Abstract
The role of normal transcription and RNA processing in maintaining genome integrity is becoming increasingly appreciated in organisms ranging from bacteria to humans. Several mutations in RNA biogenesis factors have been implicated in human cancers, but the mechanisms and potential connections to tumor genome instability are not clear. Here we discuss how RNA processing defects could destabilize genomes through mutagenic R-loop structures and by altering expression of genes required for genome stability. A compelling body of evidence now suggests that researchers should be directly testing these mechanisms in models of human cancer.
Keywords: RNA processing, R-loops, genome instability, cancer, transcriptome
Linking RNA processing and tumor genome instability
Most tumor cells exhibit some level of genome instability symptomatic of either increased rates of aneuploidy or mutation [1]. Although a genome instability phenotype has dramatic effects on oncogenesis, tumor progression, and resistance to therapy, the mechanistic basis of tumor genome instability is often unclear. Various genetic, epigenetic, and environmental factors can shape tumor genomes, manifesting their mutagenic activities by interfering with mitosis, damaging DNA, or preventing normal DNA replication or repair [2, 3].
Early studies linked transcription elongation, RNA export and splicing defects with genome instability phenotypes such as hyper-mutation and hyper-recombination [4–7]. Since then, recent functional genomic studies in mammalian cell lines and model organisms have implicated several more aspects of RNA processing in the prevention of genome instability [8–11]. These systematic studies, along with more focused work [12–18], reveal that virtually every major aspect of RNA processing is potentially mutable to a genome instability phenotype, from transcript elongation to termination, 3′-end processing, splicing, RNA transport, and RNA degradation. More than a decade of directed efforts have convincingly linked some RNA processing defects to increases in transcription-coupled DNA:RNA hybrid-mediated R-loop formation, which in turn constitute a major source of genome instability across species (Figure 1, Table 1). In addition, R-loops are known to play important roles in gene expression regulation by influencing transcription termination, DNA methylation, and chromatin modification [19–22]. Thus the formation of R-loops could play a role in genome integrity both by creating a damage-prone site on the genome and by altering the expression of key genome maintenance proteins. Significantly, regardless of R-loop formation, RNA processing defects can change gene expression patterns due to their effects on RNA levels. There is considerable support for this informational mechanism, discussed below, although it is less explored than DNA:RNA hybrid formation. Finally, a role for RNA processing in transcription-coupled nucleotide excision repair can also influence genome stability, although there are only a few direct examples of this that are reviewed elsewhere [23].
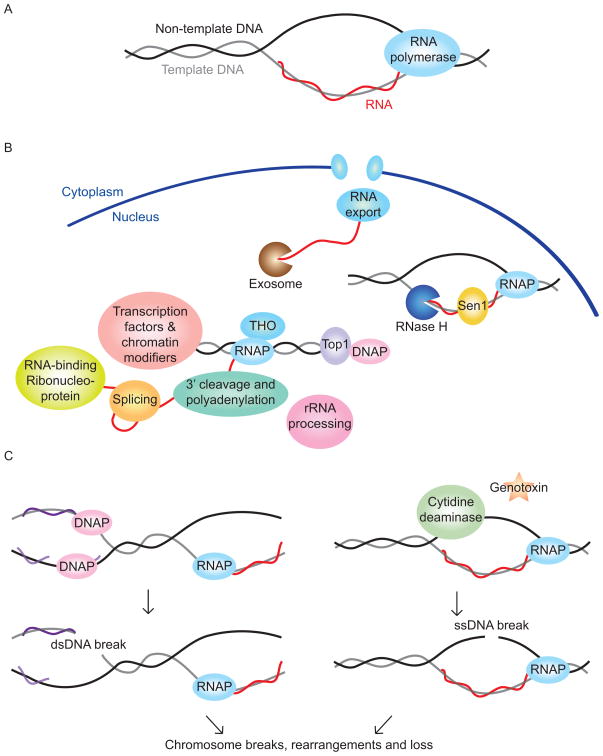
Figure 1. R-loop formation and potential genome instability outcomes.
(A) The structure of an R-loop. (B) RNA processing steps and other events that are required to prevent abnormal R-loop formation. The majority of the known factors are involved in co-transcriptional events but there are also other DNA replication and repair complexes such as DNA polymerase (DNAP) that are implicated in R-loop formation. (C) Mutational events arising from R-loops that lead to genome instability. The left diagram shows one way in which collision between replication and transcriptional machinery can result in chromosome instability. The right diagram shows how the exposed ssDNA in an R-loop can be damaged by cytidine deaminases and genotoxins to culminate in chromosome instability.
Table 1.
