Abstract
The leukocyte antigen CD38 is expressed after all-trans retinoic acid (ATRA) treatment in HL-60 myelogenous leukemia cells and promotes induced myeloid differentiation when overexpressed. We found that Vav1 and SLP-76 associate with CD38 in two cell lines, and that these proteins complex with Lyn, a Src family kinase (SFK) upregulated by ATRA. SFK inhibitors PP2 and dasatinib, which enhance ATRA-induced differentiation, were used to evaluate the involvement of Lyn kinase activity in CD38-driven signaling. Cells treated with ATRA for 48 hours followed by one hour of PP2 incubation show SFK/Lyn kinase inhibition. We observed that Lyn inhibition blocked c-Cbl and p85/p55 PI3K phosphorylation driven by the anti-CD38 agonistic mAb IB4 in ATRA-treated HL-60 cells and untreated CD38+ transfectants. In contrast, cells cultured for 48 hours following concurrent ATRA and PP2 treatment did not show Lyn inhibition, suggesting ATRA regulates the effects on Lyn. 48 hours of co-treatment preserved CD38-stimulated c-Cbl and p85/p55 PI3K phosphorylation indicating Lyn kinase activity is necessary for these events. In contrast another SFK inhibitor (dasatinib) which blocks Lyn activity with ATRA co-treatment prevented ATRA-induced c-Cbl phosphorylation and crippled p85 PI3K phosphorylation, indicating Lyn kinase activity is important for ATRA-propelled events potentially regulated by CD38. We found that loss of Lyn activity coincided with a decrease in Vav1/Lyn/CD38 and SLP-76/Lyn/CD38 interaction, suggesting these molecules form a complex that regulates CD38 signaling. Lyn inhibition also reduced Lyn and CD38 binding to p85 PI3K, indicating CD38 facilitates a complex responsible for PI3K phosphorylation. Therefore, Lyn kinase activity is important for CD38-associated signaling that may drive ATRA-induced differentiation.
Keywords: CD38 signalling, Src family kinases, Lyn, all-trans retinoic acid
1. Introduction
All-trans retinoic acid (ATRA) is used clinically to treat acute promyelocytic leukemia (APL), but is largely unsuccessful in treating other types of leukemias that are t(15,17) negative. HL-60 is a human acute myelogenous leukemia (AML) cell line that is t(15,17) negative and used as a model to study the mechanisms of ATRA-propelled myeloid differentiation in non-APL cells. Molecules and signaling pathways that confer ATRA responsiveness in HL-60 cells may be important in elucidating how a non-APL leukemia cell can be induced to differentiate by ATRA, and may ultimately provide knowledge that could expand the use of ATRA as a therapeutic agent.
CD38 is a leukocyte antigen that is an early marker of ATRA induction whose expression is mediated via retinoic acid receptor α (RARα) and drives differentiation when overexpressed [1,2]. CD38 is an ectoenzyme receptor and has enzymatic activity that generates the Ca2+ mobilizing compounds NAADP+ and cADPR. It also has receptor functions that drive cell signaling including the phosphorylation of c-Cbl, extracellular signal-regulated kinase (ERK), and the p85 PI3K regulatory subunit [2–9].
Enzymatic activity and receptor/signaling functions can operate independently [10–12]. For example, CD38 metabolic activity is unnecessary for ATRA-induced differentiation while the receptor function associated with membrane-expressed CD38 is required [13]. In addition, siRNA targeting CD38 cripples differentiation [14]. These reports suggest that CD38-driven signaling is important for ATRA-driven myeloid maturation. Therefore, it is of interest to identify CD38-associated signaling molecules and how they may regulate ATRA efficacy. Such knowledge may indicate targets for therapeutic intervention.
CD38 forms a complex with c-Cbl [15,16] and CD38 agonist ligand interaction results in c-Cbl phosphorylation [3]. c-Cbl is an E3 ubiquitin ligase and adaptor molecule that, like CD38, promotes mitogen-activated protein kinase (MAPK) signaling and ATRA-induced differentiation when overexpressed [3,15,16]. This suggests that the c-Cbl/CD38 interaction may cooperatively drive MAPK signaling and other aspects of ATRA therapy. This is consistent with a report that a c-Cbl tyrosine kinase binding domain mutant (G306E) that does not bind CD38 also fails to drive MAPK signaling and differentiation [16].
c-Cbl is known to interact with the guanine nucleotide exchange factor Vav1, the SLP-76 adaptor, and, like CD38, the p85 regulatory subunit of PI3K [15–18]. c-Cbl, SLP-76, and Vav1 protein expression and p85 PI3K activity are upregulated during granulocytic maturation [19–23]. These four proteins also form complexes in myeloid cells after ATRA treatment. For example, Vav1 associates with PI3K and may facilitate the characteristic nucleoskeleton remodeling that occurs with ATRA treatment in HL-60 and NB4 cells [24,25]. Consistent with this, downmodulation of Vav1 impedes induced myeloid maturation and nucleoskeleton remodeling, and affects differentiation-related protein expression [23]. This suggests Vav1 may be a key regulator of myeloid differentiation.
The Src homology 2 domain of Vav1 interacts with c-Cbl and SLP-76 in a differentiation-dependent manner. After ATRA treatment Vav1/c-Cbl complexes are detectable in the cytosol, while Vav1/SLP-76 interactions are predominant in nuclei [24]. SLP-76 is also upregulated after ATRA and forms a complex with c-Cbl [16]. Co-expression of SLP-76 with CSF-1/c-FMS enhances ATRA-induced ERK activation, G0 cell cycle arrest, and a number of additional differentiation markers [21].
CD38 ligation by the anti-CD38 agonistic mAbs IB4 (IB4) and T16 induces phosphorylation of the p85 regulatory subunit of PI3K and is associated with normal and leukemic B cell growth suppression [26–28]. Consistent with this, PI3K inhibitors relieved CD38-mediated growth suppression in ATRA-treated HL-60 cells [29], which suggests a PI3K-modulated CD38 feedback loop.
CD38 also drives transient MAPK activation after agonist ligation, which is orchestrated by the Raf/MEK/ERK axis [6,7]. Transient or protracted signaling from this cascade can lead to either cell proliferation or differentiation respectively [30], and sustained MAPK signaling characterizes ATRA-induced differentiation [31,32]. CD38 overexpression results in persistent ERK phosphorylation, therefore CD38 appears capable of propagating a transient or sustained MAPK signal.
Lyn and other Src family kinases (SFKs) are known to be modulated by ATRA treatment, and Lyn is linked to CD38-driven signaling events. For example, CD38 stimulation of B lymphocytes obtained from Lyn-deficient mice showed defective differentiation, and drugs interfering with PI3K or ERK decreased differentiation [33]. This suggests that Lyn may cooperate with other CD38-associated signaling molecules, such as PI3K and ERK. Other reports show that both Fyn and Lyn are required for B cell signaling after CD38 ligation [34]. Likewise, in lymphoblastoid B-cell membrane rafts CD38 is associated with Lyn and modulates cell signaling [35].
SFK inhibitors are known to enhance aspects of ATRA induction, including expression of CD11b and other myeloid maturation markers [36,37]. A recent study reported that dasatinib, which inhibits Lyn kinase activity alone and with ATRA co-adminstration, enhances differentiation [38]. However the inhibitor PP2, which inhibits Lyn alone but does not block kinase activity with ATRA co-treatment, shows a more significant enhancement of ATRA-induced differentiation than dasatinib. Therefore, Lyn kinase activity may function to drive some aspects of differentiation.
While neither inhibitor is specific for Lyn, the protein was the primary target of significance at the treatment levels used. PP2 can affect ZAP70 and JAK2, for example, but at higher concentrations; we found negligible expression of SFKs Fyn and Lck in HL-60s, and a recent study revealed that the SFK Fgr, while expressed in response to ATRA treatment, was not activated in response to ATRA, PP2, dasatinib, or ATRA and SFK inhibitor co-treatment [38,39]. HL-60s also do not express the BCR/ABL fusion protein, a target of dasatinib.
