Abstract
Cortical granules are specialized organelles whose contents interact with the extracellular matrix of the fertilized egg to form the block to polyspermy. In sea urchins, the granule contents form a fertilization envelope (FE), and this construction is critically dependent upon protease activity. An autocatalytic serine protease, cortical granule serine protease 1 (CGSP1), has been identified in the cortical granules of Strongylocentrotus purpuratus eggs, and here we examined the regulation of the protease activity and tested potential target substrates of CGSP1. We found that CGSP1 is stored in its full-length, enzymatically quiescent form in the granule, and is inactive at pH 6.5 or below. We determined the pH of the cortical granule by fluorescent indicators and micro-pH probe measurements and found the granules to be pH 5.5, a condition inhibitory to CGSP1 activity. Exposure of the protease to the pH of seawater (pH 8.0) at exocytosis immediately activates the protease. Activation of eggs at pH 6.5 or lower blocks activation of the protease and the resultant FE phenotypes are indistinguishable from a protease-null phenotype. We find that native cortical granule targets of the protease are β-1,3 glucanase, ovoperoxidase, and the protease itself, but the structural proteins of the granule are not proteolyzed by CGSP1. Whole mount immunolocalization experiments demonstrate that inhibition of CGSP1 activity affects the localization of ovoperoxidase but does not alter targeting of structural proteins to the FE. The mistargeting of ovoperoxidase may lead to spurious peroxidative cross-linking activity and contribute to the lethality observed in protease-null cells. Thus, CGSP1 is proteolytically active only when secreted, due to the low pH of the cortical granules, and it has a small population of targets for cleavage within the cortical granules.
INTRODUCTION
Polyspermy is lethal to the eggs of most species. If more than one sperm fertilizes the egg, then the newly formed zygote will cleave abnormally and die. One effective way to block polyspermy is for the egg to rapidly modify its extracellular surface after sperm activation so that subsequent sperm cannot reach the egg cell surface. A conserved mechanism for this modification is for the fertilizing sperm to induce the egg to exocytose contents from specialized organelles called cortical granules. The contents of the granule serve to modify the extracellular matrix of the egg, making it impenetrable to additional sperm fusions. The cortical granules have a specific and singular role during development: these organelles are present only in eggs and oocytes, and most of the known content proteins are expressed selectively in oocytes. Although the molecular mechanisms used to block polyspermy are different between animals, all mammals, most vertebrates, and many invertebrates, including the sea urchin share this fundamental process (Shapiro et al., 1989; Wessel et al., 2001).
Cortical granules of the sea urchin contain a diverse population of proteins, including structural molecules, enzymes, and glycosaminoglycans, which are selectively packaged into the granule. Each of the detectable content proteins is now known (Wessel et al., 2001; Wong and Wessel, 2004). After exocytosis, the granule contents interact with each other as well as the proteins of the egg extracellular matrix (the vitelline layer) to form the fertilization envelope (FE), a tough, translucent protein matrix that is completely detached from the plasma membrane and forms a protective environment for the newly formed zygote. In <1 min after insemination, the mass of proteins stored in the granule transforms itself into a new, highly organized protein matrix that blocks polyspermy.
Structural proteins make up the main scaffolding components of the fertilization envelope. These proteins include SFE1, SFE9, MGB, and proteoliasin. SFE9 is 128 kDa and has a mosaic of conserved structural domains, including low-density lipoprotein (LDL) receptor-like repeats (Wessel, 1995). SFE1 is a 175-kDa molecule that it is comprised of five LDL receptor-like repeats, 22 cysteine-rich repeats, and three serine/threonine-rich repeats (Wessel et al., 2000). Proteoliaisin is ∼235 kDa and binds to the vitelline layer, perhaps serving as scaffolding for additional fertilization envelope proteins. Proteoliaisin also binds to ovoperoxidase (see below) and targets its enzymatic activity to the envelope (Weidman et al., 1985). The final major fertilization envelope protein is MGB (Wong and Wessel, unpublished data). It is predicted to be 90 kDa with a number of cysteine-rich protein interaction domains classified as CUB domains. These domains, as for LDL receptor-like domains, are known to function in other molecules in protein–protein interactions (Bork and Beckmann, 1993). In addition, the granules secrete hyalin, a 330-kDa fibrillar protein that is not incorporated into the FE, but instead becomes the main component of the hyaline layer, a distinct extracellular matrix underneath the fertilization envelope that coats the eggs and provides support for the embryo as blastomeres cleave during development (Hylander and Summers, 1982).
The cortical granules contain diverse enzyme activities as well. Glucanase was the first molecule identified from the cortical granule (Epel et al., 1969; Talbot and Vacquier, 1982). The β-1,3 glucanase from Strongylocentrotus purpuratus eggs was cloned and shown to be a 68-kDa protein that is homologous to horseshoe crab and bacterial glucanases (Bachman and McClay, 1996), but the function of this enzyme at fertilization is yet unknown. The cross-linking activity of the ovoperoxidase enzyme is also present in the granule. After granule exocytosis, ovoperoxidase interacts specifically with proteoliaisin, which effectively tethers ovoperoxidase to the vitelline layer. In the presence of H2O2 generated by an egg oxidase, ovoperoxidase catalyzes the formation of covalent dityrosine bonds between proteins (Foerder and Shapiro, 1977; Hall and Vacquier, 1982), including SFE1, SFE9, and itself (Shapiro et al., 1989; Somers and Shapiro, 1991; Wessel et al., 2001), and thereby creates a highly cross-linked or hardened FE (Klebanoff et al., 1979; Somers et al., 1989). In the absence of ovoperoxidase activity, the FE still forms and rises off the egg plasma membrane, but it does not become stabilized like a normal envelope.
The third enzyme identified in the cortical granule is a trypsin-like protease, cortical granule serine protease 1 (CGSP1). CGSP1 is 61 kDa and contains two LDL receptorlike repeats at its N terminus and a trypsin-like serine protease domain in its C terminus (Haley and Wessel, 1999). An activation sequence separates the N- and C-terminal domains, and the protease undergoes autocatalysis at this position. Sea urchin eggs fertilized in the presence of serine protease inhibitors exhibit an abnormal fertilization envelope that does not fully detach from the plasma membrane and the eggs become highly polyspermic (Hagström, 1956; Vacquier et al., 1972b; Schuel et al., 1973). These observations led to the conclusion that serine protease activity is necessary for the permanent block to polyspermy in sea urchins. CGSP1 is believed to hydrolyze a number of proteins on the egg surface, including proteinaceous connections of the vitelline layer to the egg plasma membrane, the putative sperm binding protein (Carroll and Epel, 1975; Hirohashi and Lennarz, 1998), and an integrin (Murray et al., 2000). We have recently shown that an extracellular membrane-bound protein, p160, is also proteolyzed from the egg surface at fertilization (Haley and Wessel, unpublished data). The coordinated proteolysis of these proteins is believed to be responsible for the rapid separation of the forming FE from the cell surface.
