Abstract
Background
Serum and glucocorticoid regulated kinase (SGK) plays a critical role in the regulation of renal sodium transport. We examined the association between SGK genes and salt sensitivity of blood pressure (BP) using single-marker and gene-based association analysis.
Methods
A 7-day low-sodium (51.3 mmol sodium/day) followed by a 7-day high-sodium intervention (307.8 mmol sodium/day) was conducted among 1,906 Chinese participants. BP measurements were obtained at baseline and each intervention using a random-zero sphygmomanometer. Additive associations between each SNP and salt-sensitivity phenotypes were assessed using a mixed linear regression model to account for family dependencies. Gene-based analyses were conducted using the truncated p-value method. The Bonferroni-method was used to adjust for multiple testing in all analyses.
Results
In single-marker association analyses, SGK1 marker rs2758151 was significantly associated with diastolic BP (DBP) response to high-sodium intervention (P = 0.0010). DBP responses (95% confidence interval) to high-sodium intervention for genotypes C/C, C/T, and T/T were 2.04 (1.57 to 2.52), 1.79 (1.42 to 2.16), and 0.85 (0.30 to 1.41) mmHg, respectively. Similar trends were observed for SBP and MAP responses although not significant (P = 0.15 and 0.0026, respectively). In addition, gene-based analyses demonstrated significant associations between SGK1 and SBP, DBP and MAP responses to high sodium intervention (P = 0.0002, 0.0076, and 0.00001, respectively). Neither SGK2 nor SGK3 were associated with the salt-sensitivity phenotypes in single-maker or gene-based analyses.
Conclusions
The current study identified association of the SGK1 gene and BP salt-sensitivity in the Han Chinese population. Further studies are warranted to identify causal SGK1 gene variants.
Introduction
High salt intake has been associated with an elevated risk of high blood pressure (BP) and cardiovascular disease [1], [2]. BP responses to dietary sodium intake vary considerably among individuals, a phenomenon described as salt sensitivity of BP [3]. Previous family studies have documented a moderate to high heritability of salt sensitivity, generally ranging from 22% to 84% [4]–[6]. Likewise, linkage analyses and genetic association studies have suggested that genetic mechanisms may play a pivotal role in BP salt sensitivity [7]. The identification of genetic variants for salt sensitivity will help elucidate how genes interact with dietary factors to influence the regulation of BP.
The serum and glucocorticoid regulated kinases (SGK) play important roles in regulating Na+ transport in both proximal and distal elements of the kidney tubule [8], [9]. The SGK family of protein kinases have three isoforms, SGK1, SGK2 and SGK3 [9]. All of the three isoforms are highly expressed in kidney [10]–[12]. SGK1 stimulates the epithelial Na+ channel (ENaC), Na+/H+ exchanger 3 (NHE3) and many other renal tubular ion channels to regulate renal Na+ reabsorption [9], [13], [14]. SGK1 also contributes to the stimulation of mineralocorticoid-induced salt intake [14]. Recent studies showed increased salt concentration induces SGK1 expression, and increased SGK1 subsequently enhances TH17 cell differentiation and promotes tissue inflammation [15]. These findings support the opinion that the intensity of immune cell infiltration in the kidney is correlated with the severity of salt-sensitive hypertension [16]. Thus the effect of SGK1 on sodium homeostasis and inflammation is expected to have an important impact on blood pressure control. Similar to SGK1, both SGK2 and SGK3 can influence NHE3 activities and potentially regulate Na+ transport [9]. SGK’s biological relevance combined with evidence in vitro and in vivo make the genes encoding SGK interesting candidates for genetic study of BP response to sodium intake. However, only a few studies have reported associations of variants in the SGK gene family with BP phenotypes in human samples [17]–[19]. None have investigated the joint effect of multiple variants within single genes, and none of them were conducted in the Chinese population. The aim of the current study was to assess the single and joint effects of genetic variants in SGK genes with BP salt sensitivity among Han Chinese participants of the Genetic Epidemiology Network of Salt-Sensitivity (GenSalt) study.
