Abstract
Gene inactivation is an important tool for correlation of phenotypic and genomic data, allowing researchers to infer normal gene function based on the phenotype when the gene function is impaired. New and better approaches are needed to overcome the shortfalls of existing methods for any significant acceleration of scientific progress. We have adapted the CRISPR/Cas system for use in Xenopus tropicalis and report on the efficient creation of mutations in the gene encoding the enzyme tyrosinase, which is responsible for oculocutaneous albinism. Biallelic mutation of this gene was detected in the F0 generation, suggesting targeting efficiencies similar to that of TALENs. We also find that off-target mutagenesis appears to be negligible and therefore CRISPR/Cas may be a useful system for creating genome modifications in this important model organism.
Keywords: TALENs, CRISPR/Cas, zinc finger nucleases, gene knockouts, gene targeting
INTRODUCTION
The frog Xenopus has long been an engine for elucidating the mechanisms underlying human disease and many reverse genetic methodologies have been pioneered in the frog system. Methods have evolved from the use of simple dominant-negative proteins and inhibitory antibodies, to various forms of antisense oligonucleotide-mediated knockdowns, RNAi, and small molecule inhibitors of signaling pathways. Recently there has been rapid progress in creating targeted gene mutations through the use of zinc finger and transcription activator-like effector nucleases (ZFNs and TALENs, respectively) and these tools have been applied to Xenopus and, like in other systems, with varying efficiencies (Bibikova et al., 2001; Ishibashi et al., 2012; Lei et al., 2012; Lei et al., 2013; Nakajima et al., 2012; Suzuki et al., 2013; Young et al., 2011). Both ZFNs and TALENs function as dimers to bind targeted sites in genes and elicit double-strand breaks. These breaks are then repaired through either non-homologous end joining (NHEJ) or homology-directed repair. The predominant mechanism is NHEJ, an error-prone repair mechanism that primarily creates small deletions centered at the site of the double-strand break.
Both ZFNs and TALENs function as dimers to create double-strand breaks (Segal and Meckler, 2013). They share an architecture composed of an array of DNA-binding domains fused to the nuclease domain from the Fok1 restriction enzyme. In the case of ZFNs, the target site for cleavage is recognized by a series of zinc finger DNA-binding domains that each bind three nucleotide bases. In the case of TALENs, the DNA-binding array is composed of TALE domains that each recognize a single base (Boch et al., 2009; Segal and Meckler, 2013). The active enzyme is created through the dimerization of Fok1 domains between two DNA-bound ZFN or TALEN monomers, thereby producing double-strand breaks between the two half sites. ZFNs have been problematic largely because (1) the combinations of nucleotide triplets capable of being targeted is dependent upon on a limited library of previously characterized zinc finger domains and (2) non-native arrangements of adjacent zinc fingers don't always efficiently function in the manner predicted. Therefore, much effort is expended selecting ZFN combinations with strong specific DNA-binding interactions to ensure success using this approach. The strength of the TALEN strategy lies in the biochemistry of the TALE DNA-binding domain. TALE domains are 33–34 amino acids in length and their recognition of single base pairs occurs via two amino acids called repeat variable diresidues (RVD), which confer specificity for binding. Thus a DNA binding code for TALE RVDs has been elucidated in which each domain interacts with a single nucleotide and fabrication of a series of such repeats creates a DNA binding domain with high specificity for interaction with a targeted site (Boch et al., 2009).
