Abstract
Lactobacillus casei has traditionally been recognized as a probiotic, thus needing to survive the industrial production processes and transit through the gastrointestinal tract before providing benefit to human health. The two-component signal transduction system (TCS) plays important roles in sensing and reacting to environmental changes, which consists of a histidine kinase (HK) and a response regulator (RR). In this study we identified HKs and RRs of six sequenced L. casei strains. Ortholog analysis revealed 15 TCS clusters (HK–RR pairs), one orphan HKs and three orphan RRs, of which 12 TCS clusters were common to all six strains, three were absent in one strain. Further classification of the predicted HKs and RRs revealed interesting aspects of their putative functions. Some TCS clusters are involved with the response under the stress of the bile salts, acid, or oxidative, which contribute to survive the difficult journey through the human gastrointestinal tract. Computational predictions of 15 TCSs were verified by PCR experiments. This genomic level study of TCSs should provide valuable insights into the conservation and divergence of TCS proteins in the L. casei strains.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-014-0456-x) contains supplementary material, which is available to authorized users.
Keywords: Comparative genomics, Lactobacillus casei, Probiotic, Two-component signal transduction system
Introduction
Lactobacillus spp. have achieved popularity in the manufacturing of probiotic products because of their convincing beneficial effects on human health. However, before providing benefit to human health, probiotic bacteria must survive the industrial production processes and transit through the gastrointestinal tract [1, 2]. Bacterial two-component signal transduction systems (TCSs) play important roles for many bacteria by enabling them to detect and respond to diverse changes/stresses in the environment [3]. TCS genes are typically located within the same operon encoding two signalling proteins: a transmembrane sensor histidine kinase (HK) and a cytoplasmic response regulator (RR), which may sometimes be carried by a single polypeptide to form the hybrid HKs [3].
Individual HKs contain a conserved kinase core and respond to environmental signals by autophosphorylation of a histidine residue, creating a high-energy phosphoryl group, which is then transferred to an aspartate residue in the RRs. The RRs, which are usually transcriptional regulators, contain a conserved regulatory domain. Phosphorylated RRs then activate downstream specific responses [3]. In most HKs, the transmitter domain shows high sequence conservation, especially within a set of six recognizable motifs or boxes designated H, N, F, G1, G2, and G3. In particular, the H box contains an invariant H residue that is autophosphorylated in an ATP-dependent manner [4]. RRs generally contain at least two functional domains: a conserved N-terminal receiver domain (REC domain) that is phosphorylated by the HK at a strictly conserved D residue, and one or more variable C-terminal output domains [5]. Modulation of the phosphorylated state of the RR controls either expression of the target genes or cellular behaviour.
Lactobacillus casei is a facultative heterofermentative lactic acid bacterium. It has traditionally been recognized as a probiotic and used in commercial products for its health-promoting and nutritional properties [6–8]. L. casei requires a complex array of TCS proteins to cope with diverse human hosts, host responses, and environmental conditions. The TCS MaeKR belonging to the citrate family is essential for the expression of malic enzyme of L. casei strains BL23 and ATCC 334, and MaeKR expression was induced by l-malic acid [9]. The genome sequences of L. casei strains BL23 and ATCC 334 harbor 17 putative TCSs, among which the role of three TCSs involved in bile response, cell envelope stress response, oxidative stress tolerance, and acid tolerance [10]. However, the role of TCSs in L. casei is not still well understood.
With the advance of large scale sequencing technologies and bioinformatics tools, it has become possible to computationally predict TCS proteins and their putative functions from the whole genome of an organism. The availability of complete genome sequences of six L. casei strains enables a more comprehensive study of the role of TCS in the stress response of this organism. In this study, we conducted a thorough comparative analysis of the identified TCS proteins which provides valuable insights into the conservation and divergence of TCS proteins in the L. casei strains studied here.
Materials and Methods
Data Collection
Complete genome sequences of the L. casei strains ATCC 334, LC2W, BD-II, BL23, W56 and str. Zhang were collected from the National Center for Biotechnology Information (NCBI) (ftp.ncbi.nih.gov/genomes/Bacteria/). The genomes used in this study was listed in Table 1.
Table 1.
