Abstract
Background
The DEAD-box helicases are required mostly in all aspects of RNA and DNA metabolism and they play a significant role in various abiotic stresses, including salinity. The p68 is an important member of the DEAD-box proteins family and, in animal system, it is involved in RNA metabolism including pre-RNA processing and splicing. In plant system, it has not been well characterized. Here we report the cloning and characterization of p68 from pea (Pisum sativum) and its novel function in salinity stress tolerance in plant.
Results
The pea p68 protein self-interacts and is localized in the cytosol as well as the surrounding of cell nucleus. The transcript of pea p68 is upregulated in response to high salinity stress in pea. Overexpression of p68 driven by constitutive cauliflower mosaic virus-35S promoter in tobacco transgenic plants confers enhanced tolerances to salinity stress by improving the growth, photosynthesis and antioxidant machinery. Under stress treatment, pea p68 overexpressing tobacco accumulated higher K+ and lower Na+ level than the wild-type plants. Reactive oxygen species (ROS) accumulation was remarkably regulated by the overexpression of pea p68 under salinity stress conditions, as shown from TBARS content, electrolyte leakage, hydrogen peroxide accumulation and 8-OHdG content and antioxidant enzyme activities.
Conclusions
To the best of our knowledge this is the first direct report, which provides the novel function of pea p68 helicase in salinity stress tolerance. The results suggest that p68 can also be exploited for engineering abiotic stress tolerance in crop plants of economic importance.
Introduction
The DEAD-box families of proteins are helicases conserved from bacteria to humans and are involved in a variety of nucleic acid metabolic processes such as replication, repair, recombination, transcription, pre-mRNA processing, RNA degradation, RNA export, ribosome assembly and translation [1]–[3]. At the sequence level, helicases have been classified into five superfamilies (SF1–SF5). The largest of these groups are SF1 and SF2. Most of these contain nine conserved helicase motifs, Q, I, Ia, Ib, II, III, IV, V and VI. All the helicases exhibit nucleic acid dependent ATPase activity which provides energy for the helicase action [3], [4].
The p68 is a prototype member of DEAD-box family and is one of the best characterized helicases and it plays a very important role in cell/organ development and participates in a variety of biological processes including pre-mRNA and pre-rRNA processing [5]–[6], rearrangement of RNA secondary structures [6], RNA splicing [7]–[8] and gene transcription. In human, p68 is a RNA-binding protein endowed with an ATP-dependent RNA helicase, RNA-dependent ATPase and RNA-protein complex remodeling activities. In human malaria parasite Plasmodium falciparum, p68 has also been reported as a dual helicase and its helicase and ATPase activities are stimulated after phosphorylation with protein kinase C [9]–[10].
The DEAD-box RNA helicases are becoming a subject of attention as they play a significant role during development and stress responses in plants [11]–[14]. Each DEAD box RNA helicase is thought to be differentially regulated during development and in response to environmental stresses in plants [2], [13]. Arabidopsis LOS4 and RCF1 (a DEAD-box RNA helicase) were reported to regulate gene expression in response to chilling stress [8], [15]. The overexpression of pea DNA helicase (PDH45) was shown to confer salt tolerance in tobacco and rice [12], [14]. A rice DEAD-box RNA helicase (OsBIRH1) has been reported to function in defense responses against pathogen and oxidative stresses [16]. OsSUV3, a member of DEAD-box RNA helicase, has been shown to be involved in salinity stress tolerance recently [17]. Splicing factors are also known to affect the alternative splicing of many genes which lead to alter the gene expression through splicing factor networks and thereby involving in plant stress tolerance [18]. The plant homologue of animal splicing factor p68 has not been characterized in detail till date. The transcript of Arabibopsis thaliana p68 DEAD-box RNA helicase (AtDRH1) was reported to be accumulated at a high level and almost equally in every part of the Arabidopsis plant [19]. Studies on MA16 (maize RNA-binding protein) and ZmDRH1 (Z. mays DEAD-box RNA helicase 1) revealed that these proteins might be part of a ribonucleoproteins complex involved in ribosomal RNA (rRNA) metabolism [20]. However, the precise role of DEAD-box RNA helicase, especially the role of p68 in abiotic stress tolerance, has not been reported so far. Here, we report detailed characterization of p68 from pea (Pisum sativum). Our results show that pea p68 self-interacts and the overexpression of pea p68 enhances the salinity stress tolerance in tobacco by controlling the generation of stress-induced reactive oxygen species (ROS) through modulating antioxidative defence machinery thereby protecting the photosynthesis and yield.
Materials and Methods
Construction of Plasmid for Tobacco Transformation
The pea p68 gene was isolated by screening of pea cDNA library using a heterologous probe from Arabidopsis thaliana. The complete ORF of p68 was cloned into pGEMT easy vector and sequenced (Accession number: AF271892.1). The insert (1.8 kb) was release from pGEMT-p68 by digesting with EcoRI and BamHI restriction enzymes and ligated into the MCS of pRT101. The CaMV35S-p68-polyA cassette generated in pRT101 was then cut with Hind III and ligated to the MCS of pCAMBIA-1301 binary vector containing GUS as the reporter and hygromycin phosphotransferase as the selection marker gene. The pCAMBIA1301-p68 clone was then transformed into Agrobacterium LBA4404 strain for generation of transgenic tobacco using standard protocol [21]. Positive transgenic lines were confirmed by PCR using gene specific primers (FP: 5′- GAATTCATGTCGTATGTTCCTCCACAC-3/EcoRI site bold) and (RP: 5′- GGATCCCCATTACCTACAAACATGACTGAT-3/BamHI site bold). The transgenic tobacco plants were analyzed in two independent experiments with three technical replicates.
Transcript Analysis and Yeast-two-hybrid Assay
To analyze the expression of pea p68 transcripts, northern blots analysis was performed as described earlier [11]. The pGBKT7-p68 and pGADT7-p68 were generated by inserting a PCR fragment encoding the complete ORF of p68. The empty yeast strain AH109, pGADT7, pGBKT7, pGADT7/pGBKT7, pGADT7/pGBKT7-p68, pGADT7-p68/pGBKT7 and pGBKT7-p68/pGADT7-p68 were transformed separately on yeast cells. Yeast cells carrying all the plasmids were selected on the synthetic medium lacking Leu and Trp (SD-Leu-Trp-). The yeast cells were then streaked on SD medium [(Leu), (Trp), (His)] containing 30 mM 3-AT (3-Amino-1, 2, 4-triazole) to determine the expression of HIS3 nutritional reporter. The β-galactosidase expression of the fusion proteins encoded by pGBKT7-p68, pGADT7-p68 constructs was assayed by colony filter lift assays as per manufacturer instruction (Clontech).
In vivo Localization by Immunofluorescence Staining and Confocal Microscopy
Polyclonal antibody against pea p68 was raised in rabbit as per the standard procedure. For in vivo localization, exponentially growing tobacco BY2 [11] suspension cells were fixed in 4% formaldehyde, permeabilized by cellulase and layered onto poly-L-lysine-coated cover slips. The cells were immunostained with p68-specific primary rabbit antibody in 1∶2000 dilutions and Alexafluor 488-labeled goat antirabbit secondary antibody (Molecular Probes, Eugene, OR, USA) in 1∶1000 dilutions. Counter staining of the cells with DAPI (4/, 6-diamidino-2-phenylindole), confocal laser scanning microscopy and image processing was carried out in a method described earlier [11].
Southern and Western Blots Analysis
Genomic DNA was isolated by CTAB method from PCR positive p68 tobacco transgenic lines and WT. For southern blots analysis, ∼20 µg of genomic DNA was digested with HindIII and resolved on agarose gel. The DNA was transferred to nylon membrane (Hybond N, Amersham Pharmacia, http://www.gelifesciences.com/), and hybridized with radiolabelled p68 cDNA as described previously [12]. For western blot analysis, the total soluble proteins were isolated from the unstressed tissue samples of transgenic lines as well as WT plants and separated on 12% SDS-PAGE. Western blot analysis was performed by using anti-p68 (1∶5,000 dilutions) as primary and anti-rabbit (alkaline phosphatase conjugated antirabbit antibody-Sigma) as secondary antibody (1∶12,500 dilutions). The blot was developed as per manufacturer’s protocol (Sigma, USA).