Methods
Study Population
The GenSalt study is a unique dietary feeding study examining gene-dietary sodium and potassium interaction on blood pressure among a rural Han population in north China with habitual high salt intake. Probands were defined as having mean SBP between 130 and 160 mm Hg and/or mean DBP between 85 and 100 mm Hg and no use of antihypertension medications. The probands were identified through a community-based BP screening carried out among adults aged 18–60 years in the study villages. Probands along with their spouses, siblings, and offspring were recruited as volunteers for the dietary intervention study. Detailed eligibility criteria for the probands and siblings/spouses/offspring have been presented elsewhere [20]. Briefly, individuals with stage-2 hypertension (≥160/100 mmHg), current or recent use of antihypertension medications, secondary hypertension, history of clinical CVD, diabetes, chronic kidney failure, liver disease or peptic ulcer disease requiring treatment during the previous two years, along with pregnant women, heavy alcohol drinkers, and those currently adhering to a low-sodium diet or unable to sign the informed consent were excluded from the study. The study recruited 1,906 eligible participants from 633 families. A total of 1,871 (98.2%) and 1,860 (97.6%) participants who completed the low-sodium and high-sodium dietary interventions, respectively, were included in the current analysis.
Ethnics Statement
Institutional review boards at the Tulane University Health Sciences Center, Washington University School of Medicine, University of Texas School of Public Health, Fu Wai Hospital and Chinese National Human Genome Center at Beijing, and Chinese Academy of Medical Sciences approved the GenSalt study. Written informed consents for the baseline observation and for the intervention program were obtained from each participant.
Dietary Intervention
After a 3 day baseline examination period, the study participants underwent a 7 day low-sodium dietary intervention (51.3-mmol of sodium/day) followed by a 7 day high-sodium dietary intervention (307.8-mmol of sodium/day). Dietary potassium intake remained unchanged during the intervention phases. Total energy intake was varied according to each participant’s baseline energy intake. All study foods were cooked without salt, and pre-packaged salt was added to the individual study participant’s meal when it was served by the study staff. To ensure compliance with the intervention program, participants were required to have their breakfast, lunch and dinner in the study kitchen under the supervision of the study staff during the entire intervention period. The study participants were also instructed to avoid consuming any foods that were not provided by study personnel. Three timed urinary specimens were collected at baseline and at the end of each intervention phase (days 5, 6 and 7) to monitor the compliance with the intervention. The results of 24-hour urinary excretion of sodium showed excellent compliance with the study diet among all participants. The respective mean (SD) of 24-hour urinary excretions at baseline, during the low-sodium intervention, and during the high-sodium intervention were 242.4 (66.7), 47.5 (16.0) and 244.3 (37.7) mmol for sodium, and 36.9 (9.6), 31.4 (7.7), and 35.7 (7.5) mmol for potassium.
Phenotype Measurement
During the 3 days baseline examination, trained staff collected information on family structure, demographic characteristics, personal and family medical history, and lifestyle risk factors using a standard questionnaire. BP was measured 3 times at the same time each morning during the 3-day baseline and days 5, 6, and 7 during each intervention period according to a standard protocol. All BP was measured by trained and certified observers using a random zero sphygmomanometer with the participants in the sitting position after resting for 5 minutes [21]. Participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes before the BP measurements. All BP observers were unaware of the dietary intervention stages. In addition, body weight and height were measured twice during the baseline examination with the participants in light indoor clothing without shoes. Body mass index (BMI) was calculated as kilograms per meters squared.
Salt sensitivity phenotypes were defined as the absolute changes in SBP, DBP, and MAP from baseline to low-sodium intervention and from low-sodium intervention to high-sodium intervention. The means of 9 BP measures at baseline and on days 5, 6, and 7 of each intervention stage were calculated. Mean BP responses to low-sodium intake were calculated as the mean BP measures during low-sodium intervention minus the mean BP at baseline, and mean BP responses to high-sodium intake as mean BP during high-sodium intervention minus that during low-sodium intervention.