The protease is also hypothesized to cleave content proteins of the cortical granule, because limited proteolysis might be necessary for proper formation and elevation of the fertilization envelope. This premise is based on the observation that the structural proteins are highly fragmented and may have to be cleaved before they can be properly arranged in the envelope (Wessel et al., 2001). The enzymes ovoperoxidase and β-1,3 glucanase might also require proteolysis, either to become active, or to limit their activity once secreted from the egg.
In this report, we demonstrate that CGSP1 cleaves a subpopulation of the granule content proteins: the enzymes ovoperoxidase and β-1,3 glucanase as well as the protease itself are cleaved in a CGSP1-dependent manner. Surprisingly, the structural proteins do not seem to be cleaved by CGSP1. Instead, processing by a secretory pathway protease must explain their diverse fragmentation. We conclude that the cleavage of the granule enzymes by CGSP1 takes place after exocytosis at fertilization, and not while stored in the granule, and that a change in the pH resulting from release by an acidic organelle activates this processing step.
MATERIALS AND METHODS
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Handling of Eggs
Adult S. purpuratus were obtained from Charles Hollahan (tidalflux@yahoo.com; Santa Barbara, CA), and gametes were shed by intercoelomic injection of 0.5 M KCl. Eggs were dejellied by passing the eggs through cheesecloth twice and then incubating the eggs in Instant Ocean artificial sea water (ASW; Aquarium Systems, Mentor, OH) adjusted to pH 5.2 for 2 min and then washed twice in ASW, pH 8.0. To disrupt the vitelline layer, the dejellied eggs were incubated in 10 mM dithiothreitol (DTT) in pH 9.2 ASW for 10 min at room temperature and then washed three times in ASW (Epel et al., 1970).
Cortical granule exudate (Vacquier et al., 1972b) was obtained as follows: a 10% suspension of dejellied DTT-treated eggs was activated with calcium ionophore A23187 (10 mM final concentration). After 5 min, the eggs were removed by centrifugation (5 min, 500 × g) and the supernatant centrifuged again at 10,000 × g for 15 min. The supernatant was dialyzed against distilled water for 12 h at 4°C and then lyophilized before analysis. To make protease-deficient exudate, soybean trypsin inhibitor (SBTI) was added to the egg suspension before activation at a final concentration of 100 μg/ml.
To observe pH effects on FE formation, dejellied eggs (10% suspension) were placed in ASW at various pH levels and then activated with A23187 (10 mM final concentration). FE formation was observed over time with an Axioplan microscope (Carl Zeiss, Thornwood, NY), and images were recorded with a SpotCam 2000 digital recorder.
Western Blot Analysis
For immunoblot analysis, exudate samples (10 μg of total protein/lane) were subjected to SDS-PAGE and immunoblotting essentially as described previously (Towbin et al., 1979). The exudate samples for analysis were lyophilized and resuspended in 2× sample buffer (20% sucrose, 5 mM Tris-Cl, pH 6.8, 2% SDS), 1 mM DTT, and 0.1 mM phenylmethylsulfonyl fluoride and denatured for 5 min at 100°C. The proteins were then resolved by SDS-PAGE on 4–20% and 10–20% gradient gels and electroblotted. Blots were washed twice for 30 min each in blotto (50 mM Tris-Cl, pH 7.5, 0.18 M NaCl, 0.05% Tween 20, 3% nonfat dry milk) and then incubated for 3 h in blotto containing antibodies of interest. Rabbit polyclonal antibodies against various cortical granule proteins and their dilutions were as follows: anti-CGSP1, 1:500 (Haley and Wessel, 1999); anti-ovoperoxidase, 1:500 (LaFleur et al., 1998); anti-β-1,3 glucanase, 1:50,000 (Peeler et al., 1987); anti-SFE9, 1:1500 (Wessel, 1995); and anti-MGB, 1:1000 (Conner, unpublished data). In addition, a monoclonal antibody was used to hyalin at 1:1000 (Wessel et al., 1998). After primary antibody incubation, the blots were then washed three more times >1 h and incubated with either goat anti-rabbit antibodies conjugated to alkaline phosphatase diluted to 1:30,000 or goat anti-mouse antibodies conjugated to alkaline phosphatase diluted to 1:10,000. Blots were washed in blotto three more times >30 min and then washed twice in blotto without milk, and immunolabel signals were detected by bromo-chloro-indoryl phosphate/nitro blue tetrazolium colorimetric development (Promega, Madison, WI) as described previously (Harlow and Lane, 1988).
Ovoperoxidase Activity Assay
Ovoperoxidase activity secreted by the egg was detected by using the fluorogenic substrate Amplex Red (10-acetyl 1-3,7-dihydroxyphenoxazine; Molecular Probes, Eugene, OR). In the presence of peroxidase, Amplex Red reacts with H2O2 to produce the highly fluorescent product resorufin (530-nm excitation and 590-nm emission; Zhou et al., 1997). To perform real-time activity assays, eggs were placed in 96-well microtiter plates, activated with A23187, and read on a Fluoroskan Ascent FL type 374 fluorimeter; Labsystems, Rochester, NY). Assays were performed in triplicate, by using 100 eggs/well in 200-μl final volume. The reaction buffer included 200 μM Amplex Red in calcium-free seawater (used to prevent precipitation of calcium in the assay buffer), 0.075% H2O2, and 10 μg/ml A23187 in DMSO. In no-activation control experiments, A23187 was omitted, and only DMSO was used. Benzamidine at 10 mM was included to inhibit protease activity when needed. Fluorescence intensity of the resorufin product was quantitated in the wells by reference to known horseradish peroxidase standards.
Glucanase Activity Assay
β-1,3 Glucanase activity is measured by digestion of laminarin, a branched β-1,3 glucose polymer, and free glucose is then detected fluorometrically. We are unable to read glucanase activity in real time in vivo as in the ovoperoxidase assays because the activated egg produces prodigious amounts of H2O2, a complication for sensitive glucose detection. We therefore had to rely on the conventional glucanase determination by first isolating cortical granule exudate, as described above. Cortical granule exudate was collected from a 30% suspension of dejellied, DTT-treated eggs in ASW buffered with 10 mM Tris, pH 8.0, after activation in 10 μg/ml A23187. Inhibitor-treated samples were first preincubated in 10 mM benzamidine. After activation, the samples were incubated for 5 min at 23°C, and protease activity was then stopped by adding 1 mM final phenylmethylsulfonyl fluoride, and the eggs were rapidly removed by hand centrifugation. Remaining particulate matter was removed by centrifugation at 5000 × g for 1 min at 4°C, and the clarified supernatant was then dialyzed overnight at 4°C against distilled H2O and 10 mM benzamidine. Glucanase activity was measured essentially as described by Peeler et al. (1987), but using Amplex Red as the fluorogenic agent. The protein levels of various samples were normalized, and the exudate samples were assayed in triplicate in 96-well plates at 200-μl final volume. The reaction buffer was as follows: 250 μg (0.125%) of laminarin, 200 μM Amplex Red (Molecular Probes), 3 U of glucose oxidase, and 1 U of horseradish peroxidase. Samples were read at 530-nm excitation and 590-nm emission, and time points were taken every 30 s for 10 min in a Fluoroskan Ascent FL type 374 fluorimeter.