Since membrane-expressed CD38 has a role in differentiation, signaling that may involve c-Cbl, SLP-76, Vav1, PI3K, and Lyn are important in understanding how ATRA provides therapeutic benefit. Clarification of pathways that confer ATRA responsiveness in t(15,17) negative HL-60 cells could lead to new treatment targets in a larger array of leukemias, as well as other types of cancers. For example, ATRA has shown some promise in treating reproductive leiomyomas by modulating the PI3K signaling cascade [40].
Our results show that CD38, SLP-76, and Vav1 were able to interact together in two cell lines, HL-60 and NB4. Lyn also complexed with these molecules and we evaluated if Lyn kinase activity had an effect on CD38 ligand-induced signaling, including phosphorylation of c-Cbl, ERK, and p85 PI3K. Using the SFK inhibitor PP2 we found that blocking Lyn kinase activity had modest effects on ERK phosphorylation, but was able to completely abrogate c-Cbl and p85 PI3K phosphorylation driven by IB4 [28,41]. We used CD38+ stable transfectants and ATRA-treated HL-60 cells to evaluate if the effects of the inhibitor were associated with either ATRA or CD38 expression alone, and found that PP2 blocked pY-c-Cbl and pY-p85 PI3K in both cell lines. A previous report showed that co-treatment with ATRA and PP2 followed by 48 hours of culture protects Lyn kinase activity from PP2 inhibition and significantly enhances differentiation [38]. Protecting Lyn kinase activity using ATRA/PP2 co-treatment also permitted CD38 ligand-induced phosphorylation of c-Cbl and p85 PI3K. We also observed that dasatinib, which unlike PP2 blocks Lyn activity with ATRA co-treatment, prevented ATRA-propelled c-Cbl phosphorylation and crippled p-p85 PI3K. Therefore Lyn activity appears needed for two signaling events that are downstream of CD38 and induced by retinoic acid.
Finally, we observed that inhibition of Lyn decreased interactions among Vav1/Lyn/CD38, SLP-76/Lyn/CD38, and p85 PI3K/Lyn/CD38. Importantly, the p85 PI3K/Lyn/CD38 association correlates with p85 phosphorylation. These results suggest that the loss of Lyn activity interrupts interactions in a proposed CD38-facilitated signaling complex that involves SLP-76, Vav1, and Lyn, and that this complex regulates the downstream phosphorylation of c-Cbl and p85 PI3K. The observed interaction between p85 PI3K, Lyn, and CD38 suggests that CD38 has direct role in assembling a Lyn kinase-containing complex that phosphorylates p85 PI3K. Together these results indicate that Lyn kinase activity regulates CD38 signaling which results in pY-c-Cbl and pY-p85 PI3K, which is associated with ATRA induction and PP2-enhanced differentiation. These outcomes are important in understanding how the CD38 receptor functions during differentiation, and how it may contribute to the effects of PP2/ATRA co-treatment that result in differentiation enhancement.
2. Materials and methods
2.1 Cell culture
HL-60 and NB4 cells were grown in 5% serum-supplemented RPMI 1640 with 1% antibiotic/antimycotic from Invitrogen (Carlsbad, CA) and treated with 1 –M ATRA as previously described [13]. PP2 from EMD Chemicals (Gibbstown, NJ) was solubilized in dimethyl sulfoxide (DMSO) at 10 mM. Cells were treated with a final concentration of 10 –M with a 0.1% concentration of carrier DMSO. Dasatinib from Santa Cruz Biotechnology (Santa Cruz, CA) was solubilized in DMSO at 5 mM. Cells were treated with a final concentration of 300 nM. SFK activity inhibition was confirmed by Western blot. The concentrations of drugs were approximately 3–4 fold less than that found to cause overt toxicity in titrations monitoring cell growth with a hemacytometer and trypan blue exclusion.
2.2 Antibodies and reagents
Protein A/G beads used for immunoprecipitation, rabbit anti-c-Cbl, rabbit anti-Vav1, and p-Tyr antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). PureProteome Protein G Magnetic Beads were from Millipore (Billerica, MA). Antibodies for GAPDH, beta-actin, p-Erk1/2, ERK1/2 (rabbit), pan-SFK416, Lyn, Fgr, Lck, Fyn, Vav1, SLP-76, pY-p55/p85 PI3K, total p85 PI3K, HRP anti-mouse, and HRP anti-rabbit were from Cell Signaling (Danvers, MA). CD38 antibody was purchased from BD Pharmingen (San Jose, CA). M-PER Mammalian Protein Extraction Reagent lysis buffer was from Pierce (Rockford, IL). Propidium iodide, protease and phosphatase inhibitors, and DMSO were purchased from Sigma (St. Louis, MO).
2.3 Construction of CD38+ stable transfectants
CD38 knock-in plasmid construction and transfection were performed as previously described [2]. To maintain CD38 high expression in stable transfectants, the cells were stained with APC-conjugated anti-CD38 antibody (BD Biosciences, San Jose, CA) and sorted based on high expression of CD38 using a fluorescence activated cell sorter (FACS) flow cytometer (FACS Aria BD Biosciences). Western blotting confirmed CD38 expression.
2.4 Western blot analysis and immunoprecipitation
For immunoprecipitation experiments, cells were lysed as previously described [13]. Equal amounts of protein were pre-cleared with either Protein A/G beads or PureProteome Protein G Magnetic Beads. The beads were pelleted and supernatant was incubated with appropriate antibodies and fresh beads overnight. All incubations included protease and phosphatase inhibitors used for lysis with constant rotation at 4°C. Bead/antibody/protein slurries were then washed and subjected to standard SDS-PAGE analysis as previously described [13].
2.5 Fluorescence resonance energy transfer (FRET)
Cells were harvested, fixed, and permeabilized as previously described [16]. Cells were resuspended in 200 –l of PBS containing 5 –l of primary rabbit anti-SLP-76 or rabbit anti-Vav1 and mouse anti-CD38 antibodies and then stained with Alexa-350 and 430-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies, respectively, from Invitrogen. The immunocomplexes were analyzed using flow cytometry (LSR II, BD Biosciences). The FRET signal was measured as previously described [16]. The Alexa 350 emission from 325 excitation was measured reflected from a 505 longpass dichroic through a 440/40 bandpass filter. Alexa 430 emission from 488 nm excitation (from an argon ion laser) was collected through a 505 longpass dichroic and 530/30 bandpass filter. Controls with secondary antibody(s) only or secondary(s) plus donor or acceptor primary antibody were included. Cells stained with just SLP-76 or Vav1 or CD38 primary antibody and Alexa 350 or 430 respectively were used for compensation controls for spillover into all fluorescence collection channels. Timing gates on the collected fluorescence defined acceptor emission synchronized to donor excitation. FRET signals were corrected by subtraction of background fluorescence of negative controls with just secondary antibodies and compensation controls.
2.6 Signaling experiments
For signaling experiments with Lyn kinase inhibition cells were cultured for 48 hours with 1 –M ATRA, and then washed twice with serum-free RPMI 1640 media. Appropriate samples were incubated with 10 –M PP2 for one hour and all samples were incubated at 37°C with constant rotation. The indicated samples were then treated with 5 –M IB4, which was graciously provided by Fabio Malavasi, for the time points as shown [9]. For samples with ATRA/PP2 co-administration cells were cultured for 48 hours with 1 –M ATRA and 10 –M PP2. Signaling experiments were performed as described above for IB4 treatment, but the one hour PP2 pre-incubation was omitted.
2.7 Statistics
Three independent repeats were conducted in all experiments. Error bars represent the standard error. The student’s t-test function in Microsoft Excel was used to analyze the data.