Single Nucleotide Polymorphism (SNP) Genotyping of SGK Genes
SNPs located within the SGK genes and their ±5,000 base-pair flanking regions were genotyped among all participants using chip based hybridization assays (Affymetrix 6.0, Santa Clara, CA). SNPs were excluded if they had a call rate less than 95%, were not in Hardy-Weinberg Equilibrium after adjustment for multiple comparisons (Bonferroni correction), or had a minor allele frequency (MAF) less than 1%. MapMaker/Sibs and PedCheck were used to check for Mendelian inconsistencies within families for each marker. Among the 71 SNPs within SGK genes, 56 met the quality control criteria, and 39 were tagged (r2<0.9) using Haploview software for inclusion in the current analysis [22]. Detailed information about the SGK genes and the 39 SNPs including their genomic locations, MAFs, call rates and P values for HWE tests are presented in Table 1 and Table S1.
Table 1. Characteristics of SGK1, SGK2 and SGK3 genes.
| Genesymbol | Gene name | Locus | Physical position±5,000 bp | Tag-SNPs | Function |
| SGK1 | serum/glucocorticoid regulated kinase 1 | 6q23 | (134485384,134644196) | 29 | Contributes to Na+ retention and K+elimination of the kidney. |
| SGK2 | serum/glucocorticoid regulated kinase 2 | 20q13.2 | (42182635,42219273) | 8 | Encodes a serine/threonine protein kinase. |
| SGK3 | serum/glucocorticoid regulated kinasefamily, member 3 | 8q12 | (67619653,67779257) | 2 | Involved in neutral amino acid transport andactivation of potassium and chloride channels. |
Chr, chromosome; SNP, single-nucleotide polymorphism.
Statistical Analysis
The means or percent of baseline characteristics and BP response variables were calculated for all participants. Additive associations between single SNPs and BP responses to each sodium intervention were assessed using a mixed linear regression model to account for familial correlations. Age, gender, field center, and BMI were controlled in all the association analyses. To correct for multiple testing with the 39 variants, the Bonferroni method was used (α-threshold = 0.05/39 = 1.28×10−3). For significant SNPs, we estimated the mean BP responses and 95% confidence interval (CI) for each genotype using a mixed linear regression model. The association analyses were conducted using SAS software (version 9.2; SAS Institute, Inc., Cary, North Carolina).
The joint effect of variants in the SGK genes on BP responses to sodium intervention was evaluated by combining P values from single SNP association analyses using the truncated product method (TPM) [23], [24]. Truncation point was set as τ = 0.10, and the P value for TPM was estimated by 10,000 simulations. If 10,000 simulations failed to generate a P value, simulations were increased up to 1,000,000. Sensitivity analyses were conducted using TPM after excluding significant SNPs within a gene to examine their influence on the gene-based analysis. Bonferroni correction was applied to account for multiple testing with 3 genes (α-threshold = 0.05/3 = 0.017). Gene-based analysis was performed using R software (Version 2.15.2; http://www.r-project.org).
Results
The GenSalt participant’s baseline characteristics and BP responses to sodium interventions are shown in Table 2 . Blood pressure significantly decreased in response to the low-sodium dietary intervention and increased in response to the high-sodium dietary intervention (all P<0.0001). In response to the low-sodium intervention, decreases of 5.5 mmHg, 2.8 mmHg, and 3.7 mmHg were observed for SBP, DBP and MAP, respectively. SBP, DBP and MAP increased 4.9 mmHg, 1.9 mmHg, and 2.9 mmHg, respectively, in response to high-sodium intervention.
