Abstract
The difference of gene expression between sclerotia-producing and non-sclerotia-producing single spore isolates from Morchella conica were preliminary analyzed by mRNA differential display reverse transcription-polymerase chain reaction (RT-PCR) technique and 67 differential gene fragments were obtained. Fifty-eight of their second PCR products were cloned and sequenced. Thirteen special differential gene fragments related to sclerotial formation were validated by semi-quantitative RT-PCR. Some gene fragments had certain homologies with lipoprotein, cyclin-dependent kinase C-3, glycerophosphoryl diester phosphodiesterase, Rho GDP-dissociation inhibitor, gamma-aminobutyrate permease, OmpA family protein, Transcript antisense to ribosomal RNA protein, sodium–calcium exchange protein and keratin-associated proteins 5, 6. In addition, the putative protein of some DNA fragments had higher similarity with hypothetical protein-coding gene in NCBI database, as well as some were only putative gene fragments. All these fragments were speculated to be the functional gene associated with sclerotial formation in morel.
Keywords: mRNA differential display, Morchella conica, Sclerotia, Semi-quantitative RT-PCR
Introduction
The fungal sclerotia, which were composed of aggregated mycelium-large cells with thick wall, were crucial for storing lipid and polysaccharides making the fungus survive on the adverse natural conditions and formation of fruiting bodies. It had been demonstrated that the sclerotia were important in morel life cycle [1]. The specific conditions of nutrition, humidity, pH, CO2, light and temperature should be met for sclerotial formation [2–5] and the morphological polymorphism (i.e. size, color, quantity and microstructure) of morel sclerotia were frequently found in different species, different strains of the same species and different single spore isolates from the same ascocarp [6–9].
The mRNA differential display technique established by Liang and Pardee [10] was a fast and efficient method for characterizing and isolating differential expression genes. Until now, no information on the genes related with sclerotial formation in Morchella has been reported. The sclerotia-producing and non-sclerotia-producing isolations from single ascocarp of M. conica were obtained in our previous research [9] which were important materials used for comparative and functional genomic studies. Some genes or fragments may be found by performing the differential display polymerase chain reaction (PCR) from these special isolations. In this study, 72 primer combinations were used to perform the differential display PCR in sclerotia-producing and non-sclerotia-producing single spore isolations for intending to get some gene fragments related to sclerotia formation in morel.
Materials and Methods
Isolates and Mycelium Inoculation
Three hundred and eighteen single spore isolates from ascocarps of M. conica YAASM582 were collected [9]. Four sclerotia-producing and four non-sclerotia-producing single spore isolations were selected as test strains in this study (Table 1). All strains were inoculated on YPD agar media (0.2 % yeast extract, 0.2 % peptone, 2 % glucose and 1.2 % agar) in darkness at 25 °C for 7–9 days.
Table 1.
Strains used in this study
| Strains | Sources | Sclerotia |
|---|---|---|
| S1 | ZD-23 | + |
| S2 | ZD-23 | + |
| S3 | ZD-23 | − |
| S4 | ZD-23 | − |
| S5 | ZD-18 | + |
| S6 | ZD-18 | + |
| S7 | ZD-18 | − |
| S8 | ZD-18 | − |
“+” Sclerotia formation, “−” no sclerotia formation
Total RNA Extraction and Purification
About 1 g mycelia was harvested and ground in liquid nitrogen using a mortar. Total RNA was extracted by TaKaRa RNAiso Reagent Kit [Takara Biotechnology (Dalian) Co., Ltd., China, Code: D9108A] and DNase I [Takara Biotechnology (Dalian) Co., Ltd.] was used to remove any traces of residual genomic DNA in a 25 μL reaction volume at 37 °C for 30 min. The RNA integrity and purity was checked by using 1.0 % agarose gel electrophoresis. The quantification and concentration of RNA samples were assessed by using Protein and Nucleic Acid Analytic Apparatus 6400 (Beckmann, USA).
mRNA Differential Play RT-PCR
About 750 ng RNA sample was reversely transcribed using the primers H-T11G (5′-AAGCTTTTTTTT TTTG-3′), H-T11A (5′-AAGCTTTTTTTT TTTA-3′) and H-T11C (5′-AAGCTTTTTTTTTTTC-3′) by Reverse Transcriptase M-MLV (RHase H-) Reagent Kit [Takara Biotechnology (Dalian) Co., Ltd.]. The cDNA was diluted to 10 % for subsequent PCR reaction. The modification of the originally developed differential display reverse transcription-PCR (DDRT-PCR) technique was performed. The PCR amplification was carried out using 72 primer combinations including 3 anchored primers and 24 arbitrary primers (Table 2). The reactions were carried out in Bio-Rad C1000 Thermal Cycler (Bio-Rad, USA) with the following programs: 1 cycle of 94 °C for 5 min; 30 s at 94 °C, 60 s at 40 °C and 60 s at 72 °C for 25 cycles; 30 s at 94 °C, 60 s at 53 °C and 60 s at 72 °C for 15 cycles. This was followed by a final extension of 10 min at 72 °C.
Table 2.