The information of six sequenced L. casei genomes
| Organism | Assembly | Chrs | Plasmids | Size (Mb) | GC, % | Gene | Protein | Origin |
|---|---|---|---|---|---|---|---|---|
| L. casei ATCC 334 | ASM1452v1 | 1 | 1 | 2.92 | 46.6 | 2,922 | 2,764 | Emmental cheese |
| L. casei LC2W | ASM19478v1 | 1 | 1 | 3.08 | 46.4 | 3,264 | 3,164 | Dairy products |
| L. casei BD-II | ASM19476v1 | 1 | 1 | 3.13 | 46.3 | 3,300 | 3,199 | Homemade koumiss |
| L. casei BL23 | ASM2648v1 | 1 | – | 3.08 | 46.3 | 3,072 | 2,997 | Human Feces |
| L. casei str. Zhang | ASM1924v1 | 1 | 1 | 2.90 | 46.4 | 2,949 | 2,847 | Homemade koumiss |
| L. casei W56 | ASM31803v1 | 1 | 1 | 3.13 | 46.3 | 3,234 | 3,126 | Dairy products |
Identification of HKs and RRs
The approach used to identify putative HKs and RRs from the complete genome sequences of L. casei strains ATCC 334, LC2W, BD-II, BL23, W56 and str. Zhang was similar to that described previously [11]. Briefly, the HMM profile (Accession numbers PF00512) was found in Pfam database that targets the HisKA family of HKs, which was used to recognize the HKs in the L. casei genomes. A profile HMM downloaded from Pfam protein families database [12], which targets the RR REC domain (Accession number PF00072), was used to recognize the RRs in each L. casei genome. Recovered sequences were further scrutinized according to the following criteria: (i) the HATPase domain had to be located in the C-terminus (last 2/3) of the encoded protein and (ii) a putative H-box had to precede the HATPase domain. Detection of HK–RR gene pairs and ‘orphan’ HK and RR genes was similar to that described previously [11].
Identification of Common and Unique TCS Proteins
TCS proteins that are common or unique among L. casei strains were identified through ortholog analysis. The ortholog groups were constructed by using the OrthoMCL-DB tool (http://www.orthomcl.org) [13]. Briefly, HK protein sequences of L. casei strains were assigned to OrthoMCL-DB for the ortholog group identification, and HK proteins belong to the same orthomcl_group were recognized as a common TCS protein.
Bioinformatic Analysis
Protein domain organizations of the HKs and RRs were identified using SMART (smart.embl-heidelberg.de) [14]. Domain limits for proteins were also derived from the graphical output of the SMART web interface. Transmembrane helices of HKs were predicted by the TMHMM2 program (http://www.cbs.dtu.dk/services/TMHMM/) [15]. Phylogenetic trees of the HKs and RRs were built by the software MEGA version 4 [16].
PCR Verification
To verify the presence of 15 TCSs in L. casei, PCR amplification with original DNA from two L. casei strains ATCC334, LC2W and five isolated strains was performed. The primers were designed using the PRIMER-BLAST at online NCBI. Conditions for this conventional PCR were: 94 °C, 2 min; followed by 30 cycles of 94 °C for 30 s; annealing temperature 58 °C for 30 s; and 72 °C for 30 s; final extension at 72 °C for 5 min. The amplified PCR products were resolved in a 1.5 % agarose gel.
Results and Discussion
Identification of TCS Proteins of L. casei Strains
The putative HKs and RRs in the six L. casei strains were identified by searching the complete genome sequences for proteins containing HK and RR domains using Pfam HMM profiles. The repertoires of potential TCS proteins (HKs and RRs) were obtained, as shown in Table 2. By analyzing the putative operon organizations of genes encoding the identified TCS proteins 98.9 % of the total putative HKs and 93.1 % of the total putative RR were found to constitute HK–RR pairs. No hybrid HKs could be detected in all the genomes of the six L. casei strains compared in this study.
Table 2.