Histochemical GUS Staining and Morphological Characterization of Transgenic Plants
The seedlings of pea p68 overexpressing tobacco transgenic lines and WT were vacuum infiltrated for 10 m and histochemical GUS staining was performed by a method described earlier [22]. For in vitro pollen germination, mature pollen was isolated aseptically and cultured on pollen germination media (SMM: 0.3 M sucrose, 1.6 mM H3BO3, 3 mM Ca(NO3)2 4H2O, 0.8 mM MgSO4.7H2O, 1 mM KNO3) supplemented with 100 and 200 mM NaCl and incubated at 26°C in the dark. The germination status of the pollens was monitored and scored during a 7 d experimentation period. The sensitivity of seed germination to NaCl was assayed on MS agar plates saturated with 200 mM NaCl and incubated at 26°C under cool-white light for germination. Leaf disks assays, measurement of growth characteristics like shoot length, root length, leaf area and plant dry weight, measurement of tolerance index, determination of the total chlorophyll content and yield characteristics of transgenic and wild-type (WT) plants were performed as described earlier [14].
Measurement of Photosynthetic Characteristics and Photosystem II Activity (Fv/Fm)
The photosynthetic characteristics like net photosynthetic rate (PN), stomatal conductance (gs), intercellular CO2 concentration (Ci) and chlorophyll fluorescence (Fv/Fm) of transgenic lines and WT plants were recorded on the fully expanded leaves using infra–red gas analyzer (Li–6400, Li–COR, Lincoln, NE, USA) between 11∶00 and 12∶00 h. The conditions during the measurement were photosynthetically active radiation (PAR) 945±8 µmol m– 2 s– 1, relative humidity 75±6%, temperature 28±2°C and an ambient CO2 concentration of 350 µmol mol– 1. The chlorophyll fluorescence i.e. maximal efficiency of PSII photochemistry (Fv/Fm) was also determined on the same leaves used for photosynthetic measurements after dark adaptation for 30 min.
Measurement of Ion Content
The Na+ and K+ ion content was measured as described earlier [23]. Salinity treated (0, 100 or 200 mM NaCl) leaves of the transgenic lines and WT plants were collected and rinsed with deionised water thoroughly. The fresh weight was determined for each sample. After drying (70°C for 48 h), dry weight was also measured. The samples are then subjected to an overnight digestion with HNO3/H2O2. The materials were picked in 2 M HCl, and Na+ and K+ ion content was analyzed by using simultaneous inductively coupled plasma emission spectrometry (ICP trace analyzer, Labtam, Braeside, Australia).
Measurement of Oxidative Stress, Enzymatic Antioxidants (SOD, CAT, APX, GR) and Non-enzymatic Antioxidants (AsA and GSH) in p68 Transgenic Lines and WT
Oxidative stress was detected by measuring thiobarbituric acid reactive substances (TBARS), hydrogen peroxide (H2O2) content, electrolyte leakage and oxidative DNA damage (8-OHdG) in the leaves of p68 overexpressing transgenic lines and WT plants by a previously described method [14]. The salinity stressed (0, 100 or 200 mM NaCl) and fully expanded leaves from transgenic and WT plants were used for the measurement of enzymatic antioxidants (superoxide dismutase, catalase, ascorbate peroxidase, glutathione reductase) and non-enzymatic antioxidants (ascorbate and glutathione). Leaf samples (transgenic lines and WT) were homogenized with an extraction buffer containing 100 mM potassium phosphate buffer (pH 7.0), 0.5% Triton X–100 and 1% polyvinylpyrrolidone (PVP) using pre-chilled mortar and pestle. The homogenate was centrifuged at 15,000×g for 20 min at 4°C. The supernatant obtained after centrifugation was used for enzyme assays. Measurement of enzymatic antioxidants (SOD, CAT, APX, and GR) and non-enzymatic antioxidants (AsA and GSH) was performed as described previously [14]. All the measurements were carried out 3 weeks after initiating the NaCl treatment.
Transgenic Plants and Salinity Stress Tolerance
For the assay of sensitivity to salinity stress, 14 d-old seedlings of WT and the transgenic plants (grown on vermiculite pots) were transferred to nutrient solutions containing 200 mM NaCl. After 2 d, growth status of the transgenic lines was observed. In another experiment, 40 plants of each line and WT were grown in vermiculite pots and watered for 10 d and then 200 mM NaCl solution was irrigated for every 3 d interval up to 12 d. After treatments, morphological changes were observed. In order to investigate the effect of salinity stress, 35-day-old plants were also subjected to salt stress for 28 d and growth was observed till maturity and photographs were taken.
Statistical Analysis
All the treatments were performed in three independent trials with consistent results. The results from only one representative experiment are shown, expressed as means± standard errors. Analysis of one-way variance (ANOVA) was performed on the data using SPSS (12.0 Inc., USA) to determine the least significant difference (LSD) for the significant data to identify the differences in the mean among the treatments. The means were separated by Duncan’s multiple range tests (DMRT). The graphs were prepared using Sigmaplot Ver. 11. Different letters indicate significant difference at P<0.05.
Results
Isolation and Sequence Analysis of Pea p68 cDNA
The pea cDNA library was screened using a 1.9 kb cDNA fragment of p68 from Arabidopsis thaliana (kindly provided by Tetsuo Meshi of Kyoto University, Japan) as a probe. This resulted in the isolation of a positive clone of pea p68 (Figure S1A). The sequence analysis shows that the open reading frame (ORF) of pea p68 cDNA (1.8 kb) encodes a protein of 622 amino acid residues, with a calculated molecular mass of 67.65 kDa and a pI of 6.46. It also exhibits all the known canonical helicase motifs (Q, I, Ia, Ib, II-VI) (Figure S1B). Phylogenetic analysis identified the closest orthologs of pea p68 as Arabidopsis thaliana and Saccharomyces cerevisiae p68 followed by Oryza sativa p68 (Figure S1C).
Tissue Specific Distribution and Regulation of Pea p68 Transcript in Response to Abiotic Stresses
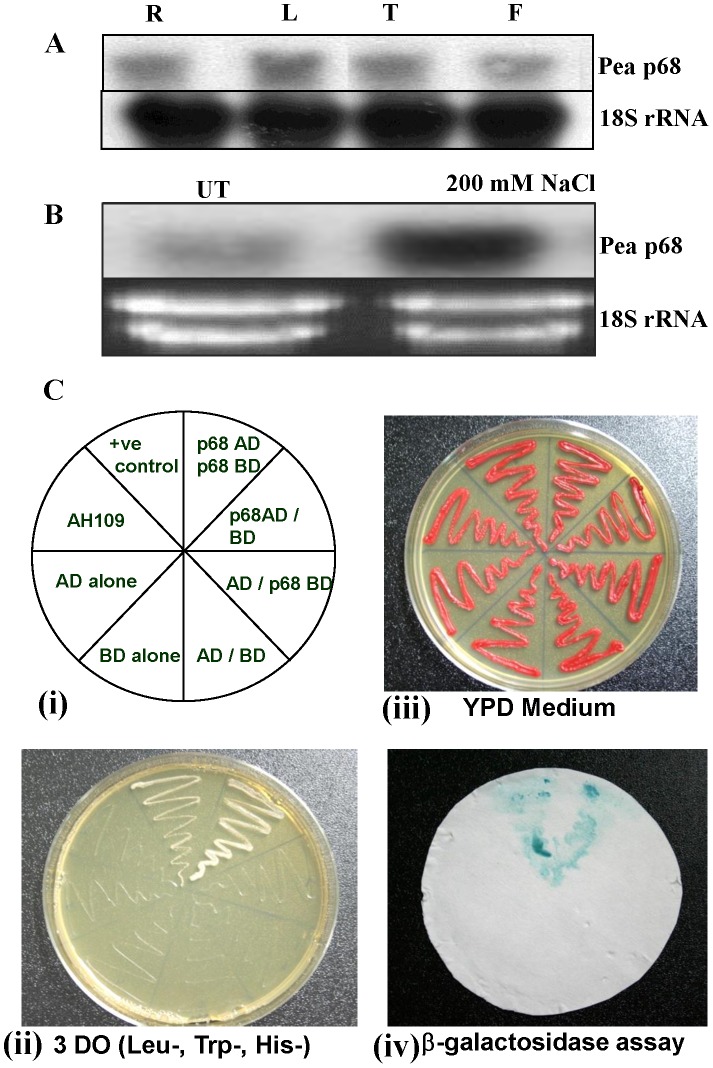
To investigate the expression pattern of the transcript level of pea p68 in different parts of pea plant, the total RNA was isolated from root, leaf, tendril and flower and was subjected to northern blot analysis using 1.8 kb pea p68 cDNA as the probe. For the internal control, 18S ribosome probe was used to show the equal loading. A single transcript of the expected size for pea p68 mRNAs was present in all the tissues (Figure 1A). Further, to analyze the expression of pea p68 under salinity stress, 7-day old pea seedlings were treated with 200 mM NaCl for 24 h and used for northern blot analysis. The results show that the pea p68 transcript was dramatically up-regulated (∼4.8 fold) in response to salinity stress (Figure 1B).