Sequences of the arbitrary primers used in this study
| Codes | Sequences | Codes | Sequences |
|---|---|---|---|
| H-AP1 | 5′-AAGCTTGATTGCC-3′ | H-AP13 | 5′-AAGCTTCGGCATA-3′ |
| H-AP2 | 5′-AAGCTTCGACTGT-3′ | H-AP14 | 5′-AAGCTTGGAGCTT-3′ |
| H-AP3 | 5′-AAGCTTTGCTCAG-3′ | H-AP15 | 5′-AAGCTTACGCAAC-3′ |
| H-AP4 | 5′-AAGCTTCTCAACG-3′ | H-AP16 | 5′-AAGCTTTAGAGCG-3′ |
| H-AP5 | 5′-AAGCTTAGTAGGC-3′ | H-AP17 | 5′-AAGCTTACCAGGT-3′ |
| H-AP6 | 5′-AAGCTTGCACCAT-3′ | H-AP18 | 5′-AAGCTTAGAGGCA-3′ |
| H-AP7 | 5′-AAGCTTAACGAGG-3′ | H-AP19 | 5′-AAGCTTATCGCTC-3′ |
| H-AP8 | 5′-AAGCTTTTACCGC-3′ | H-AP20 | 5′-AAGCTTGTTGTGC-3′ |
| H-AP9 | 5′-AAGCTTCATTCCG-3′ | H-AP21 | 5′-AAGCTTTCTCTGG-3′ |
| H-AP10 | 5′-AAGCTTCCACGTA-3′ | H-AP22 | 5′-AAGCTTTTGATCC-3′ |
| H-AP11 | 5′-AAGCTTCGGGTAA-3′ | H-AP23 | 5′-AAGCTTGGCTATG-3′ |
| H-AP12 | 5′-AAGCTTGAGTGCT-3′ | H-AP24 | 5′-AAGCTTCACTAGC-3′ |
Gel Electrophoresis, Separation of Differential Bands and Re-amplification
Each of 7 μL differential display products were size-fractionated in parallel on a 6 % polyacrylamide gel at 100 V for 2.5 h. Then silver staining method was employed for showing DNA bands as described as follow: the gel was fixed with 250 mL solution containing 1 % acetic acid and 10 % ethyl alcohol for 30 min; rinsed with ddH2O for two times; dyed with 250 mL dying solution (0.2 % silver nitrate, 1 % acetic acid and 10 % ethyl alcohol) for 20 min; washed with water for 30 s, and oxidation was performed with 3 % sodium hydroxide and 0.7 % formaldehyde for 15 min. Obvious different bands were considered to be the differential expression genes between two kinds of strains. These bands were cut down and recuperated by soaking in 100 μL ddH2O after the gel was crushed. The gel pieces were boiled in water for 15 min, and the sediment was incubated overnight at 4 °C. Amplification reaction by PCR was performed using 5 μL product as template with the same primer combination. Reaction system and parameters were same to the DDRT-PCR. The re-amplified products were visualized on 1 % agarose gels and the positive bands was purified using the DNA Gel Extraction Kit (Axygen, Union City, CA, USA).
Cloning and Identification of Reampilfied Product of the Differential Fragments
The eluted DNA fragments ligated into the pMD18-T Vector [Takara Biotechnology (Dalian) Co., Ltd., Code: D101A] were transferred into Escherichia coli DH5α competent cells, and three clones were sequenced for each sample (Sangon Biotech Corporation Limited, China). Homology analysis was conducted with the BLASTN and/or BLASTX algorithms in GenBank database.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed to identify the gene fragments described in DDRT-PCR [11]. The housekeeping gene 18S rRNA was selected as the internal control. Amplification was conducted according to the method described below: pre-denaturation at 94 °C for 3 min, denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min. The number of cycles and annealing temperature in the specific DNA PCR varied depending on the sequences and the original amount of target mRNA. Detection and analysis of the PCR products was performed using 1 % agarose gels.
Results and Discussion
DDRT-PCR Screenings and Identification of Differentially Expressed Genes
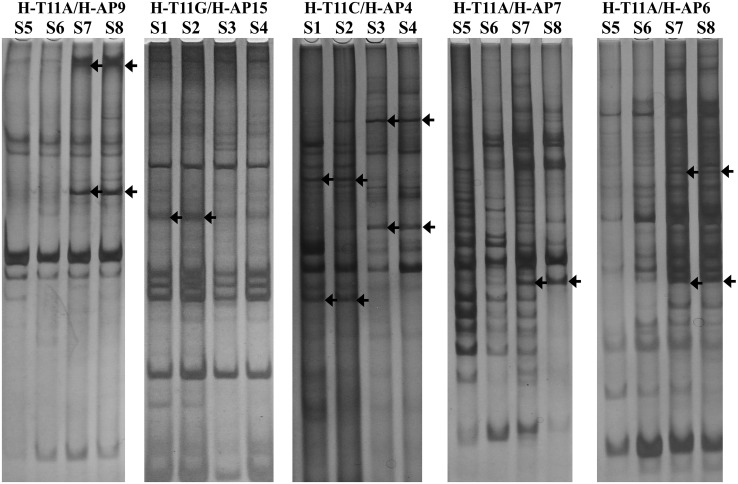
The DDRT-PCR method could reveal the expressive differences of many genes [12]. Sixty-seven different bands were obtained by 72 primer combinations in our study. The size of these fragments were ranged from 250 to 2,000 bp, and mainly centralized on the range of 300–800 bp (Fig. 1).