Identification of putative two component systems in the six sequenced L. casei strains
| Strains | L. casei ATCC 334 | L. casei BD-II | L. casei BL23 | L. casei LC2W | L. casei str. Zhang | L. casei W56 |
|---|---|---|---|---|---|---|
| Total TCS proteins | 31 | 29 | 31 | 31 | 31 | 30 |
| Total paired HKs | 15 | 14 | 15 | 15 | 15 | 13 |
| Orphan HKs | 0 | 0 | 0 | 0 | 0 | 1 |
| Total paired RRs | 15 | 14 | 15 | 15 | 15 | 13 |
| Orphan RRs | 1 | 1 | 1 | 1 | 1 | 3 |
Ortholog Analysis of TCS Proteins
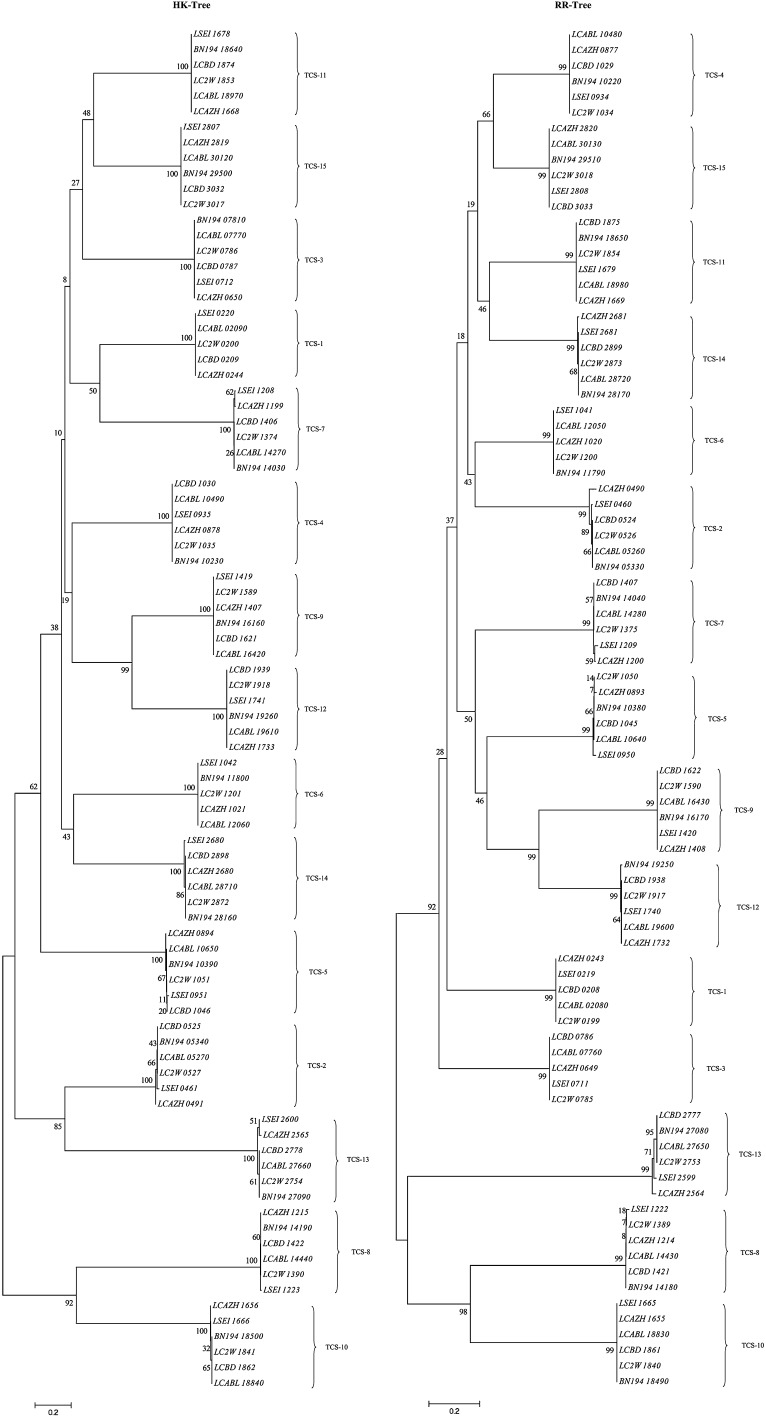
Ortholog analysis of the paired or non-paired TCS proteins among the six L. casei strains revealed a total of 15 different TCS clusters, one orphan HKs and three orphan RRs (Table 3). Co-evolution of TCS proteins could be clearly observed. This means, HKs and RRs which belong to a particular TCS cluster are usually co-present or co-absent in a specific strain. Twelve of the 15 TCS clusters (designated as TCS-2, TCS-4, TCS-5, TCS-7, TCS-8, TCS-9, TCS-10, TCS-11, TCS-12, TCS-13, TCS-14, TCS-15) were common to all the strains. Three clusters (TCS-1, TCS-3, TCS-6) were observed to be absent in one or several strains. One orphan HK was identified as uniquely present in L. casei W56 (BN19407810, named as orphan HK1). In contrast, an orphan RR (orphan RR1, LSEI2389 in L. casei ATCC 334) was found to be common to all strains except for L. casei BD-II. L. casei W56 harbored an additional unique orphan RR (BN19402120, orphan RR3). In addition, a clear clustering of HK and RR orthologs can be visualized in the phylogenetic tree shown in Fig. 1, which additionally illustrates the relationships between the different TCS clusters.
Table 3.