Figure 1. Expression analysis and self-interaction assay for pea p68.
(A). Transcript analysis of pea p68 by Northern blot analysis (R: root, S: shoot, T: tendril and F: flower tissue respectively). (B) Transcript level in response 200 mM NaCl stress respectively (UT: untreated samples). About 30 µg of total was separated by electrophoresis, blotted and hybridized with the 32P-labeled ORF of pea p68 cDNA (1.8 kb). For equal loading of RNA in each lane, the same blot was hybridized with the 18S rRNA, as shown in bottom of the each panel. (C) Yeast two-hybrid system-based self interaction showing pea p68 interacts with pea p68 in vivo (i) Template showing the organization of Y2H experiment (ii) Showing phenotypes on a YPD plate (iii) Yeast growing on a synthetic dextrose plate lacking leucine, tryptophan and histidine (3 DO) and (iv) β-galactosidase filter lift assay showing positive pea p68 self interaction. Yeast strain (AH109) carrying and Gβ-AD+Gγ-BD used as a positive control for yeast two-hybrid assay.
Pea p68 Interacts with Itself in the Yeast Two-hybrid System
To verify the self-interaction, we co-expressed BD-pea p68 with a construct encoding the full ORF of pea p68 fused to the GAL4 activation domain (AD-Pea p68). Yeast cells carrying both the plasmids of BD-pea p68+ AD-Pea p68 were able to grow on 3DO media [Figure 1C (ii)]. All the clones including negative and positive control transformed in yeast cells were grown in YPD medium [Figure 1C (iii)]. Yeast cell carrying the plasmids of BD-pea p68+ AD-Pea p68 and Gβ-pGADT7+Gβ-pGBKT7 exhibited β-galactosidase activity, indicating the expression of reporter genes [Figure 1C (iv)]. These results confirm that the full-length pea p68 is able to self-interact in the yeast two-hybrid system.
In vivo Localization of p68
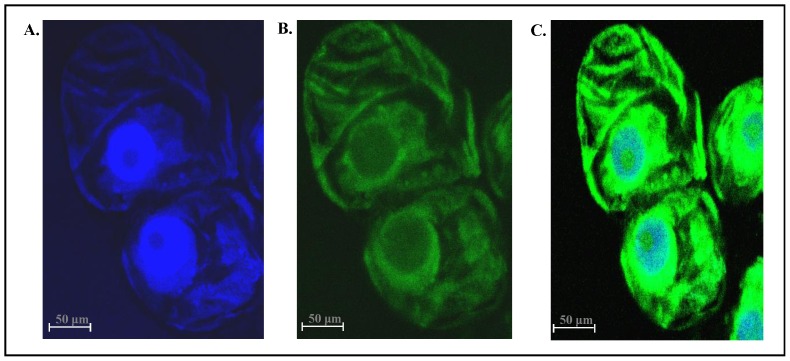
The in vivo localization of p68 was also analyzed by immunofluorescence labeling and observed under confocal microscopy. Immunofluorescent labeling of tobacco BY2 cells with anti-pea p68 antibodies showed p68 protein exclusively localized in the cytoplasm as well as in the surrounding of cell nucleus (Figure 2).
Figure 2. In vivo localization of the p68 in tobacco BY2 cells.
The tobacco BY2 cells were fixed, permeabilized and immunostained with primary antibodies against p68 followed by Alexafluor 488-labeled secondary antibody and then counterstained with DAPI. A single confocal image is shown. (A) Image of cell stained with DAPI (blue). (B) Immunofluorescently stained cell (green). Anti-p68 labeling is restricted to the nucleus and cytosol. (C) Superimposed image of cell.
Overexpression of p68 and Molecular Analysis of Transgenic Tobacco Lines
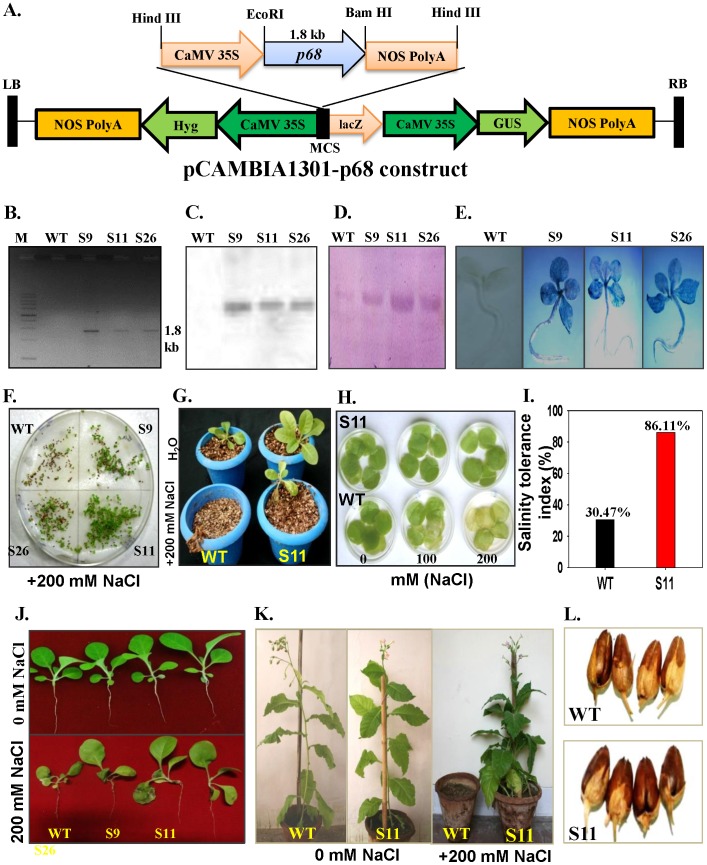
To establish the functional significance of the p68 gene, the complete ORF of the gene was cloned in Hind III site of pCAMBIA1301 (Figure 3A). Pea p68-pCAMBIA-1301 construct was overexpressed in tobacco plants by using Agrobacterium-mediated transformation. The integration of the transgene was confirmed by PCR (Figure 3B). The stable integration was also confirmed by Southern blot analysis (Figure 3C). Western blot detected ∼68 kDa band in transgenic lines (Figure 3D). The GUS expression was positive for all the three transgenic lines while no GUS expression was observed in WT plant (Figure 3E). The germination efficiency of the seeds was also tested. The seeds of pea p68 transgenic lines and WT were grown on MS media containing 200 mM NaCl (Figure 3F). In NaCl containing medium, the seeds of pea p68 transgenic plants started to germinate at 3 d with germination efficiency of more than 70% (data not shown). In contrast, the germination of WT seeds was observed at 9 d on the NaCl-containing medium. The highest germination rates of WT seeds were 35.6% on the NaCl-containing MS medium (data not shown), which was lower than those of pea p68 transgenic seeds, suggesting the susceptibility of WT plants to 200 mM NaCl. The seedlings of p68 transgenic S11 and WT were transferred to pots supplemented with 200 mM NaCl, WT plants died after sometime whereas S11 survived and resumed the growth (Figure 3G). Salinity stress tolerance was also tested by leaf disk senescence assay. Leaf disks of p68 transgenic line S11 and WT were floated separately on 0 (H2O only), 100 or 200 mM NaCl for 72 h. The salinity induced damage was reflected in the degree of bleaching observed in the leaf tissue after 72 h. The leaves of WT plants were bleached, whereas, the leaf disks of S11 retained chlorophyll (Figure 3H). The measurement of salinity stress tolerance index of the 200 mM NaCl treated p68 transgenic line S11 and WT plants was made using the data of plant dry weight and it was noted that the tolerance potential of p68 transgenic line S11 was 86.11%, whereas, it was only 30.47% in WT plants (Figure 3I). The performance of p68 transgenic lines in the presence or absence of salt was similar to the WT plants, which revealed the potential of p68 in salinity stress tolerance (Figure 3J).