Fig. 1.
Differential display polymerase chain reaction of total RNA from hyphae of non-sclerotia producing strains versus, sclerotia producing strains, Morchella conica. Total RNAs of non-sclerotia producing strains (S1, S2, S5, S6) and sclerotia producing strains (S3, S4, S7, S8) were reverse transcribed into cDNA, and then were amplified. The differential display PCR products were run on to 6 % naturing polyacrylamide gel in pairs(non-sclerotia producing strains S1, S2 and sclerotia producing strains S3, S4 must be next to each other, non-sclerotia producing strains S5, S6 and sclerotia producing strains S7, S8 can be done in the same manner). The representative differential display RT-PCR banding patterns were showed, which had been obtained from amplification with the H-T11A/H-AP9, H-T11G/H-AP5, H-T11C/H-AP4, H-T11A/H-AP7, H-T11A/H-AP6 primer combinations. Arrows indicate differentially expressed cDNA fragments that were recovered from gel and analyzed further
The polymorphisms of DDRT-PCR product not only related with primer combination but also with DNA template (Table 3). The statistic result suggested that most of the primers combinations were suitable for screening difference segment except primer combinations with primers AP6, -7, -16 and -22. It was also considered that polyAG and polyAC structure existed more frequently in M. conica genome based on the more differentially expressed bands obtained from the amplification of primer combinations with anchored primer HT11C or HT11G. Furthermore, the specific bands in non-sclerotia-producing isolations were more than ones in sclerotia-producing isolations by the anchored amplification of HT11A and HT11C, and just the opposite for anchored amplification of HT11G.
Table 3.
Differential fragments amplified by primer combinations
| Anchored primers | Group | Arbitrary primers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-AP1 | H-AP2 | H-AP3 | H-AP4 | H-AP5 | H-AP6 | H-AP7 | H-AP8 | H-AP9 | H-AP10 | H-AP11 | H-AP12 | H-AP13 | ||
| H-T11A | S | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 1 | 1 | |
| H-T11C | S | 0 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 1 | |
| H-T11G | S | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 1 | 2 | 1 | 2 |
| NS | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total | 4 | 12 | 1 | 3 | 1 | 0 | 0 | 7 | 6 | 2 | 2 | 2 | 4 | |
| Anchored primers | Group | Arbitrary primers | Sum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H-AP14 | H-AP15 | H-AP16 | H-AP17 | H-AP18 | H-AP19 | H-AP20 | H-AP21 | H-AP22 | H-AP23 | H-AP24 | |||
| H-T11A | S | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| NS | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 15 | |
| H-T11C | S | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 7 |
| NS | 3 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 4 | 22 | |
| H-T11G | S | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 12 |
| NS | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 10 | |
| Total | 6 | 2 | 0 | 1 | 2 | 2 | 2 | 1 | 0 | 2 | 5 | 67 | |
The Group S was consist of sclerotia-producing strains S1, S2, S5, S6; the Group NS was consist of non-sclerotia-producing strains S3, S4, S7, S8
Reamplification and Semi-quantitative RT-PCR
False positive could affect the determination on differential band, and the good way was to PCR amplify again and semi-quantitative RT-PCR [10, 13].
Sixty-seven different bands were recovered as template for re-amplification. Only 58 samples could repeated first amplification results (Fig. 2) and the these 58 s PCR products were cloned and sequenced. The sequence analysis showed that 26 segments had corresponding similar function genes, 9 segments hit hypothetical protein coding fragments of known species. Twenty-three putative protein sequences could be found.
Fig. 2.
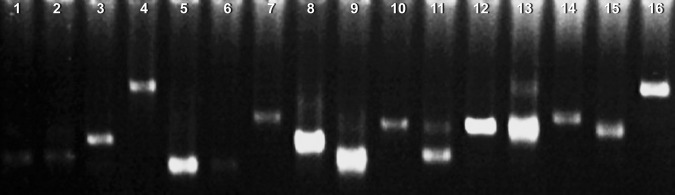
The differential fragments were amplificated in the same way. The putative bands should be repeatable single one showing in lanes 1–6, 10, 12 and 14–16, and the false positive bands were display in lanes 8, 9, 11 and 13
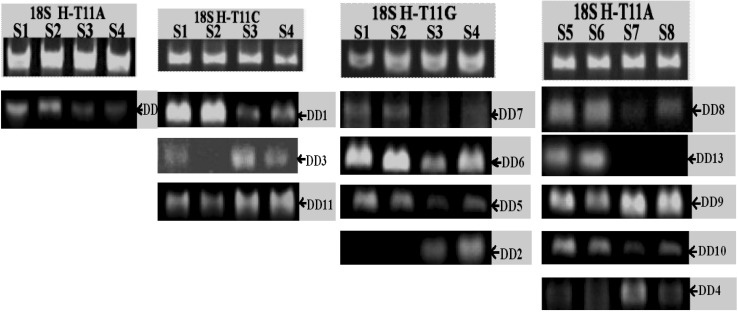
Fifty-eight primer pairs were designed for semi-quantitative RT-PCR according to sequenced results. Only 13 (denoted as DD1–13) in 58 fragments were identified as positive by testing their expression at two types of strains compared with housekeeping gene 18S rRNA (Fig. 3), and the expression level these fragments were consistent with the result displayed on 6 % polyacrylamide gel.
