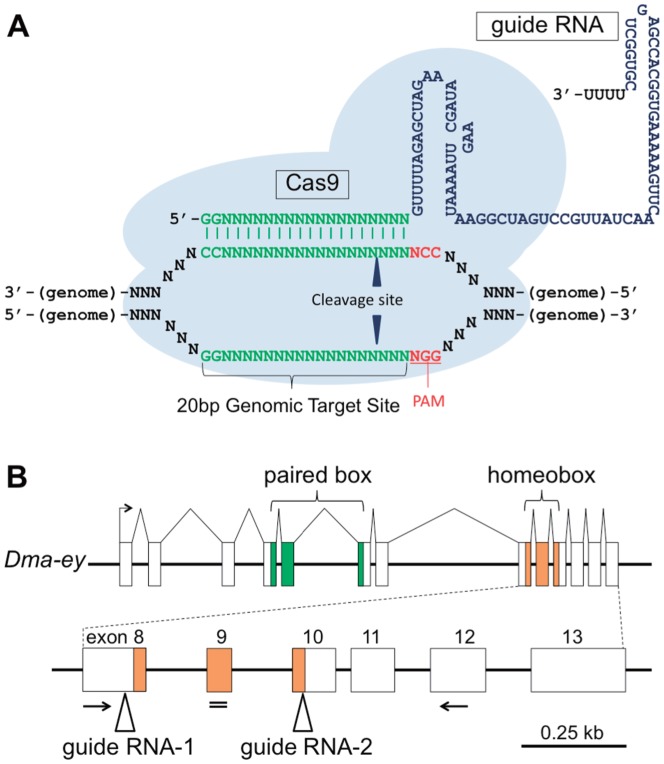
Figure 1. A target gene for the CRISPR/Cas-based targeted mutagenesis: D. magna eyeless (Dma-ey) gene.
(A) Schematic illustration of the CRISPR/Cas system. The guide RNA (gRNA) binds to the 20-bp genomic target site with its complementary sequences (green). The genomic target site requires the PAM sequence (NGG; red) immediately downstream of its 3′ side. The latter sequences of gRNA (blue) interact with Cas9 nuclease (light blue spheres). Cleavage sites are shown by triangles. (B) Schematic illustration of Dma-ey locus. Dma-ey gene putatively consists of 13 exons (shown by open boxes). DNA-binding domain-encoding regions are colored in green (paired box) and orange (homeobox). Primers designed for RT-PCR are indicated by arrows. siRNA was targeted in a site shown by a double line. gRNA-targeted sites are depicted by triangles.
The mammalian pax6 gene ortholog is a functionally conserved regulator for eye development that has been conserved from invertebrates to vertebrates. It encodes a transcription factor with two DNA-binding domains: paired box and homeobox domains. In Drosophila, a mammalian pax6 homolog was originally mapped in the eyeless (ey) locus [27], whose mutants show abnormal eye morphogenesis, resulting in complete or partial loss of eye as well as mutations in the mouse pax6 gene [27]–[31]. Animals with abnormal eyes could be easily distinguished from normal ones in appearance, suggesting that pax6 ortholog would be a useful model target gene for targeted mutagenesis as previously used for TALEN-mediated mutagenesis [32].
In the present study, D. magna ey gene was partially cloned and its role in eye development was confirmed by RNAi, thus prompting us to use ey as a target for CRISPR/Cas-mediated mutagenesis. We showed that the CRISPR/Cas system efficiently introduces heritable in-del mutations into the ey loci of the D. magna genome, resulting in deformations of the compound eye. In addition, undesired mutations (off-target mutations) were not detected in the genomes of the ey-deficient daphniids. Thus, we concluded that the CRISPR/Cas system is a powerful tool for modifying targeted genomic sites and can facilitate studies on the functional genomics of D. magna.
Results
Functional analyses of D. magna eyeless (Dma-ey) gene
To utilize the ey gene as a marker gene for targeted mutagenesis, we searched for mammalian pax6 orthologs in the D. magna genome and found the ey gene in addition to its paralog, twin of eyeless (toy) gene, both of which are conserved among several arthropods [33]. We named D. magna ey as Dma-ey in this study. By using the D. magna genome database, the Dma-ey gene was predicted to consist of 13 exons (Figure 1B). Reverse transcription-PCR (RT-PCR) of the Dma-ey gene using a primer set encompassing the homeobox as well as sequencing of the PCR fragments revealed that Dma-ey is expressed in D. magna (Figure S1).