In the past year an even simpler technology has emerged. The prokaryotic CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas (CRISPR-associated) genes comprise an immune system for exclusion of incoming infective DNA (Bhaya et al., 2011; Westra et al., 2012; Segal and Meckler, 2013). Components of the type 2 CRISPR/Cas system from Streptococcus pyogenes have been successfully deployed in eukaryote cells for targeted genome modification (Jinek et al., 2012). The enzymatic portion responsible for creating double strand breaks is encoded by the Cas9 endonuclease, containing two separate nuclease domains each responsible for cleaving one of the two strands of DNA in the target site. Thus, unlike ZFNs and TALENs, the Cas9 nuclease creates double-strand breaks as a monomer. In the form most commonly employed in eukaryotes the targeting portion of the CRISPR/Cas system is a short RNA artificially created by fusing two different S. pyogenes RNAs into a single ~100 nucleotide guide RNA (gRNA; Jinek et al., 2012). The 5' end of a gRNA contains 20 nucleotides that are complementary to the target site in a DNA molecule and these appear to displace the non-complementary strand in the target site to form a gRNA:DNA heteroduplex with the complementary strand. The remaining 80 nucleotides of the gRNA is suggested to form a stem-loop structure that interacts with Cas9 to recruit it to the target site to commence cleavage. The double strand break occurs several base pairs upstream of one end of the target site within the heteroduplex (Gasiunas et al., 2012; Jinek et al., 2012). Therefore creation of a CRISPR/Cas-mediated mutation requires delivery of two molecules into cells: the Cas9 enzyme and a gRNA specific to the sequence being targeted. A three nucleotide sequence is required immediately downstream of the target site, referred to as a PAM (for protospacer adjacent motif) sequence which consists most often of the sequence NGG. Delivery of gRNA can be accomplished either by driving its transcription from polIII promoters like the U6 promoter, or by in vitro transcription using bacteriophage RNA polymerases. The ease of creating gRNAs for gene targeting is an attractive feature of this method and explains the great interest in adapting the CRISPR/Cas system to a variety of biological systems.
Our long-term goal is to generate mutants for analysis of early embryogenesis and we sought to adapt the CRISPR/Cas system for use in Xenopus tropicalis. Two advantages of this organism are its diploidy, with a gene content similar to that of humans, and its relatively fast generation time (Hirsch et al., 2002; Harland and Grainger, 2011; Hellsten et al., 2010). Furthermore, the full genome sequence for X. tropicalis is readily available (Hellsten et al., 2010), making genome-wide searches for unique target sites more straightforward.
In this study we use the gene tyrosinase as a test case for the CRISPR/Cas system in Xenopus. Albino embryos can be efficiently generated in the F0 generation implying that mutation of the tyr locus in individual cells is biallelic. Examination of the tyr target site reveals small indels typical of NHEJ repair. The issue of off-target mutagenesis was also investigated and no evidence of such undesirable mutations was found. We conclude that the CRISPR/Cas system is an efficient means of genome modification in Xenopus that will add to the toolbox of methods available for research in this important disease model.
RESULTS AND DISCUSSION
CRISPR/Cas-mediated knockout of the tyrosinase gene
We wished to determine whether the CRISPR/Cas system is capable of efficiently creating gene mutations in the F0 generation and sought a phenotypic marker that could screened morphologically. tyr encodes the enzyme tyrosinase and is required for the biosynthesis of melanin during tailbud and early tadpole stages of early Xenopus development. Pigment begins to appear in the early stage 30s, approximately 1.5–2 days post-fertilization in X. tropicalis. Oculocutaneous albinism is caused by homozygous loss of try function in mammals and biallelic knockout of this gene using either TALENs or ZFNs similarly results in albinism in Xenopus (Ishibashi et al., 2012; Nakajima et al., 2012; Suzuki et al., 2013). Therefore if mutation of the tyr gene occurs efficiently enough to knockout both alleles in a subset of cells in the embryo, this can be simply detected by visually inspecting injected embryos for the relative abundance of albino tissue at the expense of otherwise pigmented melanocytes and retinal pigmented epithelium (RPE).
To efficiently drive expression of Cas9 protein and a tyr guide RNA in Xenopus embryos we created expression vectors designed for in vitro transcription of these RNAs to permit microinjection into early cleavage stage Xenopus embryos. A human codon-optimized Streptococcus pyogenes Cas9 coding sequence with a C-terminal nuclear localization signal (hCas9; Mali et al., 2013) was cloned downstream of the bacteriophage T7 RNA polymerase promoter in pCDG1 (Blumberg et al., 1989). The resulting plasmid, pCasX, permits the synthesis of a 5'-capped ~5.5kb hCas9 RNA containing Xenopus 5' and 3' β-globin sequences to enhance translatability and an SV40 intron and polyA signal for high RNA stability upon processing in microinjected Xenopus embryos. To create a tyr-targeting guide RNA we scanned the tyr coding sequence for potential sites of the form GGN18NGG using the ZiFiT Targeter webtool (Hwang et al., 2013) and found a candidate site that overlaps well the region targeted by TALENs in Ishibashi et al. (2012). This site (Figure 1A) lies in the 5' end of the ORF within sequence encoding an N-terminal predicted 22-amino acid signal peptide that is responsible for directing tyrosinase protein into membrane-bound melanosomes. Therefore, we surmise that disruption of this sequence even by single codon in-frame deletions might result in a loss of proper signal peptide function and impairment of melanin synthesis. A double-stranded oligonucleotide containing the target sequence was cloned into the plasmid DR274 (Hwang et al., 2013) to permit in vitro synthesis of 100nt tyr gRNAs using T7 RNA polymerase.