RNA processing factors whose mutation elevates R-loops and genome instability phenotypes.
| Genes | Organism |
|---|---|
| RNA Polymerase II transcription regulation and chromatin modification | |
| BRE1 [68], CDC36 [30], LEO1 [38], MED12 [38], MED13 [11], MOT1 [30], RTT103 [18], SDS3 [30], SIN3 [11, 38], SPT2 [17], TAF5 [30] | S. cerevisiae |
| THO complex | |
| HPR1 [69]c [28]bc [12]b [32]c, MFT1 [32]c, THO2 [69]a [28]a, THP2 [70]c |
C. elegansa Homo sapienb S. cerevisiaec |
| mRNA cleavage and polyadenylation, transcription termination | |
| CLP1 [18]c, CFT2 [18]c, FIP1 [18]c, PCF11 [18]c, RNA14 [6]c, RNA15 [18]c, SEN1[71]b [33]c [22]a |
H. sapiena Mus musculusb S. cerevisiaec |
| Splicing | |
| ASF/SF2 [13]b [5] ab, MUD2 [30]c, NPL372c, PRP31 [30]c, SNU114 [30]c, YHC1 [30]c |
Gallus gallusa H. sapienb S. cerevisiaec |
| rRNA processing | |
| DBP6 [30], DBP7 [30], IMP4 [30], RPF1 [30], SNU13 [30], SNU66 [30] | S. cerevisiae |
| Exosome and RNA degradation | |
| DIS3 [30], KEM1/XRN1 [6, 38], RNH1 [11, 18, 30], RNH201 [6, 18], RRP6 [38], TRF4 [35] | S. cerevisiae |
| Nuclear export | |
| CRM1 [6], MEX67 [4], MTR2 [4], MTR3 [6], NAB2 [73], NUP60 [6], NUP133 [30], RNA1 [30], SAC3 [73], SRM1 [18], STS1 [30], SUB2 [4, 14, 30], THP1 [73], YRA1 [4] | S. cerevisiae |
Despite our mechanistic understanding of how RNA processing defects could cause genome instability, their importance in oncogenesis is virtually unknown. It has become increasingly clear that some RNA processing factors are prevalently mutated in cancers and likely act as oncogenes or tumor suppressors (Table 2). However, the mechanistic link is usually unclear and the biological consequences of mutations across different RNA processing pathways are expected to vary. For example, defects in RNA processing components such as Senataxin helicase and RNase H2 cause neurological diseases, whereas splicing factor mutations are strongly cancer-associated [24–26]. Here we discuss the major candidate mechanisms for genome instability caused by RNA processing mutations in the context of tumor genomes and review recent data describing how R-loops regulate DNA damage and gene expression.
Table 2.
Representative RNA processing and maturation factors mutated in cancer.
| Gene | Predominant mutation | Tumor Type(s) | Reference |
|---|---|---|---|
| mRNA 3′-end processing | |||
|
| |||
| FIP1L1 | Translocation (PDGFRα) | Eosinophilic leukemia | [74] |
| PCF11 | Missense | Clear-cell renal cell carcinoma (ccRCC) | [75]* |
|
| |||
| mRNA splicing | |||
|
| |||
| DHX9 | Missense/deletion | ccRCC | [75] |
| DHX38 | Missense | ccRCC | [75] |
| HNRNPA1 | Missense | ccRCC | [75] |
| PRPF40B | Missense | Lung Adenocarcinoma | [76] |
| SFRS1 | Missense | Chronic lymphocytic leukemia (CLL) | [77] |
| SFRS2 | Missense | Chronic myelomonocytic leukemia (CMML) | [78] |
| SFRS6 | Amplified | Colon, Lung | [54] |
| SFRS7 | Missense | CLL | [77] |
| SF3A1 | Missense | Myelodysplastic syndromes (MDS) | [79] |
| SF3B1 | Missense | Breast, CLL, CMML, Lung adenocarcinoma, MDS, Pancreatic, Uveal Melanoma, | [76–78, 80–82] |
| U2AF1 | Missense | MDS, Lung adenocarcinoma, CMML | [76, 78, 79] |
| U2AF2 | Missense | CLL, Lung adenocarcinoma | [76, 83] |
| ZRSR2 | Nonsense/frameshift | MDS | [79, 84] |
|
| |||
| Nuclear transport | |||
|
| |||
| RANBP2 | Missense | ccRCC | [75] |
| SEH1L | Missense | ccRCC | [75] |
| XPO1 | Missense | CLL | [83] |
|
| |||
| RNA deadenylation and degradation | |||
|
| |||
| CNOT3 | Missense/frameshift | T-Acute lymphoblastic leukemia | [51] |
| DIS3 | Missense | Multiple myeloma, AML | [45, 49] |
Sato et al.[75] Identified 31 mutations in RNA processing factors in 106 exomes, highlighting the pathway as a significant class of mutations, those cited here were found in >1 sample.
