Abstract
In ciliary and flagellar axonemes, various discrete structures such as inner and outer dynein arms are regularly arranged on the outer doublet microtubules. Little is known about the basis for their regular arrangement. In this study, proteins involved in the attachment of inner-arm dyneins were searched by a microtubule overlay assay on Chlamydomonas mutant axonemes. A 58-kDa protein (p58) was found ∼80% diminished in the mutants ida6 and pf3, both lacking one (species e) of the seven inner-arm species (a–g). Analysis of its cDNA indicated that p58 is homologous to tektin, a protein that was originally found in sea urchin and thought to be crucial for the longitudinal periodicity of the doublet microtubule. Unlike sea urchin tektin, which is a component of protofilament ribbons that occur after Sarkosyl treatment of axonemes, p58 was not contained in similar Sarkosyl-resistant ribbons from Chlamydomonas axonemes. Immunofluorescence microscopy showed that p58 was localized uniformly along the axoneme and on the basal body. The p58 signal was reduced in ida6 and pf3. These results suggest that a reduced amount of p58 is sufficient for the production of outer doublets, whereas an additional amount of it is involved in inner-arm dynein attachment.
INTRODUCTION
Eukaryotic cilia and flagella are elaborate molecular machines constructed of >250 proteins (Dutcher, 1995). Their core structure, the axoneme, displays an evolutionarily conserved, ninefold symmetrical arrangement of doublet microtubules, with two singlet microtubules at the center (Gibbons, 1981). Outer-arm and inner-arm dyneins, radial spokes, and nexin links are attached at precise positions on the A-tubule of the outer doublet, with longitudinal periodicity characteristic of each component (Goodenough and Heuser, 1985; Mastronarde et al., 1992). In Chlamydomonas axonemes, outer-arm dynein, a large protein complex composed of three heavy chains and ∼10 associated proteins, is arranged in a single row along the length of the outer doublet microtubules with a 24-nm spacing. In contrast, inner-arm dynein exists as seven discrete species, each of which is composed of one or two heavy chains and two to five associated proteins (Kamiya, 2002) and localized at a specific locus within the 96-nm repeat length of the outer doublet (Goodenough and Heuser, 1985; Mastronarde et al., 1992). Because the spacings of inner-arm and outer-arm dynein arrangement are much longer than the 8-nm spacing of the microtubule surface lattice, some proteins may possibly function as rulers to determine their arrangement. In fact, a protein complex (ODA-DC) has been found that may potentially function as such a ruler for the arrangement of outer-arm dynein (Takada and Kamiya, 1994; Koutoulis et al., 1997; Wakabayashi et al., 2001; Takada et al., 2002; Casey et al., 2003); its two major coiled-coil subunits seem to produce a heterodimer, whose total length can be as long as the 24-nm spacing of the outer arm arrangement.
In contrast to the progress in the studies on the outer-arm arrangement, almost nothing is known about how inner-arm dyneins are attached to specific loci on the outer doublet. Regarding the 96-nm repeat length, however, a protein called tektin has been suggested to play a crucial role. It was first discovered as a component of stable protofilaments in detergent-fractionated sea urchin sperm flagella (Linck, 1976). Those protofilaments (called “the protofilament ribbons”) were thought to be derived from the junctional region between A- and B-tubules of the outer doublet (Linck, 1976; Stephens et al., 1989). In sea urchin sperm flagella, tektin comprises three isoforms, tektin A, B, and C (Linck and Langevin, 1982; Linck and Stephens, 1987), each with structural features common to intermediate filament proteins (Norrander et al., 1992, 1996; Chen et al., 1993). Experiments using Sarkosyl and urea extraction suggested that tektin A and B form a fiber that runs along axonemes, and tektin C is attached to it (Linck and Stephens, 1987; Pirner and Linck, 1994). Their structural properties and putative location in the A-tubule suggest that tektin may function as the ruler that specifies the complex spacing of axonemal components (Nojima et al., 1995; Norrander et al., 1996). In experiments with sea urchin embryos grown under various conditions, tektin A was found to be synthesized in proportion with the ciliary length (Stephens, 1989; Norrander et al., 1995). This observation supports that a fixed amount of tektin is incorporated into the unit length of axoneme as an integral component. However, it is unknown whether tektin is actually involved in the periodic attachment of particular axonemal components.