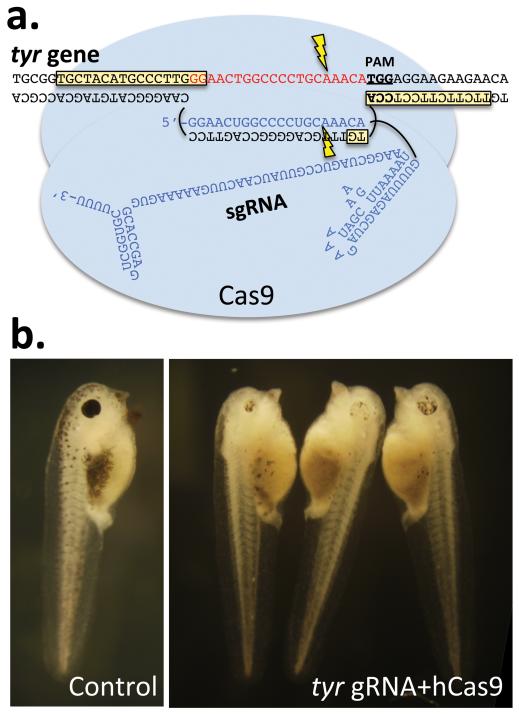
Figure 1. CRISPR/Cas targeting of the tyrosinase locus in Xenopus tropicalis.
A) Cas9-bound gRNA:DNA target region on the tyr gene. The 5'→3' orientation of the transcription unit is from right to left. Overlap with the TALEN binding sites (boxed in yellow) from Ishibashi et al. (2012) is shown. B) Right panel contains examples of stage 41 embryos injected with hCas9 and tyr gRNA. These tadpoles exhibiting typical oculocutaneous albinism found in most of the embryos. A subset retained more pigment but significantly less than wild-type levels (not shown). Left panel is a wild-type control sibling from the same clutch.
hCas9 mRNA and tyr gRNA were coinjected into 1–2 cell stage X. tropicalis embryos and these were cultured at 24–25°C until they reached early tadpole stages. We scored the embryos at stages 41–42 where, in wild types, not only are the eyes well developed and the RPE is strongly melanized, but pigmented melanocytes are also in great abundance. Of 139 injected embryos that survived to stage 25, forty (28.8%) were discarded as being abnormal (cyclopic, strongly bent axes, edema). By stage 41, of the 99 remaining injected embryos, 27 were phenotypically abnormal (defective gut and/or head development) and 72 were normally developing, and the latter were scored for albinism. Among uninjected control embryos 49% were similarly malformed suggesting that developmental abnormalities were not the result of the injected RNAs. Similar observations have been made by Nakayama et al. (manuscript submitted). All 72 embryos showed various degrees in reduction in melanocyte and RPE pigmentation from intermediate to near complete loss (Figure 1B). These observations suggest that, in all the scored embryos, large numbers of cells contain biallelic loss of tyr gene function. Twenty three tadpoles survived through metamorphosis and at the time of manuscript submission are 17 weeks of age (Figure 2). A wide range of pigmentation can be seen in these immature adults. The size differences reflect the typical variation seen within any population of sibling frogs, with some individuals traversing metamorphosis faster than others over the course of about one month.
Figure 2. The range of albino phenotypes in froglets after targeting tyrosinase.
Top left panel: A wild-type frog pigmentation pattern as comparison. The extent of mosaicism is highly variable. Scale bar in lower right panel indicates 1cm.