3. Results
3.1 CD38 interacts with the ATRA-regulated proteins Vav1 and SLP-76
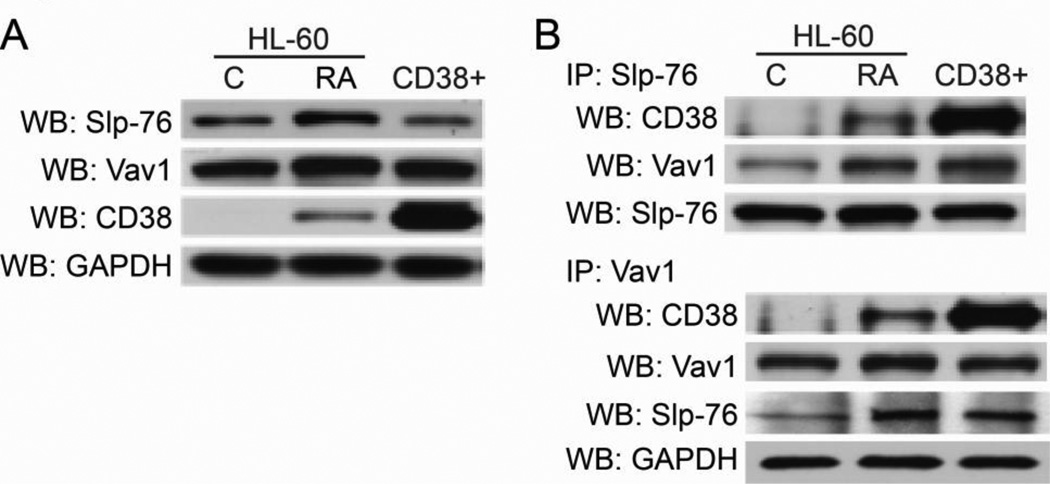
We first determined if there was interaction between CD38 and SLP-76 or Vav1, and investigated whether or not ectopically expressing CD38 in the absence of ATRA affected these interactions. Therefore we could compare effects that were dependent on ATRA treatment versus CD38 expression alone. Western blotting for total protein showed that SLP-76, Vav1, and CD38 were all upregulated by ATRA, with CD38 expression showing dependence on ATRA treatment (Figure 1a). CD38 overexpression in stable transfectants (CD38+) did not significantly increase Vav1 or SLP-76, indicating that upregulated expression of these proteins was dependent on ATRA.
Figure 1. CD38 interacts with SLP-76 and Vav1.
A: Western blots (WB) for protein expression of SLP-76, Vav1, and CD38 in HL-60 cells after 48 hours of ATRA treatment, or in untreated CD38+ transfectants. B: Immunoprecipitation (IP) of either SLP-76 or Vav1 shows interaction among CD38/SLP-76/Vav1.The IP:SLP-76/WB:SLP-76 blot shows protein loading for CD38/Vav1/SLP-76 interaction (top). The IP:Vav1/WB:Vav1 shows protein loading for CD38/Vav1 interaction, and WB for GAPDH shows total protein input for the Vav1/SLP-76 interaction (bottom IP).
Immunoprecipitation experiments show that ATRA-induced CD38 was able to complex with SLP-76 and Vav1, and CD38+ transfectants showed increased interaction (Figure 1b). ATRA treatment also increased interaction between Vav1 and SLP-76. Untreated CD38+ cells also showed increased Vav1/SLP-76 interaction as well, suggesting that CD38 expression regulates binding between these two proteins and facilitates a CD38/SLP-76/Vav1 signaling complex. GAPDH was used as a loading control for protein input for the SLP-76 probed membrane.
3.2 FRET corroborates the CD38/SLP-76/Vav1 protein complex
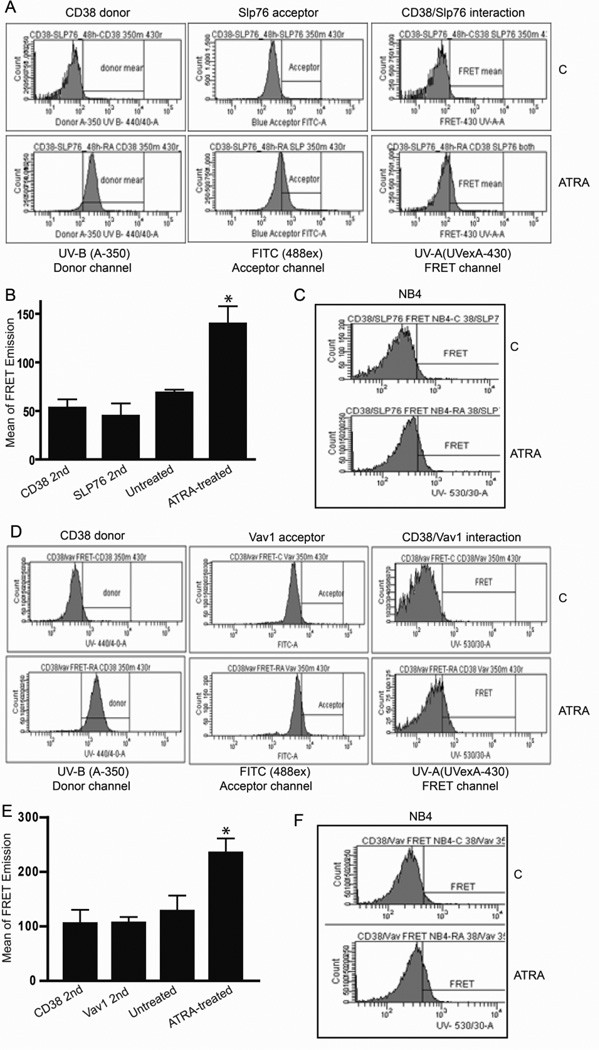
We used FRET to confirm the interaction between CD38 and SLP-76 or Vav1. We included an additional myeloid leukemia cell line (NB4) which bears the characteristic APL t(15,17) translocation to show that these interactions are not specific to HL-60 cells [42,43]. As expected, CD38, SLP-76, and Vav1 were ATRA-upregulated as indicated in donor and acceptor channels (Figures 2a&d). FRET signals between CD38 and SLP-76 were observed in HL-60 and NB4 cells after 48 hours of ATRA treatment (Figures 2a,b,c) but not in untreated cells that did not express CD38. Likewise we detected interaction between and CD38 and Vav1 in ATRA-treated HL-60 and NB4 cells (Figures 2d,e,f).
Figure 2. FRET corroborates the CD38/Vav1/SLP-76 interaction.
A: Flow cytometry FRET histograms confirm 48 hours of ATRA upregulates CD38 and SLP-76 expression and their interaction in HL-60 cells. B: Graph showing means of CD38/SLP-76 FRET emission in HL-60 cells. C: CD38/SLP-76 FRET interaction in NB4 cells after 48 hours of ATRA treatment. D: Flow cytometry FRET histograms confirm ATRA-upregulated CD38 and Vav1 expression and interaction in HL-60 cells. E: Graph showing means of CD38/Vav1 FRET emission in HL-60 cells. F: CD38/Vav1 FRET interaction in NB4 cells after 48 hours of ATRA treatment.
Together, the results from co-immunoprecipitation and FRET experiments demonstrated that ATRA-treated HL-60 and NB4 cells showed interaction among CD38, SLP-76, and Vav1, and indicated that CD38 facilitated a SLP-76/Vav1/CD38 complex. These results prompted interest in CD38/Vav1/SLP-76 interactions with other signaling molecules that are regulated by both ATRA and CD38, specifically the SFK Lyn.