In the present study, we aimed to identify proteins involved in the attachment of inner-arm dyneins to the outer doublet microtubules in Chlamydomonas axonemes. We searched for such proteins assuming that those proteins can bind to microtubules and may be diminished in mutant axonemes that lack inner-arm dyneins. We identified a 58-kDa protein (p58) that satisfies those conditions. Strikingly, p58 was found to be a tektin homologue. We report here the cloning and characterization of p58 and discuss the potential function of p58 in inner-arm dynein assembly.
MATERIALS AND METHODS
Strains and Culture
The strains used in this study are listed in Table 1. Cells were grown at 25°C in Tris-acetate/phosphate medium with aeration on a 12-h/12-h light/dark cycle.
Table 1.
Mutants used in this study
| Strain | Protein | Motility phenotype | Flagellar lengtha (μm) | Missing structuresb | References |
|---|---|---|---|---|---|
| Wild type | 11.1±1.9 | ||||
| oda1 | DC2 | Slow swimming | 11.4±2.0 | Outer arm | Kamiya, 2002 |
| ida1 | 1αHC | Slow swimming | 10.9±1.6 | f | Kamiya, 2002, Kamiya et al., 1991a |
| ida2 | 1βHC | Slow swimming | 11.0±1.2 | f | Kamiya, 2002, Kamiya et al., 1991a |
| ida3 | Slow swimming | 10.2±1.8 | f | Kamiya, 2002, Kamiya et al., 1991a | |
| ida4 | p28 | Slow swimming | 9.4±1.6 | acd | Kamiya, 2002, Kamiya et al., 1991a |
| ida5 | Actin | Slow swimming | 12.4±1.2 | acde | Kamiya, 2002, Kato et al., 1993a |
| ida6 | Slow swimming | 8.1±1.4 | ec | Kamiya, 2002, Kato et al., 1993a | |
| pf2 | DRC4 | Slow swimming | 8.9±1.3 | be (reduced), DRC (partial) | Rupp and Porter, 2003; Piperno et al., 1992a; Piperno et al., 1994; Gardner et al., 1994 |
| pf3 | DRC1d | Slow swimming | 9.1±1.4 | e, DRC (partial) | Kamiya, 2002; Piperno et al., 1992a; Piperno et al., 1994; Gardner et al., 1994 |
| pf13 | Paralyzed | c and outer arm | Kamiya, 2002 | ||
| pf14 | Rsp3 | Paralyzed | Radial spoke | Harris, 1989; Huang et al., 1981 | |
| pf18 | Paralyzed | Central pair | Harris, 1989 | ||
| pf22 | Paralyzed | 4.0±1.2 | Inner arms, outer arm | Kamiya, 2002; Piperno et al., 1992 | |
| pf23 | Paralyzed | 4.0±1.0 | acdf | Kamiya, 2002; Piperno et al., 1992 | |
| sup-pf-3 | Wild-type motility | 12.6±1.6 | e (reduced), DRC (partial) | Piperno et al., 1992; Piperno et al., 1994; Gardner et al., 1994 | |
| sup-pf-4 | DRC5d | Wild-type motility | 12.2±1.5 | DRC (partial) | Piperno et al., 1992; Piperno et al., 1994; Gardner et al., 1994 |
Mean ± SD.
Italic characters (abcdefg) indicate inner-arm subspecies.
Defects in DRC have not been investigated.
Putative gene products based on dikaryon rescue.
Isolation of Axonemes
Flagellar axonemes were isolated by the method of Witman et al. (1978). Flagella were suspended in HMDE buffer (30 mM HEPES, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM EGTA, pH 7.4) and extracted with 0.2% Nonidet P-40 in HMDE buffer. The protein concentration of the axonemal samples was determined using Advanced Protein Assay Reagent (Cytoskeleton, Denver, CO). SDS-PAGE of axonemal proteins was performed with 7.5 or 8% acrylamide gels by the method of Laemmli (1970). Gels were stained with Coomassie Brilliant Blue (CBB) or silver.