Protease Activity Assay
The cortical granule protease was purified by SBTI affinity chromatography (Fodor et al., 1975), and column fractions were concentrated and washed in 10 mM Tris, pH 4.0, buffer in Centricon-10 centrifugal filter devices (Millipore, Beverly, MA). Samples were then diluted in 10 mM Tris, pH 8.0, buffer and protease activity was measured by benzoyl-l-arginine ethylester (BAEE) as described previously (Schwert and Takenaka, 1955). To measure activity in different buffer conditions, the protease was diluted in buffers of defined pH. Activity was assayed by BAEE hydrolysis as an increase in absorbance at 253 nm, recorded every 30 s over 10 min.
Whole Mount Immunolocalizations
Immunofluorescence localization of cortical granule content proteins was performed on activated eggs, either under control conditions or treated with 100 μg/ml soybean trypsin inhibitor, and fixed for whole-mount immunolabeling (Wessel et al., 1998). The embryos were then stained for the following fertilization envelope proteins: proteoliaisin (Somers et al., 1989), SFE1 (Wessel et al., 2000), SFE9 (Wessel, 1995), and MGB (Conner, unpublished data), each at 1:125 dilution; ovoperoxidase (LaFleur et al., 1998) at 1:50 dilution; and glucanase at 1:500 dilution. The samples were then incubated in CY3-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) at 1:100 dilution, for 30 min, washed three times in 10 mM Tris, pH 7.6, 0.15 M NaCl, 0.01% Tween 20. The samples were then visualized by fluorescence staining on an Axioplan microscope (Carl Zeiss) equipped with an ORCA ER camera (Hamamatsu, Bridgewater, NJ).
pH Determination of Cortical Granules
The pH of the cortical granules was determined in three ways, either using LysoSensor or by microelectrode measurement of the exudates of activated eggs. For the micro-pH electrode measurements, eggs were resuspended to 50% by volume in nonbuffered sea water, 0.5 M NaCl, or Na+-free sea water and activated by 10 μg/ml A23187 either in the presence or absence of 0.5 mM amiloride to inhibit Na+/H+ exchange. Because cortical granules are the major, if not only, vesicle exocytosis to occur in the time frame of this experiment, it enables us to measure directly the protons liberated from the cortical granules. The pH was monitored over 10 min, and the extent of acid efflux was calculated using the following values: 15,000 cortical granules per cell, each of 1.5-μm diameter in an 80-μm-diameter egg (Wessel et al., 2001).
For in situ measurements of cortical granule pH, LysoSensor Yellow/Blue DND-160 (Molecular Probes) was used at 10 μM on intact eggs or cell surface complex isolated as in Kinsey (1986). The LysoSensor Yellow/Blue type spectral profile changes depending on the protonation state of the dye, thus accurately reflecting the environment in which the dye is evaluated. Standard pH curves were determined on a Zeiss 510 equipped with META imaging by using a 405 diode excitation and collecting emissions from 420 to 750 nm. The profiles of the emission spectrum at known pH were calculated and stored for later comparison. Eggs or cell surface complex were immersed in LysoSensor and immediately recorded as in the control, focusing on either the cortex of the intact egg, or on sheets of cell surface complex. These emission spectra were then compared with the calculated standards and assigned pseudocolor values for display. The META spectrum was also reproduced for single cortical granules and found to replicate the standards accurately (data not shown). We found that LysoSensor protonation would basify the pH of the organelle after several minutes of incubation, so the samples were recorded within 2 min.
LysoSensor pH based determinations were also made fluorometrically in a fluoro Max-2 fluorimeter (Jobin Yvon-Spex Instruments S.A., Edison, NJ) equipped with Datamax for Windows software. In these recordings, the excitation wavelength was set at 350 nm, the scans were recorded from 365 to 700 nm, at 0.5-nm intervals with an integration time of 0.5 s. The pH standards were recorded in duplicate without detectable variation between replicates. Isolated cell surface complex was then added to the LysoSensor under the same conditions and assayed. Each of these methods of pH determination of cortical granules resulted in the same general values, although the fluorimeter determinations give the greatest resolution.
RESULTS
The Cortical Granule Is Acidic
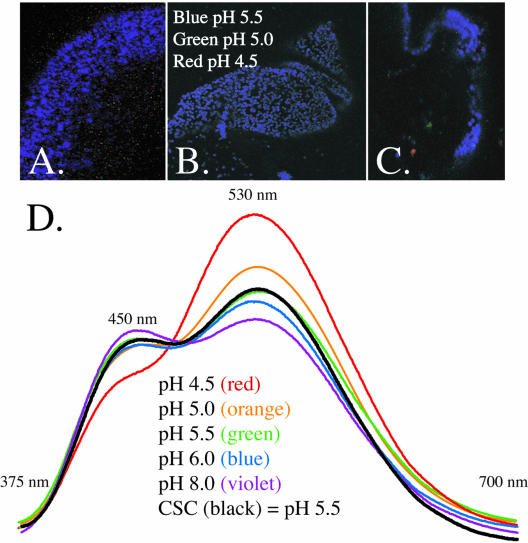
Three methods of determining the pH of the cortical granules converged on the same value of pH 5.5 (Figure 1). Location of LysoSensor in situ enabled definitive identification of the cortical granules and by use of the Zeiss 510 META system of imaging, enabled an accurate spectral array determination (Figure 1, A–C). Almost all of the detectable cortical granules in the intact egg are approximately pH 5.5 (±0.25 U), and the majority of cortical granules in isolated cell surface complex also fall in the pH 5.5 spectral range. On a population basis in the fluorimeter, the cortical granules most accurately followed the biphasic spectrum of pH 5.5 as well (Figure 1D). The least accurate method of pH determination was by micro-pH electrode. In these experiments, eggs were activated in nonbuffered salts, in the presence of amiloride, or in Na+-free seawater to block Na+/H+ exchange. The pH of the exudate was then monitored and the proton concentration calculated based on known organelle dimensions. This value consistently yielded a value of pH 5.3 (data not shown), approximating the pH 5.5 value of the LysoSensor.
Figure 1.
Spectral profile of LysoSensor Yellow/Blue DND-160 was determined in situ by a Zeiss 510 equipped for META spectral imaging (A–C) and by a fluorimeter (D). After standard recordings at various pHs, the LysoSensor spectrum was determined in situ in whole eggs slightly compressed to reveal the cortical granules at the surface of the cell (A), or in isolated cell surface complex consisting of cortical granules attached to the plasma membrane (B and C). Pseudocolor was assigned to pixels based on an overlay with standards. (D) Isolated cell surface complex was also examined in a fluorimeter after treatment with LysoSensor. In comparison with standards of defined pH, the cortical granules closely overlay the pH 5.5 standard.