Next, we tested whether the Dma-ey gene is required for compound eye development by RNAi. We designed siRNA within the homeobox of Dma-ey. A compound eye in a wild-type daphniid is located at the anterior parietal edge and roughly spherical in shape (Figure 2A, left), whereas daphniids injected with 100 µM siRNA had deformed compound eyes, which were located at the inner side and could not build a precise spherical shape (Figure 2A, right). Afterward, we call this phenotype as deformed eye in this study. Taken together, our results indicated that the eyeless gene is functionally conserved in D. magna, suggesting that it would be a useful marker gene for subsequent knockout experiments using the CRISPR/Cas system.
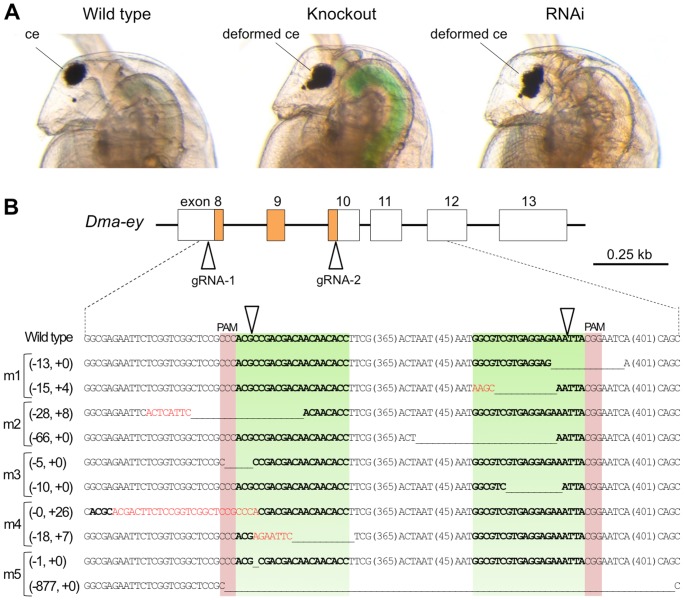
Figure 2. Knockout of Dma-eye gene.
(A) Typical phenotypes of Dma-ey deficient daphniids. The images to the left, center and right show the lateral head parts of the wild-type daphniid, Dma-ey knocked-out daphniid by the CRISPR/Cas system, and Dma-ey RNAi daphniid, respectively. The knocked-out individual is “mutant #5 (m5)” described in Figure 2B. Ventral side is left. ce: compound eye. (B) Genome sequences of established Dma-ey-knocked out mutant lines around gRNA-targeted sites. A part of the exon-intron structure of the Dma-ey gene including the homeobox colored in orange is shown above. In the alignment, the top line in the below alignment represents the wild-type Dma-ey sequence, and subsequent lines show sequences of five mutant alleles (see Table 1). The target sites for gRNAs are indicated in green, PAM in red, and cleavage site by triangles. The length (base pairs) of each in-del mutation is written in the left of each sequence (-, deletions; +, insertion). In each mutant sequence, deletion is indicated by underbars, insertions by red letters, sequences corresponding to the wild-type targeted sequences by bold letters, and the length (base pairs) of an abbreviated sequence within a parenthesis.
Dma-ey gene disruption by CRISPR/Cas system
To test whether the CRISPR/Cas system could induce targeted mutagenesis in D. magna, we attempted to introduce in-del mutations into the homeobox domain of the Dma-ey locus. Previous reports described ey-deficient Drosophila flies whose mutated ey allele lacked all C-terminal domains including the homeobox domain [31]. Thus, we hypothesized that Dma-ey-deficient daphniids can be generated by inducing frameshifts at an earlier part of the homeobox domain. To target the homeobox region, we used two gRNAs, gRNA-1 and gRNA-2, which were designed to bind the sense strand from exon 8 and the anti-sense strand from exon 10, respectively (Figures 1B and 2B). Because a previous report suggested that co-injection of multiple gRNAs increased mutation efficiency [17], we coinjected two gRNAs together with the Cas9 mRNA that contained the untranslated regions (UTRs) of D. magna vasa (Dmavas) gene, which is exclusively expressed in D. magna germ cells [34]. We tested three and two different concentrations of Cas9 mRNA (500; 1,000; 2,000 ng/µL) and gRNAs (50; 150 ng/µL each), respectively (Table 1). At the first instar juvenile stage, 61–78% of the injected embryos survived. In addition, as expected, 18–47% of the surviving juveniles developed deformed eye phenotypes without significant dose dependency, suggesting that the CRISPR/Cas system worked in D. magna (Table 1).