An RNA roadblock: R-loops as a structural source of genome instability
Biological functions and genomic sites of R-loop formation
R-loop structures consist of a DNA:RNA hybrid and a displaced single DNA strand (Figure 1A). R-loops have been ascribed numerous regulated biological functions in cells such as facilitating somatic hyper-mutation at immunoglobulin genes, mitochondrial DNA replication, alternative mRNA degradation, and pause site-dependent transcription termination [22] (reviewed [27]). Moreover, roles for R-loop formation in the genome continue to be identified in chromatin condensation [28], recruitment of chromatin remodelers [20], telomere length regulation [29], and antisense transcription regulation [21].
Perturbation of R-loop functions may impact genome integrity but it is excessive or misregulated R-loop formation at sites in the genome (Box 1) that has been commonly associated with DNA damage and genome instability [27]. Importantly, DRIP profiling in yeast mutants of the Sen1 helicase or RNase H showed mutant-specific differences from wild type (Box 1), providing a generalizable tool for linking R-loop forming mutants with specific sensitized loci and thereby suggesting testable mechanisms [30]. For instance, the Sen1 mutant accumulates R-loops in genes encoding various snoRNA and tRNA, which are known to rely on Sen1 for maturation and transcription termination [30, 31]. It remains to be understood whether these site-specific R-loops or their effect on the expression of snoRNA and tRNA play a role in neurological diseases caused by mutations in Senataxin helicase. Ideally, future studies will determine R-loop binding profiles in a greater variety of RNA processing mutants, as well as in different environments, to provide further insights into the various influencing factors and potential consequences of unchecked R-loop formation.
BOX 1. DNA:RNA hybrid prone loci in the eukaryotic genome.
Certain genomic loci have been reported to be prone to DNA:RNA hybrid formation, either because of innate properties or because R-loops play a regulated biological function at those sites. The most universal properties that promote R-loop formation seem to be high transcription frequency and high GC content [30, 49, 64]. In addition, properties that promote collision with the DNA replication machinery also predispose loci to R-loop formation, for example very long genes or highly transcribed genes oriented to collide with replication forks are DNA:RNA hybrid prone [65–67]. Site-specific studies and, more recently, genome-wide studies using antibodies to precipitate DNA:RNA hybrids (i.e. DRIP, DNA:RNA immunoprecipitation) have mapped many specific examples of hybrid prone loci [19, 20, 30]. In yeast, the ribosomal DNA locus and retrotransposons are hybrid prone and DNA damage prone genomic sites [30, 67] likely due in part to their transcription frequency, while sub-telomeric regions that are transcribed into telomeric-repeat containing RNA (TERRA) accumulate hybrids that seem to play a regulatory role in telomere maintenance by recombination [29]. In the yeast genome-wide analysis, a set of genes with generally high GC content and transcription-frequency were also identified [30]. Interestingly, genes associated with overlapping antisense transcripts were also implicated as R-loop prone, raising the possibility that transcriptional collisions are inducing R-loops in vivo[30]. Novel insights to R-loop prone regions of the human genome have arisen from using a suite of genomic technologies, including DRIP [19, 20]. Analysis of R-loops in the human genome identify GC skew (i.e. regional strand asymmetry in the distribution of G and C bases) as an important enabling characteristic of DNA:RNA hybrid formation due to the thermodynamic stability of G-rich RNA and C-rich DNA duplexes [20]. GC-skew induced R-loops at CpG island promoters were shown to regulate the recruitment of DNA methyltransferases [20]. In subsequent work, the authors extended these observations to functions for 3′-end R-loops in transcription termination [20]. Together we are beginning to understand the ways in which genomic loci are sensitized to R-loop formation incidentally and for normal cellular functions. A challenge will be to determine how cells differentiate potentially genotoxic versus functional R-loops and prevent each from causing genome instability.
Genetic enhancers of R-loop formation
The mutagenic and hyper-recombinogenic potential of R-loops has been directly demonstrated in mutants of Saccharomyces cerevisiae, Caenorhabditis elegans, mammalian cells and other systems to generate a picture of the various processes that prevent anomalous R-loop formation (Figure 1B). The extent of involved cellular pathways remains to be fully investigated but the list of genes is growing (Table 1). It appears that mutations in nearly every RNA processing function can lead to DNA:RNA hybrids, although not all mutants exhibit equally strong defects and, indeed, some alleles show no effect (Table 1; [11, 18, 30]). The prevailing model of R-loop formation relies on failed RNA transcript sequestration due to the absence of RNA binding proteins (e.g. in Hpr1 mutants [32]). It is worth noting two special cases, Senataxin helicase that unwinds hybrids and participates in transcription termination [22, 33], and RNase H that degrades RNA in DNA:RNA hybrids and functions in DNA replication. Unlike other RNA processing factors, Senataxin helicase and RNase H may operate as cellular surveillance for inappropriate R-loop structures and could regulate biological R-loop functions because of their ability to recognize and reverse R-loop structures. Interestingly, sumo-modified Senataxin has recently been physically linked to the exosome at nuclear stress foci [34]. In combination with observations linking exosome mutants to DNA:RNA hybrids [6, 30, 35], these data raise the possibility that DNA:RNA unwinding by Senataxin is coordinated with RNA degradation by the exosome [34]. Indeed the network of cellular regulation of Senataxin is of intense interest as it also functions actively to coordinate transcription with DNA replication and repair activities [36, 37].