Microtubule Overlay Assay
Microtubule overlay assay was modified from Nakaseko et al. (1996). Tubulin was purified from porcine brain by cycles of temperature-dependent polymerization and phosphocellulose chromatography (Shelanski et al., 1973). The purified tubulin was labeled with 5-[5-(N-succinimidyloxycarbonyl)pentylamido]hexyl d-biotinamide (Dojindo Laboratories, Kumamoto, Japan) following the method of Hyman et al. (1991). The biotin-labeled tubulin was polymerized by incubation in the presence of 10% dimethyl sulfoxide in PEMG buffer (100 mM PIPES, 2 mM EGTA, 1 mM MgSO4, 1 mM GTP, pH 7.0) at 37°C for 15 min at 37°C. After taxol was added to the final concentration of 20 μM, the polymerization mixture was incubated for additional 15 min. The microtubules were pelleted by centrifugation at 350,000 × g for 12 min, resuspended in PEMG buffer, and stored on ice. The microtubules were used without shearing. Protein samples separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes and incubated in PEMT buffer (100 mM PIPES, 2 mM EGTA, 1 mM MgSO4, 0.1% Tween 20, pH 7.0) containing 5% skim milk at room temperature for 1 h. After three 5-min washes with PEMT buffer alone, the membranes were incubated with the biotin-labeled microtubules (50 μg/ml) in the same buffer at room temperature for 1 h. After three 10-min washes with the same buffer, microtubule-binding bands were detected using a VECTASTAIN ABC kit (Vector Laboratories, Burlingame, CA) and a TMB Peroxidase Substrate kit (Vector Laboratories).
Extraction of Axonemes
Extraction of axonemes with KCl of various concentrations was performed in HMDE buffer at 0°C for 30 min. The extracted axonemes were centrifuged at 14,000 × g for 10 min. Urea extraction was performed in MESH buffer (50 mM Mes, 1 mM EDTA, 0.1% 2-mercaptoethanol, pH 6.8) or in TED buffer (10 mM Tris-HCl, 0.1 mM EDTA, 1 mM DTT, pH 8.0) at 15°C for 1 h. Sarkosyl extraction was performed in TED buffer at 0°C for 1 h. The extracted axonemes were centrifuged at 100,000 × g for 1 h, and the resultant pellet was suspended in MESH or TED buffer.
Isolation of p58 and Peptide Sequencing
Wild-type flagellar axonemes were extracted twice with 0.6 M KCl in HMDE buffer and centrifuged as described above. The pellet was then extracted with 2 M urea in MESH buffer. The insoluble fraction was pelleted and solubilized in 8 M urea in MESH buffer. After debris in the sample were removed by centrifugation at 100,000 × g for 30 min, the sample was applied to a Hitrap SP FF column (Amersham Biosciences, Piscataway, NJ) equilibrated with MESH buffer containing 8 M urea. The bound proteins were eluted with a 0 to 0.5 M NaCl gradient produced in the same buffer. p58 was eluted at ∼0.15 M NaCl. The fractions containing p58 were subjected to SDS-PAGE. The 58-kDa polypeptide band was excised and in-gel digested with modified trypsin (Promega, Madison, WI) according to the method of Rosenfeld et al. (1992). The reaction was stopped by addition of 10% trifluoroacetic acid. The polypeptides were eluted from gels by soaking twice with 0.1% trifluoroacetic acid in 60% acetonitrile for 40 min at room temperature, followed by fractionation by reverse-phase chromatography on an μRPC C2/C18 SC 2.1/10 column (Amersham Biosciences). Fractions of four discrete peaks were collected and subjected to sequencing on an ABI 494 instrument (Applied Biosystems, Foster City, CA) at the National Institute for Basic Biology Center for Analytical Instruments (Okazaki, Japan).