Ortholog analysis and classifications of the putative TCS proteins in the six sequenced L. casei strains
| TCS cluster | TCS protein | Family | L. casei ATCC 334 | L. casei BD-II | L. casei BL23 | L. casei LC2W | L. casei str. Zhang | L. casei W56 |
|---|---|---|---|---|---|---|---|---|
| TCS-1 | HK | IIIA | LSEI0220 | LCBD0209 | LCABL02090 | LC2W0200 | LCAZH0244 | Absent |
| RR | OmpR | LSEI0219 | LCBD0208 | LCABL02080 | LC2W0199 | LCAZH0243 | Absent | |
| TCS-2 | HK | IIIA | LSEI0461 | LCBD0525 | LCABL05270 | LC2W0527 | LCAZH0491 | BN19405340 |
| RR | OmpR | LSEI0460 | LCBD0524 | LCABL05260 | LC2W0526 | LCAZH0490 | BN19405330 | |
| TCS-3 | HK | IIIA | LSEI0712 | LCBD0787 | LCABL07770 | LC2W0786 | LCAZH0650 | Absent |
| RR | OmpR | LSEI0711 | LCBD0786 | LCABL07760 | LC2W0785 | LCAZH0649 | Absent | |
| TCS-4 | HK | IIIA | LSEI0935 | LCBD1030 | LCABL10490 | LC2W1035 | LCAZH0878 | BN19410230 |
| RR | OmpR | LSEI0934 | LCBD1029 | LCABL10480 | LC2W1034 | LCAZH0877 | BN19410220 | |
| TCS-5 | HK | IIIA | LSEI0951 | LCBD1046 | LCABL10650 | LC2W1051 | LCAZH0894 | BN19410390 |
| RR | OmpR | LSEI0950 | LCBD1045 | LCABL10640 | LC2W1050 | LCAZH0893 | BN19410380 | |
| TCS-6 | HK | IIIA | LSEI1042 | Absent | LCABL12060 | LC2W1201 | LCAZH1021 | BN19411800 |
| RR | OmpR | LSEI1041 | Absent | LCABL12050 | LC2W1200 | LCAZH1020 | BN19411790 | |
| TCS-7 | HK | IIIA | LSEI1208 | LCBD1406 | LCABL14270 | LC2W1374 | LCAZH1199 | BN19414030 |
| RR | OmpR | LSEI1209 | LCBD1407 | LCABL14280 | LC2W1375 | LCAZH1200 | BN19414040 | |
| TCS-8 | HK | II | LSEI1223 | LCBD1422 | LCABL14440 | LC2W1390 | LCAZH1215 | BN19414190 |
| RR | CitB | LSEI1222 | LCBD1421 | LCABL14430 | LC2W1389 | LCAZH1214 | BN19414180 | |
| TCS-9 | HK | IIIA | LSEI1419 | LCBD1621 | LCABL16420 | LC2W1589 | LCAZH1407 | BN19416160 |
| RR | OmpR | LSEI1420 | LCBD1622 | LCABL16430 | LC2W1590 | LCAZH1408 | BN19416170 | |
| TCS-10 | HK | II | LSEI1666 | LCBD1862 | LCABL18840 | LC2W1841 | LCAZH1656 | BN19418500 |
| RR | CitB | LSEI1665 | LCBD1861 | LCABL18830 | LC2W1840 | LCAZH1655 | BN19418490 | |
| TCS-11 | HK | IIIA | LSEI1678 | LCBD1874 | LCABL18970 | LC2W1853 | LCAZH1668 | BN19418640 |
| RR | OmpR | LSEI1679 | LCBD1875 | LCABL18980 | LC2W1854 | LCAZH1669 | BN19418650 | |
| TCS-12 | HK | IIIA | LSEI1741 | LCBD1939 | LCABL19610 | LC2W1918 | LCAZH1733 | BN19419260 |
| RR | OmpR | LSEI1740 | LCBD1938 | LCABL19600 | LC2W1917 | LCAZH1732 | BN19419250 | |
| TCS-13 | HK | ? | LSEI2600 | LCBD2778 | LCABL27660 | LC2W2754 | LCAZH2565 | BN19427090 |
| RR | LytT | LSEI2599 | LCBD2777 | LCABL27650 | LC2W2753 | LCAZH2564 | BN19427080 | |
| TCS-14 | HK | IIIA | LSEI2680 | LCBD2898 | LCABL28710 | LC2W2872 | LCAZH2680 | BN19428160 |
| RR | OmpR | LSEI2681 | LCBD2899 | LCABL28720 | LC2W2873 | LCAZH2681 | BN19428170 | |
| TCS-15 | HK | IIIA | LSEI2807 | LCBD3032 | LCABL30120 | LC2W3017 | LCAZH2819 | BN19429500 |
| RR | OmpR | LSEI2808 | LCBD3033 | LCABL30130 | LC2W3018 | LCAZH2820 | BN19429510 | |
| Orphan HK1 | HK | IIIA | Absent | Absent | Absent | Absent | Absent | BN19407810 |
| Orphan RR1 | RR | LytT | LSEI2389 | Absent | LCABL25620 | LC2W2549 | LCAZH2351 | BN19425170 |
| Orphan RR2 | RR | OmpR | Absent | LCBD1189 | Absent | Absent | Absent | BN19407790 |
| Orphan RR3 | RR | OmpR | Absent | Absent | Absent | Absent | Absent | BN19402120 |
Fig. 1.