A fascinating recent addition to this field was the report of a mechanism for R-loop formation in trans involving the homologous recombination (HR) repair machinery in yeast [38]. It was demonstrated that Rad51-coated ssRNA can catalyze R-loop formation in trans in an analogous fashion to ssDNA invasion of a dsDNA duplex during homology-directed repair. In this scenario, removal of Rad51 prevents R-loop formation in trans and stabilizes the genome to this type of R-loop-mediated instability [38]. In many R-loop producing mutants, removal of DNA:RNA hybrids by overexpression of RNase H has been found to suppress genome instability such as chromosome loss in yeast [18] and double stranded break (DSB) formation in mammalian cells [9]. These observations provide causal links between unchecked R-loop formation and genome instability through several mechanisms discussed below.
R-loop-associated genome instability
R-loops that form due to abnormalities in RNA processing are recognized sources of transcription-associated mutagenesis (TAM) and transcription-associated recombination (TAR) (reviewed [27]). One popular model of R-loop-mediated DNA damage involves collisions between the transcription and DNA replication machinery [13, 27] (Figure 1C). In this scenario, the stalled replication fork occasionally collapses and the resulting repair of the DSB by HR is mutagenic and contributes to TAM. Another mechanism of R-loop-mediated genome instability, relevant to mammalian cells, involves the displaced ssDNA serving as a substrate for cytidine deaminases, which can result in mutagenic repair of the deaminated base [27] (Figure 1C). On a related note, overexpression of the cytidine deaminase APOBEC3B is known to cause genome instability in cancer, although it is not known whether R-loops play a role in targeting APOBEC3B to sites of mutagenesis in cancer [39]. Generally, it is unknown whether R-loop formation is upregulated in cancer or if R-loops are a common cause of genome instability in tumor evolution. Studies in model organisms and tissue culture strongly suggest that, in the event of a mutation in RNA metabolism or DNA repair that stimulates R-loop formation, the cell becomes predisposed to genome instability.
Staying on message: an altered transcriptome as a source of genome instability
One of the most obvious consequences of perturbed RNA processing is changes in gene expression. Producing more, less, or altered sets of mRNAs will influence the amount of protein produced and ultimately control the fate of the cell and the potential progression into disease [40, 41]. Cancer is often caused by changes in expression and activity of oncogenes and tumor suppressors. Mutations and mis-regulation of transcription factors are well documented in tumors and can lead to pleiotropic effects on cell biology [41, 42]. Recent examples have linked altered chromatin or transcription specifically to expression of genome stability factors (e.g. polycomb modifiers of Aurora A expression [43, 44]). In addition to these traditional modifiers of gene expression, co- and post-transcriptional activities like splicing and RNA degradation also control gene expression and have been linked to genome instability and cancer [45–47].
RNA turnover
In yeast, defects in both major RNA degradation pathways, the exosome and Processing (P)-bodies, have been implicated in regulating genome maintenance through altered gene expression (Figure 2). The exosome is a nuclear and cytoplasmic exonuclease complex involved in degradation or processing of various RNA. P-bodies are cytoplasmic ribonucleoprotein aggregates containing multiple enzymatic activities involved in RNA turnover (e.g. deadenylation, decapping, exonucleolytic activity). Analysis of mutants lacking the core P-body component LSM1 revealed a defect in histone mRNA degradation in response to DNA replication stress [15]. Maintaining a dynamic pool of histones is essential for cells to effect changes in their chromatin state in response to genotoxic stress and to transit S-phase. In lsm1Δ mutants, increased levels of histones caused destabilization of stalled DNA replication forks leading to genome instability [15]. The observations at the histone-level indicate redundancy with functions associated with the exosome, which had previously been shown to regulate histone levels and genome stability [16]. In another study, mutation of several exosome subunits, including the catalytic subunit DIS3, were shown to dramatically affect the function of microtubules, probably through changes in gene expression, which would impact chromosome segregation fidelity [48]. These data show that different perturbations of RNA degradation can elicit genome instability phenotypes by altering turnover of specific transcripts in various pathways (Figure 2). Perhaps most importantly, mutations in human DIS3 occur at a significant frequency in multiple myeloma and acute myeloid leukemia [45, 49] and the paralogous gene DIS3L2 is mutated in Wilms tumor susceptibility [50]. Even more recently, mutations in CNOT3, a gene coding for a component of the CCR4-NOT deadenylase complex found in P-bodies, were identified in T-cell acute lymphoblastic leukemia [51]. Thus RNA turnover is an emerging player in the biology of some cancers and may have unappreciated connections to genome integrity in these tumors. Interestingly, mutations in several RNA degradation proteins such as Xrn1, Dis3 and Rrp6 have also been found to increase R-loop formation [6, 30, 38]. This raises the intriguing possibility of both direct (via R-loops) and indirect (via changed gene expression) causes of DNA damage, and complicates interpretation of genome instability phenotypes in these mutants.