Fig. 3.
Positive results getting from semi-quantitative PCR 18S rRNA was the internal standard showing in the each first line and the test segment can be determined to positive different fragment corresponding to the fragment brightness of internal standard. The measured positive fragment should conform to phenotype of the source strains
Analysis of Positive Fragments
Most of these positive fragments were cloned from sclerotia-producing isolates except fragments DD1–4 (Table 4). All fragments hit the functional gens or hypothetical protein (Tables 4, 5).
DD5 was similar to the gene encoding gamma-aminobutyrate (GABA) permease of Shigella dysenteriae. It was known that the level of GABA transporter gene expression could be increased with the invasion of pathogens and insect, physiological stress difference and even anyone else adversity conditions. GABA also participated in nitrogen storage. Morel sclerotia were hard-surfaced resting organisms that act as a nutrient reservoir, and resistant to unfavorable environmental conditions [14], so GABA may regulate more substances transportation to mycelia for promoting the sclerotia formation. More attention had paid to the nutrition conditions of sclerotia formation in Morchella sp., and nitrate and composite nitrogen such as yeast extract were useful for the formation of sclerotia, but the contribution of GABA permease for nitrogen metabolic processes remained unproven.
DD10 was similar to the gene encoding glycerophosphoryl diester phosphodiesterase, which was involved in process of lecithin’s metabolism of E. coli. The phosphatide was the important nutrient in fungi, but no research was reported on the physiological function in fungi itself. The inflation pressure and oxidative stress response had a great contribution to morel sclerotia formation [15] and glycerophosphoryl diester phosphodiesterase may participate in two courses above, so further research should be done on the relationship between them.
DD11 had 69 % similarity with fragment coding transcript antisense to ribosomal RNA protein of Ajellomyces capsulatus. In the yeast, this protein was the constitute of transcription subunit of nuclear rDNA repeat sequence [16], and was useful for the selective expression of repeat sequence. The parallel clips in Morchella may be a part of duplicated gene sequences which were variable in quantity and quality of units related to sclerotia.
Table 4.
DDRT-PCR transcripts identified by sequence homologies found in the NCBI sequence database
| Numbers | Primer pairs | Sources | Size (bp) | Blast similarity/species/accessions numbers | E-value | Max ident (%) |
|---|---|---|---|---|---|---|
| DD1 | H-T11C/H-AP8 | NS | 350 | Rho guanidine dissociation inhibitor/Tuber borchii/ABW17274 | 8e−09 | 71 |
| DD2 | H-T11G/H-AP15 | NS | 295 | Similar to 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase 1/Pyronema omphalodes CBS 100304/CCX33765.1| | 7e−31 | 61 |
| DD3 | H-T11C/H-AP8 | NS | 189 | Ras superfamily GTPase/Nannochloropsis gaditana/XP_005854920.1 | 6.2 | 37 |
| DD4 | H-T11A/H-AP6 | NS | 334 | Hypothetical protein slr0639/Synechocystis sp. PCC 6803/NP_442288.1 | 0.29 | 50 |
| DD5 | H-T11G/H-AP13 | S | 261 | GABA permease/Shigella dysenteriae/EGP24113 | 6e−46 | 98 |
| DD6 | H-T11G/H-AP12 | S | 333 | Zinc finger protein ZFAT isoform X2/Falco peregrinus/XP_005237283.1 | 5.3 | 31 |
| DD7 | H-T11G/H-AP9 | S | 346 | Hypothetical protein/Actinomyces georgiae/WP_005870647.1 | 0.022 | 32 |
| DD8 | H-T11A/H-AP2 | S | 334 | Hypothetical protein slr0639/Synechocystis sp. PCC 6803/NP_442288.1 | 3.3 | 60 |
| DD9 | H-T11A/H-AP7 | S | 239 | Hypothetical protein COCC4DRAFT_155570/Bipolaris maydis ATCC 48331/ENH98510 | 2e−14 | 58 |
| DD10 | H-T11A/H-AP6 | S | 327 | Glycerophosphoryl diester phosphodiesterase/Escherichia coli/EGJ96270 | 5e−69 | 98 |
| DD11 | H-T11C/H-AP20 | S | 370 | Transcript antisense to ribosomal RNA protein/Ajellomyces capsulatus/EGC42647.1 | 7e−24 | 69 |
| DD12 | H-T11A/H-AP9 | S | 403 | Diacylglycerol kinase [Escherichia coli]/WP_001472712.1 | 4e−44 | 96 |
| DD13 | H-T11A/H-AP2 | S | 334 | Protein of unknown function [Pyronema omphalodes CBS 100304]/CCX05279.1 | 0.20 | 26 |
Table 5.