Phylogenetic trees of the paired HKs and RRs in the six sequenced L. casei strains. The trees were constructed using MEGA version 4 by applying the neighbor-joining method. The scale bar is shown above the trees and the scale is in units of “substitution/site”
Classification of HKs Based on Domain Architecture Analysis
Using the classification method as previously described [17], the putative HKs were grouped into three different groups: extracytoplasmic-sensing HKs, cytoplasmic-sensing HKs, and membrane-sensing HKs (HKs with sensing mechanisms associated with membrane-spanning helices), as shown in Fig. 2.
Fig. 2.
Domain architectures of histidine kinases representative of each TCS clusters. The pictorial depiction is based on the predictions carried out using the SMART web interface http://smart.embl-heidelberg.de/. The transmembrane helices (TMs) were predicted using the tool TMHMM. C, E and M stand for cytoplasmic, extracytoplasmic and membrane sensing, respectively
Among all the HKs identified, HKs of TCS-3 and TCS-4 were recognized as extracytoplasmic sensing HKs by displaying at the N-terminal region an extracytoplasmic putative signal perception domain, which were flanked by (at least) two transmembrane helices (TMs). The cytoplasmic part of the HK proteins harboring the transmitter domain comprised either a HisKA-HATPase_c domain (HK of TCS-3) or a PAS-HisKA-HATPase_c domain (HK of TCS-4). Per-Arnt-Sim (PAS) domains play important roles as sensory modules for sensing oxygen tension, cellular redox state, or light intensity [18]. Most PAS domain-containing proteins are intracellularly located with dual functions of monitoring both the external and internal environments by perceiving alterations in the electron transport system caused by intracellular or extracellular changes in redox potential [19]. It should be noticed that a region of low compositional complexity was found to exist between two TM regions of the TCS-3 HK. The region starts at position 121 and ends at position 135.
The HKs of seven TCS clusters, namely the soluble HK of TCS-11 and the membrane anchored HKs of TCS-2, TCS-6, TCS-7, TCS-10, TCS-14, and TCS-15, and Orphan HK1 were identified as HKs with putative cytoplasmic sensing functions.
HKs of TCS-2, TCS-6, TCS-11, TCS-14, and TCS-15 possess a histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins and phosphatases (HAMP) domain. HAMP functions as a linker to bridge the transmembrane helix and the transmitter domain [20]. HK of TCS-15 possesses the PAS and PAC domains. It has been reported that PAS domains are often associated with proxy auto-config (PAC) domains and they are directly linked and together form the conserved 3D PAS fold [21], as also exemplified by the HK of TCS-15 in this study.
HKs of TCS-1, TCS-5, TCS-8, TCS-9, TCS-12 and TCS-13 in this study were all found to belong to membrane-sensing HK group, indicating that a relatively high percentage of HKs of the L. casei strains are involved in sensing signals directly associated with the membrane.
Classification of RRs
The majority of the putative RRs identified in this study were classified into the following 3 families: CitB, LytT and OmpR, with RRs of the OmpR family constituting the largest group. The assignment of RRs of the 15 TCS clusters and the orphan RRs to the corresponding RR families is given in Table 3.
The RRs of TCS-8 and TCS-10 contain a protein of the CitB family, respectively. Members of the CitB family have been documented to control expression of the genes for citrate fermentation in response to external citrate under anaerobic conditions [22, 23], and to have an effect on the inheritance of iteron-containing plasmids and on the SOS response to β-lactam antibiotics [24, 25].
The RR of TCS-13 and Orphan RR1 contain a protein of the LytT family, respectively. RR proteins of the LytT family are characterized by having a non-HTH DNA binding domain, which modulate the expression of many genes coding for virulence factors, fimbriae, cell wall components, bacteriocins, extracellular polysaccharides etc. [26, 27].