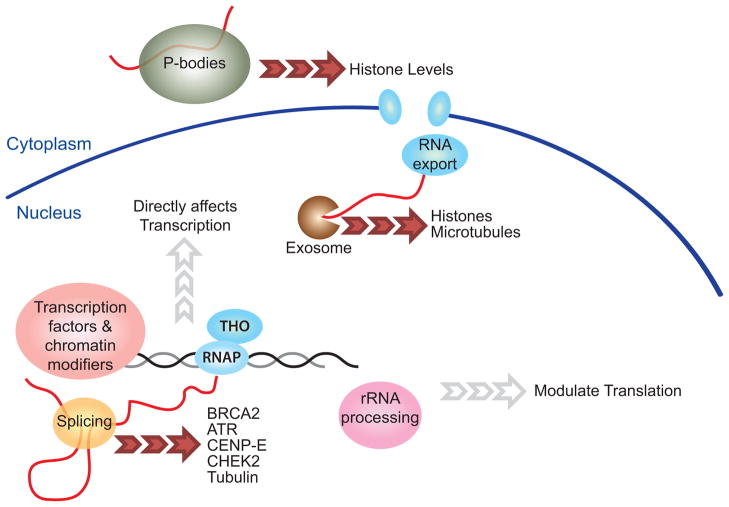
Figure 2. Altered gene expression as a cause of genome instability in RNA processing mutants.
A cellular schematic highlights examples from the main text where (I) disruption of yeast RNA-degrading P-bodies reduces histone levels, (II) disruption of the yeast exosome alters histones and microtubules and (III) disruption of splicing proteins lead to reduced expression of many important genome stability proteins in yeast and humans. Mutations of transcription factors and chromatin modifiers clearly impact gene expression and represent a large, related topic not covered here (black arrows), while a potential role for rRNA processing, and ribosome biogenesis in controlling gene expression is suspected but less well-understood (grey arrow, e.g. Rpl10 mutations in T-cell acute lymphoblastic leukemia [51]).
Splicing
An informational mechanism of genome instability has also been suggested for a variety of perturbed splicing conditions. In mammalian cells, depletion of a splicing regulator called ERH (enhancer of rudimentary homologue) leads to chromosomal instability due to failed splicing of the mitotic motor protein CENP-E and other cell cycle and DNA repair proteins [52]. The authors identified ERH as a synthetic lethal partner of mutant KRASG13D and they proposed that the mechanism of synthetic lethality relates to additional mitotic stress in cells with activating KRAS mutations [52]. Finally, genome-wide analysis of genes that regulate HR identified many splicing regulators [8]. These authors went on to show that knockdown of the splicing factors RBMX, PRPF6, SF3A2, SF3B3 and possibly others, affect HR by specific depletion of the breast cancer susceptibility gene BRCA2 and, in the case of RBMX, the checkpoint kinase ATR [4]. These effects were quite specific and did not extend to other HR proteins (i.e. BRCA1, PALB2, BARD1 were unaffected) raising the question of how to identify specific transcripts that are hypersensitive to reduced levels of core splicing proteins. Of course, not only failed splicing, but also changes in alternative splicing represent another likely effect of splicing factor mutations. For instance, the alternative splicing of CHEK2 has been linked to chromosomal instability [53] and alternative splicing patterns have been identified in tumors with spliceosomal mutations [47]. Together these data show that disruption of splicing factors can change the gene expression program in a highly specific manner that can impede genome maintenance (Figure 2).