Table 1. Mutation frequencies induced by microinjection using various concentrations of Cas9 mRNA/gRNA mix.
| RNA concentration (ng/µL) | Embryos | Juveniles | Adults | |||
| gRNA mix | Cas9 mRNA | Injected | Surviving | Deformed eye | Surviving | Founder lines |
| 50 each | 500 | 77 | 59/77 (77%) | 28/59 (47%) | 49/77 (64%) | *4/49 (8.2%) |
| 1,000 | 121 | 90/121 (74%) | 16/90 (18%) | 81/121 (67%) | 5/81 (6.2%) | |
| 2,000 | 113 | 75/113 (66%) | 29/75 (39%) | 61/113 (54%) | 0/61 (0%) | |
| 150 each | 500 | 98 | 70/98 (71%) | 19/70 (27%) | 60/98 (61%) | 2/60 (3.3%) |
| 1,000 | 86 | 67/86 (78%) | 20/67 (30%) | 60/86 (70%) | *2/60 (3.3%) | |
| 2,000 | 64 | 39/64 (61%) | 8/39 (21%) | 38/64 (59%) | *1/38 (2.6%) | |
*Mutant lines subjected to sequencing of Dma-ey loci. In Figure 2B, two mutants named m1 and m2 were from 4 mutants injected with 50 ng/µL each of gRNA and 500 ng/µL Cas9 mRNA (50 each, 500), m3 and m4 from 2 mutants (150 each, 1,000), and m5 from 1 mutant (150 each, 2,000).
After the injected animals matured, we counted the number of adults producing deformed eye G1 progenies ( = founder G0 animals) and normal eye G1 progenies, respectively, to calculate the efficiency in inducing heritable mutations. The founder G0 animals were generated at a rate of 2.6–8.2% of the surviving adults, excluding the case when 50 ng/µL each of gRNA-1 and gRNA-2 were injected with 2,000 ng/µL Cas9 mRNA (Table 1 and Figure 2A). The highest efficiency was achieved by injecting 50 ng/µL each of gRNA-1 and gRNA-2 with 500 ng/µL Cas9 mRNA. Of the 14 founder animals, 13 founders produced deformed eye progenies which died within a week. The lethality of Dma-ey knockout mutants was consistent with the previous report on Drosophila [31]. However, the remaining founder animal produced viable deformed eye progenies and the phenotype was observed over generations. The difference of viability between mutant lines is discussed in detail (see Discussion). In sum, these results suggest that the CRISPR/Cas system worked not only in somatic cells but also in germ line cells.
To investigate how in-del mutations were introduced into the Dma-ey loci, we collected deformed eye G1 progenies from 5 different founder lines and extracted their genomic DNAs respectively (Table 1). Genomic PCR products encompassing the gRNA-targeted sites were cloned and sequenced. All of the deformed eye G1 progenies had biallelic in-del mutations around gRNA-targeted sites (Figure 2B). In contrast, monoallelic in-del mutations were found in normal eye G1 progenies (data not shown). Thus, we concluded that the CRISPR/Cas system could induce heritable mutations in the endogenous Dma-ey locus of D. magna.
Evaluation of off-target effects in deformed eye mutants
We further analyzed if Dma-ey-deficient mutants had undesired mutations (off-target mutations). Pioneering works suggested that genomic sites have mismatches fewer than 5 bp with PAM (NGG at 3′ end) sequence could be cleaved ( = potential off-target sites) [19], [35]. By using a BLASTn search on the D. magna genome database, we looked for potential off-target sites and found that gRNA-1 had 4 potential off-target sites, whereas gRNA-2 did not (Table 2). To test if Dma-ey-deficient mutants have off-target in-del mutations, we designed a primer set to amplify each potential off-target site of gRNA-1 and performed PCRs using genomic DNAs from 5 different G1 mutants subjected to previous analyses of in-del mutations in the Dma-ey locus. Consequently, no off-target mutation was observed. These results suggested that the CRISPR/Cas-mediated targeted mutagenesis approach was highly specific.