Evaluation of tyrosinase gene indels
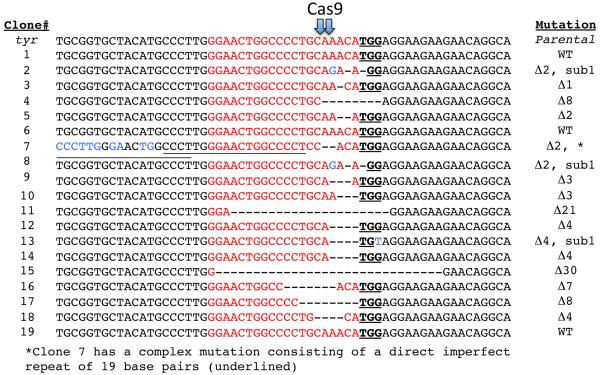
To assess whether albino embryos indeed contain indel mutations at the targeted site in the tyr, we prepared genomic DNA from injected embryos and PCR amplified a 425bp region of the tyr gene overlapping the targeted site using a high fidelity polymerase. As F0 embryos are expected to be mosaic, containing a population of cells with different mutations, we subcloned this PCR product from both wild-type and injected embryos and sequenced a small number of independent clones to sample the variety of mutations that might be present. Sequencing 19 independent clones from a single embryo, we found that 16 (84%) contain a diversity of small indels within the targeted region and 3 clones had wild-type sequence (Figure 3). Independently generated indels of a similar nature were found when targeting the same site by Nakayama et al. (manuscript submitted). Mutations were centered upstream and proximal to the PAM site, which is the expected location for the cleavage site based on the biochemistry of cleavage by Cas9 (Gasiunas et al., 2012; Jinek et al., 2012). The indel variations we observed are similar to those induced by a TALEN pair targeting this region (Ishibashi et al., 2012) and to indels produced by NHEJ repair in other Xenopus genes (Lei et al., 2012; Lei et al., 2013; Nakajima et al., 2012; Suzuki et al., 2013; Young et al., 2011) and in other systems. We conclude that CRISPR/Cas efficiently targeted the tyr gene in X. tropicalis, with apparently both alleles being mutated in most cells especially in the more strongly albino animals.
Figure 3. A catalog of the mutations found in a single albino CRISPR/Cas embryo.
The wild-type sequence (parental) is shown at the top. Nineteen PCR product sequences are shown. Red highlighted bases indicate the target site with the PAM sequence in black bold and underlined. The nature of the mutations is indicated in the right column. Δ, deletion; sub, substitutions; WT, wild-type.
Assessing the possibility of off-target mutagenesis
Numerous publications have suggested that use of the CRISPR/Cas system might be highly problematic due to unwanted targeting of the Cas9 to genomic sites containing mismatches to the 5' portion of the gRNA (Cong et al., 2013; Cradick et al., 2013; Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013; Pattanayak et al., 2013; Sapranauskas et al., 2013), with a minority of these studies suggesting off-target mutagenesis might be infrequent (Bassett et al., 2013; Cho et al., 2013; Fujii et al., 2013; Gratz et al., 2013; Wang et al., 2013; Xie and Yang, 2013; Yang et al., 2013). We wished to evaluate whether our tyr gRNA-directed Cas9 created mutations at off-target sites. To address this question, the v7.1 genome assembly was bioinformatically searched to identify all sequences similar to the targeted site in tyr. The search maintained the requirement for a PAM sequence (5'-NGG-3'), which is essential for Cas9 enzyme binding and cleavage (Gasiunas et al., 2012; Jinek et al., 2012). Table 1 catalogs the distribution of sequences containing 5 mismatched nucleotides or less. We found a total of 621 sequences with five mismatches or fewer and the locations and sequences of each site can be found in Supplementary Table 1. Initially, 4 genomic sites were chosen for inspection because they had 3 or fewer mismatches (Table 2, sites 1–4). Approximately 400bp PCR products containing these sites was amplified from the same genomic DNA used to assess mutations in tyr shown in Figure 1 and 10 independent clones from each site were sequenced. From these 40 clones, no mutations at any site were observed. This implies that the relative efficiency of cleavage at the tyr target site is significantly higher, perhaps at least 10 fold higher, than at sites containing 3 or fewer mismatches.