Mutations affecting core components of the splicing machinery are remarkably common in various cancer types including uveal melanoma, colon cancer, lung cancer, breast cancer, pancreatic cancer myelodysplastic syndromes, chronic lymphocytic leukemia, and other hematological malignancies (Table 2, references therein). In some cases there is evidence that these mutations affect gene expression [47, 54] but the ensuing cellular phenotype relevant to malignancy is not known [25]. Although the influence on gene expression is the most probable common theme among all of these cancer-associated splicing mutants, there is also precedent for splicing factors functioning outside of splicing reactions (e.g. the direct role of SFPQ/PSF in DNA repair [55], the function of SF3B1 in polycomb-mediated gene repression [56], or the role of factors like SRSF1, formerly ASF/SF2, in preventing transcription-coupled R-loops [5]). Disruption of any of these activities could lead to genome instability independent of altered gene expression due to splicing defects and only direct mechanistic analysis will differentiate these possibilities. The contributions of individual splicing mutations to tumor genome instability remains to be determined but is an area of growing interest coincident with the growing recognition of spliceosomal mutations in cancer.
Bridging the gap: R-loops as regulators of gene expression
R-loops play important roles in gene expression through functions in transcription termination, DNA methylation, and chromatin modification [19, 20, 22, 28]. For instance, R-loops at CpG island promoters were recently shown to be correlated with protection against DNA methylation and transcriptional silencing in mammalian cells [19, 20] (Box 1). Interestingly, in Schizosaccharomyces pombe, DNA:RNA hybrids formed by non-coding RNA were found to interact with the RNA-induced transcriptional silencing (RITS) complex and to play a role in RNAi-directed heterochromatin assembly [57]. There is also recent evidence that SEN1 mutations, themselves known to cause genotoxic R-loops [33], reduce the expression of the yeast ribonucleotide reductase subunit, RNR1, which is crucial to the DNA damage response [58]. Thus, single mutations can induce R-loops and alter expression, and the R-loop structure itself may play a key role in modulating gene expression in some cases.
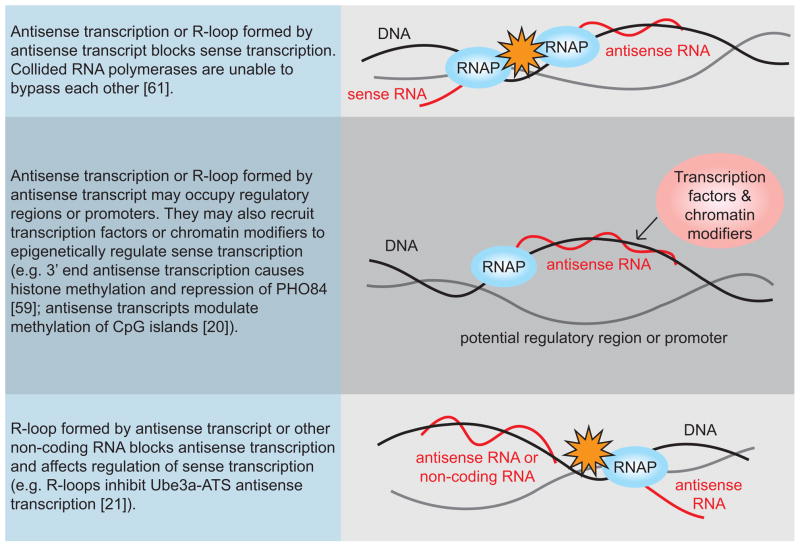
Additionally, a group of studies have demonstrated a role for R-loops in antisense transcription regulation, which suggests a mechanism by which anomalous R-loop formation could significantly alter gene expression [21, 30]. Antisense transcripts are transcribed on the opposite strand from, and in some cases have been found to play a role in the regulation of expression of, sense transcripts (e.g. antisense transcription represses PHO84 [59]). They are highly prevalent in the mammalian genome and have been observed at active promoters and most transcribed genes [60]. The mismanagement of R-loops, possibly resulting from RNA processing defects, can alter the transcriptome by perturbing various transcription regulation mechanisms (Figure 3). One model is that colliding RNA polymerases are unable to bypass each other, resulting in failure of transcription which has been demonstrated in vitro [61]. The authors showed that convergent transcription impedes transcription and there is increased RNA polymerase density in regions between convergent genes in vivo [61]. However, it remains to be determined whether RNA polymerase collisions occur at a significant rate in vivo and whether R-loops are involved. Another model links antisense transcription to chromatin modification. It has been shown that R-loop formation inhibits antisense transcription, decondenses chromatin, and removes silencing of the Ube3a gene in mammalian cells [21]. Thus, deregulated R-loop formation may lead to significant alterations in the expression of genes that are key to disease progression. The role that altered gene expression may play in the phenotype of R-loop prone cells is unknown. However, it is important to consider the possibility of both direct and indirect effects of R-loop formation as causes of genome instability.
Figure 3. R-loop formation influences antisense transcription regulation.