Cloning and Sequencing of cDNA and p58 Gene
Total RNA was isolated from wild-type cells with TRIzol reagent (Invitrogen, Carlsbad, CA) 40 min after deflagellation by pH shock. First strand cDNAs were synthesized by Super Script II reverse transcriptase (Invitrogen) with oligo d(T)12–18 primer or 3′AP (3′ rapid amplification of cDNA ends [RACE] system; Invitrogen). Two sets of degenerate primers, designed based on two peptide sequences (NAHHWLADSAR and VDEAVLSIPTD), were used to amplify the intervening cDNA fragment. For 3′RACE polymerase chain reaction (PCR), two sense primers were designed from the sequence within the obtained cDNA fragment and nested PCR was performed using the primers and AUAP (3′RACE system; Invitrogen). A 495-base pair fragment corresponding to the 3′-untranslated region (UTR) was obtained by digestion with PvuII of the 3′RACE product. This fragment was used as a probe for screening a cDNA library (a gift from Dr. P. A. Lefebvre, University of Minnesota, St. Paul, MN). To obtain a genomic clone containing the p58 gene, a genomic library constructed from the wild-type strain was screened with the same probe. Positive clones were analyzed by Southern blot. A ∼9-kb NotI fragment containing the entire p58 gene was subcloned into pBluescript and partially sequenced.
Mapping of the p58 Gene
DNA was isolated from the C. reinhardtii standard laboratory strain 21gr and a strain S1-D2. To design primers for PCR-based mapping analysis, a ∼1-kb region within the p58 3′UTR was PCR amplified using the template DNA from each strain and sequenced. Three primers were designed within that region: P58-F (GGAGGGAGATGCGGTATGAACG, a common forward primer), P58-R1 (CACGGCCCCATAAATAGCGCGA, a 21gr specific reverse primer), and P58-R2 (GCGCCAGCCCAATATTCCGCT, an S1-D2 specific reverse primer). The underlined bases indicate sites of single nucleotide polymorphism between the two strains. PCR using the three primers yielded a 575-base pair product from the 21gr template and a 432-base pair product from the S1-D2 template, respectively. PCR-based mapping was kindly performed in Dr. Pete Lefebvre's laboratory by Dr. Masafumi Hirono, by using a previously described method (Kathir et al., 2003).
Northern and Southern Blot Analyses
To determine the size of the p58 transcript and also to detect its up-regulation upon deflagellation, total RNA was isolated from wild-type cells every 15 min after deflagellation. The RNA samples were analyzed by Northern blots by using the 495-base pair PvuII fragment as a probe. The α-1 tubulin genomic clone P-151 (Chlamydomonas Genetics Center, Department of Biology, Duke University, Durham, NC) and the 16S rRNA sequence amplified by PCR from the genomic DNA were used as control probes. To determine the number of gene copies, DNA was isolated from the wild-type strain and digested with NotI, SacI, and SalI. The DNA samples were analyzed by Southern blots by using as a probe a 417-base pair HinfI fragment of cDNA, corresponding to a partial coding region of p58.
Bacterial Expression of p58
The coding region of the cDNA was amplified by PCR with primers P58-B (CGGGATCCGATGCCGTCCTATAACCAT) and P58-H (GCAAGCTTTAGAAGGCGCCACCGCCC), which contained the recognition sites for BamHI and HindIII, respectively (underlined). The PCR product was ligated to the BamHI and HindIII sites of the bacterial expression vector pProExHTa (Invitrogen). The resulting fusion protein contained a His tag sequence at its N terminal. Expression of the fusion protein was induced by addition of isopropyl-β-d-thiogalactopyranoside to a logarithmically growing culture of Escherichia coli to a final concentration of 0.6 mM. Almost all of the expressed protein was contained in inclusion bodies. To obtain the produced protein, cells were suspended in buffer A (50 mM sodium phosphate, 0.6 M NaCl, 1 mg/ml lysozyme, pH 8.0), incubated at 0°C for 30 min, and lysed by sonication. The lysate was centrifuged at 18,000 × g for 5 min at 4°C. The resultant pellet was washed three times with buffer B (50 mM Tris·HCl, 1 mM EDTA, 3% Triton X-100, 0.05% 2-mercaptoethanol, pH 8.0) and once with 50 mM Tris·HCl (pH 8.0) alone. The pellet, containing purified inclusion bodies, was suspended in buffer C (20 mM Tris·HCl, 0.5 M NaCl, 8 M urea, 1 mM 2-mercaptoethanol, pH 8.0) containing 5 mM imidazole, incubated at 15°C for 20 min, and centrifuged at 400,000 × g for 30 min. The supernatant was applied to a Ni2+-charged Hitrap chelating HP column (Amersham Biosciences) equilibrated with buffer C. After the column was washed with buffer C containing 20 mM imidazole, the recombinant protein was eluted with buffer C containing 0.5 M imidazole.