The sequences of difference cDNA fragment and their prediction protein amino acid sequences
| Numbers | cDNA sequences | Coding bases | Amino acid sequences of prediction protein |
|---|---|---|---|
| DD1 | AAGCTTTTACCGCGGACACTACGATGCCATAAGCAAGTTCATGGATGACGACCATAACACCCATTTGGAGTTTTACTGGAGCTTCAATATTTCTAAGACCTGGGAATAAGTCCTGCTTTCTCTCTGCCAATTAGTTTTGTTTCTCGGGCAGTAGTCTAGTTAAATAGCTAGCCAGCACATTATGACTCTTAATGGGCGCTCATCCTCCTTTTCGGTGTATTTCCCATTCTTTATTGAATGCTTTTTTTTTCTACCATCGGTTTTTCAAATATCATGAAGCGCATTTCTTGTCTGAGCGTAGGTTGAGGACACAATAAAGACGTTTATTTGGGAGAAAAAAAAAAAGCTT | 2–106 | SFYRGHYDAISKFMDDDHNTHLEFYWSFNISKTWE |
| DD2 | AAGCTTGGCTATGCAAGGAGAGAAGTTTGTTGATGCACATTGGAGAAATTACATGGCTACTCGAAACACTATTACGCCACCACAATCTCCAAAGGTCGAAAGACTCTTATTCGAGGATGCTGAACGCCTTTGCCGACGTCTTCATATAAATTGCTCCCGCCAGTTTCTTAAAGATCGTTTTAAGCGTGCAGATAAAGACAACTCAGGATATCTTAACTTCCCCGAGTTCCAGAAATTTGTGCAGTTATTGAAAGAGCGGGAAGAGATAAAAGATATTTGGAAAAAAAAAAAGCTT | 2–295 | SLAMQGEKFVDAHWRNYMATRNTITPPQSPKVERLLFEDAERLCRRLHINCSRQFLKDRFKRADKDNSGYLNFPEFQKFVQLLKEREEIKDIWKKKKL |
| DD3 | AAGCTTTTTTTTTTTGATGAGGGAAGGATTCTTGTAGCACGGACACTAAATACAGAGACACCGATTTGGGGATACTACATACAGACACGCCCCGAAGATCATGCACCCCTTGTAGACTACGGTAATTACCAGTCAAGACGTACTCTGTCCTTTTCTACGAGCCTGGTCTCTTCCGCCATAGCCAAGCTT | 1–189 | KLFFFDEGRILVARTLNTETPIWGYYIQTRPEDHAPLVDYGNYQSRRTLSFSTSLVSSAIAKL |
| DD4 | AAGCTTTTTTTTTTTATAATGATATCATTTGATAATATTTTCCGAGCAGTTCAGCAGCTAATCATAAGTACATCGCTAGAAATCCAATCTTAACCTGCCAAAGCAAACACCGCTATAACCGTTGCCGCGCCCGCTAACCCAGCCCTGAAAGTGCTCCTTGAGCTCCACTCTTCTAGATCACTTTTCACTTTCCGATCGTCCACAACTTGAACACCCGTTTTTCTTTTCTCGCTCGCCTCCACAGATTCCGCAGCAGCCAGTAAACTGGATTTAGCTTTCTTCAAGATTATCTGGCTATACGGAATGACAATCGCCAACAGCACAGTCGAAGCTT | 1–90 | KLFFFIMISFDNIFRAVQQLIISTSLEIQS |
| DD5 | AAGCTTTTTTTTTTTGGGTACTGGTTATCCCGCTGGAAGCCAACATCGCCGCCATGATCCTGCACTCATGGGTTCCAGGCATTCCCATCTGGTTATTTTCCCTCGTCATTACCCTCGCCTTAACTGGCAGTAACTTATTAAGCGCTAAAAACTACGGCGGATTTGAGTTCTGGCTGGCGCTGTGCAAAGTCATCGCTATCCTGGCCTTTATTTTCCTTGGTGCAGTCGCAATTAGCGGTTTTTACCCGTATGCCGAAGCTT | 3–260 | AFFFWVLVIPLEANIAAMILHSWVPGIPIWLFSLVITLALTGSNLLSAKNYGGFEFWLALCKVIAILAFIFLGAVAISGFYPYAEA |
| DD6 | AAGCTTCGGGTAACGATGAGGATGATGATTTCGACCAGCTCATTGAGCGTGAAGGAGAGGAACTCGAGGATATGCTTTCTCAGTTGGATTTAGACTTTGACCATGCCGTTGACGATTCCATTAAAGTTCAGGATCAAGTTAACGTAACCTTGCCATCCTCTGTTACTTGAACTATAACTACAGGAGTGTGTTCGGCGTCCTGGTATTAATTGTAAATTTGGTTCACTTGATGTTTTAGAGTATTTGTATTCTTTTTTCTTTCTCTCTCTTTACTTGAGGTGTATGGATAAGTGGTCCCGCGATTCCCTTTTCTTCCTCAAAAAAAAAAAGCTT | 3–167 | ASGNDEDDDFDQLIEREGEELEDMLSQLDLDFDHAVDDSIKVQDQVNVTLPSSVT |
| DD7 | AAGCTTCATTCCGAGCCGGGAGGAACTGTAGTAAAGCCAGTGCTAGTCTCATAATCACACCTCACGTCGGGTCACGATAACGATTCGTTGCTGCCGATTACCAGATACAACTACTCAACGGCAACACGCAGACAAGAAGAACACCACCCTCCGCTCTAGAGTGCACTTCTCATTTCTGGAACACCTGACTCGATTCTGGAGAGATAGGGACCAAGCGGGAATTCATTCGATCAAAGGAACGGCTTAAAAGCACGCGTTACCGTTCAAGTGGACCAAGACCAAGAGAGAAGGCAAGACAGACTTCAATAAACTTAACAACCAGCTACAGCACAAAAAAAAAAAGCTT | 3–344 | ASFRAGRNCSKASASLIITPHVGSR.RFVAADYQIQLLNGNTQTRRTPPSALECTSHFWNTCLDSGEIGTKREFIRSKERLKSTRYRSSGPRPREKARQTSINLTTSYSTKKKS |