Table 2. Potential off target sites of gRNA-1.
| On/off target sites (5′-3′) | Locations | Annotations | Mismatches | |
| On target | GGTGTTGTTGTCGTCGGCGTggg | exon | eyeless | - |
| Off target | GGTGTTGGTGTCGTCGGCGTcgg | exon | protocadherin fat 2 precursor | 1 bp |
| GTTGGTGTTGTCGTCGGCATcgg | exon | cytochrome c oxidase | 3 bp | |
| ATTGGTGTTGTCGTCGGCATcgg | exon | cytochrome c oxidase | 4 bp | |
| TGCGCCGTTGACGTCGGCGTtgg | intronic | - | 5 bp |
Genomic sequences of on/off target sites (uppercase) with PAM (NGG, lowercase), their locations, annotations, and the number of base pair differences are noted. Bold letters exhibit mismatched nucleotides. bp: base pairs.
Discussion
Here, we described a simple, rapid, and efficient technique for inducing target mutagenesis into an endogenous locus of the D. magna genome by using a CRISPR/Cas system, which will enable us to perform high-throughput functional analyses of D. magna genes. This system would contribute to overcoming two limitations in previous Daphnia reverse genetics studies using RNAi by microinjection of dsRNAs into eggs [5]: (1) incapability to induce null phenotypes and (2) transient nature of gene manipulation. The knockout Daphnia lines allow us to observe the loss-of-function phenotypes throughout life cycle, which will undoubtedly advance our understanding of D. magna gene functions.
We coinjected pairs of gRNAs, gRNA-1, and gRNA-2, which targeted exons 8 and 10 of Dma-ey, together with Cas9 mRNA. We found that all five Dma-ey G1 mutants with deformed eyes have biallelic mutations. This simple and efficient induction of biallelic mutations seems to be beneficial for researchers studying Daphnia that usually produce parthenogenetic females. For crossing to establish homozygous mutants in Daphnia, we have to induce the production of males and sexual females that lay haploid eggs by stimulating the parthenogenetic females with environmental cues such as shortened photoperiod, lack of food, and/or increased population density, which makes the crossing procedure laborious and time consuming. Ability of the CRISPR/Cas system to induce biallelic mutations would significantly improve the genetics of parthenogenetic organisms, including Daphnia.
In the current study, no founder animals were observed when we used 2,000 ng/µL of Cas9 mRNA and 50 ng/µL of each gRNA for injection. One possible explanation for this result is the aggregation of free abundant Cas9 proteins which prevents them from forming a complex with gRNAs. This interpretation seemingly corresponds with our data since we could establish one founder animal by injection of the same concentration of Cas9 mRNA with a higher concentration of gRNAs (see Table 1). Although the reason is not clear based on our current data, the proportion of Cas9/gRNA used should be important for the activity of CRISPR/Cas systems.
Of the 14 deformed eye mutants established in this study, one was viable. Biallelic Dma-ey mutations should give rise to lethality because previous reports described that homozygous mutation in mammalian pax6 and its homolog ey were lethal to mice and flies, respectively [29], [31]. Our sequence data showed that the viable mutant lost the 877 bp region including the entire homeobox domain on the Dma-ey allele but it retained the correct reading frame of the remaining C-terminal region (Figure 2B, m5), whereas coding frames of the other sequenced alleles were shifted, hence all the sequences from the homeobox domain to the C-terminal region were lost (Figure 2B. m1 to m4). This might account for the difference in the mutants' viability. Previous works described the PAX6 protein as having a conserved C-terminal domain which, follows homeobox domain, is rich in proline (P), serine (S), and threonine (T) residues, and which mediates activation of PAX6 protein via phosphorylation [36], [37], [38]. Therefore, the presence of the C-terminal activation domain on the 877 bp deleted Dma-ey allele might contribute to the viability of the deformed eye mutant D. magna.
Of the 10 alleles from five deformed eye mutants analyzed in this study, nine had indel mutations in one of the two target sites (Figure 2B). The patterns of these mutations were consistent with those induced by Cas9-based cleavage in the other animals. Interestingly, one allele had an 877-bp deletion spanning both target sites, suggesting the possibility that concurrent DSBs at two distantly target sites for gRNAs induced this large deletion, as reported in previous studies [9], [12], [17], [22].