Table 1.
Distribution of sites with 5 or fewer mismatches in the X. tropicalis genome
| Number of mismatches | Number of target sites |
|---|---|
| 0 | 1 |
| 1 | 0 |
| 2 | 1 |
| 3 | 3 |
| 4 | 55 |
| 5 | 559 |
A genome-wide search for sequences related to the tyr target site found 661 sites with 5 mismatches or fewer. The number of instances containing different mismatches is shown.
Table 2.
Sites for off-target analysis
| Site | Sequence of sites | Number of mismatches |
|---|---|---|
| tyr | GGAACTGGCCCCTGCAAACATGG | 0 |
| 1 | GGGACTGGCCCCTGGAAACAAGG | 2 |
| 2 | GGAAGTGGTCCCAGCAAACAGGG | 3 |
| 3 | GGAGATGGCCCCTTCAAACAGGG | 3 |
| 4 | GGAACTGGTCCCAGCAAGCAGGG | 3 |
| 5 | GGGAATGGGTCCTGCAAACAAGG | 4 |
| 6 | AAGAATTGCCCCTGCAAACAAGG | 5 |
| 7 | GGACAAGTTCCCTGCAAACACGG | 5 |
| 8 | CTGACTCGGCCCTGCAAACAAGG | 5 |
Seven motifs across the genome were sequenced from an albino embryo and are shown. Positions highlighted in red differ from the tyr target site shown at the top. The PAM sequence is in bold. Sites 5–7 contain an uninterrupted “seed” region of 10–13 bases. No mutations were found after sequencing a total of 95 clones. Genomic locations of each motif can be found in Supplementary Tables 1 and 2.
A number of studies have shown that off-targeting efficiency depends on the location of the mismatch within the target site (Jinek et al., 2013; Cong et al., 2013; Hsu et al., 2013; Jiang et al., 2013; Pattanayak et al., 2013; Qi et al., 2013). A “seed” sequence of nine to thirteen bases proximal to the 3' PAM sequence appear to tolerate fewer mismatches than positions further upstream. Examination of the four sites analyzed above shows that all contain at least one mismatch within this seed region. Therefore, we inspected 4- and 5-mismatch sites for examples where the mismatches are more 5' in the target sequence. Only one site could be found that contain the 3'-most 13 bases as identical matches (Table 2, site 6). Three additional sites were found with identity through at least the 3'-most 10 bases (sites, 5, 7 and 8). Of these four sites, three were similarly PCR amplified from genomic DNA and 14–16 clones from each were sequenced (sites 5–7). Again, no mutations were identified amongst these 45 clones. Using this approach unwanted mutations were not detectable at 7 different sites in the genome, strongly suggesting that off-target mutagenesis by the CRISPR-Cas system may not be a major factor in Xenopus. As the current analysis involves a single gRNA and target gene, it will be important to evaluate whether other gRNA-gene combinations in Xenopus also display low off-target mutation rates.
Many of the studies showing strong off-target mutation rates are the result of analyses being done in cell culture, whereas studies in whole animal models appear to find little evidence of such undesirable mutations. Many of the latter studies, like the current report, rely on RNA microinjection to deliver the gRNA and Cas9 expression, whereas the studies in cell culture utilize plasmid-based delivery. The stability of gRNAs is presently unclear. However expression from plasmid-based vectors would be expected to deliver gRNA continuously over several days. Therefore, plasmid-driven expression of these factors might provide a sustained period of action and/or higher levels of expression of these components for Cas9-mediated cleavage thereby resulting in higher off-target mutagenesis rates than observed following direct RNA microinjection.
The efficiency of creating mutations using the CRISPR/Cas system as reported here and elsewhere rivals that of TALENs. This begs the question of whether CRISPR/Cas will supplant TALENs as a method for generating mutants. While CRISPR/Cas has the advantage of simpler design and assembly of targeting components than TALENs, the latter has the promise of being able to target any sequence in the genome. CRISPR/Cas targeting is limited by the requirement for the downstream 5'-NGG-3' PAM sequence. Nevertheless, GG dinucleotides are prevalent enough that targeting by CRISPR/Cas is not significantly hampered. Both systems will likely be advantageous, dependent on the targeting requirements of individual experiments. Genome modification in Xenopus using both of these systems will greatly expand the use of this organism as a tool for understanding biological mechanisms and human disease.