The RRs of the others 12 TCS clusters and two orphan RRs contain a protein of the OmpR family, respectively. RRs of the OmpR family constituted the largest group. Proteins of the OmpR family have been reported to mediate a wide range of biological functions related to, for example, osmolarity, phosphate assimilation, antibiotic resistance, virulence and toxicity [28].
TCS Proteins Common to All the Six L. casei Strains
TCSs are conserved in closely related microorganisms [29–31]. Ancient TCSs, on one hand, may have maintained basic functions in different bacteria, and on the other hand, may also have evolved new functionalities in niche-specific bacteria.
Proteins of the TCS clusters 2, 4, 5, 7, 8, 9, 10, 11, 12, 13, 14, 15 are common to all the six L. casei strains compared here, indicating probably the functional importance of these TCS clusters for the adaptation and survival of these L. casei strains isolated almost from dairy products (Tables 1, 3, 4). For instance, TCS-12 is highly conserved across the six L. casei strains. TCS-12 is homologous to the three paralogous TCS of Bacillus subtilis, BceRS, YvcPQ, and YxdJK, involved in the cell envelope stress response against the nisin [10, 32]. TCS-12 of L. casei BL23 strain has been found to play a vital role in the growth under a low pH environment [10]. Therefore, it is conceivable that conservation of TCS-12 across the L. casei strains is essential for their acid tolerance. These functional similarities and differences of the core TCSs clearly indicate that although they are conserved in the L. casei, they may have developed new niche-specific functions during evolution.
Table 4.
A brief summary of known/putative functions of the TCSs identified in L. casei BL23
| TCS cluster | TCS protein | GenBank locus tag | Functions | References |
|---|---|---|---|---|
| TCS-1 | HK-hpk31 | LCABL02090 | Bile and NaCl response, antimicrobials response, cell envelope stress tolerance | [10] |
| RR-rrp11 | LCABL02080 | |||
| TCS-2 | HK-resE | LCABL05270 | Unclear | |
| RR-spaR | LCABL05260 | |||
| TCS-3 | HK-ciaH | LCABL07770 | Unclear | |
| RR-llrF | LCABL07760 | |||
| TCS-4 | HK-hpk2 | LCABL10490 | Antimicrobials resistance | [10] |
| RR-rrp2 | LCABL10480 | |||
| TCS-5 | HK-resE | LCABL10650 | Unclear | |
| RR | LCABL10640 | |||
| TCS-6 | HK-hpk7 | LCABL12060 | Cell envelope stress response, nisin resistance, bile and NaCl response, oxidative stress tolerance, H2O2 stress tolerance | [10, 35–37] |
| RR-rrp7 | LCABL12050 | |||
| TCS-7 | HK | LCABL14270 | Unclear | |
| RR | LCABL14280 | |||
| TCS-8 | HK-hpk6 | LCABL14440 | Unclear | |
| RR-rrp6 | LCABL14430 | |||
| TCS-9 | HK-hk07 | LCABL16420 | Cell envelope stress response, bacitracin and nisin resistance | [10, 32] |
| RR | LCABL16430 | |||
| TCS-10 | HK | LCABL18840 | Antimicrobials response, | [10] |
| RR | LCABL18830 | |||
| TCS-11 | HK-hpk5 | LCABL18970 | Nisin resistance | [10] |
| RR-rrp5 | LCABL18980 | |||
| TCS-12 | HK-hpk1 | LCABL19610 | Cell envelope stress response, acid tolerance, nisin resistance | [10, 32] |
| RR-rrp1 | LCABL19600 | |||
| TCS-13 | HK | LCABL27660 | Unclear | |
| RR | LCABL27650 | |||
| TCS-14 | HK-kinE | LCABL28710 | Bacitracin resistance | [10] |
| RR-rrp2 | LCABL28720 | |||
| TCS-15 | HK-hpk3 | LCABL30120 | Bacitracin resistance | [10] |
| RR-rrp3 | LCABL30130 | |||
| Orphan RR1 | RR-pltR | LCABL25620 | Unclear |
L. casei strains have achieved popularity in the manufacture of probiotic products because of their convincing beneficial effects on human health. However, before providing benefit to human health, L. casei strains have to survive the difficult journey through the human gastrointestinal tract in sufficient densities in the presence of bile salts [33]. The implicated pathways of L. casei are involved with a complex physiological response under bile salts stress, particularly including cell protection (DnaK and GroEL), modifications in cell membranes (NagA, GalU, and PyrD), and key components of central metabolism (PFK, PGM, CysK, LuxS, PepC, and EF-Tu) [34].