CRISPR-based genome engineering will also provide novel approaches to integrate foreign DNAs into the D. magna genome. DSB sites are predominantly repaired by either NHEJ or homologous recombination (HR) [39]. Double-strand cleavage has been shown to facilitate the rate of homologous gene targeting at the cleaved site [40]. Successes of targeted knock-ins have been reported through the co-introduction of CRISPR components and exogenous DNAs such as plasmids or single-strand oligoDNAs (ssODNs) that have a homologous region to the cleavage site [16], [17], [19], [25]. This approach will enable us to induce targeted knock-in of DNA fragments such as integrase-targeting sequences or epitope tag-coding sequences, even in D. magna.
Thus, CRISPR/Cas system-mediated genome editing technique described here will definitely accelerate the development of Daphnia functional genomics.
Materials and Methods
Daphnia strain and culture conditions
The D. magna strain (NIES clone) was obtained from the National Institute for Environmental Studies (NIES, Tsukuba, Japan) and cultured under laboratory conditions for many generations. To minimize variations in maternal effects that may influence microinjection, we maintained the strain under the following conditions: 120 neonates (under 24 h) were transferred to 5 L of ADaM medium [41] and cultured at 22–24°C, under a light/dark photoperiod of 16 h/8 h. The culture medium was not changed for 3 weeks. Daphniids were fed once a day with 7.5 mg of Chlorella vulgaris (Nikkai Center, Tokyo, Japan) and 7.5 mg of baker's yeast (marusanPantry, Ehime, Japan) during the first week; after they matured, their offspring was removed once per day and they were fed 15 mg of Chlorella and 15 mg of yeast daily. The addition of yeast helped in maintaining the number of juveniles per clutch constantly higher than that observed when fed with Chlorella only (data not shown).
Construction of RNA expression vectors
To generate the Cas9 expression vector pCS-Dmavas-Cas9, the Cas9 ORF was amplified using the plasmid MLM3613 (Addgene plasmid 42251, [18]) as a PCR template. DNA fragments of the 5′ and 3′ UTRs from the Dmavas gene (Accession: AB193324.1) were obtained by PCR using cDNAs of the NIES strain. These PCR products were simultaneously cloned into the downstream region of the SP6 promoter of pCS+ vector using the In-Fusion PCR cloning kit (Clontech, California, USA) [42]. To generate the gRNA expression vector pDR274-Dma-ey, the plasmid DR274 (Addgene plasmid 42250, [18]) was digested with BsaI (NEW ENGLAND BioLabs, Connecticut, USA), followed by dephosphorylation with Antarctic Phosphatase (NEW ENGLAND BioLabs, Connecticut, USA). A pair of Dma-ey targeting oligonucleotides was annealed and then ligated into the linearized pDR274 vector using a ligation mix (TaKaRa Bio, Shiga, Japan). The genomic target sites and sequences of the oligonucleotides constructed in this study are listed in Tables S1 and S2. All PCRs in this section were performed with PrimeSTAR (Takara Bio, Shiga, Japan).
In vitro RNA synthesis
siRNAs for knocking down the Dma-ey gene were designed by using Block-iT RNAi Designer (Life Technologies, California, USA) and two nucleotides dTdT were added to each 3′ end of the siRNA strand. The sequences of the siRNA are listed in Table S1.
For the syntheses of Cas9 mRNAs, templates with T7 promoter were amplified by PCR from the pCS-Dmavas-Cas9. The primer sequences for the PCR are shown in Table S2. Amplified PCR fragments were subjected to in vitro transcription with the mMessage mMachine T7 kit (Life Technologies, California, USA). Poly (A) tails were attached to capped Cas9 RNAs by using a Poly(A) Tailing Kit (Life Technologies, California, USA), following the manufacturer's instructions. The synthesized mRNAs were column purified using mini Quick Spin RNA columns (Roche diagnostics GmbH, Mannheim, Germany), followed by phenol/chloroform extraction, ethanol precipitation, and dissolution in DNase/RNase-free water (Life Technologies, California, USA).