CGSP1 Is Inactive in the Acidic Environment of the Cortical Granule
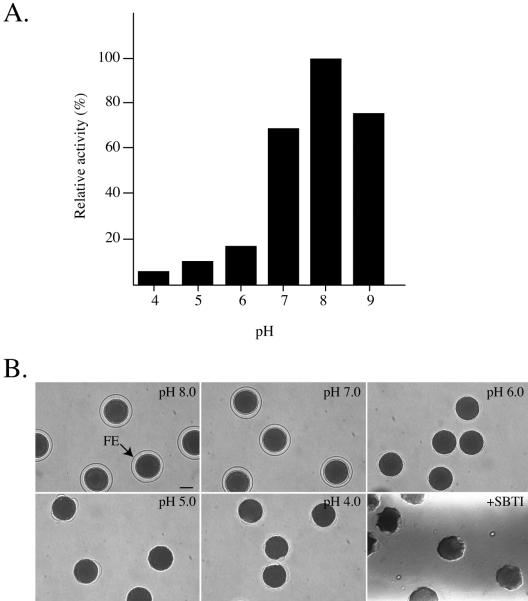
To examine pH-dependent protease activity, column-purified CGSP1 was assayed in vitro by using BAEE as a substrate. CGSP1 showed maximal activity at pH 8.0 (Figure 2A), which had been reported previously for protease activity found in cortical granule exudate as well as ammonium sulfate-precipitated protease activity (Vacquier et al., 1972a; Sawada et al., 1984; Alliegro and Schuel, 1985; Lois et al., 1986). The protease activity dropped significantly (∼25%) when placed in pH 7.0 buffer, and at pH 6.0 the protease was only 20% active compared with that assayed at pH 8.0 ASW. At pH 5.0 and pH 4.0, activity was <10%. This lack of activity in acidic buffers is replicated in the phenotype of FE formation. When eggs are activated in low pH seawater, they demonstrated a phenotype similar to eggs activated in the presence of SBTI (Figure 2B). The eggs showed a uniform level of inhibition, and the formation of abnormal envelopes occurred similarly as eggs activated in the presence of the serine protease inhibitor SBTI. Eggs activated at any pH <7.0 exhibited a similar abnormal phenotype, which suggests that low pH suppresses proteolytic activity in vivo as well. Thus, we believe the cortical granule protease is inactive when stored in the cortical granules and becomes activated by autocatalysis once exposed to the normally basic seawater. This conclusion is supported by the finding of predominantly full-length, mature protease in the cortical granules by Western blot analysis and by zymography (Figure 5; Haley and Wessel, 1999).
Figure 2.
CGSP1 is inhibited at low pH. (A) Column-purified CGSP1 was assayed in vitro at different pH values by using BAEE as a substrate. (B) Brightfield images of live eggs activated with A23187 in low pH sea water form abnormal fertilization envelopes, similar to the phenotype displayed when eggs are activated in the presence of SBTI. Bar, 40 μm.
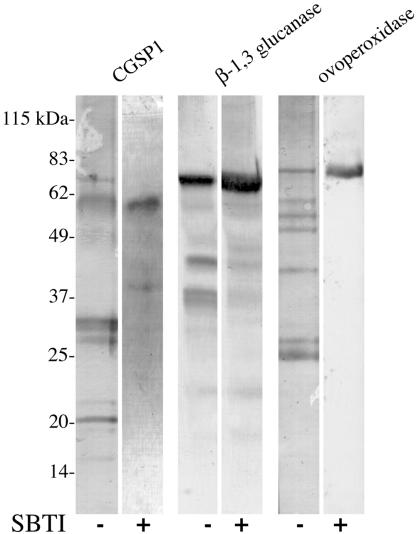
Figure 5.
Enzymatic contents of the cortical granule are cleaved at fertilization. Immunoblot analysis of cortical granule exudate using antibodies generated against the enzymes of the granule. The enzymes are cleaved in a protease-dependent manner after fertilization. SBTI concentration 100 μg/ml.
Fertilization Envelope Structural Proteins Derived from the Cortical Granules Are Not Cleaved by CGSP1
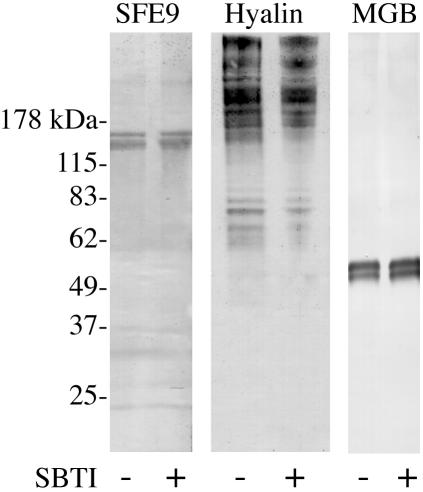
We examined the molecular constituents of the cortical granule exudate of eggs activated under normal conditions and in the presence of the trypsin inhibitor SBTI. By Western blot analysis, the structural proteins of the FE exhibit no difference in migration patterns when CGSP1 is blocked that would indicate a functional cleavage event at fertilization (Figure 3). SFE9 occurs as a doublet at approximately its full size of 127.7 kDa in both the presence and absence of trypsin inhibitor. MGB (Conner, unpublished data; Wong and Wessel, 2004) is also unaltered in the presence of SBTI as are the other known structural proteins of the granule, proteoliaisin and SFE 1 (our unpublished data). The migration pattern of hyalin, although present as a smear as reported previously (Adelson and Humphreys, 1988; Wessel et al., 1998), is also not affected by the presence of active CGSP1. Although it is possible that the cortical granules contain an alternative protease activity in addition to CGSP1 and that it is this hypothetical protease that is responsible for cleavage of the structural proteins, protease activity in the cortical granules not ascribable to CGSP1 has not been detected (Shapiro et al., 1989; Haley and Wessel, 1999).
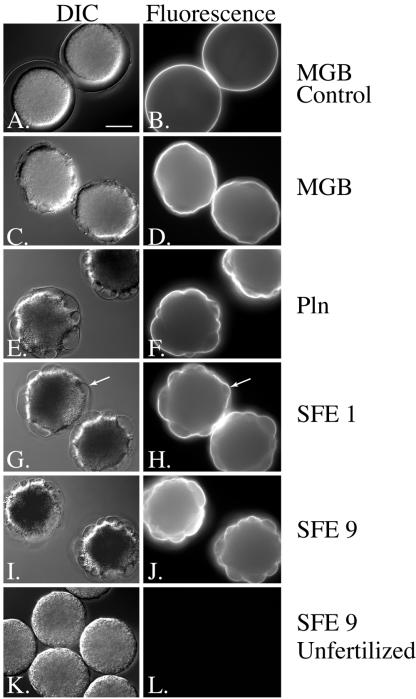
Figure 3.