In this study, we found that TCS-1 and TCS-6 clusters are involved with the response under bile salts stress, and TCS-12 is involved with the response under the acid tolerance. In addition, the TCS-6 cluster is also involved in the stress tolerance of oxidative and H2O2. These TCSs in L. casei will contribute to survive the difficult journey through the human gastrointestinal tract.
TCS Proteins Uniquely Present/Absent in One or Several Strains
The TCS-1 cluster was predicted to be absent in L. casei W56 strain, which is involved in cell envelope stress tolerance, and the response of the bile, NaCl and antimicrobials of L. casei BL23 (Table 4) [10]. The TCS-3 was also absent in L. casei W56. Taken together, these findings indicate dramatic differences in the regulation of the response of the bile, NaCl, antimicrobials and the cell envelope stress of L. casei W56 in comparison to the other L. casei strains.
The TCS-6 cluster could be not found in L. casei BD-II strain, which is involved in cell envelope stress response, nisin resistance, bile and NaCl response, oxidative stress tolerance, H2O2 stress tolerance of L. casei BL23 strain [10, 35–37]. Orphan RR1 was also absent in L. casei BL23 strain.
It has been suggested that specific TCS systems may play critical roles in microbe–host relationship, such as the HrpXY system in plant enterobacteria, which regulates type III secretion [38].
PCR Verification of Predicted TCSs
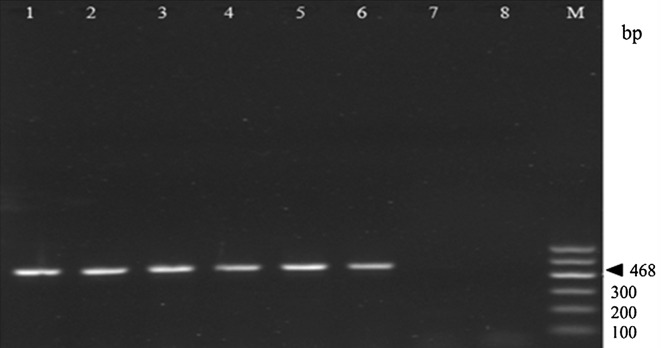
To verify the presence of predicted TCSs in L. casei, 15 primer pairs were designed based on 15 TCS genes, respectively (Supporting Information, Table S1). PCR amplifications using these primers were performed with two sequenced L. casei strains (L. casei ATCC334 and LC2W), four isolated strains (L. casei BD00054, BD00090, BD01649, and BD01803) and one L. paracasei BD03416. All primer sets exhibited 100 % inclusivity for six L. casei strains (Supporting Information, file 1). However, no clearly products were obtained from the isolated strain L. paracasei BD03416, which needs to be further clarified. Typical data is shown in Fig. 3. These results supported successfully the identification of these TCSs in L. casei strains by bioinformatics analysis.
Fig. 3.
PCR verification of the presence of TCS genes in Lactobacillus casei strains. Agarose gel electrophoresis of PCR products amplified using TCS-2 primers. Lane 1, Lactobacillus casei ATCC334; Lane 2, Lactobacillus casei LC2W; Lane 3, Lactobacillus casei BD00054; Lane 4, Lactobacillus casei BD00090; Lane 5, Lactobacillus casei BD01649; Lane 6, Lactobacillus casei BD01803; Lane 7, Lactobacillus paracasei BD03416; Lane 8, negative control (ddH2O); M 100 bp DNA Marker
Conclusion
In the present study we conducted a genome-wide identification, classification, and ortholog analysis of the TCS proteins in six sequenced L. casei strains. Totally, 15 TCS clusters comprising HK–RR pairs were identified, with 12 of them shared by all the six strains compared, three being absent in one strain. In addition, one orphan HKs and three orphan RRs were identified. We believe that the results from this genomic level study will be certainly helpful for the design of physiological studies which in turn will lead to a better understanding of response mechanisms for survival in the gastrointestinal tract of L. casei strains.
Electronic supplementary material
Acknowledgments
This work was supported by the Open Project Program of State Key Laboratory of Dairy Biotechnology, Bright Dairy & Food Co. Ltd., (No. SKLDB2012-007).