Figure 3. Molecular and morphophysiological analysis of pea p68 overexpressing transgenic tobacco plants.
(A) The map of the construct of pCAMBIA1301 containing the p68 gene (pCAMBIA1301-p68). (B) PCR analysis using gene-specific primers. (C) Southern blot analysis for the integration of pea p68 gene in tobacco genome. (D) Western blot analysis of each transgenic lines using anti-pea p68 polyclonal antibody. (E) Histochemical GUS staining of each transgenic line. (F) Comparison of seeds germination of the WT and transgenic lines in response to 200 mM NaCl. (G) Phenotypic comparison of WT and transgenic line (S11) in response to 200 mM NaCl stress. 21 d-old-seedling growing in vermiculite pots and supplied with 200 mM NaCl solution for 5 d. (H) Leaf-disk senescence assay for salinity stress tolerance in transgenic line (S11). (I) Salinity tolerance index potential of WT and transgenic line (S11). (J) Stress responses of WT and p68 overexpressing transgenic lines. 14 d-old vermiculite grown seedlings were transferred to without or with NaCl in nutrient solution (0 and 200 mM NaCl). (K) WT and p68 overexpressing plant (S11) in soil pots supplied without or with 200 mM NaCl solution. Note that the WT plant could not sustain growth under salinity stress. (L) Phenotypic comparison of pods grown in water and stress condition of WT and transgenic line (S11).
The p68 transgenic S11 line and WT plants were phenotypically similar when grown in absence of NaCl. To assess the effect of high salt (200 mM NaCl) on growth, morphology and development of p68 overexpressing and WT plants, the three-week old seedlings were grown in the presence of continuous salt (200 mM NaCl). In the presence of NaCl some stress induced symptoms appeared on the transgenic plants but it still grew normally and set viable seeds, whereas the WT plants could not survive under continuous salinity stress (Figure 3K). In response to stress, the p68 transgenic set healthy pods similar to H2O grown WT plants (Figure 3L). The results show that the p68 overexpressing transgenic lines have better ability to tolerate salinity stress. p68 overexpressing transgenic lines were further analysed for physiological and biochemical parameters to understand the mechanism of salinity stress tolerance in p68 overexpressing transgenic lines.
Segregation Ratio and Seedling Survival of Transgenic and WT Plants Under Salinity Stress
To determine whether the salinity tolerance imparted by p68 is functionally and genetically stable, the homozygous T2 progeny was analyzed. Seeds from the T0 plants, when plated onto hygromycin containing medium, segregated in 3∶1 ratio (Table S1). The percent seedlings survival of p68 overexpressing transgenic lines and WT were also observed and it was found that 200 mM NaCl did not affect the seedlings survival and there was no difference when compared with H2O grown WT plants (Table S1).
Effect of Salinity on Germination of Pollens and Seeds of Pea p68 Transgenic Tobacco Plants
To compare the germination efficiency, first pollens of WT and p68 transgenic lines were sown on pollen germination medium (PGM) alone or on medium supplemented with either 100 or 200 mM NaCl. Without addition of NaCl, the germination of the pollen of pea p68 transgenic was similar to that of WT pollen (Figure S2). The germination of the WT pollen was repressed in the medium containing 100 mM NaCl while no germination was observed in 200 mM NaCl (Figure S2 B, C).
Effect of Salinity on Growth Performance and Yield Transgenic and WT Plants
Significant growth reduction was noted for the WT plants in response to 200 mM NaCl treatment while p68 transgenic lines resist the adverse effects of stress by maintaining vigorous growth. Growth performance measured in terms of shoot length, root length, leaf area and plant dry weight remained almost similar in p68 overexpressing transgenic lines and WT plants under 0 mM NaCl (Figure S3). High concentration of salt (200 mM NaCl) significantly reduced the shoot length, root length, leaf area and plant dry weight of WT plants by 51.70, 57.31, 64.10 and 61.96%, in comparison to 0 mM NaCl. However, the decrease was 16.67, 19.55, 18.82 and 17.35% in the case of S9; 14.50, 18.04, 18.22 and 13.31% in the case of S11 and 18.85, 18.01, 16.92 and 15.62% in the case of S26 in comparison to their respective controls (Figure S3). Under salinity stress, transgenic lines maintained yield contributing parameters including time required for flowering, number of pods per plant, seed number per pod and seed weight per pod and therefore set normal seeds (Table S1).
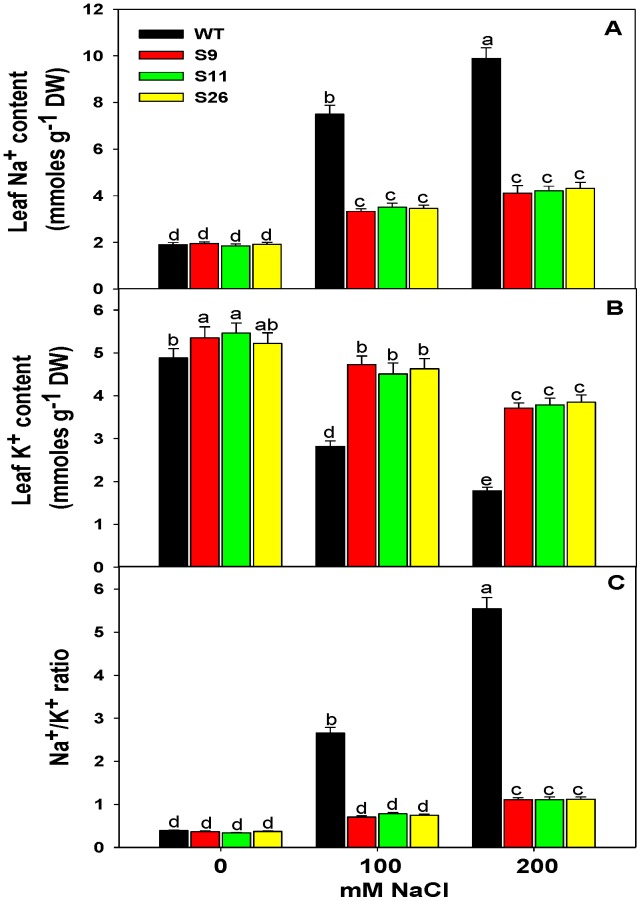
Ion (Na+ and K+) Accumulation in Transgenic and WT Plants under Salinity Stress
To observe Na+ and K+ accumulation, WT and p68 transgenic plants were exposed to salt stress. No significant difference was observed in the accumulation of Na+ in between WT and p68 overexpressing transgenic lines without NaCl treatment. However, with the increasing salt concentration (100 or 200 mM NaCl), the accumulation of Na+ was significantly increased in WT plants while p68 transformed plants accumulated less Na+ (Figure 4A). The pattern of K+ accumulation was similar in the leaves of p68 overexpressing transgenic lines and WT in response to 0 mM NaCl treatment (Figure 4B) but in response to salinity stress, p68 overexpressing transgenic lines retained more K+ compared to the WT plants (Figure 4B). Furthermore, p68 overexpressing transgenic lines showed lower Na+/K+ ratio in comparison to WT plants (Figure 4C), which reflect the potential of transgenic lines to tolerate salinity stress.
Figure 4. Analysis of ion content in transgenic and WT tobacco plants.
(A) Na+ content in the leaves of transgenic and WT plants. (B) K+ content in the leaves of transgenic and WT plants. (C) Na+/K+ ratio in the leaves of transgenic and WT plants. The leaves were exposed to 0, 100 or 200 mM NaCl for 3 weeks of salinity treatment. Values are mean ± SE (n = 3). Different letters on the top of bars indicate significant differences at P<0.05 level as determined by Duncan’s multiple range test (DMRT). The results are representative of similar results obtained from two independent experiments.