Table 2. Characteristics of 1,906 GenSalt dietary intervention participants.
| Variables | Mean SD or percentage | Median (interquartile range) |
| Age | 38.7±9.6 | 39.0 (33.0 to 46.0) |
| Men, % | 53.0 | |
| BMI, kg/m2 | 23.3±3.2 | 22.9 (21.1 to 25.2) |
| Creatinine level at baseline, mg/dL | 0.9 (0.2) | 0.9 (0.8 to 1.1) |
| Baseline blood pressure | ||
| SBP | 116.9±14.2 | 115.8 (106.4 to 127.1) |
| DBP | 73.7±10.3 | 73.3 (66.7 to 80.7) |
| MAP | 88.1±10.9 | 87.7 (80.1 to 95.4) |
| Response to low sodium intervention | ||
| SBP, mm Hg | −5.5±7.0* | −4.4 (−8.9 to −1.3) |
| DBP, mm Hg | −2.8±5.5* | −2.7 (−5.6 to 0.4) |
| MAP, mm Hg | −3.7±5.3* | −3.3 (−6.6 to −0.6) |
| Response to high sodium intervention | ||
| SBP, mm Hg | 4.9±6.0* | 4.4 (0.6 to 8.2) |
| DBP, mm Hg | 1.9±5.4* | 1.8 (−1.6 to 5.3) |
| MAP, mm Hg | 2.9±5.0* | 2.7 (−0.4 to 5.9) |
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; MAP, mean arterial pressure;
*P<0.0001 when comparing blood pressure change with 0.
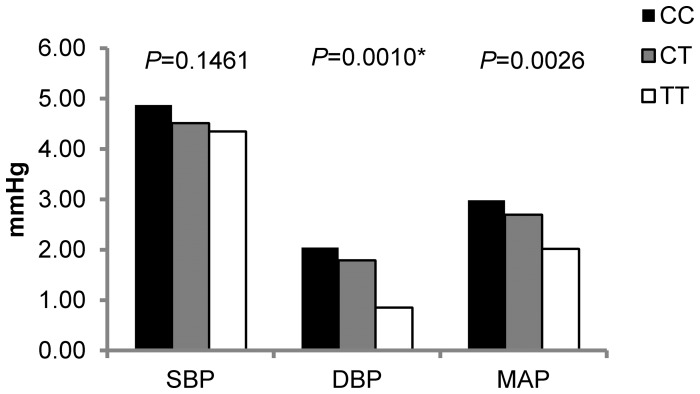
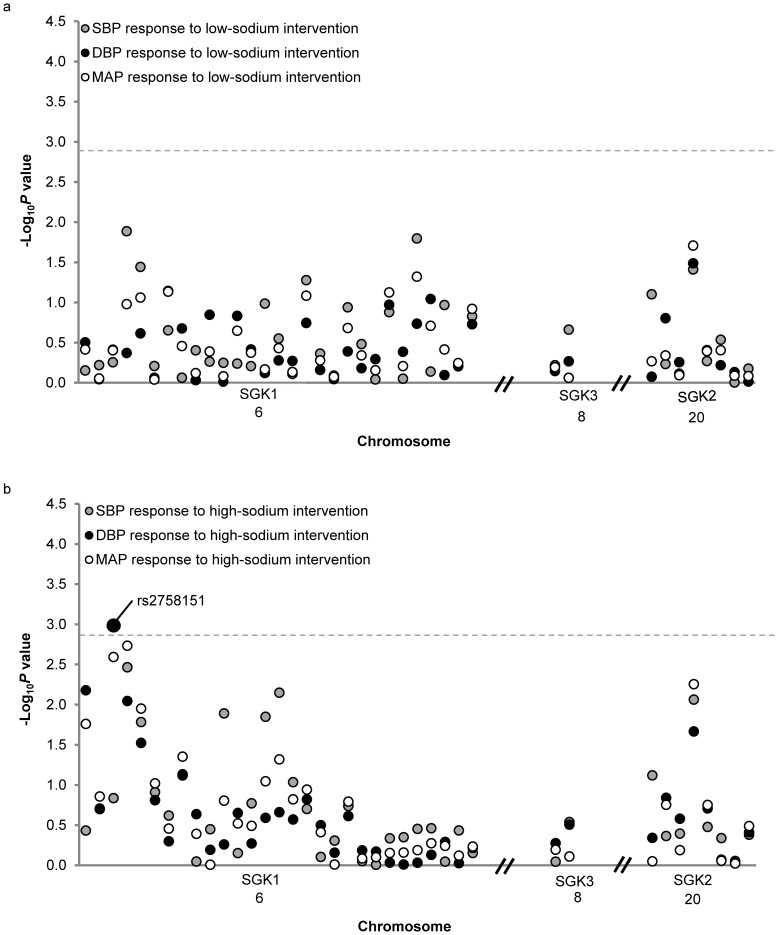
The single-marker association analyses are shown in Figure 1 . SNP, rs2758151, which is located 2.7 kbp downstream to the SGK1 gene, was significantly associated with absolute DBP responses to high-sodium intervention (P = 0.001). As shown in Figure 2 , DBP responses to high-sodium intervention decreased with the number of minor T alleles of rs2758151. Mean DBP response was 2.04 (1.57 to 2.52) mmHg for participants with CC genotype (n = 510), 1.79 (1.42 to 2.16) mmHg for those with CT genotype (n = 911), and 0.85 (0.30 to 1.41) mmHg for those with TT genotype (n = 415). Similar trends were observed for SBP and MAP response to the high sodium interventions (P = 0.146 and 0.003, respectively). Exact P values for all single SNP association tests are shown in Table S2.
Figure 1. –log10 P values for the associations of 39 SNPs in SGK1, SGK2 and SGK3 with blood pressure responses to low-sodium intervention (a) and high-sodium intervention (b).
Labeled SNP was significant after Bonferroni correction. The horizontal dashed lines indicate the Bonferroni corrected significant level.
Figure 2. Blood pressure responses to high sodium intervention among participants with different genotypes of rs2758151, respectively.
*significant after Bonferroni correction.