Antisense transcription regulation by collision of transcription machinery (orange stars) or recruitment of, or protection against, chromatin modifiers (pink circle). Specific examples with citations are stated for each model.
Concluding Remarks
RNA processing directly impacts the genome by regulating gene expression and by preventing the annealing of potentially harmful complementary RNAs to genomic DNA. DNA:RNA hybrids can lead directly to replication stress and DNA damage [27], and also have the potential to influence gene expression through epigenetic mechanisms or by transcriptional interference [19–22].
These observations highlight a number of unanswered fundamental questions. First and foremost, how do cells differentiate R-loops with normal functions from deleterious ones? In other words, how have cells evolved to prevent the activities of functional R-loops from blocking replication and causing genome instability? Moreover, how do cells distinguish and respond differently to trans versus cis R-loops, which are presumably associated with a different suite of proteins (i.e. HR proteins versus transcriptional machinery respectively)? Answering these questions will require additional mechanistic studies in models and a comparison of genomic profiles of DNA:RNA hybrid prone sites across various mutant backgrounds.
From a human health standpoint, what is the impact of RNA processing mutations that are found in cancer? If the mutations are important for tumor cell biology, do they work through promoting R-loops, by altering gene expression, or through another mechanism? We acknowledge that if the mechanism of a cancer-associated RNA processing mutant invokes R-loops, changes transcript levels, or alters epigenetic states as predicted in models, the ultimate phenotypic impact relevant to disease may or may not be genome instability. One method to probe for R-loop formation sites that is gaining in popularity and could be extended to tumor genome analysis is native bisulfite-treated genome sequencing [5, 20].
Better understanding of the role that RNA processing factors play in tumor cells will intersect with emerging prognostic and therapeutic opportunities. An excellent example of this is the SF3B complex where the frequent mutations in SF3B1 are becoming part of the prognostic criteria in chronic lymphocytic leukemia [62]. Meanwhile, candidate anti-cancer therapeutic inhibitors of the spliceosome such as Spliceostatin A target the SF3B complex [63]. The molecular mechanisms of cancer-associated mutations in SF3B components, which potentially cause genome instability, are unknown but, once elucidated these mechanisms will surely benefit the prognostic and therapeutic applications of the complex.
Highlights.
Genome instability is an established consequence of perturbations in RNA processing.
Genome instability in RNA processing mutants occurs via DNA:RNA hybrids.
Genome instability can also occur through changes in gene expression.
RNA processing genes are mutated in many cancers but mutant function is unknown.
Acknowledgments
We apologize to those colleagues whose work we could not cite due to space limitations. We thank Nigel O’Neil, Doug Koshland and Lamia Wahba for critical reading of the manuscript. P.C.S. is supported by the Cancer Research Society. P.H. is supported by the Canadian Institutes of Health Research (MOP-38096) and National Institutes of Health (R01-CA158162). Y.A.C. is supported by the Roman M Babicki fellowship.
GLOSSARY BOX
- Antisense transcription
Transcription of an (antisense) RNA from the opposite DNA strand as the template encoding a sense mRNA transcript
- DRIP
DNA, RNA immunoprecipitation; a technique to isolate DNA:RNA hybrids from cells using a specific monoclonal antibody
- R-loop
A three-stranded nucleic acid structure in which RNA is hybridized to the complementary strand of double-stranded DNA and the other DNA strand is exposed in a single-stranded loop
- RNA processing
In the context of this article, the suite of cellular activities that lead to maturation and proper localization of a nascent transcript, and to the various steps in its eventual degradation
- Genome Instability
An increase in the rate of mutations or chromosomal abnormalities, such as aneuploidy, experienced by a cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stratton MR. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 2.Burrell RA, et al. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crasta K, et al. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jimeno S, Rondon AG, Luna R, Aguilera A. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Manley JL. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Luna R, Jimeno S, Marin M, Huertas P, Garcia-Rubio M, Aguilera A. Mol Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Wellinger RE, Prado F, Aguilera A. Mol Cell Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen RD, et al. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stirling PC, et al. PLoS Genet. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahba L, Amon JD, Koshland D, Vuica-Ross M. Mol Cell. 2011;44:978–988. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Sanchez MS, Barroso S, Gomez-Gonzalez B, Luna R, Aguilera A. PLoS Genet. 2011;7:e1002386. doi: 10.1371/journal.pgen.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan W, et al. Genes Dev. 2011;25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Gonzalez B, et al. EMBO J. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero AB, Moreno S. EMBO J. 2011;30:2008–2018. doi: 10.1038/emboj.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis CC, Campbell JL. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikdar N, Banerjee S, Zhang H, Smith S, Myung K. PLoS Genet. 2008;4:e1000290. doi: 10.1371/journal.pgen.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stirling PC, et al. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. Genome Res. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell WT, et al. Proc Natl Acad Sci U S A. 2013;110:13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaillard H, Aguilera A. Biochim Biophys Acta. 2013;1829:141–150. doi: 10.1016/j.bbagrm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Crow YJ, et al. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 25.Garraway LA, Lander ES. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Suraweera A, et al. Hum Mol Genet. 2009;18:3384–3396. doi: 10.1093/hmg/ddp278. [DOI] [PubMed] [Google Scholar]