Effect of Salinity on Chlorophyll Content, Photosynthesis and Chlorophyll Fluorescence of Transgenic and WT Plants
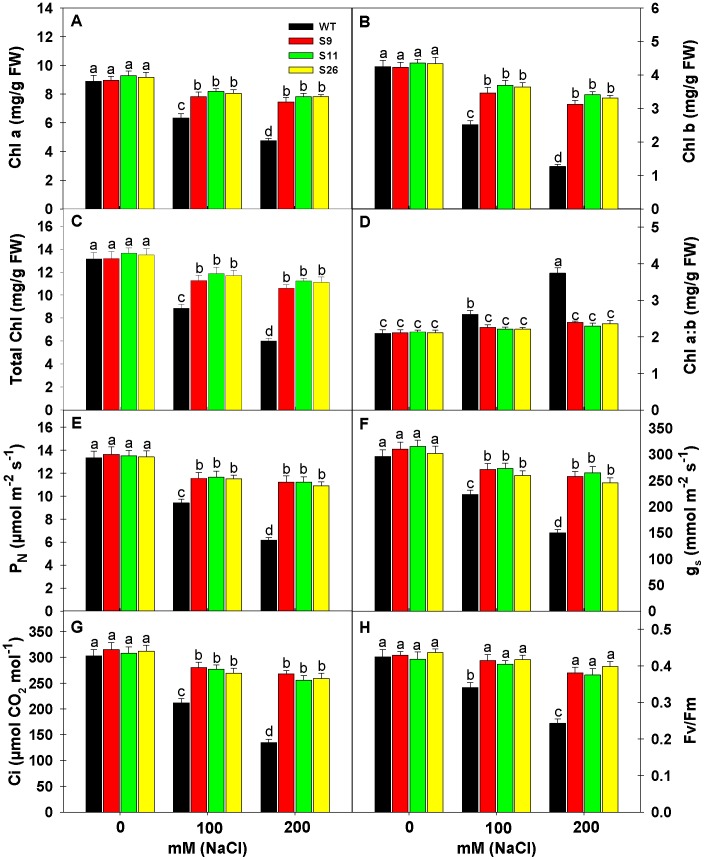
To access the effect of salinity on Chlorophyll (Chl), Chl a, Chl b, total Chl and Chl a: b was measured in p68 overexpressing transgenic lines and WT plants (Figure 5A–D). Salinity stress (100 or 200 mM NaCl) significantly reduced the Chla, Chlb and total Chl in transgenic lines and WT plants but the extent of reduction was higher in WT than p68 overexpressing transgenic lines. The reduction in Chl a, Chl b and total Chl under 200 mM NaCl in WT was 46.56, 70.12 and 54.18% in comparison to 0 mM NaCl, whereas, the reduction was 16.65, 26.24 and 19.73% in the case of S9; 15.71, 21.61 and 17.59% in the case of S11 and 14.83, 23.73 and 17.69% in the case of S26 in comparison to their respective controls (Figure 5A–C). Under salinity stress the Chl content remained significantly higher in transgenic than WT plants. In WT plants the Chl a:b ratio followed the reverse pattern as of Chl content and significantly increased with the increasing salt concentration, whereas, no significant change was noted in the case of transgenic lines (Figure 5D). It reflects that Chl b was severely affected by salinity stress than Chl a under increasing salt concentration in WT plants than transgenic lines which led to significant increase in Chla: b ratio.
Figure 5. Effects of salinity stress on chlorophyll content, photosynthesis and chlorophyll fluorescence of transgenic and WT tobacco plants.
(A) Chlorophyll a content. (B) Chlorophyll b content. (C) Total Chlorophyll content. (D) Ratio of Chlorophyll a and b. (E) Measurement of Net photosynthetic rate (PN). (F) Measurement of stomatal conductance (gs). (G) Measurement of internal CO2 concentration [(Ci)]. (H) Measurement of Chlorophyll fluorescence (Fv/Fm). Data’s were recorded from WT and p68 overexpressing transgenic tobacco lines after 3 weeks exposure to 0, 100 or 200 mM NaCl treatment. Each value represents mean of three replicates ± SE. Means were compared using ANOVA. Data followed by the same letters are not significantly different at P<0.05 as determined by least significant difference (LSD) test. a, b, c indicate significant differences at P<0.05 level as determined by Duncan’s multiple range test (DMRT).
Salinity stress affects the photosynthetic functions at various levels, therefore, the photosynthetic parameters like net photosynthetic rate (PN), stomatal conductance (gs) and internal CO2 (Ci) were measured in p68 overexpressing transgenic lines (S9, S11 & S26) and WT plants under different salinity levels (0, 100 or 200 mM NaCl) (Figure 5E–G). High level of salinity (200 mM NaCl) significantly reduced the photosynthetic parameters but the extent of reduction was several folds higher in WT plants than transgenic lines. It is interesting to note that p68 overexpressing transgenic lines maintained higher photosynthesis than WT even under 0 mM NaCl. The reduction in PN, gs and Ci of WT plants was 53.75, 49.32 and 55.44% under 200 mM NaCl in comparison to their controls, whereas, the decrease in PN, gs and Ci of S9, S11 and S26 was 17.67, 16.77, 14.92%; 16.94, 15.87, 16.88% and 18.85, 18.54, 16.99%, respectively in comparison to their controls.
Chlorophyll fluorescence measurement is one of the most commonly used parameters to study the ecophysiology of plants under salinity stress. To understand the response of salt stress, we measured maximal efficiency of PSII photochemistry (Fv/Fm) in p68 overexpressing transgenic lines and WT plants (Figure 5H). High salinity stress (100 or 200 mM NaCl) significantly reduced the Fv/Fm, whereas, it remained unaltered in transgenic lines. Statistically insignificant change in Fv/Fm in p68 overexpressing transgenic lines reflects that PSII complex did not suffer damage under NaCl stress.