3.3 Lyn interacts with CD38, SLP-76, and Vav1, and kinase inhibition affects CD38-stimulated signaling
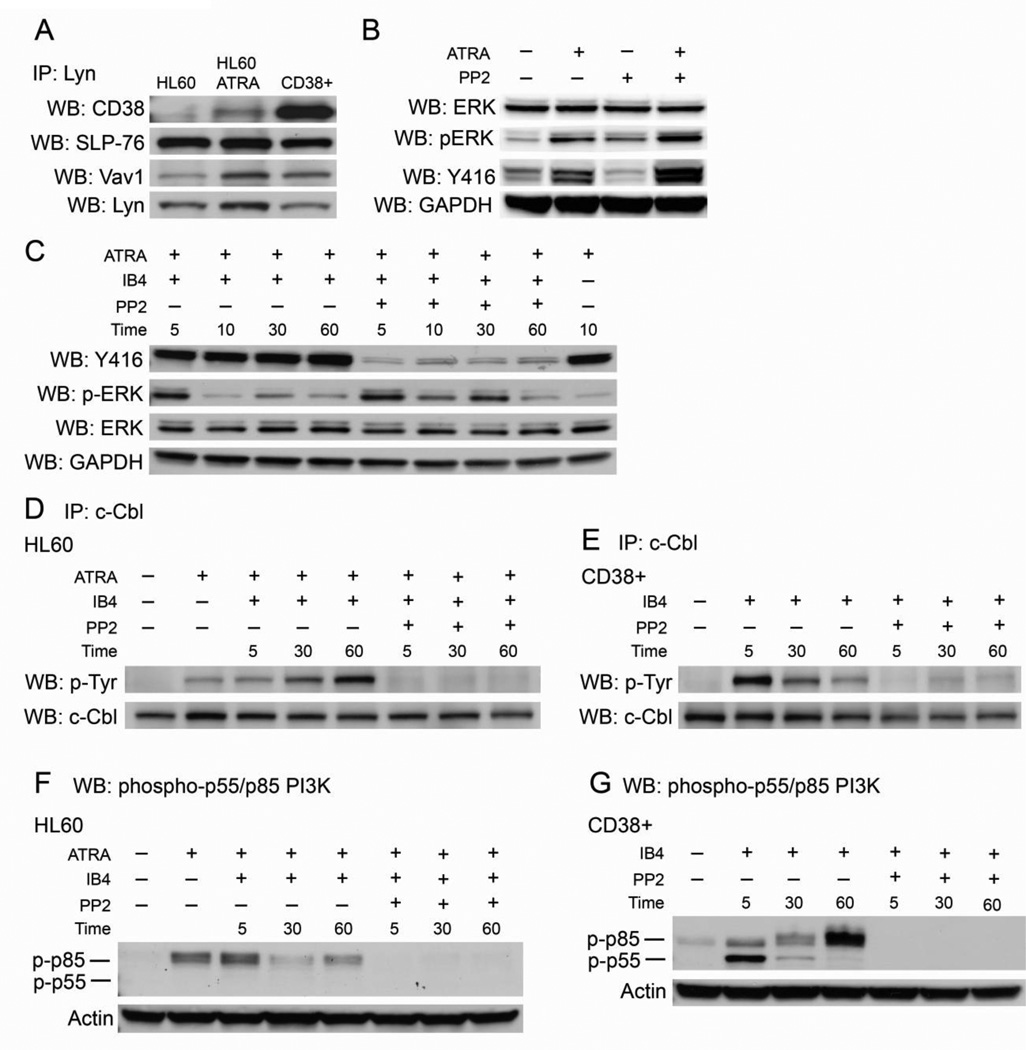
Lyn is upregulated by ATRA and may modulate induction therapy since siRNA against Lyn interferes with differentiation [44]. Lyn also interacts with CD38 to promote signaling [4,34,35]. Therefore, we investigated if Lyn participates in a potential CD38/Vav1/SLP-76 signaling complex. Immunoprecipitation experiments showed that Lyn was able to interact with CD38, Vav1, and SLP-76 (Figure 3a). ATRA-induced CD38 complexed with Lyn, and untreated CD38+ cells showed significantly increased interaction, indicating that CD38 alone facilitates binding. ATRA treatment increased interactions between Lyn/SLP-76 and Lyn/Vav1, but overexpression of CD38 in untreated cells only modestly increased Lyn/Vav1 binding. Therefore Lyn/CD38 and, to a lesser extent, Lyn/Vav1 binding is modulated by CD38 expression level but Lyn/SLP-76 binding is not.
Figure 3. Lyn binds CD38, SLP-76, and Vav1 and its kinase activity regulates CD38 signaling.
A: Immunoprecipiation shows Lyn interaction with CD38, SLP-76, and Vav1 in HL-60 cells with and without 48 hours of ATRA treatment, and in untreated CD38+ cells. The IP:Lyn/WB:Lyn blot shows protein loading. B: Western blots for SFK/Lyn kinase basal activity in untreated, ATRA-treated, PP2-treated, and co-treated cells. All treatments lasted 48 hours. C: Western blots for SFK/Lyn kinase activity in ATRA-treated HL-60 cells using a pan-Y416 that detects active site phosphorylation in all SFK members. Cells were treated with ATRA for 48 hours, incubated with PP2 for one hour, and then stimulated by IB4 for the indicated time points. p-ERK blot shows PP2 did not significantly affect ERK phosphorylation. Total ERK and GAPDH show protein loading. D: Immunoprecipitation of c-Cbl shows tyrosine phosphorylation with indicated treatments in HL-60 cells. Cells treated with ATRA were cultured for 48 hours, cells treated with PP2 were exposed to the inhibitor for one hour, and cells were stimulated by IB4 for the indicated time points E: c-Cbl tyrosine phosphorylation in CD38+ transfectants in the absence of ATRA treated as indicated with PP2 for one hour and then stimulated with IB4. F: Western blot for phospho-p55/p85 PI3K in HL-60 cells treated as indicated. G: Western blot for phospho-p55/p85 PI3K in CD38+ transfectants treated as indicated. Actin shows protein loading in pY-p55/p85 PI3K blots.
Since CD38, SLP-76, and Vav1 also interact with c-Cbl [15,16], we probed for interaction between c-Cbl and Lyn but were unable to detect any significant evidence of association. We also immunoprecipitated Fgr, another SFK upregulated by ATRA, but were only able to detect very weak binding to SLP-76 and Vav1. Western blotting for additional SFKs expressed in myeloid cells (Fyn and Lck) showed negligible protein expression (above data not shown). This is consistent with previous reports that show Lyn is the predominant SFK in myeloid leukemia cells [36,45]. Therefore, we focused our attention on the role of Lyn in CD38 signaling, which motivated interest in whether the Lyn/SFK inhibitor PP2 could modulate signaling propelled by IB4. Here, IB4 was used as a tool to stimulate CD38 signaling. As HL-60 cells express so little basal CD38, IB4 does not affect levels of the targets explored in this paper when added alone (data not shown).
We first confirmed that PP2 was effective in crippling SFK activity in HL-60 cells treated with ATRA for 48 hours. We incubated the indicated samples with PP2 for one hour followed by IB4 treatment as shown to evaluate if Lyn/SFK activity was enhanced by IB4, and if PP2 was able to inhibit Lyn signaling in the presence of IB4. We analyzed samples at various time points after IB4 treatment by probing Western blot membranes with an antibody that detects active site phosphorylation of all SFK members (Y416) (Figure 3c). We found that PP2 was able to inhibit Lyn/SFK activity after IB4 stimulation in ATRA-treated cells and that IB4-driven CD38 ligation did not increase Lyn/SFK kinase activity, consistent with a previous report [28].
Lyn may cooperate with CD38 signaling molecules such as ERK, and ligation of Lyn-associated CD38 in T cells is followed by ERK and p85 PI3K phosphorylation [33,35]. We therefore evaluated whether the inhibitor affected transient IB4-induced ERK phosphorylation (Figure 3c). Although PP2 was able to inhibit SFK/Lyn kinase activity, it had minimal effects on ERK phosphorylation after IB4 stimulation of CD38, allowing the ERK phosphorylation to last slightly longer. We then turned our attention toward two alternative CD38 signaling targets, c-Cbl and p85 PI3K.