Immunoblot analysis of cortical granule exudate in the presence and absence of protease inhibitors indicates that these proteins are not cleaved by CGSP1. SBTI concentration 100 μg/ml.
The targeting of the structural proteins to the FE is also not altered by protease activity. When eggs are activated either in the presence or absence of protease inhibitors, the structural proteins still localize to the envelope as normal. Although we cannot isolate FEs from protease-null eggs to test this conclusion biochemically, we are able to immunolocalize each FE protein in either intact live, intact fixed, or intact permeabilized cells (Figure 4). We find that after fertilization or artificial activation of the egg, each of the cortical granule-derived FE structural proteins is targeted specifically to the FE in the presence or absence of protease inhibitors. Thus, we conclude that the protein interactions required for formation of the FE work independently of the activity of the cortical granule serine protease. These data also show that the major constituents of the cortical granules, the structural proteins of the fertilization envelope, are not natural targets of CGSP1 at fertilization.
Figure 4.
Fertilization envelope forms but does not detach in protease-null cells. However, the targeting of the structural proteins of the envelope is not affected by the absence of protease activity. Arrows indicate the FE on fixed cells. Bar, 40 μm.
We were surprised that the cortical granule protease is so selective in its activity because we hypothesized that the extensive fragmentation of FE structural proteins would be the result of CGSP1. Yet, with multiple serine protease inhibitors we were unable to detect any FE protein cleavage by CGSP1. Indeed, this result is now more understandable because we find that CGSP1 is active only in basic conditions and the cortical granules are pH 5.5. We now postulate that the proprotein convertases that process proteins in the trans-Golgi network (Seidah and Chretien, 1997) are responsible for the cleavage of the structural proteins and that this cleavage would occur before entry into the cortical granule. For at least one cortical granule structural protein, MGB, this seems to be the case. One of the fragments of MGB was analyzed by N-terminal sequence (Dr. Robert Donnelly, The University of Medicine & Dentistry of New Jersey, Rutgers, NJ peptide sequencing facility), and based on the sequence flanking the cleavage site it is likely that the protein is processed by a protease of the proprotein convertase (PC1/3) family.
Cortical Granule Enzymes Are Substrates of CGSP1
Cortical granules contain three known enzyme activities, each of which seem to be cleaved by the cortical granule protease (Figure 5). The protease was previously shown by zymography to autocatalyze from its full-length, 61-kDa form to smaller, active forms of ∼35, 30, and 25 kDa (Alliegro and Schuel, 1985; Haley and Wessel, 1999). However, if the eggs are activated in the presence of SBTI, the protease does not autocatalyze and is found in the exudate only in the 61-kDa proform. This result supports the hypothesis that CGSP1 is stored in the cortical granule and is then secreted as a full-length, inactive zymogen that undergoes autocatalysis after cortical granule exocytosis. It also shows that the protease inhibitor treatment used in these experiments effectively blocked the CGSP1 activation, as does lowering the pH of the medium.
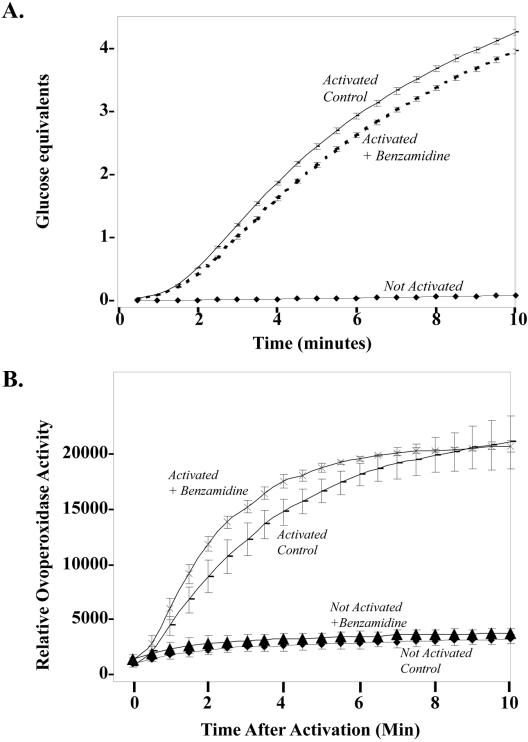
In addition to the protease, β-1,3 glucanase also seems to be proteolyzed by CGSP1 in the exudate (Figure 5). Full-length, β-1,3 glucanase is ∼68 kDa (Bachman and McClay, 1996), whereas in the exudate, several proteolytic fragments are visible, including bands at 43 kDa and a doublet at 37 kDa. If SBTI is included in the exudate, however, these proteolytic products diminish significantly with a compensatory increase in the full-length form, suggesting that CGSP1 activity is necessary for this hydrolysis to take place. In vitro assays measuring glucanase activity in cortical granule exudate demonstrate that the β-1,3 glucanase is more active by ∼15% when the protease is also active (Figure 7A). Although detailed biochemical analysis of glucanase structure and function has yet to be completed, either the proteolytic forms of glucanase have an overall 15% increase in activity, or the protease cleaves a yet undefined inhibitor of the glucanase, contributing to the activity difference observed.
Figure 7.
Protease activity has opposing effects on ovoperoxidase and β-1,3 glucanase. (A) Egg β-1,3 glucanase is more active when CGSP1 is active. In vitro glucanase activity was detected by measuring laminarin hydrolysis by β-1,3 glucanase in the presence or absence of the protease inhibitor benzamidine. (B) Protease activity suppresses ovoperoxidase activity initially after fertilization. In vivo ovoperoxidase was measured by an Amplex Red-based fluorescence assay.