Polyclonal Antibody Production
The recombinant p58 was separated by SDS-PAGE, and the corresponding band was cut out. The resultant gel piece was crushed, dialyzed against phosphate-buffered saline, and used as the antigen. Two rabbits were immunized, and antisera were obtained using standard procedures. Antibody was affinity purified using recombinant p58 blotted on polyvinylidene difluoride membranes.
Immunoblotting
Immunoblot procedures were modified from Towbin et al. (1979). Immunoreactive bands were detected using alkaline phosphatase-conjugated second antibody and either BCIP-NBT solution kit (Nacalai Tesque, Kyoto, Japan) or CSPD (Tropix, Bedford, MA). The p58 antiserum was used at 1:20,000 dilution, and the affinity-purified antibody was used at 1:2000 dilution. mAb against α-tubulin (T5168; Sigma-Aldrich, St. Louis, MO) was used at 1:5000 dilution.
Chemical Cross-linking
As cross-linkers, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), an amine to carboxyl zero-length cross-linker, and disuccinimidyl substrate (DSS), a amine to amine cross-linker, were used. All were purchased from Pierce Chemical (Rockford, IL). Axonemal samples prepared in HME buffer (HMDE minus DTT) were treated with the cross-linkers for 0.5–1 h at room temperature (King et al., 1991). Reactions were terminated by the addition of SDS-PAGE sample solution containing 2-mercaptoethanol and Tris. The resultant samples were analyzed by Western blot.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed according to Sanders and Salisbury (1995). Whole cells or nucleoflagellar apparatuses prepared by the method of Taillon and Jarvik (1995) were fixed with 2% formaldehyde for 10 min at room temperature, followed by treatment with cold methanol and acetone (–20°C). Fixed samples were stained with either preimmune serum or the p58 antiserum diluted 1:500 in immunofluorescence blocking buffer and fluorescein isothiocyanate-labeled anti-rabbit IgG antibody F0382 (Sigma-Aldrich) diluted 1:40 in immunofluorescence blocking buffer.
Electron Microscopy
For thin-section electron microscopy, axonemal samples were fixed with 2% glutaraldehyde and 1% tannic acids. Specimens were postfixed with 1% OsO4, dehydrated through a series of ethanol solutions, and embedded in Epon 812. Sections were stained with uranyl acetate and lead citrate.
Immunoelectron microscopy of detergent/urea-treated axonemes was performed according to Linck et al. (1985) with some modifications. Axonemes were adsorbed onto carbon-coated grids and extracted in situ. After two 15-min incubations with blocking buffer (0.5% bovine serum albumin, 1% fish gelatin in Tris-buffered saline [TBS]), the grids were incubated for 1 h with either preimmune serum or anti-p58 serum diluted 1:300–1:1000 in the blocking buffer. After washes with TBS, the grids were incubated for 1 h with 5-nm gold colloid-conjugated anti-rabbit IgG antibody (BBInternational, Cardiff, England) diluted 1:40 in the blocking buffer. After washes with TBS and TED buffer alone, the grids were negatively stained with 1% uranyl acetate. The specimens were observed using a JEM-1011 electron microscope (JEOL, Tokyo, Japan).
RESULTS
Mutants Lacking the Inner-Arm “e” Species Lack a 58-kDa Protein, p58
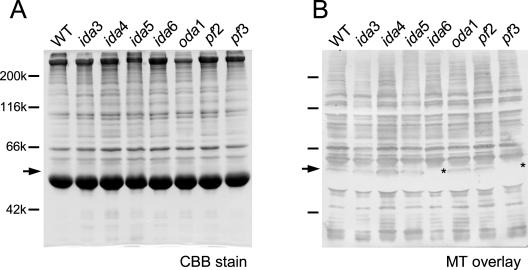
To search for microtubule binding proteins diminished or absent in mutant axonemes lacking inner-arm dyneins, we performed a microtubule overlay assay (Nakaseko et al., 1996). Axonemal proteins from various inner-arm dynein mutants were separated by SDS-PAGE (Figure 1A), transferred to polyvinylidene difluoride membranes, and incubated with microtubules polymerized from biotinylated porcine tubulin. Numerous bands occurred upon detection with avidin-conjugated horseradish peroxidase (Figure 1B), whereas only a faint ∼90-kDa band occurred in a control experiment in which biotinylated microtubules were omitted (our unpublished data). This was as expected, because axonemes must contain a large number of microtubule-associated proteins. Specificity of the method was confirmed by a control experiment in which outer-arm dynein complexes were analyzed in the same way; of the two intermediate chains, IC69 and IC78, only IC78 known as a microtubule-binding protein (King et al., 1991) was detected (our unpublished data). However, we cannot rule out the possibility that some of the detected proteins were nonspecifically bound to the microtubules because of their positive charges, and, conversely, some microtubule-binding proteins were not detected because of imperfect refolding on the membrane.