Less Oxidative Stress in p68 Transgenic Lines than WT under Salinity Stress
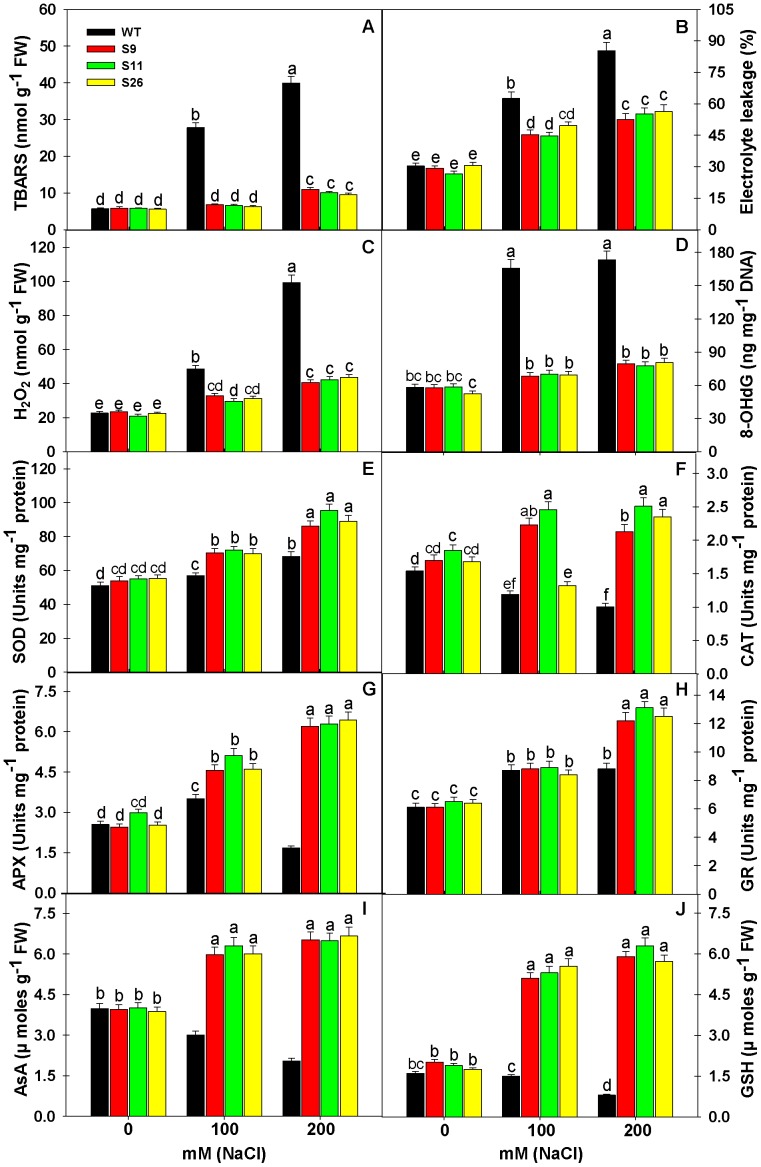
Abiotic stresses including salinity cause overproduction of ROS, which leads to oxidative stress in plants. Therefore, the indicators of oxidative stress such as lipid peroxidation (TBARS content), H2O2 content, electrolyte leakage and oxidative DNA damage were studied in p68 overexpressing transgenic lines and WT plants (Figure 6A–D). High concentration of salt (100 or 200 mM NaCl) significantly increased the extent of oxidative damage and it was significantly higher in WT as compared to p68 transgenic lines. The response of oxidative stress parameters reflects non-significant difference in the values of TBARS content, H2O2 content, electrolyte leakage and oxidative DNA damage (8-OHdG) between p68 overexpressing transgenic lines and WT plants under no salt i.e. 0 mM NaCl. The increase in TBARS, H2O2, electrolyte leakage and 8-OHdG under 200 mM NaCl in WT was 600.00, 337.44, 181.03 and 197.59% in comparison to 0 mM NaCl, whereas, the increase was 86.61, 72.76, 79.53 and 37.67% in the case of S9; 74.48, 102.39, 107.08 and 32.48% in the case of S11 and 70.20, 93.78, 84.48 and 53.62% in the case of S26 in comparison to their respective controls (Figure 6A–D). Overall, it is noted that WT plants suffered maximum oxidative damage reflected in terms of peroxidation of lipids, nucleic acid and electrolyte leakage, whereas, p68 overexpressing transgenic lines experienced less oxidative stress, therefore less oxidative damage.
Figure 6. Overexpression of pea p68 showed less oxidative damage by modulating the ROS machinery under salinity stress.
Measurement thiobarbituric acid reactive substance (A), Electrolyte leakage (B), hydrogen peroxide content (C) and oxidative DNA damage in terms of 8-OhdG (D) in the leaves of WT and p68 overexpressing transgenic tobacco exposed to 0, 100 or 200 mM NaCl after 3 weeks of salinity treatment. Measurement of the activities of superoxide dismutase (E), Catalase (F), ascorbate peroxidase (G) guaiacol peroxidase (H) ascorbate content (I) and glutathione content (J) in the leaves of WT and p68 overexpressing transgenic tobacco exposed to 0, 100 or 200 mM NaCl after 3 weeks of salinity treatment. Values are mean ± SE (n = 3). Different letters on the top of bars indicate significant differences at P<0.05 level as determined by Duncan’s multiple range test (DMRT). The results are representative of similar results obtained from two independent experiments.
Overexpression of p68 Enhances ROS Scavenging Capacity in p68 Transgenic Lines
Salinity stress is known to cause ROS induced oxidative damage in plant cells. Therefore, we analyzed the response of enzymatic (SOD, CAT, APX and GR) and non-enzymatic antioxidants (AsA and GSH) in p68 overexpressing transgenic lines and WT plants under salinity stress. Antioxidant defense machinery protects the plant cells from ROS induced oxidative damage. SOD constitutes the primary step of cellular defense, where SOD dismutates O2 • ¯ to H2O2 and O2. The increase in SOD activity was noted in both p68 transgenic lines and WT plants but activity of SOD was several folds higher as compared to WT plants. Significant increases in SOD activity in p68 overexpressing transgenic lines under 200 mM NaCl was 59.92, 73.13, 61.12%, whereas, it was just 33.92% in WT in comparison to their respective controls (Figure 6E). The H2O2 generated by the action of SOD is restricted through the action of CAT and APX, which reduces H2O2 to water. In the present study, reverse to SOD activity, the CAT and APX activity decreased significantly in WT plants but CAT and APX activity showed significant increase in p68 overexpressing transgenic lines under 200 mM NaCl (Figure 6F–G). The decrease in CAT and APX activity was 34.80 and 34.12% in WT plants under 200 mM NaCl. The activity of CAT and APX increased in p68 overexpressing transgenic lines under 200 mM NaCl by 25.29, 153.06%; 35.68, 111.07% and 39.88, 154.19%, respectively, in comparison to their respective controls (Figure 6F–G). GR catalyzes the NADPH-dependent reduction of oxidized GSSG to the reduced GSH. Here, GR activity showed an increasing trend under salinity stress in both p68 overexpressing transgenic lines and WT plants but maximum significant increase was seen in p68 transgenic lines (Figure 6H). Increase in GR activity in p68 overexpressing transgenic lines under 200 mM NaCl was 100.00, 101.54 and 95.31%, respectively, in comparison to their respective controls, whereas, it was just 44.26% in WT plants. Overall, the upregulation of ROS scavenging antioxidant enzymes in p68 overexpressing transgenic lines protected the plant cells from the damaging effect of ROS generated by salinity stress.
AsA is the only antioxidant buffer in the apoplast and a key antioxidant that reacts with superoxide and hydroxyl radicals, whereas, GSH is a major non-enzymatic scavenger of ROS. The efficient coordination of AsA and GSH in AsA-GSH cycle can protect the plants from salinity induced oxidative damage. Under control condition (0 mM NaCl), ASC and GSH contents were same in control and transformed plants. However, p68 overexpression enhances the pool of AsA and GSH in the transgenic lines. The decrease in AsA and GSH content was 48.74 and 50.00% in WT plants under 200 mM NaCl. The content of AsA and GSH increased in p68 overexpressing transgenic lines under 200 mM NaCl by 65.32, 193.53%; 61.84, 233.33% and 72.35, 226.86%, respectively, in comparison to their respective controls (Figure 6F–G).
Discussion
Abiotic stress is known to affect the cellular gene-expression that limits crop productivity worldwide. So the molecules that are involved in nucleic acid processing, such as helicases, are expected to be affected in response to stress as well. It is evident that stress triggers the expression of many genes including DEAD-box helicases, which play a crucial role in various abiotic stresses [8], [11]–[12], [14], [17], [24]. In this study, a novel DEAD-box helicase gene (p68) was isolated from pea plant which is specifically upregulated in response to salinity. The transcript of pea p68 is also accumulated at a high level and almost equally in every part (roots, leaves, tendrils and flowers) of the pea plant. This result is consistent with the earlier report of transcript analysis of AtDRH1 gene expression in A. thaliana [19]. Therefore, this gene could be a potential candidate for developing stress-tolerant transgenic plants.
The pea p68 protein contains all conserved domains that are characteristic of the DEAD-box proteins including ‘Q’ and ‘GG’ motifs [25]. The ability of p68 to interact with itself indicated that the oligomerization of p68 may be essential for its proper functioning. The p68 is exclusively localized to the cytoplasm and also seems to surround the nucleus of cell. Similar results were reported for another pea helicase (PDH45) [26]. Previously nuclear localization of some DEAD-box helicases (eg. mammalian eIF4AIII and A. thaliana UAP56) showed the involvement in nonsense-mediated mRNA decay, mRNA splicing and export [27]–[28]. RNA splicing process involves the association and dissociation of the pre-mRNA with snRNAs, which may be facilitated by RNA helicases. Previous report in animal system has shown that p68 and p72 RNA helicases are the crucial factors required for efficient RNA splicing [29]–[30]. Both p68 and p72 interact with the U1 small nuclear ribonucleoprotein that recognizes the 5′ splice site [31]. The p68 protein, devoid of RNA helicase or ATPase activity also inhibited the dissociation of U1 from 5′ splice site, and downregulation of DDX5 resulted in the accumulation of unspliced RNA [29]. The role of plant p68 in splicing has not been yet reported. However, there may be possibility that the p68 is sustaining the splicing activity during the stress condition and therefore allowing the p68 overexpressing tobacco plant to survive under the salinity stress condition.