Table 3 shows the results of the gene-based analyses. SGK1 was significantly associated with SBP, DBP and MAP responses to high-sodium intervention. In sensitivity analyses excluding significant SGK1 marker rs2758151, all statistically significant SGK1 results remained significant with the exception of DBP response to high-sodium. P-values for the association between SGK1 and SBP, DBP, and MAP responses in the sensitivity analysis were 9.0×10−5, 0.09 and 5.2×10−4, respectively.
Table 3. Gene-based associations of SGK1, SGK2 and SGK3 with blood pressure responses to dietary sodium intervention.
| SGK1 | SGK2 | SGK3 | |
| Response to low-sodium intervention | |||
| SBP | 0.1541 | 0.1037 | 0.1708 |
| DBP | 0.6240 | 0.3212 | 0.1326 |
| MAP | 0.2474 | 0.2589 | 0.1505 |
| Response to high-sodium intervention | |||
| SBP | 0.0002* | 0.0422 | 0.2008 |
| DBP | 0.0076* | 0.2669 | 0.1388 |
| MAP | 1.00E-05* | 0.1822 | 0.1309 |
BP, blood pressure; SBP, systolic blood pressure; DBP, diastolic blood pressure; MAP, mean arterial pressure;
*Significant P values after Bonferroni correction.
Discussion
In the first study to examine the association between the SGK gene pathway and salt-sensitivity of BP in an East Asian population, we identified a significant association between SGK1 variant rs2758151 and DBP response to high-sodium intervention. Each copy of the rs27858151 minor T allele predicted smaller DBP responses to the high-sodium diet. Similar trends were observed for the SBP and MAP responses to high sodium phenotypes although not significant. In addition, gene-based analysis revealed that the SGK1 gene was significantly associated with all BP responses to the high-sodium intervention. While it appeared that the gene-based association of the DBP response phenotype could be explained by significant marker rs27858151, the SBP and MAP response phenotypes remained significant after sensitivity analyses excluding this SNP. Neither the gene-based nor single-marker analyses identified significant association of the SGK2 and SGK3 genes with the BP response phenotypes. To the best of our knowledge, this is the first study examining the joint effect of SNPs in genes of the SGK family on the blood pressure salt-sensitivity phenotype. In aggregate, these findings could provide valuable information towards delineating the genomic mechanisms underlying human BP salt-sensitivity.