We immunoprecipitated c-Cbl and compared tyrosine phosphorylation status after the indicated treatments (Figure 3d). c-Cbl became phosphorylated after ATRA, and IB4 further stimulated phosphorylation. However, pre-incubation with PP2 for one hour was able to abrogate c-Cbl phoshphorylation during all time points, indicating that Lyn activity is necessary for ATRA- and IB4-induced c-Cbl phosphorylation. We also evaluated c-Cbl phosphorylation in untreated CD38+ cells to investigate if the effects of IB4 and PP2 were dependent on ATRA or CD38 expression alone (Fig. 3d). We found that PP2 was still able to block c-Cbl phosphorylation induced by IB4, showing the inhibitor blocked CD38-driven signaling independent of ATRA. It is noteworthy that the kinetics of c-Cbl phosphorylation induced by IB4 were different in CD38+ transfectants compared to ATRA-treated HL-60 cells, suggesting ATRA may temporally regulate modification of c-Cbl.
We also evaluated the effects of PP2 on p85 PI3K regulatory subunit phosphorylation. In HL-60 cells ATRA induced phosphorylation of the p85 PI3K subunit after 48 hours (Figure 3f). IB4 treatment did not result in a significant increase in phosphorylation but this may be because ATRA is already stimulating p-p85 PI3K, and additional phosphorylation could not be achieved. Like pY-c-Cbl, PP2 was able to block ATRA-induced p85 phosphorylation. Phosphorylation of the p55 isoform was not detectable. In untreated CD38+ cells, IB4 was able to stimulate the phosphorylation of the p85 PI3K subunit in the absence of ATRA (Figure 3g). We also detected an IB4-induced transient phosphorylation of the p55 PI3K isoform that was undetectable in HL-60 cells. This indicates that CD38 ectopic expression is able to regulate PI3K proteins differently than ATRA-induced CD38 in HL-60 cells. PP2 was able to abrogate phosphorylation of both the p55 and p85 subunit proteins, similar to its ability to block PI3K activity in HL-60 cells. Together this data suggests that Lyn kinase activity regulates CD38-stimulated phosphorylation of two downstream targets, c-Cbl and PI3K, in ATRA-treated HL-60 cells and CD38+ transfectants. PP2 had minimal effects on CD38-propelled ERK phosphorylation, indicating that the signaling cascade that results in p-ERK is independent of Lyn kinase activity.
3.4 Early PP2 treatment co-administered with ATRA protects Lyn kinase activity and permits CD38-driven c-Cbl and p85 PI3K phosphorylation
After 48 hours of ATRA, one hour of PP2 incubation inhibits Lyn activity (Figure 3). However, if PP2 treatment occurs concurrently with ATRA and cells are then cultured for 48 hours, Lyn kinase activity is protected [38]. Though we cannot rule out some PP2 degradation over the 48 hour treatment, based on inhibition of Y416 phosphorylation, PP2 is still effective at 48 hours [38]. The mechanism by which ATRA protects Lyn kinase activity from inhibition by PP2 is not known. Since one-hour PP2 incubation with ATRA-treated cells blocks SFK/Lyn kinase activity but PP2/ATRA co-treatment for 48 hours protects it, we used this strategy to evaluate if preserving Lyn activity would permit CD38 ligand-induced phosphorylation of c-Cbl and p85 PI3K.
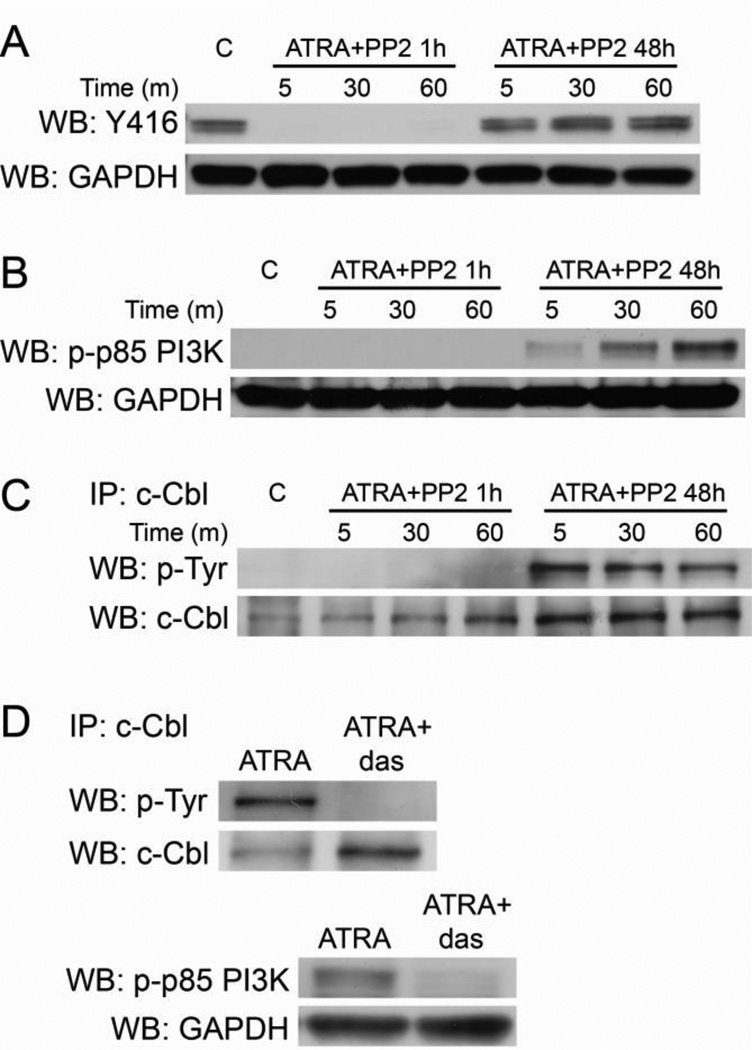
First we confirmed that 48 hours of co-administered PP2 and ATRA were able to protect Lyn activity. We compared Y416 phosphorylation in cells that were co-treated with ATRA and PP2 simultaneously and then cultured for 48 hours to cells that received only ATRA for 48 hours followed by PP2 incubation for one hour (Figure 4a). All ATRA-treated samples were treated with IB4 for the indicated time points to stimulate CD38. As expected, cells treated with ATRA and then later incubated with PP2 showed no Y416 phosphorylation, while cells co-treated with ATRA and PP2 for 48 hours permitted SFK/Lyn kinase activity.
Figure 4. Protecting Lyn kinase activity preserves CD38-stimulated c-Cbl and p85 PI3K phosphorylation.
A: Cells were treated with ATRA for 48 hours, incubated with PP2 for one hour or co-treated for 48 hours as indicated, and then stimulated by IB4 for the indicated time points. Western blotting shows Y416 inhibition in HL-60 cells treated with ATRA for 48 hours followed by one hour of PP2 incubation, but activation after 48 hours of ATRA/PP2 co-treatment in culture. GAPDH shows protein loading. B: Western blot for p85 PI3K phosphorylation in HL-60 cells. Samples were either untreated (C), treated with ATRA for 48 hours followed by one hour of incubation with PP2 to inhibit Lyn activity (ATRA+PP2 1h), or co-treated with ATRA and PP2 and then cultured for 48 hours (ATRA+PP2 48h) which protects Lyn kinase activity. C: c-Cbl phosphorylation in HL-60 cells after the treatments described in B. D: Cells were treated with ATRA or a combination of ATRA and dasatinib for 48 hours. c-Cbl was immunoprecipitated and membranes were probed for tyrosine phosphorylation (top). Western blotting was performed for phosphorylated p85 PI3K (bottom).
We then evaluated if protecting Lyn activity correlated with the ability of ATRA and IB4 to stimulate CD38-driven c-Cbl and p85 PI3K phosphorylation. Confirming the results in Figure 3, one hour of PP2 incubation in ATRA-treated cells blocked p85 PI3K and c-Cbl phosphorylation in IB4-stimulated cells (Figures 4b&c). However, 48 hour co-treatment with ATRA and PP2 permitted ATRA- and IB4-induced c-Cbl and p85 PI3K phosphorylation.
Finally we used dasatinib, which unlike PP2 abrogates Lyn activity with ATRA co-administration [38], to evaluate if kinase inhibition blocked ATRA-induced c-Cbl and p85 PI3K phosphorylation (Figure 4d). We found that dasatinib blocked c-Cbl phosphorylation and significantly decreased p85 PI3K phosphorylation, suggesting that ATRA-induced pY-c-Cbl requires Lyn activity and ATRA-upregulated p85 PI3K activity is largely dependent on Lyn.