Previously a number of studies demonstrated that DEAD-box RNA helicases are involved in regulating many stages of plant development processes including plant morphogenesis, embryogenesis, pollen tube guidance, floral meristems, flowering, plastids and seed development [32]–[34]. In plant the first report of stress induced helicase gene came by cDNA microarray analysis of 1300 Arabidopsis genes where the authors reported a DEAD-box helicase gene (accession number AB050574) as a cold stress-inducible gene suggesting a new role of helicases in stress signaling [35]. Later, many plant DEAD-box helicases were identified and found to be activated in response to changing environmental conditions [2], [13]–[14], [36]. Evidence is accumulating that transcript of PDH45 and PDH47 was found to be induced in response to high salt, dehydration and cold stresses [11], [13]–[14], [17], [26]. In barley, a salt-responsive transcript HVD1 is induced under salt stress, cold stress, and ABA treatment [37]. AvDH1 is another DEAD-box helicase gene from the halophyte dogbane plant that also strongly upregulated in response to salinity and low temperature [24]. Under normal growth conditions relatively high level of basal expression of the pea p68 gene in different plant parts implies its function in growth and/or development processes. Under salt treatment, a single species of pea p68 mRNA was detected abundantly and constitutively in the tissues examined. This indicated that basic activity of cells might be regulated by pea p68 under salt stress.
Genome-wide expression analysis of many DEAD-box helicase genes have been identified and suggested that these genes might be stress regulated [38]. Overexpression analysis in different DEAD-box helicases has been shown to provide multiple abiotic stress tolerance in crop plants by regulating different signalling pathways [11]–[12], [17]. For example, overexpression of PDH45 and OsSUV3 gene provided salinity stress tolerance in tobacco and rice respectively [12], [17]. LOS4 and RCF1 mutant analysis in Arabidopsis was found to play an important role in response to cold and heat stress [8], [15]. Our study showed that overexpression of pea p68 provides salinity stress tolerance in tobacco.
The reduction in leaf chlorophyll content under abiotic stress has been attributed to the destruction of chlorophyll pigments in various crop plants [3], [39]–[40]. We observed that stress-induced chlorophyll loss was enhanced in WT plant while transgenic lines retained more chlorophyll. This finding has strong correlation with the previous studies in other DEAD-box helicases [12], [41]–[42]. Hence it indicated that overexpression of pea p68 could have positive effects on the growth and photosynthetic metabolism process. Under stress condition, maintenance of the Na+, K+ levels and Na+/K+ ratio are important indicators for plant stress tolerance [43]–[45]. The pea p68 overexpressing tobacco plants accumulated less Na+ and more K+ as compared to the WT plants. Higher K+ content implies delayed leaf senescence in the transgenic lines. Previously it was reported that lower cytosolic K+ content controlled endonuclease and caspase-like proteases activity in the cells causing leaf senescence under stress conditions [40], [46]. Transgenic tobacco plants also extruded more Na+ from cells and, as a result, low Na+ content was detected which indicated that overexpression of pea p68 enhances stress tolerance in transgenic plants. In wheat increased K+ uptake and Na+ extrusion was reported in response to salinity stress [47]. The pea p68 overexpressing transgenic plants are also capable of absorbing more water and diluting the Na+ content. Earlier transgenic approaches with transporter proteins and DESD-box helicase (PDH45) showed that lower Na+/K+ ratio helps plants to respond to salinity stress tolerance [14], [45], [48]. We found lower Na+/K+ ratio in transgenic lines which suggested overexpression of pea p68 might restrict the entry of Na+ ions into the cells thereby protecting photosynthetic machinery from abiotic stresses.
Stress also leads to the rapid production of ROS including H2O2 in plant tissues that ultimately cause damages to the cell membrane and other cellular components such as plasma membrane, mitochondria and chloroplasts [14], [40]. Hence, to avoid any stress-induced injuries plant needs to develop efficient mechanism to remove excess ROS from cells. Enzymatic ROS-scavenging and non-enzymatic antioxidants system are such mechanisms in the plant cells that prevent ROS induced oxidative damage [49]–[51]. In the present study, stress-induced high H2O2 accumulation was noted more in WT plants in comparison to the transgenic lines. Therefore, we speculated that transgenic plants are capable of removing excess H2O2 from the cells, hence preventing cellular damages. Catalase, ascorbate and peroxidase are the major enzymes that are known to be involved in scavenging of cellular production of H2O2 [52]–[53]. Interestingly in this study the activity of these enzymes increased in transgenic lines in response to stress treatment. This indicated that overexpressing lines could readily scavenge H2O2 either decomposing it through increased activity of catalase or by ascorbate through the ascorbate/glutathione cycle. Previously, a number of overexpression studies have shown an increased activity of catalase, ascorbate and peroxidase in response to abiotic stress treatment [14], [54]–[56].
The involvement of DEAD-box helicases in various metabolic processes in plant cells might have general implications. The present study provides new insights into the novel function of p68 DEAD-box protein in conferring salinity stress tolerance in transgenic tobacco plants without affecting yield. The overexpression of stress-induced DEAD-box p68 can also provide a good example of the exploitation of factors of RNA metabolism pathways including splicing factor for enhanced agricultural production of economically important crops under stress conditions.
Supporting Information
Cloning and sequence analysis of p68 .
(TIF)
Percent pollen germination of WT and transgenic lines under salinity stress.
(TIF)
Effects of salinity stress on growth parameter of transgenic and WT tobacco plants.
(TIF)
Comparison of segregation ratio, plant seedlings survival and various yield parameters of the WT and transgenic plants.
(DOCX)
Acknowledgments
We thank Mr Kazi Mostaque Ahmed for helpful corrections.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are included in the manuscript.
Funding Statement
Work on RNA/DNA metabolism and plant abiotic stress tolerance in N.T.’s laboratory is partially supported by Department of Biotechnology (DBT), Government of India and Department of Science and Technology (DST), Government of India. S.S.G. acknowledges the receipt of research grants from CSIR, New Delhi for helicase work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tanner NK, Linder P (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8: 251–262. [DOI] [PubMed] [Google Scholar]
- 2. Owttrim GW (2006) RNA helicases and abiotic stress. Nucleic Acids Res 34: 3220–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuteja N, Gill SS, Tuteja R (2012) Helicases in Improving Abiotic Stress Tolerance in Crop Plants. In: Tuteja N, Gill SS, Tiburcio AF, Tuteja R (eds) Improving crop resistance to abiotic stress. Wiley-VCH Verlag GmbH and Co, KGaA, Germany, 433–445.