SGK1 variant rs2758151 showed evidence of association with BP salt-sensitivity in the current analysis. It is of interest to note that this variant was implicated in the only other candidate gene study to examine the relationship between SGK1 and BP salt-sensitivity to date [17]. In the previous report, Rao and colleagues identified a significant association between rs2758151 and BP response to high sodium intervention in a population of European ancestry [17]. However, their study reported an opposite effect direction of the T allele [17]. A potential reason for this discrepancy could be that rs2758151 is not the causal locus for the salt-sensitivity phenotype but is in linkage disequilibrium (LD) with the causal variant [25]. The causal variant may be on different haplotypes in populations of distinct ethnicities. In support of this hypothesis, post-hoc comparison of LD patterns between European and Chinese populations at the SGK1 locus showed distinct differences in LD structure between these populations (P = 0.01 for differences in LD pattern; see Figure S1 ) [26], [27]. In addition, differences in direction of association could be due to interactive effects with other variants or environmental factors that vary between populations [28]. Lin et al used theoretical modeling to demonstrate that such “flip-flop” associations may indeed represent confirmations of true associations due to interactive effects or LD with a causal variant at another locus [29]. Marker rs2758151 is located downstream of the SGK1 gene and is unlikely to play a causal role in altering the synthesis or structure of the SGK1 protein. Future studies will be needed to pinpoint the true functional variant responsible for the strong signal reported by both us and Rao et al. Such work could leverage LD information in East Asians and Caucasians to help localize the region for sequencing study.
Our study also represents the first report of gene-based associations of SGK1 and BP salt-sensitivity phenotypes. We identified significant associations of SGK1 with SBP, DBP and MAP responses to high-sodium intervention. Although sensitivity analyses revealed that the association between SGK1 and DBP response was largely driven by marker rs2758151, this variant could not explain the gene-based relation of SGK1 with both SBP and MAP responses to the high-sodium dietary intervention. Such findings indicate that there might be multiple independent loci within SGK1 that are related to these phenotypes. Furthermore, our findings highlight the importance of considering both singular and joint effects of variants to elucidate the genetic architecture of complex phenotypes like BP response to dietary sodium intervention.
SGK2 and SGK3 were not associated with BP salt-sensitivity in the current analysis, nor were any single variants within the two genes. Although very few studies have been conducted to examine the association of the SGK2 and SGK3 genes with BP phenotypes [8], [30], our results are consistent with animal models. For example, Schnackenberg and colleagues showed that SGK2 knockout mice did not exhibit urinary Na+ wasting or low blood pressure [31]. Similarly, SGK3 knockout mice were not found to have obvious defects in Na+ handling in a manuscript by McCormick and collagues [32].
Our study has several important strengths. GenSalt is the largest dietary intervention study to examine the associations between the SGK gene pathway and BP responses to dietary sodium intervention. A high proportion of study participants completed the dietary intervention (97.6%), and compliance with the study interventions was excellent, as confirmed by urinary excretion of sodium and potassium during each intervention period. Measurement error was reduced and power enhanced by the large number of BP measures that were collected for each participant. Furthermore, stringent quality control methods were used in measuring BP and other study covariables, genotyping, and marker data cleaning. Finally, the recruitment of only Han Chinese participants should make the analysis robust to even fine levels of population stratification. Although the Affymetrix 6.0 platform generally provides good genomic coverage of common variants in the Han Chinese population [33], coverage of SGK3 was limited, with only two tag SNPs in the SGK3 gene. Therefore, common SGK3 variants associated with BP salt-sensitivity may have been missed by the current study. Further, the gene-based analysis for SGK3 must be interpreted with caution. Finally, due to the uniqueness of salt-sensitivity phenotype, our findings could not be replicated in an independent East Asian sample.