References
- 1.Corcoran BM, Stanton C, Fitzgerald G, Ross RP. Life under stress: the probiotic stress response and how it may be manipulated. Curr Pharm Des. 2008;14:1382–1399. doi: 10.2174/138161208784480225. [DOI] [PubMed] [Google Scholar]
- 2.Santivarangkna C, Kulozik U, Foerst P. Inactivation mechanisms of lactic acid starter cultures preserved by drying processes. J Appl Microbiol. 2008;105:1–13. doi: 10.1111/j.1365-2672.2008.03744.x. [DOI] [PubMed] [Google Scholar]
- 3.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 4.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17:204–210. doi: 10.1016/j.copbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Food Biotechnol. 2008;111:1–66. doi: 10.1007/10_2008_097. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Y, Zhang YP, Li Y. Understanding the industrial application potential of lactic acid bacteria through genomics. Appl Microbiol Biotechnol. 2009;83:597–610. doi: 10.1007/s00253-009-2034-4. [DOI] [PubMed] [Google Scholar]
- 9.Landete JM, Garcia-Haro L, Blasco A, Manzanares P, Berbegal C, Monedero V, Zuniga M. Requirement of the Lactobacillus casei MaeKR two-component system for l-malic acid utilization via a malic enzyme pathway. Appl Environ Microbiol. 2010;76:84–95. doi: 10.1128/AEM.02145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcantara C, Revilla-Guarinos A, Zuniga M. Influence of two-component signal transduction systems of Lactobacillus casei BL23 on tolerance to stress conditions. Appl Environ Microbiol. 2011;77:1516–1519. doi: 10.1128/AEM.02176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Been M, Francke C, Moezelaar R, Abeel T, Siezen RJ. Comparative analysis of two-component signal transduction systems of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis. Microbiology. 2006;152:3035–3048. doi: 10.1099/mic.0.29137-0. [DOI] [PubMed] [Google Scholar]
- 12.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ., Jr Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;12:11–19. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 16.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 17.Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhulin IB, Taylor BL. PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/S0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 21.Hefti MH, Francoijs KJ, de Vries SC, Dixon R, Vervoort J. The PAS fold: a redefinition of the PAS domain based upon structural prediction. Eur J Biochem. 2004;271:1198–1208. doi: 10.1111/j.1432-1033.2004.04023.x. [DOI] [PubMed] [Google Scholar]
- 22.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto K, Matsumoto F, Oshima T, Fujita N, Ogasawara N, Ishihama A. Anaerobic regulation of citrate fermentation by CitAB in Escherichia coli. Biosci Biotechnol Biochem. 2008;72:3011–3014. doi: 10.1271/bbb.80301. [DOI] [PubMed] [Google Scholar]
- 24.Ingmer H, Miller CA, Cohen SN. Destabilized inheritance of pSC101 and other Escherichia coli plasmids by DpiA, a novel two-component system regulator. Mol Microbiol. 1998;29:49–59. doi: 10.1046/j.1365-2958.1998.00895.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- 26.Nikolskaya AN, Galperin MY. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 2002;30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galperin MY. Telling bacteria: do not LytTR. Structure. 2008;16:657–659. doi: 10.1016/j.str.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MartinezHackert E, Stock AM. The DNA-binding domain of OmpR: crystal structure of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/S0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 29.Lavin JL, Kiil K, Resano O, Ussery DW, Oguiza JA. Comparative genomic analysis of two-component regulatory proteins in Pseudomonas syringae. BMC Genomics. 2007;8:397. doi: 10.1186/1471-2164-8-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian W, Han ZJ, He CZ. Two-component signal transduction systems of Xanthomonas spp.: a lesson from genomics. Mol Plant Microbe Interact. 2008;21:151–161. doi: 10.1094/MPMI-21-2-0151. [DOI] [PubMed] [Google Scholar]
- 31.Zhao YF, Wang DP, Nakka S, Sundin GW, Korban SS. Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics. 2009;10:245. doi: 10.1186/1471-2164-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- 33.Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr. 2001;73:386s–392s. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 34.Wu R, Sun Z, Wu J, Meng H, Zhang H. Effect of bile salts stress on protein synthesis of Lactobacillus casei Zhang revealed by 2-dimensional gel electrophoresis. J Dairy Sci. 2010;93:3858–3868. doi: 10.3168/jds.2009-2967. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi K, Ogura M, Yamaguchi H, Yoshida KI, Ogasawara N, Tanaka T, Fujita Y. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J Bacteriol. 2001;183:7365–7370. doi: 10.1128/JB.183.24.7365-7370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller C, Sanguinetti M, Riboulet E, Hebert L, Posteraro B, Fadda G, Auffray Y, Rince A. Characterization of two signal transduction systems involved in intracellular macrophage survival and environmental stress response in Enterococcus faecalis. J Mol Microbiol Biotechnol. 2008;14:59–66. doi: 10.1159/000106083. [DOI] [PubMed] [Google Scholar]
- 37.Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J Bacteriol. 2010;192:59–67. doi: 10.1128/JB.00873-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heermann R, Fuchs TM. Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics. 2008;9:40. doi: 10.1186/1471-2164-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.