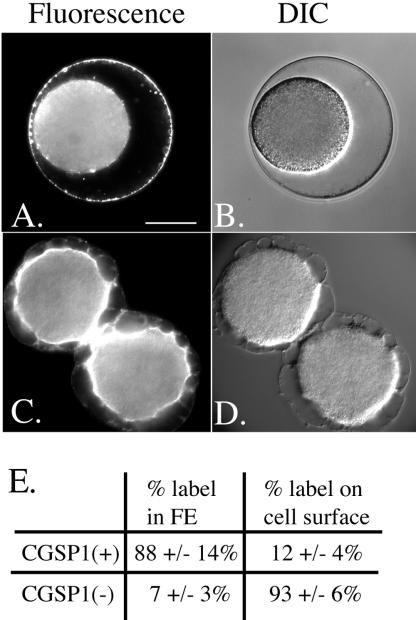
Another substrate of CGSP1 is ovoperoxidase. The observed molecular mass of S. purpuratus ovoperoxidase stored in the granule is ∼70 and 50 kDa (Deits et al., 1984; LaFleur et al., 1998). In the exudate of activated eggs, the 70-kDa form as well as six proteolytic fragments are observed at 60, 55, 50, 40, and a doublet at 25 kDa (Figure 5). If protease activity is inhibited, however, only the 70-kDa form is detected, suggesting that like the other enzymes of the cortical granule, ovoperoxidase is normally hydrolyzed by CGSP1 after exocytosis. One function of the protease activity may be to delineate the site of ovoperoxidase activity in vivo. After fertilization, control eggs have nearly 90% of their ovoperoxidase in the FE, as determined by in situ immunolocalization. In protease null cells, however, only 7% of the detectable ovoperoxidase is in the FE, and the remainder of the detectable ovoperoxidase is present on the surface of the fertilized egg (Figure 6). Thus, either the protease cleaves a cell surface protein that ovoperoxidase adheres to, or cleavage of ovoperoxidase enables it to incorporate into the FE. Under normal conditions, ovoperoxidase is specifically targeted to the FE via a calcium-dependent interaction with proteoliaisin (Weidman et al., 1987). Because proteoliaisin is not detectably cleaved by CGSP1 and is targeted to the FE specifically, even in the absence of protease activity, we can exclude an interpretation that misdirection of ovoperoxidase is a result of alteration of proteoliaisin function directly (Figure 4F).
Figure 6.
Protease activity is necessary for targeting ovoperoxidase to the fertilization envelope. Ovoperoxidase localizes to the fertilization envelope after cortical granule exocytosis in fixed embryos (A and B). However, when CGSP1 is inhibited, the ovoperoxidase is mistargeted to the egg plasma membrane (C and D). (E) Quantification of ovoperoxidase in the fertilization versus the plasma membrane. Bar, 40 μm.
To determine the effect proteolysis has on ovoperoxidase activity, eggs were incubated in Amplex reagent and then activated in the presence or absence of SBTI (Figure 7B). Ovoperoxidase activity was measured in real time by the production of a fluorogenic product, resorufin, for 10 min, a time frame that encompasses all the natural ovoperoxidase activity in FE processing. When CGSP1 activity is blocked, the overall ovoperoxidase activity is greater than in control eggs, suggesting that the protease directly limits ovoperoxidase activity at fertilization.
DISCUSSION
At fertilization, the proteins of the cortical granule are rapidly arranged into a new and highly organized matrix. We have investigated what role, if any, the cortical granule protease has in the molecular interactions that take place during this process. The results shown here demonstrate that the cortical granule protease has a very limited target population and does not cleave all the proteins of the granule. The protease does not cleave the structural proteins that are stored in the granule before fertilization, and targeting of the structural proteins to the envelope is independent of protease activity. Electron microscopic analysis of eggs fertilized in the presence of SBTI shows that although the envelope still forms, it lacks the multilayered organization typical of mature envelopes (Longo and Schuel, 1973). Perhaps this substructural difference reflects the lack of ovoperoxidase incorporation into the FE.
Although the structural proteins each contain multiple recognition sites for protease cleavage, their conformation or their complexing with other proteins must make them inaccessible to CGSP1 cleavage. Although we cannot exclude the possibility that the cortical granules contain another protease activity in addition to CGSP1 that is responsible for the structural protein cleavage, neither we nor other investigators have detected protease activity not ascribable to CGSP1 (Shapiro et al., 1989; Haley and Wessel, 1999). The extreme fragmentation seen in structural proteins of the cortical granule must instead result from protease activity elsewhere in the secretory pathway. One such candidate mechanism is the proprotein convertases that process proteins in the trans-Golgi network (Seidah and Chretien, 1997) and for at least one cortical granule structural protein, MGB, the data support this contention. We analyzed one of the fragments of MGB by N-terminal sequence (Dr. Robert Donnelly, The University of Medicine & Dentistry of New Jersey peptide sequencing facility), and the sequence flanking the cleavage site is a conserved site for the proprotein convertase (PC1/3) family. The function of this processing has yet to be determined, but given the numerous repeated motifs that characterize all of the structural proteins, variable processing might serve to enhance the organization of the envelope.
In contrast to the structural proteins, each of the known enzymes of the cortical granule are cleaved by CGSP1. By analysis of ovoperoxidase activity released from the egg in the presence of protease inhibitors, it seems that ovoperoxidase is negatively regulated by CGSP1. Negative regulation may be important to protect the egg surface from nonspecific, potentially harmful cross-linking activity, until ovoperoxidase is tethered to proteoliaisin. The regulation of ovoperoxidase activity seems to be important, because proteoliaisin seems to specifically target cross-linking activity of ovoperoxidase to the proteins of the FE (Somers and Shapiro, 1991). Proteoliaisin has also been suggested to protect ovoperoxidase from proteolytic digestion, by envelysin, the hatching enzyme (Nomura et al., 1997). It has not yet been determined whether ovoperoxidase is also protected by proteoliaisin from cleavage by CGSP1, but one of the cleavage sites in ovoperoxidase is a conserved serine protease site (LaFleur et al., 1998), likely that of CGSP1 because of its arginyl preference (Green, 1986). Such a scenario would suggest that the protease inhibits the activity of only free, and potentially harmful, ovoperoxidase. Indeed, inhibition of protease activity results in the mistargeting of ovoperoxidase to the fertilized egg surface.
The β-1,3 glucanase enzyme is also a CGSP1 target, but in this case overall enzymatic activity is increased. Based on the Western blot analysis in which the glucanase is not completely cleaved, we believe that proteolysis of the enzyme seems to enhance the β-1,3 glucanase activity. It has been reported that an extracellular glucanase of the cellulolytic bacterium Cellulomonas fimi is cleaved by a secreted serine protease, and proteolysis impairs the ability of the glucanase to bind to its substrate (Gilkes et al., 1989). Likewise, CGSP1 may have a role in modulating glucanase association with its substrate.
The enzymatic function of β-1,3 glucanase at fertilization is poorly understood at present. The glucanase could cleave sugars from extracellular glycoproteins upon exocytosis to release free glucose, or it could be involved in the elevation of the FE. Because many CUB domains are found in the FE structural proteins, and these domains function as a lectin upon homodimerizing, perhaps glucanase cleaves the CUB ligand and enables FE detachment from the cell surface. Alternatively, it may function in a pathogen defense system, as has been reported in other invertebrates (Seki et al., 1994). A β-1,3 glucanase the bears close similarity to the S. purpuratus glucanase has been identified in the earthworm Eisenia foetida (Beschin et al., 1998). This glucanase is involved in the activation of the prophenoloxidase cascade, which is a defense mechanism that recognizes microbes, including yeast and Gram-negative bacteria. In addition, the proprophenoloxidase cascade is activated by proteolysis by serine proteases. The in vitro substrate of β-1,3 glucanase is long-chain oligosaccharides, which are cleaved at internal and external β-1,3 linkages.