The current study identified a significant association of SGK1 marker rs2758151 with BP salt-sensitivity. Having previously been reported to associate with BP responses to dietary sodium in a population of European ancestry, these findings provide some evidence of trans-ethnic replication in the Han Chinese population. Furthermore, our gene-based analysis revealed potentially important joint actions of SGK1 SNPs on the salt-sensitivity phenotypes. These findings contribute important information towards elucidating the genomic mechanisms underlying blood pressure regulation. Still, further studies will be necessary to localize the reported SGK1 signals and identify the causal variants for BP salt sensitivity.
Supporting Information
(TIF)
Quality control information of the tagged 39 SNPs in SGK1 , SGK2 and SGK3.
(DOC)
P -Values of single SNP association analysis of the 39 SNPs in SGK1 , SGK2 and SGK3 with blood pressure responses to dietary sodium intervention.
(DOCX)
Acknowledgments
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD.
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Identifying data could not be made freely available, but GenSalt may provide the association result for certain SNP and phenotypes. Requests for this data may be sent to Dr. Tanika N. Kelly.
Funding Statement
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD. https://www.nhlbi.nih.gov/Drs Tanika Kelly and Changwei Li are supported by American Heart Association award number 11SDG5130026. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1. Elliott P, Stamler J, Nichols R, Dyer AR, Stamler R, et al. (1996) Intersalt revisited: further analyses of 24 hour sodium excretion and blood pressure within and across populations. Intersalt Cooperative Research Group. BMJ 312: 1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strazzullo P, D’Elia L, Kandala NB, Cappuccio FP (2009) Salt intake, stroke, and cardiovascular disease: meta-analysis of prospective studies. BMJ 339: b4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luft FC, Weinberger MH (1997) Heterogeneous responses to changes in dietary salt intake: the salt-sensitivity paradigm. Am J Clin Nutr 65: 612S–617S. [DOI] [PubMed] [Google Scholar]
- 4. Gu D, Rice T, Wang S, Yang W, Gu C, et al. (2007) Heritability of blood pressure responses to dietary sodium and potassium intake in a Chinese population. Hypertension 50: 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller JZ, Weinberger MH, Christian JC, Daugherty SA (1987) Familial resemblance in the blood pressure response to sodium restriction. Am J Epidemiol 126: 822–830. [DOI] [PubMed] [Google Scholar]
- 6. Svetkey LP, McKeown SP, Wilson AF (1996) Heritability of salt sensitivity in black Americans. Hypertension 28: 854–858. [DOI] [PubMed] [Google Scholar]
- 7. Kelly TN, He J (2012) Genomic epidemiology of blood pressure salt sensitivity. J Hypertens 30: 861–873. [DOI] [PubMed] [Google Scholar]
- 8. Lang F, Böhmer C, Palmada M, Seebohm G, Strutz-Seebohm N, et al. (2006) (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev 86: 1151–1178. [DOI] [PubMed] [Google Scholar]
- 9. Pao AC (2012) SGK regulation of renal sodium transport. Curr Opin Nephrol Hypertens 21: 534–540. [DOI] [PubMed] [Google Scholar]
- 10. Raikwar NS, Snyder PM, Thomas CP (2008) An evolutionarily conserved N-terminal Sgk1 variant with enhanced stability and improved function. Am J Physiol Renal Physiol 295: F1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi T, Deak M, Morrice N, Cohen P (1999) Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J 344 Pt 1: 189–197. [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Zhou D, Phung S, Masri S, Smith D, et al. (2011) SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Mol Endocrinol 25: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faresse N, Lagnaz D, Debonneville A, Ismailji A, Maillard M, et al. (2012) Inducible kidney-specific Sgk1 knockout mice show a salt-losing phenotype. Am J Physiol Renal Physiol 302: F977–985. [DOI] [PubMed] [Google Scholar]