METHODS
Plasmid construction
To create pCasX, a 4.2kb NcoI-PmeI restriction fragment containing human codonoptimized S. pyogenes Cas9 coding sequence was first liberated from pcDNA3.3-TOPO (Mali et al., 2013; obtained from Addgene). This fragment was then ligated between the NcoI and EcoRV sites in pCDG1 (Blumberg et al., 1998). To create DR274tyr we first used the ZiFiT Targeter webtool (Hwang et al., 2013) to scan the tyrosinase ORF (NM_001103048). The target sequence 5' GGAACTGGCCCCTGCAAACATGG-3' was chosen that contains a PAM sequence NGG at its 3' end, is uninterrupted by introns and appears to be unique to in the X. tropicalis genome based on blastN search. Oligonucleotides, 5'-TAGGAACTGGCCCCTGCAAACA-3' and 5'-AAACTGTTTGCAGGGGCCAGTT-3' were annealed and contained 5' overhangs (underlined) compatible with the BsaI sites in DR274 (Hwang et al., 2013; obtained from Addgene). Following ligation into BsaI-digested DR274, individual clones were sequenced (GeneWiz, Inc.) using the oligonucleotide 5'-AACGACGGCCAGTTTATCTAGTC-3' as primer to verify correct oligo subcloning into the 5' end of the gRNA coding sequence.
RNA synthesis, embryo microinjection and rearing
hCas9 mRNA was prepared by linearizing pCasX with Asp718I, which digests downstream of the SV40 polyadenylation cleavage signal. The Ambion T7 mMessage Machine kit was used to synthesize mRNA, which was subsequently DNAsed to remove plasmid DNA, phenol/CHCl3 extracted and NH4OAc/isopropanol precipitated following manufacturers recommendations. The RNA pellet was washed twice with 70% ethanol/30% DEPC-treated water to remove residual NH4OAc. gRNAs were prepared from DR274 plasmid templates by digesting the plasmids with DraI, which creates a blunt 3' end immediately downstream of the 100bp gRNA coding sequence. One microgram of each linearized template was used in 20 μl reactions employing the Ambion T7 Megascript kit. gRNA synthesis reactions were incubated for 4hrs and similarly deproteinized as described above. All RNA concentrations were quantified using A260 spectrophotometer readings and integrity was confirmed by agarose gel electrophoresis.
For microinjection, X. tropicalis eggs were obtained following injection of females with 100U human chorionic gonadotropin. Eggs laid in 1X MMR were collected and in vitro fertilized using testis macerated in 1X MMR containing 1mg BSA/ml. Sperm suspension was diluted with 1/9thX MMR to activate the sperm and thirty minutes after sperm addition the fertilized eggs were dejellied using 3% cysteine, pH 7.6, prepared in 1/9thX MMR. Embryos were microinjected in agarose-coated plates containing 1X MMR at room temperature. The dose range injected was 3–4ng hCas9 mRNA/embryo and 150pg tyr gRNA/embryo. Each embryo received this cocktail in 4nl of total injection volume, delivered into 4 sites per embryo above the equator, and embryos were subsequently cultured in agarose-coated plates in 1/9thX MMR at 24–25°C until desired stages. Staging was according to the Xenopus laevis normal table of Nieuwkoop and Faber (1967).
Genomic PCR and sequencing
DNA was prepared from single embryos at tailbud to early tadpole stages by homogenizing embryos in 100ul of 50mM Tris-HCl (pH8.8), 1mM EDTA, 0.5% Tween-20 and 200ug/ml proteinase K. Homogenates were incubated for 6hrs to overnight at 56°C followed by inactivation of the proteinase K by heating to 95°C for 10 minutes. Samples were stored at −80°C. To amplify genomic regions for sequencing, the high fidelity polymerase Platinum Pfx (Invitrogen) was used in PCR reactions according to manufacturer's specifications. Reactions were 25ul in volume and used 1ul of genomic DNA. Amplification consisted of a 5 minute step at 94°C during which 0.5ul of Pfx polymerase was added (“hot start”), followed by 13 cycles of “touchdown” PCR, where cycles consisted of 94°C for 20 seconds, 65°C for 20 seconds and 68°C for 30 seconds, with each subsequent cycles dropping the annealing temperature by 0.5 Celsius degrees. This segment was typically followed by 20–30 cycles of 94°C for 20 seconds, 58°C for 20 seconds and 68°C for 30 seconds, and a final 68°C step for 5 minutes. PCR oligonucleotide primers used are indicated in Supplementary Table 2. PCR products were subcloned into pCR-Blunt II-Topo (Invitrogen) according to manufacturer's recommendations. Minipreps from single colonies were sequenced (GeneWiz, Inc.) using the standard T7 promoter oligonucleotide primer.