Therefore, SFK/Lyn kinase activity regulated ATRA-propelled and CD38-stimulated signaling that results in phosphorylation of c-Cbl and p85 PI3K. This motivated interest in whether or not CD38 associations with other ATRA-induced molecules, specifically SLP-76 and Vav1, were affected by intact or lost Lyn kinase activity.
3.5 PP2 inhibition of c-Cbl and p85 PI3K phosphorylation coincides with decreased CD38/Lyn/SLP-76, CD38/Lyn/Vav1, and CD38/Lyn/p85 PI3K interactions
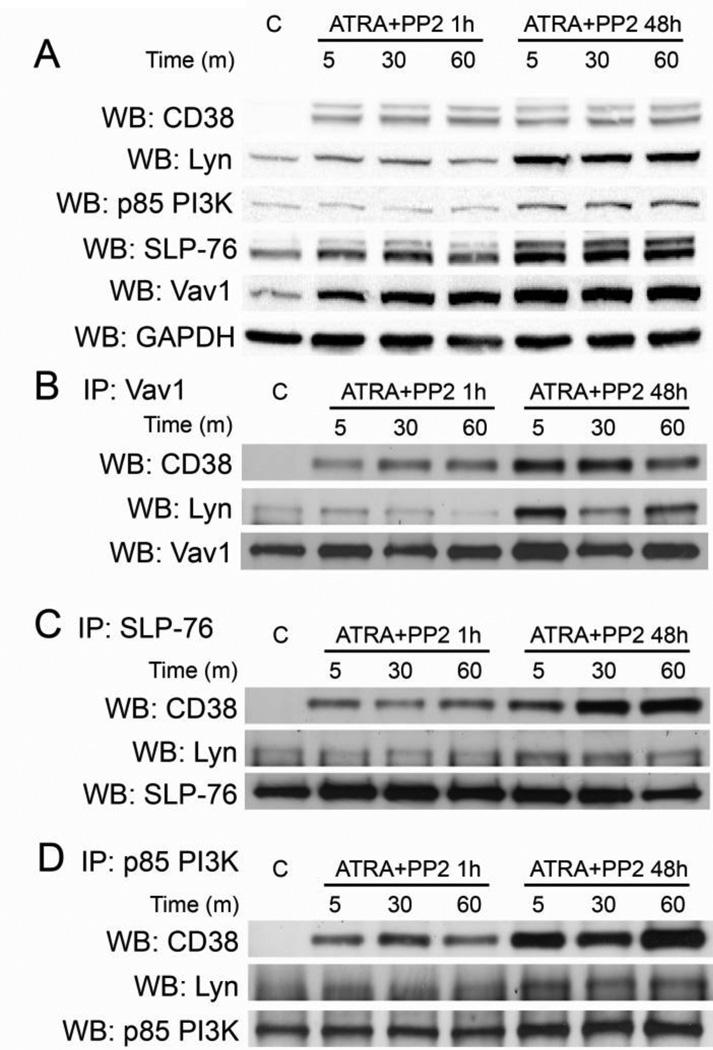
Since SLP-76 and Vav1 complex with both CD38 and Lyn, we investigated if those interactions were affected by the loss of Lyn activity. We immunoprecipitated Vav1 in samples that were either incubated with PP2 for one hour after 48 hours of ATRA treatment or received simultaneous PP2/ATRA treatment for 48 hours. All ATRA-treated samples were stimulated by IB4 to elicit CD38 signaling. We anticipated that an ATRA-induced effect seminal to protecting Lyn kinase activity from PP2 inhibition may also be relevant to interactions between CD38 and its related signaling molecules. We found that the loss of Lyn activity corresponded with a significant decrease in CD38/Vav1 and Lyn/Vav1 interaction (Figure 5b). This suggests that these three molecules constitute part of a signaling complex that regulates an ATRA-induced and CD38-stimulated modification of p85 PI3K and c-Cbl.
Figure 5. Lyn inhibition disrupts interactions among CD38-associated proteins.
A: Cells were treated with ATRA for 48 hours, incubated with PP2 for one hour or co-treated for 48 hours as indicated, and then stimulated by IB4 for the indicated time points. Western blots show basal protein expression levels of target proteins. B: Immunoprecipitation of Vav1 shows changes in CD38 and Lyn interaction after treatments as indicated in HL-60 cells. The Vav1 blot shows protein loading. C: Immunoprecipitation of SLP-76 shows changes in CD38 and Lyn interaction after treatments as indicated. The SLP-76 blot shows protein loading. D: Immunoprecipitation of p85 PI3K shows changes in CD38 and Lyn interaction after treatments as indicated. The p85 PI3K blot shows protein loading.
We also immunoprecipitated SLP-76 and probed for CD38 and Lyn. Similar to Vav1, loss of Lyn kinase activity resulted in decreased CD38/SLP-76 interaction (Figure 5c). Lyn/SLP-76 interaction was modestly decreased, yet the effects were not as significant as observed with Vav1 experiments. These results show that loss of c-Cbl and p85 PI3K phosphorylation, which corresponded to the inhibition of SFK/Lyn kinase activity, is associated with decreased interaction among CD38/Lyn/Vav1 and CD38/Lyn/SLP-76. This suggests that a putative signaling complex including CD38, Vav1, SLP-76, and active Lyn regulates CD38-driven signaling which characterizes ATRA-induced differentiation. Finally we observed that Lyn inhibition decreased CD38/Lyn/p85 PI3K interaction (Figure 5d), suggesting that CD38 facilitates a Lyn kinase-containing complex that is directly responsible for p85 PI3K phosphorylation.
4. Discussion
ATRA therapy is successful in treating t(15,17) positive APL, yet it has shown little efficacy in the treatment of other types of leukemias and cancers. The t(15,17) negative HL-60 cell line shows ATRA responsiveness and is used as a model to investigate how a non-APL leukemic cell can be induced to differentiate. Therefore, elucidation of signaling pathways that may confer ATRA responsiveness could broaden its usefulness in other diseases and aid in identifying new therapeutic molecular targets [46].
CD38 is a leukocyte enzyme and receptor that drives MAPK signaling and differentiation when overexpressed [2]. It has important functions in ATRA induction, since siRNA targeting CD38 interferes with differentiation, and a CD38 mutant (CD38 Δ11–20) that is not membrane-expressed also cripples ATRA effectiveness [13,14]. Identifying signaling pathways orchestrated by CD38 that are involved in myeloid maturation is important in understanding how ATRA works.
4.1 CD38 may direct ATRA-induced differentiation via c-Cbl, Vav1, and SLP-76
In this report, we found that CD38 was able to interact with SLP-76 and Vav1, which regulate differentiation. Therefore, signaling pathways modulated by these proteins may also be propelled by the CD38 receptor. For example downregulation of Vav1 prevents ATRA-induced differentiation, as evidenced by loss of nucleoskeleton remodeling and maturation markers [22–24]. These reports also show that ATRA induction is characterized by the association of Vav1/PI3K and Vav1/SLP-76 in the nucleus and Vav1/c-Cbl in the cytosol. Those complexes could modulate signaling cascades that are important for neutrophil differentiation, and may be coordinated by CD38. CD38 also associates with c-Cbl and interruption of this interaction by a c-Cbl mutation (G306E) results in loss of MAPK signaling and ATRA efficacy [15,16]. These studies report that like CD38, c-Cbl binds Vav1 and SLP-76. This supports our results and suggests that membrane-expressed CD38 coordinates a putative cytosolic signaling complex involving c-Cbl, Vav1, and SLP-76, which could regulate associations with effectors including PI3K.