| DD8 | AAGCTTTTTTTTTTTATAATGATATCATTTGATAATATTTTCCGAGCAGTTCAGCAGCTAATCATAAGTACATCGCTAGAAATCCAATCTTAACCTGCCAAAGCAAACACCGCTATAACCGTTGCCGCGCCCGCTAACCCAGCCCTGAAAGTGCTCCTTGAGCTCCACTCCTCTAGATCACTTTTCACTTTCCGATCGTCCACAACTTGAACACCCGTTTTTCTTTTCTCGCTCACCTCCACAGATTCCGCAGCAGCCAGTAAACTGGATTTAGCTTTCTTCAAGATTATCTGGCTATACGGAATGGCAATCGCCAACAGCACAGTCGAAGCTT | 1–90 | KLFFFIMISFDNIFRAVQQLIISTSLEIQS |
| DD9 | AAGCTTTTTTTTTTTAAAGTAAAAGTCCTGGTTCCCCGACACACCCAGTAAAGGGCATGCGGCTAGCCAGAAGGAAAGGTCCGGCCGGATCAGTACACTCGGTGAGGAGGACCGACCAGCCAGACCCAAGGTTCAACTACGAGCTTTTAACTGCAACAACTTTAATATACGCTACTGGAGCTGGAATTACCGCGGCTGCTGGCACCAGACTTGCCCTCCAATTGTTCCTCGTTAAGCTT | 21–239 | KSPGSPTHPVKGMRLARRKGPAGSVHSVRRTDQPDPRFNYELLTATTLIYATGAGITAAAGTRLALQLFLVKL |
| DD10 | AAGCTTGCACCATGCCAGTGAGTTTGATATTACCCGGCTGCGATGTCTCCTCAATCAACATATGGTAATCCGGACCAATACCATCTGCATATTCCGCCACCTGTTTCATGGCACCCGGCTTAAACATCCAGTCGTAGTTGTAGTTAACCCAGCTTCCATCCGGCTGTTCCTGCTGCGTTTCATTCCAGTCGGTATAGGCAATCAGCTGTACCAGATTGAGCTCCATGCCCATTTTGGGTTCCAGCTCATTCTTAATACGCTTCAGCTCATCAGCATCAGAACATTGCAAATAAACTTTATCGTCTTTACCGGTGATGGTGCAAGCTT | 3–326 | ACTITGKDDKVYLQCSDADELKRIKNELEPKMGMELNLVQLIAYTDWNETQQEQPDGSWVNYNYDWMFKPGAMKQVAEYADGIGPDYHMLIEETSQPGNIKLTGMVQA |
| DD11 | AAGCTTTTTTTTTTTCCCCGCTGAACTTAAGCATATCAATAAGCGGAGGAAAAGAAACCAACAGGGATTGCCTCAGTAACAGCGAGTGAAGCGGCAAAAGCTCAAATTTTAAAGCTGACATCTTCGGTGTCCGCGTTGTAATTTTGTGAGGCATCTCCGGGTAGGGCGCTGGCCTAAGTTCCTTGGAACAGGACGCCACAGAGGGTGAGAATCCCGTGCTTGGCTGGCTGTCCCCGTCCATGTGAGGTGTCTTCGACGTGTCGAGTTGTTTGGGAATGCAGCTCAAAATGGGTGGTAAATTTCATCTAAAGCTAAATATTGGTGGGAGACCGATAGCGCACAAGTAGAGTGATCGAAAAAAAAAAAGCTT | 2–259 | SFFFFDHSTCALSVSHQYLALDEIYHPFCAAFPNNSTRRRHLTWTGTASQARDSHPLWRPVPRNLGQRPTRRCLTKLQRGHRRCQL |
| DD12 | AAGCTTCATTCCGTCAGGAAGGCGCAGCGGTATTGTTGGCGGTGGTCATCGCCTGCTGGCTGGATGTGGACGCGATTACCCGCGTGCTGCTTATCAGCTCCGTGATGCTGGAGATGATTGTGGAAATCCTCAATAGCGCCATCGAAGCAGTGGTTGACCGAATTGGCTCTGAATACCATGAGCTTTCCGGACGCGCAAAAGATATGGGATCCGCTGCGGTGCTGATTGCCACTATCGTCGCCGTGATTACCTGGTGCATTCTGTTATGGTCGCATTTTGGATACCCCTTCCAGAATTCGATACAATCTCTGGTTTATTGTGCAGTTTATGGTTCCAAAATCGCCTTTTGCTGTATATACTCACAGCATAACTGTATATACACCCAGGGGGCGGAATGAAGCTT | 3–395 | ASFRQEGAAVLLAVVIACWLDVDAITRVLLISSVMLEMIVEILNSAIEAVVDRIGSEYHELSGRAKDMGSAAVLIATIVAVITWCILLWSHFGYPFQNSIQSLVYCAVYGSKIAFCCIYSQHNCIYTQGAE |
| DD13 | AAGCTTCGACTGTGCTGTTGGCGATTGTCATTCCGTATAGCCAGATAATCTTGAAGAAAGCTAAATCCAGTTTACTGGCTGCTGCGGAATCTGTGGAGGCGAGCGAGAAAAGAAAAACGGGTGTTCAAGTTGTGGACGATCGGAAAGTGAAAAGTGATCTAGAAGAGTGGAGCTCAAGGAGCACTTTCAGGGCTGGGTTAGCGGGCGCGGCAACGGTTATAGCGGTGTTTGCTTTGGCAGGTTAAGATTGGATTTCTAGCGATGTACTTATGATTAGCTGCTGAACTGCTCGGAAAATATTATCAAATGATATCATTATAAAAAAAAAAAGCTT | 3–282 | ASTVLLAIVIPYSQIILKKAKSSLLAAAESVEASEKRKTGVQVVDDRKVKSDLEEWSSRSTFRAGLAGAATVIAVFALAG |
Other fragments were similar to the gene coding hypothetical protein in NCBI database, and to certain protein coding genes on other position of the cloning, except clone DD2. DD1 was in common with Rho GDP-dissociation inhibitor coding part in Tuber melanosporum, DD7 was similar to Schizosaccharomyces japonicus cyclin-dependent kinase C-3 coding part. DD9 was similar to lipoprotein in Corynebacterium striatum. The morel sclerotia contain a lot of lipid, Papapostolou which was thought that differentiation of sclerotia in plant filamentous pathogenic fungi demanding superoxide dismutase (SOD), which increased with the decreases of oxygen pressure and lipid superoxide, so the lipid may be hoarding up in sclerotia in the process of sclerotia differentiation induced by SOD [17, 18].