Analysis of putative off-target sites
We scanned the version 7.1 Xenopus tropicalis genome assembly (available at ftp://ftp.xenbase.org/pub/Genomics/JGI/Xentr7.1/) for matches to the tyr target sequence GGAACTGGCCCCTGCAAACANGG, allowing up to 5 mismatches except in the last two letters of the PAM sequence (bold GG), where no mismatches were allowed. A Python script used to run this search, with instructions for its use, is included in Supplementary materials as Supplementary_file_consensus_search.py. The full list of putative off-target sequences related to the tyr guide RNA are available in Supplementary Table 1. We PCR amplified regions containing possible off-target sites of interest using Pfx polymerase, subcloned products into pCR-Blunt II-Topo and sequenced as described in the previous section. Oligonucleotides used in these amplifications are indicated in Supplementary Table 2.
Supplementary Material
Genome-wide search results for motif instances containing 5 or fewer mismatches to the tyr target site.
Off-target sites chosen for sequencing analysis including PCR primers.
ACKNOWLEDGMENTS
The authors would like to thank Drs. Takuya Nakayama and Robert Grainger for generously sharing results prior to publication. We also thank Drs. Shoko Ishibashi and Enrique Amaya for communicating their pre-publication TALEN knockdown results and for sharing their TALEN plasmids. Thanks to Drs. Amanda Janesick and Bruce Blumberg for the expression vector pCDG1 and to Drs. Nakayama and Lyle Zimmerman for many conversations on X. tropicalis techniques and animal husbandry. This work was supported by NSF grant 1147270 and NIH grant HD073179 awarded to K.W.Y.C.
REFERENCES
- Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–97. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Kang H, Bolado J, Jr., Chen H, Craig AG, Moreno TA, Umesono K, Perlmann T, De Robertis EM, Evans RM. BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites. Genes Dev. 1998;12:1269–77. doi: 10.1101/gad.12.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt714. Advanced access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–6. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–35. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. Advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Guo X, Liu Y, Cao Y, Deng Y, Chen X, Cheng CH, Dawid IB, Chen Y, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci. 2012;109:17484–9. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Guo X, Deng Y, Chen Y, Zhao H. Generation of gene disruptions by transcription activator-like effector nucleases (TALENs) in Xenopus tropicalis embryos. Cell Biosci. 2013;3:21. doi: 10.1186/2045-3701-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;9:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Nakajima T, Takase M, Yaoita Y. Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev Growth Differ. 2012;54:777–84. doi: 10.1111/dgd.12006. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) North Holland; Amsterdam: 1967. [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–83. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Ann. Rev. Genom. Human Genet. 2013;14:5.1–5.24. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- Suzuki KT, Isoyama Y, Kashiwagi K, Sakuma T, Ochiai H, Sakamoto N, Furuno N, Kashiwagi A, Yamamoto T. High efficiency TALENs enable F0 functional analysis by targeted gene disruption in Xenopus laevis embryos. Biol Open. 2013;2:448–52. doi: 10.1242/bio.20133855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Harland RM, Zeitler B. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci. 2011;108:7052–7. doi: 10.1073/pnas.1102030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Yang Y. RNA-guided Genome Editing in Plants Using A CRISPR-Cas System. Mol Plant. 2013 doi: 10.1093/mp/sst119. Advanced online publication. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome-wide search results for motif instances containing 5 or fewer mismatches to the tyr target site.
Off-target sites chosen for sequencing analysis including PCR primers.