The cortical granule protease also cleaves itself. Our data suggests that CGSP1 is inactive during storage in the granule until fertilization and then autocatalyzes to become active upon exocytosis. Western blot analysis shows that the protease is secreted in its full-length, 61-kDa form, so it has not been cleaved at its predicted activation site between the residues R333 and I334 to become proteolytically active. What keeps CGSP1 quiescent until fertilization? Previous studies have reported that pH optima of the cortical granule protease is 8.0, the same pH as sea water (Sawada et al., 1984; Alliegro and Schuel, 1985; Lois et al., 1986), whereas the pH of the granule was thought to be acidic. When eggs are activated in a low pH environment (<pH 7.0 ASW), the eggs exhibit the same phenotype as if protease activity had been blocked by SBTI. At low pH, the ionization state of the protease is likely disrupted so that it cannot cleave its substrates. A similar situation has been reported for other serine proteases, including acrosin and chymase, proteases that are kept inactive by the low pH of their granules during storage and then become active upon exocytosis (McEuen et al., 1995). Therefore, it is likely that the protease is secreted in its full-length form and then autoactivates to cleave its biological substrates, including proteins on the cell surface and in the granules. Showing here that the pH of the cortical granule is uniformly at pH 5.5 enables a single regulatory step that inhibits protease activity until its release to the extracellular environment by secretion.
Acknowledgments
We thank Craig Lassey and Doug Fishkind for generous assistance in META spectral imaging for in situ LysoSensor determinations on the Zeiss 510 META confocal microscope. In addition, we thank colleagues at the Providence Institute of Molecular Oogenesis (Providence, RI) for helpful discussions, especially Judith Nathanson for contributions in figure preparation. This work was supported by grants from the National Institutes of Health and National Science Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–11–0843. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–11–0843.
Abbreviations used: ASW, artificial sea water; CGSP1, cortical granule serine protease 1; DTT, dithiothreitol; FE, fertilization envelope; SBTI, soybean trypsin inhibitor.
References
- Adelson, D.L., and Humphreys, T. (1988). Sea urchin morphogenesis and cell-hyalin adhesion are perturbed by a monoclonal antibody specific for hyalin. Development 104, 391–402. [DOI] [PubMed] [Google Scholar]
- Alliegro, M.C., and Schuel, H. (1985). Characterization of soybean trypsin inhibitor sensitive protease from unfertilized sea urchin eggs. Biochemistry 24, 3926–31. [DOI] [PubMed] [Google Scholar]
- Bachman, E.S., and McClay, D.R. (1996). Molecular cloning of the first metazoan β-1,3 glucanase from eggs of the sea urchin Strongylocentrotus purpuratus. Proc. Natl. Acad. Sci. USA 93, 6808–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beschin, A., Bilej, M., Hanssens, F., Raymakers, J., Van Dyck, E., Revets, H., Brys, L., Gomez, J., De Baetselier, P., and Timmermans, M. (1998). Identification and cloning of a glucan- and lipopolysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J. Biol. Chem. 273, 24948–24954. [DOI] [PubMed] [Google Scholar]
- Bork, P., and Beckmann, G. (1993). The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol. 231, 539–545. [DOI] [PubMed] [Google Scholar]
- Carroll, E.J., Jr., and Epel, D. (1975). Isolation and biological activity of the proteases released by sea urchin eggs following fertilization. Dev. Biol. 44, 22–32. [DOI] [PubMed] [Google Scholar]
- Deits, T., Farrance, M., Kay, E.S., Medill, L., Turner, E.E., Weidman, P.J., and Shapiro, B.M. (1984). Purification and properties of ovoperoxidase, the enzyme responsible for hardening the fertilization membrane of the sea urchin egg. J. Biol. Chem. 259, 13525–13233. [PubMed] [Google Scholar]
- Epel, D., Weaver, A.M., and Mazia, D. (1970). Methods for removal of the vitelline membrane of sea urchin eggs. I. Use of dithiothreitol (Cleland Reagent). Exp. Cell Res. 61, 64–68. [DOI] [PubMed] [Google Scholar]
- Epel, D., Weaver, A.M., Muchmore, A.V., and Schimke, R.T. (1969). β-1,3-Glucanase of sea urchin eggs: release from particles at fertilization. Science 163, 294–296. [DOI] [PubMed] [Google Scholar]
- Fodor, E.J., Ako, H., and Walsh, K.A. (1975). Isolation of a protease from sea urchin eggs before and after fertilization. Biochemistry 14, 4923–4927. [DOI] [PubMed] [Google Scholar]
- Foerder, C.A., and Shapiro, B.M. (1977). Release of ovoperoxidase from sea urchin eggs hardens the fertilization membrane with tyrosine crosslinks. Proc. Natl. Acad. Sci. USA 74, 4214–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes, N.R., Kilburn, D.G., Miller, R.C., Jr., and Warren, R.A. (1989). Structural and functional analysis of a bacterial cellulase by proteolysis. J. Biol. chem. 264, 17802–17808. [PubMed] [Google Scholar]
- Green, J.D. (1986). Sea urchin egg-cortical granule protease has arginyl proteolytic specificity. Gamete Res. 15, 227–236. [Google Scholar]
- Hagström, B.E. (1956). Studies on polyspermy in sea urchins. Arkiv. Fortschr. Zool. 10, 307–315. [Google Scholar]
- Haley, S.A., and Wessel, G.M. (1999). The cortical granule serine protease CGSP1 of the sea urchin, Strongylocentrotus purpuratus, is autocatalytic and contains a low-density lipoprotein receptor-like domain. Dev. Biol. 211, 1–10. [DOI] [PubMed] [Google Scholar]
- Hall, H.G., and Vacquier, V.D. (1982). The apical lamina of the sea urchin embryo: major glycoproteins associated with the hyaline layer. Dev. Biol. 89, 168–78. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Hirohashi, N., and Lennarz, W.J. (1998). The 350-kDa sea urchin egg receptor for sperm is localized in the vitelline layer. Dev. Biol. 204, 305–15. [DOI] [PubMed] [Google Scholar]
- Hylander, B.L., and Summers, R.G. (1982). An ultrastructural immunocytochemical localization of hyalin in the sea urchin egg. Dev. Biol. 93, 368–380. [DOI] [PubMed] [Google Scholar]
- Kinsey, W.H. (1986). Pureification and properties of the egg plasma membrane. Methods Cell Biol. 27, 139–152. [DOI] [PubMed] [Google Scholar]