Sclerotial formation was a complex and key stage for the development of Morchella, and the research on the specific gene expression in this process is very important. The differentiation and difference of organ can be checked by the gene expression in time or space, and precise regulatory mechanism of developmental gene expression is required. The establishment and utilization of mRNA DDRT-PCR technique could help breaking the restrictions such as complex genetic background of materials [19]. The obvious advantages of this techniques is simple, sensitive and with high efficiency, but its tendency to high copy of mRNA amplification and high frequency of false positive bands production have to be avoided. In this study, the fragments obtained were expressed sequence tags in 250–2,000 bp, and most of them were located between 300 and 800 bp, which turned shorter after amplification again.
Morel sclerotium is a kind of dense structure of differentiated and systematized hyphae. The structure can help fungi to against the adverse environmental conditions and survived as hypogeous, which can revive in the cold winter and germinate to second hyphae even fruiting. Many non-nutritive factors have influences on sclerotial formation, including illumination, temperature, pH, chemicals (i.e. organic acids, phenolic compounds, and polyphenol oxidase), accumulated environmental corruption and mechanical barrier, sulfhydryl modification factor and osmotic pressure, and the factors that increase or reduce oxidation stress may play role on promoting or inhibiting sclerotial differentiation [20–24]. The positive fragments obtained in this experiment were similar to genes participating in lipid metabolism, nitrogen metabolism, adversity stress and cell cycle regulation, the genes corresponding these fragments may participate in oxidation stress reaction in M. conica, working as the upstream or downstream regulative factors. The fragments similar to S. dysenteriae GABA permease, OmpA family protein and Homo sapiens keratin-associated protein 5, 6 coding gene were important for studying the protection mechanism of sclerotium. These genes may be peculiar to process of adversity stress resistance and structure adaptive change in M. conica, and have similar structure or function to item produced by these three genes below.
In this experiment, we also found some positive fragment coding hypothetical protein and even can not be found the corresponding result in NCBI, and these fragments may be the specific genes controlling the formation or differentiation of sclerotia in M. conica. It provided important information for genetic and gene regulation on sclerotium formation of M. conica.
Acknowledgments
This work was supported by a Grant from the China Agricultural Research Systems (CARS-24).
Footnotes
Li-Jiao Chen and Hong-Mei Chai have contributed equally to this work.