- 4. Tuteja N (2000) Plant cell and viral helicases: essential enzymes for nucleic acid transactions. Cri Rev Plant Sci 19: 449–478. [Google Scholar]
- 5. Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, et al. (2005) The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J 24: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fuller-Pace FV (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34: 4206–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishizuka A, Siomi MC, Siomi H (2002) A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16: 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guan Q, Wu J, Zhang Y, Jiang C, Liu R, et al. (2013) A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis . Plant Cell 25: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pradhan A, Chauhan VS, Tuteja R (2005a) A novel ‘DEAD box’ DNA helicase from Plasmodium falciparum is homologous to p68. Mol Biochem Parasitol 140: 55–60. [DOI] [PubMed] [Google Scholar]
- 10. Pradhan A, Chauhan VS, Tuteja R (2005b) Plasmodium falciparum DNA helicase 60 is a schizont stage specific, bipolar and dual helicase stimulated by PKC phosphorylation. Mol Biochem Parasitol 144: 133–141. [DOI] [PubMed] [Google Scholar]
- 11. Vashisht A, Pradhan A, Tuteja R, Tuteja N (2005) Cold and salinity stress-induced pea bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C. Plant J. 44: 76–87. [DOI] [PubMed] [Google Scholar]
- 12. Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc. Natl Acad. Sci. USA 102: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vashisht A, Tuteja N (2006) Stress responsive DEAD box helicases: a new pathway to engineer plant stress tolerance. J Photochem Photobiol 84: 150–160. [DOI] [PubMed] [Google Scholar]
- 14. Gill SS, Tajrishi M, Madan M, Tuteja N (2013) A DESD-box helicase functions in salinity stress tolerance by improving photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. PB1). Plant Mol Biol 82: 1–22. [DOI] [PubMed] [Google Scholar]
- 15. Gong Z, Dong CH, Lee H, Zhu J, Xiong L, et al. (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis . Plant Cell 17: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li D, Zhang H, Wang X, Song F (2008) OsBIRH1, a DEAD box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J Exp Bot 59: 2133–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tuteja N, Sahoo RK, Garg B, Tuteja R (2013) OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). Plant J 76: 115–127. [DOI] [PubMed] [Google Scholar]
- 18. Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okanami M, Meshi T, Iwabuchi M (1998) Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana . Nucleic Acids Res 26: 2638–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gendra E, Moreno A, Alba MM, Pages M (2004) Interaction of the plant glycine-rich RNA-binding protein MA16 with a novel nucleolar DEAD box RNA helicase protein from Zea mays. Plant J 38: 875–886. [DOI] [PubMed] [Google Scholar]
- 21. Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, et al. (1985) A Simple and General Method for Transferring Genes into Plants. Science 227: 1229–1231. [DOI] [PubMed] [Google Scholar]
- 22. Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munns R, Wallace PA, Teakle NL, Colmer TD (2010) Measuring soluble ion concentrations (Na+, K+, Cl-) in salt-treated plants. In: Sunkar R (ed) Methods in molecular biology, vol 639. Plant stress tolerance: methods and protocols. Humana Press. Springer, New York, 371–382. [DOI] [PubMed]
- 24. Liu HH, Liu J, Fan SL, Song MZ, Han XL, et al. (2008) Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum. J Exp Bot 59: 633–644. [DOI] [PubMed] [Google Scholar]
- 25. Tanner NK, Cordin O, Banroques J, Doe’re M, Linder P (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell 11: 127–138. [DOI] [PubMed] [Google Scholar]
- 26. Pham XH, Reddy MK, Ehtesham NZ, Matta B, Tuteja N (2000) A DNA helicase from Pisum sativum is homologous to translation initiation factor and stimulates topoisomerase I activity. Plant J 24: 219–229. [DOI] [PubMed] [Google Scholar]
- 27. Ferraiuolo MA, Lee C, Ler LW, Hsu JL, Costa-Mattioli M, et al. (2004) A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA 101: 4118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kammel C, Thomaier M, Sørensen BB, Schubert T, Längst G, et al. (2013) Arabidopsis DEAD-Box RNA Helicase UAP56 Interacts with Both RNA and DNA as well as with mRNA Export Factors. PLoS ONE 8: e60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin C, Yang L, Yang JJ, Huang Y, Liu ZR (2005) ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell Biol 25: 7484–7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janknecht R (2010) Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). American J Transl Res 2: 223–234. [PMC free article] [PubMed] [Google Scholar]
- 31. Liu ZR (2002) p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA- 5′ splice site duplex. Mol Cell Biol 22: 5443–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shimizu KK, Ito T, Ishiguro S, Okada K (2008) MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana . Plant Cell Physiol 49: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohtani M, Demura T, Sugiyama M (2013) Arabidopsis root initiation defective1, a DEAH-box RNA helicase involved in pre-mRNA splicing, is essential for plant development. Plant Cell 25: 2056–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanai M, Hayashi M, Kondo M, Nishimura M (2013) The plastidic DEAD-box RNA helicase 22, HS3, is essential for plastid functions both in seed development and in seedling growth. Plant Cell Physiol 54: 1431–1440. [DOI] [PubMed] [Google Scholar]
- 35. Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, et al. (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444: 139–158. [DOI] [PubMed] [Google Scholar]
- 37. Nakamura T, Muramoto Y, Takabe T (2004) Structural and transcriptional characterization of a salt-responsive gene encoding putative ATP-dependent RNA helicase in barley. Plant Sci 167: 63–70. [Google Scholar]
- 38. Kant P, Kant S, Gordon M, Shaked R, Barak S (2007) STRS1 and STRS2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol 145: 814–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang ZH, Liu Q, Song HX, Rong XM, Ismail AM (2012) Responses of different rice (Oryza sativa L.) genotypes to salt stress and relation to carbohydrate metabolism and chlorophyll content. Afr J Agric Res 7: 19–27. [Google Scholar]
- 40. Huda KM, Banu MS, Garg B, Tula S, Tuteja R, et al. (2013) OsACA6, a P-type IIB Ca2+ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J 76: 997–1015. [DOI] [PubMed] [Google Scholar]
- 41. Dang HQ, Tran NQ, Gill SS, Tuteja R, Tuteja N (2011) A single subunit MCM6 from pea promotes salinity stress tolerance without affecting yield. Plant Mol Biol 76: 19–34. [DOI] [PubMed] [Google Scholar]
- 42. Sahoo RK, Gill SS, Tuteja N (2012) Pea DNA helicase 45 promotes salinity stress tolerance in IR64 rice with improved yield. Plant Signal Behav 7: 1037–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cuin TA, Betts SA, Chalmandrier R, Shabala S (20080 A root’s ability to retain K+ correlates with salt tolerance in wheat. J Exp Bot 59: 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao Z, He X, Zhao B, Zhou C, Liang Y, et al. (2010) Overexpressing a putative aquaporin gene from wheat, TaNIP, enhances salt tolerance in transgenic Arabidopsis . Plant Cell Physiol 51: 767–775. [DOI] [PubMed] [Google Scholar]
- 45. Hill CB, Jha D, Bacic A, Tester M, Roessner U (2012) Characterization of ion contents and metabolic responses to salt stress of different Arabidopsis AtHKT1;1 genotypes and their parental strains. Mol Plant 6: 350–368. [DOI] [PubMed] [Google Scholar]
- 46. Shabala S (2009) Salinity and programmed cell death: unraveling mechanisms for ion specific signalling. J Exp Bot 60: 709–712. [DOI] [PubMed] [Google Scholar]
- 47. Munns R, James RA, Lauchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Ex Bot 57: 1025–1043. [DOI] [PubMed] [Google Scholar]
- 48. Rajagopal D, Agarwal P, Tyagi W, Singla-Pareek SL, Reddy MK, et al. (2007) Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol. Breed 19: 137–151. [Google Scholar]
- 49. Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–939. [DOI] [PubMed] [Google Scholar]
- 50.Gill SS, Singh LP, Gill R, Tuteja N (2012) Generation and scavenging of reactive oxygen species in plants under stress. In: Tuteja N, Gill SS, Tiburcio AF, Tuteja R (eds) Improving crop resistance to abiotic stress. Wiley-VCH Verlag GmbH and Co. KGaA, Germany, 49–70.
- 51. Bhattacharjee S (2012) The language of reactive oxygen species signaling in plants. J Bot. doi:10.1155/2012/985298 [Google Scholar]
- 52. Willekens H, Langebartels C, Tire C, Van Montagu M, Inze D, et al. (1994) Differential expression of catalase genes in Nicotiana plumbaginifolia (L.). Proc. Natl Acad. Sci. USA 91: 10450–10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Noctor G, Foyer CH (1998) Ascorbate glutathione: Keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279. [DOI] [PubMed] [Google Scholar]
- 54. Jiang M, Zhang J (2002) Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and upregulates the activities of antioxidant enzymes in maize leaves. J Exp Bot 53: 2401–2410. [DOI] [PubMed] [Google Scholar]
- 55. Luna CM, Pastori GM, Driscoll S, Groten K, Bernard S, et al. (2004) Drought controls on H2O2 accumulation, catalase (CAT) activity and CAT gene expression in wheat. J Exp Bot 56: 417–423. [DOI] [PubMed] [Google Scholar]
- 56. Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, et al. (2010) Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61: 4107–4320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cloning and sequence analysis of p68 .
(TIF)
Percent pollen germination of WT and transgenic lines under salinity stress.
(TIF)
Effects of salinity stress on growth parameter of transgenic and WT tobacco plants.
(TIF)
Comparison of segregation ratio, plant seedlings survival and various yield parameters of the WT and transgenic plants.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are included in the manuscript.