- Klebanoff, S.J., Foerder, C.A., Eddy, E.M., and Shapiro, B.M. (1979). Metabolic similarities between fertilization and phagocytosis. Conservation of a peroxidatic mechanism. J. Exp. Med. 149, 938–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur, G.J., Jr., Horiuchi, Y., and Wessel, G.M. (1998). Sea urchin ovoperoxidase: oocyte-specific member of a heme-dependent peroxidase superfamily that functions in the block to polyspermy. Mech. Dev. 70, 77–89. [DOI] [PubMed] [Google Scholar]
- Lois, A.F., Lackey, D.A., and Carroll, E.J., Jr. (1986). Parital purification and characterization of sperm receptor hydrolase, a cortical granule proteoesterase, from eggs of the sea urchin Strongylocentrotus purpuratus. Gamete Res. 14, 307–321. [Google Scholar]
- Longo, F.J., and Schuel, H. (1973). An ultrastructural examination of polyspermy induced by soybean trypsin inhibitor in the sea urchin Arbacia punctulata. Dev. Biol. 34, 187–199. [DOI] [PubMed] [Google Scholar]
- McEuen, A.R., Sharma, B., and Walls, A.F. (1995). Regulation of the activity of human chymase during storage and release from mast cells: the contributions of inorganic cations, pH, heparin and histamine. Biochim. Biophys. Acta 1267, 115–121. [DOI] [PubMed] [Google Scholar]
- Murray, G., Reed, C., Marsden, M., Rise, M., Wang, D., and Burke, R.D. (2000). The αBβC integrin is expressed on the surface of the sea urchin egg and removed at fertilization. Dev. Biol. 227, 633–647. [DOI] [PubMed] [Google Scholar]
- Nomura, K., Shimizu, T., Kinoh, H., Sendai, Y., Inomata, M., and Suzuki, N. (1997). Sea urchin hatching enzyme (envelysin): cDNA cloning and deprivation of protein substrate specificity by autolytic degradation. Biochemistry 36, 7225–7238. [DOI] [PubMed] [Google Scholar]
- Peeler, M. T., Chambers, S. A., Wang, C. Y., and D. R., M. (1987). Stage-specific expression of beta-1, 3-glucanase in sea urchin embryos and hybrids. J. Exp. Zool. 244, 215–222. [DOI] [PubMed] [Google Scholar]
- Sawada, H., Miura, M., Yokosawa, H., and Ishii, S. (1984). Purification and characterization of trypsin-like enzyme from sea urchin eggs: substrate specificity and physiological role. Biochem. Biophys. Res. Commun. 121, 598–604. [DOI] [PubMed] [Google Scholar]
- Schuel, H., Wilson, W.L., Chen, K., and Lorand, L. (1973). A trypsin-like proteinase localized in cortical granules isolated from unfertilized sea urchin eggs by zonal centrifugation. Role of the enzyme in fertilization. Dev. Biol. 34, 175–186. [DOI] [PubMed] [Google Scholar]
- Schwert, G.W., and Takenaka, Y. (1955). A spectrophotometric determination of trypsin and chymotrypsin. Biochim. Biophys. Acta 16, 570–575. [DOI] [PubMed] [Google Scholar]
- Seidah, N.G., and Chretien, M. (1997). Eukaryotic protein processing: endoproteolysis of precursor proteins. Curr. Opin. Biotechnol. 8, 602–607. [DOI] [PubMed] [Google Scholar]
- Seki, N., Muta, T., Oda, T., Iwaki, D., Kuma, K., Miyata, T., and Iwanaga, S. (1994). Horseshoe crab (1,3)-β-D-glucan-sensitive coagulation factor G. A serine protease zymogen heterodimer with similarities to β-glucan-binding proteins. J. Biol. Chem. 269, 1370–1374. [PubMed] [Google Scholar]
- Shapiro, B.M., Somers, C., and Weidman, P.J. (1989). Extracellular remodeling during fertilization. In: Cell Biology of Fertilization (ed. H. Schatten and G. Schatten), San Diego: Academic Press, 251–276.
- Somers, C.E., Battaglia, D.E., and Shapiro, B.M. (1989). Localization and developmental fate of ovoperoxidase and proteoliaisin, two proteins involved in fertilization envelope assembly. Dev. Biol. 131, 226–235. [DOI] [PubMed] [Google Scholar]
- Somers, C.E., and Shapiro, B.M. (1991). Functional domains of proteoliaisin, the adhesive protein that orchestrates fertilization envelope assembly. J. Biol. Chem. 266, 16870–16875. [PubMed] [Google Scholar]
- Talbot, C.F., and Vacquier, V.D. (1982). The purification and characterization of an exo-β (1 going to 3)-glucanohydrolase from sea urchin eggs. J. Biol. Chem. 257, 742–746. [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier, V.D., Epel, D., and Douglas, L.A. (1972a). Sea urchin eggs release protease activity at fertilization. Nature 237, 34–36. [DOI] [PubMed] [Google Scholar]
- Vacquier, V.D., Tegner, M.J., and Epel, D. (1972b). Protease activity establishes the block against polyspermy in sea urchin eggs. Nature 240, 352–353. [DOI] [PubMed] [Google Scholar]
- Weidman, P.J., Kay, E.S., and Shapiro, B.M. (1985). Assembly of the sea urchin fertilization membrane: isolation of proteoliaisin, a calcium-dependent ovoperoxidase binding protein. J. Cell Biol. 100, 938–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidman, P.J., Teller, D.C., and Shapiro, B.M. (1987). Purification and characterization of proteoliaisin, a coordinating protein in fertilization envelope assembly. J. Biol. Chem. 262, 15076–15084. [PubMed] [Google Scholar]
- Wessel, G.M. (1995). A protein of the sea urchin cortical granules is targeted to the fertilization envelope and contains an LDL-receptor-like motif. Dev. Biol. 167, 388–397. [DOI] [PubMed] [Google Scholar]
- Wessel, G.M., Berg, L., Adelson, D.L., Cannon, G., and McClay, D.R. (1998). A molecular analysis of hyalin–a substrate for cell adhesion in the hyaline layer of the sea urchin embryo. Dev. Biol. 193, 115–126. [DOI] [PubMed] [Google Scholar]
- Wessel, G.M., Brooks, J.M., Green, E., Haley, S., Voronina, E., Wong, J., Zaydfudim, V., and Conner, S. (2001). The biology of cortical granules, vol 209. In: International Review of Cytology (ed. K.W. Jeon), San Diego: Academic Press, 117–206. [DOI] [PubMed] [Google Scholar]
- Wessel, G.M., Conner, S., Laidlaw, M., Harrison, J., and LaFleur, G.J., Jr. (2000). SFE1, a constituent of the fertilization envelope in the sea urchin is made by oocytes and contains low-density lipoprotein-receptor-like repeats. Biol. Reprod. 63, 1706–1712. [DOI] [PubMed] [Google Scholar]
- Wong, J., and Wessel, G.M. (2004). Major components in a sea urchin block to polyspermy are structurally and functionally conserved. Evol. Dev. 6, 134–153. [DOI] [PubMed] [Google Scholar]
- Zhou, M., Diwu, Z., Panchuk-Voloshina, N., and Haugland, R.P. (1997). A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 253, 162–168. [DOI] [PubMed] [Google Scholar]