4.2 Lyn is present in a putative ATRA-induced signaling complex
We also report the involvement of Lyn in a proposed CD38-coordinated complex including SLP-76 and Vav1, which is significant because SFK inhibitors appear to regulate ATRA-induced differentiation [38]. This motivated interest in whether or not Lyn kinase activity was able to modulate signaling driven by a CD38 agonist, IB4. We evaluated three targets of CD38: ERK, c-Cbl, and the p85/p55 PI3K regulatory subunit. Cells were treated with ATRA for 48 hours to induce CD38, and then treated with the SFK inhibitor PP2 for one hour followed by IB4. While PP2 is not specific to Lyn, many of the other SFKs that are affected at the concentration used do not appear to play important roles in RA-induced differentiation of HL-60 cells; Fyn and Lck levels were negligible and activated Fgr was not detected regardless of treatment with RA, PP2, dasatinib, or cotreatment with RA plus either inhibitor [38]. We also used CD38+ transfectants in these experiments to evaluate if there were signaling effects that were specific to ATRA or CD38 expression alone.
4.3 Lyn kinase activity is necessary for ATRA-induced c-Cbl and p55/p85 PI3K, but not ERK, phosphorylation
Since CD38 and c-Cbl interact and both drive MAPK signaling when overexpressed, it is possible that they cooperatively contribute to the persistent ERK phosphorylation that is characteristic of ATRA treatment [15,16,31,47]. However it appears that CD38-propelled ERK phosphorylation is not mediated by Lyn kinase activity since PP2 failed to affect MAPK signaling in ATRA-treated HL-60 cells. In addition, we failed to detect interaction between Lyn and c-Cbl, suggesting that CD38 and c-Cbl may cooperate to propel MAPK signaling independent of Lyn, or that Lyn/c-Cbl interaction is labile. Alternatively, it is possible that Lyn participates in CD38-driven MAPK signaling by serving as a scaffold or by facilitating signaling complex assembly. For example PP2/ATRA co-treatment for 48 hours enhances Lyn expression along with Lyn/c-Raf and c-Raf/ERK interaction, and c-Raf C-terminal domain phosphorylation. These events may be facilitated by Lyn [38,48,49]. Therefore the role of Lyn in CD38-driven MAPK signaling and orchestration may involve the potential for Lyn to act as a scaffold, which appears to be separate from kinase activity.
In contrast, Lyn inhibition completely abrogated ATRA- and CD38-driven c-Cbl and p55/p85 PI3K phosphorylation, showing that Lyn regulates these events in both ATRA-treated HL-60 cells and CD38+ transfectants. It is interesting that co-administration of ATRA and PP2 followed by 48 hours of culture protects Lyn from the effects of PP2. Protecting Lyn kinase activity permitted CD38 ligand-induced pY-c-Cbl and pY-p85 PI3K in HL-60 cells. Dasatinib, which unlike PP2 inhibits Lyn when co-administered with ATRA, blocked c-Cbl phosphorylation and impeded pY-p85 PI3K. Therefore, Lyn kinase activity regulates CD38-stimulated signaling and ATRA-induced phosphorylation events that may be driven by CD38.
4.4 Lyn kinase activity is necessary for the formation of several ATRA-induced CD38-associated complexes
We also show that the loss of Lyn kinase activity coincided with a loss in interaction among CD38/Lyn/SLP-76, CD38/Lyn/Vav1, and CD38/Lyn/p85 PI3K. This suggests that the assembly of these CD38-associated complexes, which are likely involved in CD38 effector signaling after ATRA treatment, is partially dependent on Lyn kinase activity. It also indicates that CD38 participates in the assembly of a Lyn kinase-containing complex that may result in the direct phosphorylation of p85 PI3K.
It is interesting that co-treatment with PP2 and ATRA, which significantly enhances differentiation, preserves Lyn kinase activity [38]. The mechanism by which ATRA protects Lyn from PP2 inhibition is not known. In contrast dasatinib is able to inhibit Lyn in the presence of ATRA and also abrogates c-Cbl tyrosine phosphorylation and significantly decreases p85 PI3K subunit activity driven by ATRA. Dasatinib still enhances differentiation with ATRA co-treatment, but to a significantly lesser extent than PP2. This suggests signaling that characterizes normal ATRA induction in HL-60 cells that is mediated by Lyn kinase activity may help drive differentiation and confer the increased effectiveness of PP2 compared to dasatinib.
Conclusions
We report a CD38/SLP-76/Vav1 interaction and found that Lyn also interacts with these proteins. We found that CD38-propelled p85 PI3K and c-Cbl phosphorylation, which is characteristic of ATRA-induced differentiation, is mediated by Lyn kinase activity. To our knowledge, this is the first report that Lyn regulates ATRA- and CD38-transduced signaling in myeloid leukemia cells, and elucidates how CD38 partner molecules including SLP-76 and Vav1 may regulate differentiation during ATRA or ATRA/PP2 co-treatment.
Highlights.
CD38, Lyn, SLP-76, and Vav1 interact
Lyn inhibition blocks CD38-stimulated c-Cbl and p85/p55 PI3K phosphorylation
Lyn inhibition decreases CD38/Lyn/Vav1 and CD38/Lyn/SLP-76 interaction
Acknowledgements
This work was supported by grants R01 CA033505 and R01 CA152870 from the National Institutes of Health (Yen), U54 CA143876 from the National Institutes of Health/Physical Sciences-Oncology (Shuler), the NYSTEM New York Department of Health (Yen), and the Fondo Ricerca Investimenti di Base (FIRB, Rome, Italy; Malavasi). We are very grateful to Fabio Malavasi for graciously providing the IB4 used in our experiments.
Abbreviations
- AML
acute myelocytic leukemia
- APL
acute promyelocytic leukemia
- ATRA
all-trans retinoic acid
- DMSO
dimethyl sulfoxide
- ERK
extracellular signal-regulated kinase
- FACS
fluorescence activated cell sorter
- FRET
fluorescence resonance energy transfer
- MAPK
mitogen-activated protein kinase
- RARα
retinoic acid receptor α
- SFK
Src family kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Drach J, McQueen T, Engel H, Andreef M, Robertson KA, Collins SJ, Malavasi F, Metha K. Cancer Res. 1994;54:1746–1752. [PubMed] [Google Scholar]
- 2.Lamkin TJ, Chin V, Varvayanis S, Smith JL, Sramkoski RM, Jacobberger JW, Yen A. J Cell Biochem. 2006;97:1328–1338. doi: 10.1002/jcb.20745. [DOI] [PubMed] [Google Scholar]
- 3.Kontani K, Kukimoto I, Nishina H, Hoshino S, Hazeki O, Kanaho Y, Katada T. J Biol Chem. 1996;271:1534–1537. doi: 10.1074/jbc.271.3.1534. [DOI] [PubMed] [Google Scholar]
- 4.Kitanaka A, Ito C, Coustan-Smith E, Campana D. J Immunol. 1997;159:184–192. [PubMed] [Google Scholar]
- 5.Kitanaka A, Suzuki T, Ito C, Nishigaki H, Coustan-Smith E, Tanaka T, Kubota Y, Campana D. J Immunol. 1999;162:1952–1958. [PubMed] [Google Scholar]
- 6.Zubiaur M, Izquierdo M, Terhorst C, Malavasi F, Sancho J. J Immunol. 1997;159:193–205. [PubMed] [Google Scholar]