- 27.Aguilera A, Garcia-Muse T. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Castellano-Pozo M, et al. Mol Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Balk B, et al. Nat Struct Mol Biol. 2013;20:1199–1205. doi: 10.1038/nsmb.2662. [DOI] [PubMed] [Google Scholar]
- 30.Chan YA, et al. PLoS Genet. 2014:10. doi: 10.1371/journal.pgen.1004288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huertas P, Aguilera A. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Mischo HE, et al. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard P, Feng S, Manley JL. Genes Dev. 2013;27:2227–2232. doi: 10.1101/gad.224923.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavalda S, Gallardo M, Luna R, Aguilera A. PLoS One. 2013;8:e65541. doi: 10.1371/journal.pone.0065541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alzu A, et al. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuce O, West SC. Mol Cell Biol. 2013;33:406–417. doi: 10.1128/MCB.01195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahba L, Gore SK, Koshland D. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns MB, et al. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chi P, Allis CD, Wang GG. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee TI, Young RA. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bywater MJ, Pearson RB, McArthur GA, Hannan RD. Nat Rev Cancer. 2013;13:299–314. doi: 10.1038/nrc3496. [DOI] [PubMed] [Google Scholar]
- 43.Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Genes Cancer. 2012;3:649–657. doi: 10.1177/1947601913479022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou CH, et al. Cancer Res. 2013;73:953–966. doi: 10.1158/0008-5472.CAN-12-2397. [DOI] [PubMed] [Google Scholar]
- 45.Chapman MA, et al. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David CJ, Manley JL. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furney SJ, et al. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith SB, Kiss DL, Turk E, Tartakoff AM, Andrulis ED. Yeast. 2011;28:755–769. doi: 10.1002/yea.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding L, et al. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Astuti D, et al. Nat Genet. 2012;44:277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 51.De Keersmaecker K, et al. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng MT, et al. Proc Natl Acad Sci U S A. 2012;109:E3659–67. doi: 10.1073/pnas.1207673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang HW, et al. Neoplasia. 2012;14:20–28. doi: 10.1593/neo.111574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen-Eliav M, et al. J Pathol. 2013;229:630–639. doi: 10.1002/path.4129. [DOI] [PubMed] [Google Scholar]
- 55.Rajesh C, Baker DK, Pierce AJ, Pittman DL. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Genes Dev. 2005;19:536–541. doi: 10.1101/gad.1284605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. Genes Cells. 2012;17:218–233. doi: 10.1111/j.1365-2443.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 58.Golla U, et al. PLoS One. 2013;8:e64798. doi: 10.1371/journal.pone.0064798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margaritis T, et al. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faghihi MA, Wahlestedt C. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cortese D, et al. Leukemia 2013 [Google Scholar]
- 63.Corrionero A, Minana B, Valcarcel J. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 65.Helmrich A, Ballarino M, Tora L. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 66.Tuduri S, et al. Nat Cell Biol. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El Hage A, French SL, Beyer AL, Tollervey D. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chernikova SB, et al. Cancer Res. 2012;72:2111–2119. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castellano-Pozo M, Garcia-Muse T, Aguilera A. PLoS One. 2012;7:e52447. doi: 10.1371/journal.pone.0052447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfeiffer V, Crittin J, Grolimund L, Lingner J. EMBO J. 2013 doi: 10.1038/emboj.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becherel OJ, et al. PLoS Genet. 2013;9:e1003435. doi: 10.1371/journal.pgen.1003435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Santos-Pereira JM, Herrero AB, Garcia-Rubio ML, Marin A, Moreno S, Aguilera A. Genes Dev. 2013;27:2445–2458. doi: 10.1101/gad.229880.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A. J Biol Chem. 2003;278:24225–24232. doi: 10.1074/jbc.M302900200. [DOI] [PubMed] [Google Scholar]
- 74.Cools J, et al. N Engl J Med. 2003;348:1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 75.Sato Y, et al. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 76.Imielinski M, et al. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quesada V, et al. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 78.Patnaik MM, et al. Am J Hematol. 2013;88:201–206. doi: 10.1002/ajh.23373. [DOI] [PubMed] [Google Scholar]
- 79.Papaemmanuil E, et al. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Biankin AV, et al. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ellis MJ, et al. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Puente XS, et al. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]