For the syntheses of gRNAs, pDR274-Dma-ey vectors were digested by DraI and purified by phenol/chloroform extraction. DraI-digested DNA fragments were used as templates for in vitro transcription with the mMessage mMachine T7 kit, followed by column purification with mini Quick Spin RNA columns, phenol/chloroform extraction, ethanol precipitation, and dissolution in DNase/RNase-free water.
Microinjection
In vitro synthesized RNAs were injected into Daphnia eggs according to established procedures [5]. Briefly, eggs were collected from daphniids within 2–3 weeks of age just after ovulation and placed in ice-chilled M4 medium containing 80 mM sucrose (M4-sucrose). The synthesized RNAs were injected through a glass needle with N2 gas pressure. The injection volume was approximately 0.2 nL. Finally, an injected egg was transferred into each well of a 96-well plate filled with 100 µL of M4-sucrose. Microinjections were carried out within an hour after ovulation.
PCR amplification of target loci
To characterize Cas9-induced mutations at the molecular level, target loci were amplified by PCR using genomic DNA extracted from deformed eye daphniids. Genomic DNA was extracted from single daphniids by homogenization in 90 µL of 50 mM NaOH with zirconia beads. The lysate was heated at 95°C for 10 min and then neutralized with 10 µL of 1 M Tris-HCl (pH 7.5). This crude DNA extract was centrifuged at 12,000 rpm for 5 min prior to being used as a template for genomic PCR. The targeted genomic region was amplified by PCR with KOD plus (TOYOBO, Osaka, Japan). The PCR products were analyzed by agarose gel electrophoresis and DNA sequencing. The primers used for PCR and DNA sequencing are listed in Tables S3 and S4.
Supporting Information
Results of RT-PCR for Dma-ey and cloned partial cDNA sequence. (A) Electrophoresis of PCR products. M: 100-bp ladder marker (TOYOBO, Osaka, Japan), G: genomic PCR product as positive control of PCR, RT+: RT-PCR product amplified from reverse-transcribed cDNAs, RT-: RT-PCR product amplified from total RNAs without reverse transcription. (B) Sequence of partially cloned Dma-ey cDNA.
(TIF)
Oligonucleotides for gRNAs and siRNAs. Lowercase “tt” in sense and antisense oligonucleotides for siRNA means dTdT (see Materials and Methods).
(DOCX)
Oligonucleotides used as in vitro transcription templates.
(DOCX)
Oligonucleotides used for the verification of cutting sites by sequencing.
(DOCX)
Oligonucleotides used for verification of off-target mutations by sequencing.
(DOCX)
Funding Statement
This study was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and partially supported by the Asahi Glass Foundation. One of the authors (Y.K.) would like to acknowledge the Frontier Research Base for Global Young Researchers, Osaka University, based on the Program of MEXT, for providing financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ebert D (2005) Ecology, Epidemiology, and Evolution of Parasitism in Daphnia [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information.
- 2. Hebert PDN (1978) Population Biology of Daphnia (Crustacea, Daphnidae). Biol Rev 53: 387–426. [Google Scholar]
- 3. Watanabe H, Tatarazako N, Oda S, Nishide H, Uchiyama I, et al. (2005) Analysis of expressed sequence tags of the water flea Daphnia magna . Genome 48: 606–609. [DOI] [PubMed] [Google Scholar]
- 4. Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex . Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kato Y, Shiga Y, Kobayashi K, Tokishita S, Yamagata H, et al. (2011) Development of an RNA interference method in the cladoceran crustacean Daphnia magna . Dev Genes Evol 220(11–12): 337–45. [DOI] [PubMed] [Google Scholar]
- 6. Kato Y, Matsuura T, Watanabe H (2012) Genomic integration and germline transmission of plasmid injected into crustacean Daphnia magna eggs. PLoS One 7(9): e45318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962): 167–70. [DOI] [PubMed] [Google Scholar]
- 8. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cradick TJ, Fine EJ, Antico CJ, Bao G (2013) CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41(20): 9584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, et al. (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154(6): 1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho SW, Kim S, Kim Y, Kweon J, Kim HS, et al. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24(1): 132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121): 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. (2013) RNA-guided human genome engineering via Cas9. Science 339(6121): 823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bassett AR, Tibbit C, Ponting CP, Liu JL (2013) Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4(1): 220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, et al. (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10(8): 741–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen C, Fenk LA, de Bono M (2013) Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res 41(20): e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, et al. (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194(4): 1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, et al. (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31(3): 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31(3): 233–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, et al. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4): 910–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu Z, Ren M, Wang Z, Zhang B, Rong YS, et al. (2013) Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila . Genetics 195(1): 289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ren X, Sun J, Housden BE, Hu Y, Roesel C, et al. (2013) Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A 110(47): 19012–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blitz IL, Biesinger J, Xie X, Cho KW (2013) Biallelic genome modification in F0 Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51(12): 827–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, et al. (2013) Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis . Genesis 51(12): 835–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao P, Zhang Z, Ke H, Yue Y, Xue D (2014) Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res 24(2): 247–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaj T, Gersbach CA, Barbas CF III (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7): 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quiring R, Walldorf U, Kloter U, Gehring WJ (1994) Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science 265(5173): 785–9. [DOI] [PubMed] [Google Scholar]
- 28. Walther C, Gruss P (1991) Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113(4): 1435–49. [DOI] [PubMed] [Google Scholar]
- 29. Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, et al. (1991) Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354(6354): 522–5. [DOI] [PubMed] [Google Scholar]
- 30. Ton CC, Hirvonen H, Miwa H, Weil MM, Monaghan P, et al. (1991) Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell 67(6): 1059–74. [DOI] [PubMed] [Google Scholar]
- 31. Kronhamn J, Frei E, Daube M, Jiao R, Shi Y, et al. (2002) Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless . Development 129(4): 1015–26. [DOI] [PubMed] [Google Scholar]
- 32. Suzuki KT, Isoyama Y, Kashiwagi K, Sakuma T, Ochiai H, et al. (2013) High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol Open 2(5): 448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Callaerts P, Clements J, Francis C, Hens K (2006) Pax6 and eye development in Arthropoda. Arthropod Struct Dev 35(4): 379–91. [DOI] [PubMed] [Google Scholar]
- 34. Sagawa K, Yamagata H, Shiga Y (2005) Exploring embryonic germ line development in the water flea, Daphnia magna, by zinc-finger-containing VASA as a marker. Gene Expr Patterns 5(5): 669–78. [DOI] [PubMed] [Google Scholar]
- 35. Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, et al. (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31(9): 822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mikkola I, Bruun JA, Bjorkoy G, Holm T, Johansen T (1999) Phosphorylation of the transactivation domain of Pax6 by extracellular signal-regulated kinase and p38 mitogen-activated protein kinase. J Biol Chem 274(21): 15115–26. [DOI] [PubMed] [Google Scholar]
- 37. Kim EA, Noh YT, Ryu MJ, Kim HT, Lee SE, et al. (2006) Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J Biol Chem 281(11): 7489–97. [DOI] [PubMed] [Google Scholar]
- 38. Yan Q, Gong L, Deng M, Zhang L, Sun S, et al. (2010) Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc Natl Acad Sci U S A 107(49): 21034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chapman JR, Taylor MR, Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47(4): 497–510. [DOI] [PubMed] [Google Scholar]
- 40. Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR (1991) Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253(5024): 1110–7. [DOI] [PubMed] [Google Scholar]
- 41. Kluttgen B, Dulmer U, Engels M, Ratte HT (1994) ADaM, an artificial freshwater for the culture of zooplankton. Water Res 28: 743–746. [Google Scholar]
- 42. Zhu BG, Cai GF, Hall EO, Freeman GJ (2007) In-Fusion (TM) assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43(3): 354–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of RT-PCR for Dma-ey and cloned partial cDNA sequence. (A) Electrophoresis of PCR products. M: 100-bp ladder marker (TOYOBO, Osaka, Japan), G: genomic PCR product as positive control of PCR, RT+: RT-PCR product amplified from reverse-transcribed cDNAs, RT-: RT-PCR product amplified from total RNAs without reverse transcription. (B) Sequence of partially cloned Dma-ey cDNA.
(TIF)
Oligonucleotides for gRNAs and siRNAs. Lowercase “tt” in sense and antisense oligonucleotides for siRNA means dTdT (see Materials and Methods).
(DOCX)
Oligonucleotides used as in vitro transcription templates.
(DOCX)
Oligonucleotides used for the verification of cutting sites by sequencing.
(DOCX)
Oligonucleotides used for verification of off-target mutations by sequencing.
(DOCX)