References
- 1.Ower R. Notes on the development of the “Morchella” ascocarp: “Morchella esculenta”. Mycologia. 1982;74:142–144. doi: 10.2307/3792639. [DOI] [Google Scholar]
- 2.Zhao YC, Wang F, Wu YX. Study on the formation of morel. Edible Fungi China. 1998;17:5–7. [Google Scholar]
- 3.Volk TJ, Leonard TJ. Cytology of the life-cycle of Morchella. Mycol Res. 1990;94:399–406. doi: 10.1016/S0953-7562(09)80365-1. [DOI] [Google Scholar]
- 4.Stott K, Mohammed C (2004) Specialty mushroom production systems: maitake and morels: a report for the Rural Industries Research and Development Corporation. RIRDC publication no. 04/024. ISSN 1440-6845
- 5.Kanwal HK, Reddy MS. The effect of carbon and nitrogen sources on the formation of sclerotia in Morchella spp. Ann Microbiol. 2012;62:165–168. doi: 10.1007/s13213-011-0241-6. [DOI] [Google Scholar]
- 6.Hervey AG, Bistis G, Leong I. Cultural studies of single ascospore isolates of Morchella esculenta. Mycologia. 1978;70:1269–1274. doi: 10.2307/3759331. [DOI] [Google Scholar]
- 7.Leonard TJ, Volk TJ. Production of specialty mushrooms in North America: shiitake and morels. In: Leatham G, editor. Frontiers in industrial mycology. York: Chapman and Hall; 1992. pp. 1–22. [Google Scholar]
- 8.Stamets P (1993) Growth parameters. In: Growing gourmet and medicinal mushroom. Ten Speed Press and Mycomedia, Olympia, pp 370–379
- 9.Chen LJ, Chai HMi, Huang XQ, Zhao YC. Study on cultural characteristics of single spore isolation population from Morchella conica. Biotechnology. 2011;21:63–70. [Google Scholar]
- 10.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 11.Daigo Y, Takayama I, Ponder BAJ, Caldas C, Ward SM, Sanders KM, Fujino MA. Differential gene expression in the murine gastric fundus lacking interstitial cells of Cajal. BMC Gastroenterol. 2003;3:14. doi: 10.1186/1471-230X-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voelckel C, Baldwin IT. Detecting herbivore/specific transcriptional responses in plants with multiple DDRT/PCR and subtractive library procedures. Physiol Plant. 2003;118:240–252. doi: 10.1034/j.1399-3054.2003.00105.x. [DOI] [Google Scholar]
- 13.Liang P, Averboukh L, Pardee AB. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding C, Cui JL, Liu L, Fang L. Research status of Morchellasclerotium and nutritional type. Edible Fungi. 2008;1:1–3. [Google Scholar]
- 15.Amir R, Levanon D, Hadar Y, Chet I. Factors affecting translocation and sclerotial formation in Morchellaesculenta. Exp Mycol. 1995;19:61–70. doi: 10.1006/emyc.1995.1007. [DOI] [Google Scholar]
- 16.Galopier A, Hermann-Le Denmat S. Mitochondria of the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis contain nuclear rDNA-encoded proteins. PLoS ONE. 2011;6:e16325. doi: 10.1371/journal.pone.0016325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgiou CD. Lipid peroxidation in Sclerotium rolfsii: a new look into the mechanism of sclerotial biogenesis in fungi. Mycol Res. 1997;101:460–464. doi: 10.1017/S0953756296002882. [DOI] [Google Scholar]
- 18.Papapostolou I, Georgiou CD. Superoxide radical induces sclerotial differentiation in filamentous phytopathogenic fungi: a superoxide dismutase mimetics study. Microbiology. 2010;156:960–966. doi: 10.1099/mic.0.034579-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang YC, Fang ZH, Li DW. Application of differential display techniques to the cloning of resistant gene fragment to phytophthora blight. North Hortic. 2008;12:149–150. [Google Scholar]
- 20.Christias C, Lockwood JL. Conversion of mycelical constituents in four sclerotium-forming fungi in nutrient deprived conditions. Phytopathology. 1973;63:602–605. doi: 10.1094/Phyto-63-602. [DOI] [Google Scholar]
- 21.Chet I, Henis Y. Sclerotial morphogenesis in fungi. Annu Rev Phytopathol. 1975;13:169–192. doi: 10.1146/annurev.py.13.090175.001125. [DOI] [Google Scholar]
- 22.Le Tourneau D. Morphology cytology and physiology of sclerotinia species in culture. Phytopathology. 1979;69:887–890. doi: 10.1094/Phyto-69-887. [DOI] [Google Scholar]
- 23.Willetts HJ, Wong JAL. The biology of Sclerotiniasclerotionum, S. trifoliorum, and S. minor with emphasis on specific nomenclature. Bot Rev. 1980;46:101–165. doi: 10.1007/BF02860868. [DOI] [Google Scholar]
- 24.Georgiou CD, Patsoukis N, Papapostolou I, Zervoudakis G. Sclerotial metamorphosis in filamentous fungi is induced by oxidative stress. Integr Comp Biol. 2006;46:691–712. doi: 10.1093/icb/icj034. [DOI] [PubMed] [Google Scholar]