- 7.Zubiaur M, Guirado M, Terhorst C, Malavasi F, Sancho J. J Biol Chem. 1999;274:20633–20642. doi: 10.1074/jbc.274.29.20633. [DOI] [PubMed] [Google Scholar]
- 8.Zubiaur M, Fernandez O, Ferrero E, Salmeron J, Malissen B, Malavasi F, Sancho J. J Biol Chem. 2002;277:13–22. doi: 10.1074/jbc.M107474200. [DOI] [PubMed] [Google Scholar]
- 9.Funaro A, Spagnoli GC, Ausiello CM, Alessio M, Rogerro S, Delia D, Zaccolo M, Malavasi F. J Immunol. 1990;145:2390–2396. [PubMed] [Google Scholar]
- 10.Lund FE, Muller-Steffner H, Romero-Ramirez H, Moreno-Garcia ME, Partida-Sanchez S, Makris M, Oppenheimer NJ, Santos-Argumedo L, Schuber F. Int Immunol. 2006;18:1029–1042. doi: 10.1093/intimm/dxl037. [DOI] [PubMed] [Google Scholar]
- 11.Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S. Physiol Rev. 2008;88:841–886. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 12.Kumagai M, Coustan-Smith E, Murray DJ, Silvennoinen O, Murti KG, Evans WE, Malavasi F, Campana D. J Exp Med. 1995;181:1101–1110. doi: 10.1084/jem.181.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Congleton J, Jiang H, Malavasi F, Lin H, Yen A. Exp Cell Res. 2011;317:910–919. doi: 10.1016/j.yexcr.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munshi CB, Graeff R, Lee HC. J Biol Chem. 2002;277:49453–49458. doi: 10.1074/jbc.M209313200. [DOI] [PubMed] [Google Scholar]
- 15.Shen M, Yen A. Cancer Res. 2008;68:8761–8769. doi: 10.1158/0008-5472.CAN-08-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen M, Yen A. J Biol Chem. 2009;284:25664–25677. doi: 10.1074/jbc.M109.014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adapala NS, Barbe MF, Langdon WY, Nakamura MC, Tsygankov AY, Sanjay A. J Biol Chem. 2010;285:36745–36758. doi: 10.1074/jbc.M110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adapala NS, Barbe MF, Langdon WY, Tsygankov AY, Sanjay A. Ann N Y Acad Sci. 2010;1192:376–384. doi: 10.1111/j.1749-6632.2009.05346.x. [DOI] [PubMed] [Google Scholar]
- 19.Ohashi E, Kogai T, Kagechika H, Brent GA. Cancer Res. 2009;69:3443–3450. doi: 10.1158/0008-5472.CAN-08-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholl S, Bondeva T, Liu Y, Clement JH, Hoffken K, Wetzker R. J Cancer Res Clin Oncol. 2008;134:861–872. doi: 10.1007/s00432-008-0356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen A, Varvayanis S, Smith JL, Lamkin TJ. Eur J Cell Biol. 2006;85:117–132. doi: 10.1016/j.ejcb.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 22.Bertagnolo V, Brugnoli F, Mischiati C, Sereni A, Bavelloni A, Carini C, Capitani S. Exp Cell Res. 2005;306:56–63. doi: 10.1016/j.yexcr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Bertagnolo V, Grassilli S, Bavelloni A, Brugnoli F, Piazzi M, Candiano G, Petretto A, Benedusi M, Capitani S. J Proteome Res. 2008;7:3729–3736. doi: 10.1021/pr7008719. [DOI] [PubMed] [Google Scholar]
- 24.Bertagnolo V, Marchisio M, Brugnoli F, Bavelloni A, Boccafogli L, Colamussi ML, Capitani S. Cell Growth Differ. 2001;12:193–200. [PubMed] [Google Scholar]
- 25.Bertagnolo V, Brugnoli F, Marchisio M, Celeghini C, Carini C, Capitani S. Cell Signal. 2004;16:423–433. doi: 10.1016/j.cellsig.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Kitanaka A, Ito C, Nishigaki H, Campana D. Blood. 1996;88:590–598. [PubMed] [Google Scholar]
- 27.Matsuo T, Hazeki K, Tsujimoto N, Inoue S, Kurosu H, Kontani K, Hazeki O, Ui M, Katada T. FEBS Lett. 1996;397:113–116. doi: 10.1016/s0014-5793(96)01151-9. [DOI] [PubMed] [Google Scholar]
- 28.Silvennoinen O, Nishigaki H, Kitanaka A, Kumagai M, Ito C, Malavasi F, Lin Q, Conley ME, Campana D. J Immunol. 1996;156:100–107. [PubMed] [Google Scholar]
- 29.Lewandowski D, Linassier C, Iochmann S, Degenne M, Domenech J, Colombat P, Binet C, Herault O. Br J Haematol. 2002;118:535–544. doi: 10.1046/j.1365-2141.2002.03601.x. [DOI] [PubMed] [Google Scholar]
- 30.Traverse S, Gomez N, Paterson H, Marshall C, Cohen P. Biochem J. 1992;288(Pt 2):351–355. doi: 10.1042/bj2880351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen A, Roberson MS, Varvayanis S, Lee AT. Cancer Res. 1998;58:3163–3172. [PubMed] [Google Scholar]
- 32.Wang J, Yen A. J Biol Chem. 2008;283:4375–4386. doi: 10.1074/jbc.M708471200. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Alba JC, Moreno-Garcia ME, Sandoval-Montes C, Rosales-Garcia VH, Santos- Argumedo L. Blood. 2008;111:3644–3652. doi: 10.1182/blood-2007-08-107714. [DOI] [PubMed] [Google Scholar]
- 34.Yasue T, Nishizumi H, Aizawa S, Yamamoto T, Miyake K, Mizoguchi C, Uehara S, Kikuchi Y, Takatsu K. Proc Natl Acad Sci U S A. 1997;94:10307–10312. doi: 10.1073/pnas.94.19.10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zumaquero E, Munoz P, Cobo M, Lucena G, Pavon EJ, Martin A, Navarro P, Garcia-Perez A, Ariza-Veguillas A, Malavasi F, Sancho J, Zubiaur M. Exp Cell Res. 2010;316:2692–2706. doi: 10.1016/j.yexcr.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 36.Kropf PL, Wang L, Zang Y, Redner RL, Johnson DE. Leukemia. 2010;24:663–665. doi: 10.1038/leu.2009.267. [DOI] [PubMed] [Google Scholar]
- 37.Miranda MB, Redner RL, Johnson DE. Mol Cancer Ther. 2007;6:3081–3090. doi: 10.1158/1535-7163.MCT-07-0514. [DOI] [PubMed] [Google Scholar]
- 38.Congleton J, MacDonald R, Yen A. Leukemia. 2012;26:1180–1188. doi: 10.1038/leu.2011.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollock BA, Connelly PA. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Sasson H, Ben-Meir A, Shushan A, Karra L, Rojansky N, Klein BY, Levitzki R, Ben-Bassat H. Fertil Steril. 2011;95:2080–2086. doi: 10.1016/j.fertnstert.2011.01.155. [DOI] [PubMed] [Google Scholar]
- 41.Lande R, Urbani F, Di Carlo B, Sconocchia G, Deaglio S, Funaro A, Malavasi F, Ausiello CM. Cell Immunol. 2002;220:30–38. doi: 10.1016/s0008-8749(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 42.Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- 43.Biondi A, Rambaldi A, Alcalay M, Pandolfi PP, Lo Coco F, Diverio D, Rossi V, Mencarelli A, Longo L, Zangrilli D, et al. Blood. 1991;77:1418–1422. [PubMed] [Google Scholar]
- 44.Katagiri K, Yokoyama KK, Yamamoto T, Omura S, Irie S, Katagiri T. J Biol Chem. 1996;271:11557–11562. doi: 10.1074/jbc.271.19.11557. [DOI] [PubMed] [Google Scholar]
- 45.Dos Santos C, Demur C, Bardet V, Prade-Houdellier N, Payrastre B, Recher C. Blood. 2008;111:2269–2279. doi: 10.1182/blood-2007-04-082099. [DOI] [PubMed] [Google Scholar]
- 46.Malavasi F. J Leukoc Biol. 2011;90:217–219. doi: 10.1189/jlb.0211069. [DOI] [PubMed] [Google Scholar]
- 47.Lamkin TJ, Chin V, Yen A. Am J Hematol. 2006;81:603–615. doi: 10.1002/ajh.20667. [DOI] [PubMed] [Google Scholar]
- 48.Brummer T, Naegele H, Reth M, Misawa Y. Oncogene. 2003;22:8823–8834. doi: 10.1038/sj.onc.1207185. [DOI] [PubMed] [Google Scholar]
- 49.Dougherty MK, Müller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. Mol Cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]