- 14. Lang F, Artunc F, Vallon V (2009) The physiological impact of the serum and glucocorticoid-inducible kinase SGK1. Curr Opin Nephrol Hypertens 18: 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, et al. (2013) Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quiroz Y, Johnson RJ, Rodriguez-Iturbe B (2012) The role of T cells in the pathogenesis of primary hypertension. Nephrol Dial Transplant 27 Suppl 4: iv2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao AD, Sun B, Saxena A, Hopkins PN, Jeunemaitre X, et al. (2013) Polymorphisms in the serum- and glucocorticoid-inducible kinase 1 gene are associated with blood pressure and renin response to dietary salt intake. J Hum Hypertens 27: 176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. von Wowern F, Berglund G, Carlson J, Månsson H, Hedblad B, et al. (2005) Genetic variance of SGK-1 is associated with blood pressure, blood pressure change over time and strength of the insulin-diastolic blood pressure relationship. Kidney Int 68: 2164–2172. [DOI] [PubMed] [Google Scholar]
- 19. Busjahn A, Aydin A, Uhlmann R, Krasko C, Bähring S, et al. (2002) Serum- and glucocorticoid-regulated kinase (SGK1) gene and blood pressure. Hypertension 40: 256–260. [DOI] [PubMed] [Google Scholar]
- 20. Group GCR (2007) GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 21: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perloff D, Grim C, Flack J, Frohlich ED, Hill M, et al. (1993) Human blood pressure determination by sphygmomanometry. Circulation 88: 2460–2470. [DOI] [PubMed] [Google Scholar]
- 22. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 23.Sheng X, Yang J (2012) Truncated Product Methods for Panel Unit Root Tests. Oxf Bull Econ Stat In press. [DOI] [PMC free article] [PubMed]
- 24. Yang J, Zhu Y, Cole SA, Haack K, Zhang Y, et al. (2012) A gene-family analysis of 61 genetic variants in the nicotinic acetylcholine receptor genes for insulin resistance and type 2 diabetes in American Indians. Diabetes 61: 1888–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perkins EA, Landis D, Causey ZL, Edberg Y, Reynolds RJ, et al. (2012) Association of single-nucleotide polymorphisms in CCR6, TAGAP, and TNFAIP3 with rheumatoid arthritis in African Americans. Arthritis Rheum 64: 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ong RT, Teo YY (2010) varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics 26: 1269–1270. [DOI] [PubMed] [Google Scholar]
- 27. Teo YY, Fry AE, Bhattacharya K, Small KS, Kwiatkowski DP, et al. (2009) Genome-wide comparisons of variation in linkage disequilibrium. Genome Res 19: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiedmann S, Fischer M, Koehler M, Neureuther K, Riegger G, et al. (2008) Genetic variants within the LPIN1 gene, encoding lipin, are influencing phenotypes of the metabolic syndrome in humans. Diabetes 57: 209–217. [DOI] [PubMed] [Google Scholar]
- 29. Lin PI, Vance JM, Pericak-Vance MA, Martin ER (2007) No gene is an island: the flip-flop phenomenon. Am J Hum Genet 80: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Loffing J, Flores SY, Staub O (2006) Sgk kinases and their role in epithelial transport. Annu Rev Physiol 68: 461–490. [DOI] [PubMed] [Google Scholar]
- 31. Schnackenberg CG, Costell MH, Bernard RE, Minuti KK, Grygielko ET, et al. (2007) Compensatory role for Sgk2 mediated sodium reabsorption during salt deprivation in Sgk1 knockout mice[abstract]. The FASEB Journal 21. [Google Scholar]
- 32. McCormick JA, Feng Y, Dawson K, Behne MJ, Yu B, et al. (2004) Targeted disruption of the protein kinase SGK3/CISK impairs postnatal hair follicle development. Mol Biol Cell 15: 4278–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, et al. (2008) Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics 9: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Quality control information of the tagged 39 SNPs in SGK1 , SGK2 and SGK3.
(DOC)
P -Values of single SNP association analysis of the 39 SNPs in SGK1 , SGK2 and SGK3 with blood pressure responses to dietary sodium intervention.
(DOCX)
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Identifying data could not be made freely available, but GenSalt may provide the association result for certain SNP and phenotypes. Requests for this data may be sent to Dr. Tanika N. Kelly.