Figure 1.
Microtubule overlay analysis of axonemes isolated from wild-type (WT) and mutants lacking inner-arm or outer-arm dyneins. Mutants used are listed in Table 1. (A) SDS-PAGE of axonemes stained with CBB. (B) Same samples detected with biotinylated microtubules and avidin-conjugated horseradish peroxidase. A 58-kDa band is missing in pf3 and ida6 (asterisks).
A 58-kDa band was found missing in the mutants pf3 and ida6 (Figure 1B). Both mutants are known to lack inner-arm dynein subspecies “e” (Table 1). Therefore, we thought that the 58-kDa protein (p58) is directly or indirectly involved in the attachment of inner-arm e subspecies.
Isolation and Cloning of p58
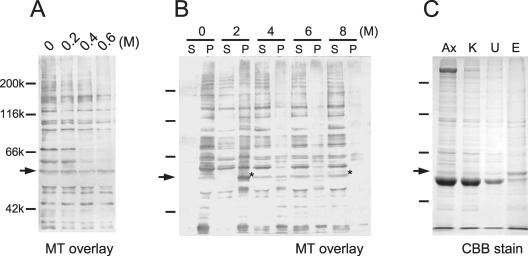
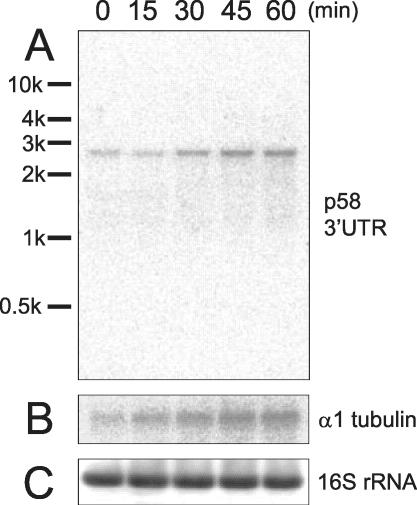
For cDNA cloning, p58 was purified from wild-type axonemes by biochemical methods. Extraction of axonemes with 0.6 M KCl removed most of the inner- and outer-arm dyneins, whereas almost fully leaving p58 in the axoneme (Figure 2A). We found that p58 was insoluble in 2 M urea, but almost completely soluble in 8 M urea (Figure 2B). For p58 purification, wild-type axonemes were first extracted with 0.6 M KCl and then with 2 M urea. The remnants were finally solubilized with 8 M urea and applied to cation-exchange column chromatography, which efficiently removed tubulin contained in a large amount. The resultant sample, containing p58 and small amounts of tubulin and other polypeptides, was resolved by SDS-PAGE (Figure 2C). This p58 protein yielded a strong band when examined by the microtubule-overlay assay described above (our unpublished data). The 58-kDa band was cut out, digested with trypsin, and separated by reverse-phase chromatography. Four major peaks were chosen, and their amino acid sequences were determined. Degenerate PCR was carried out to amplify cDNA fragments corresponding to two of these sequences. This yielded a 155-base pairs product. After 3′ RACE, PCR yielded a ∼1.5-kbp product, containing a partial coding sequence and a 3′-UTR sequence. This product was digested with PvuII, and a 495-base pair product corresponding to the 3′-UTR was obtained. Screening of a cDNA library with the 3′-UTR sequence yielded three clones, two of which contained ∼2.7-kbp, full-length p58 cDNA. Northern blots of total RNA with the probe detected an ∼2.7-kbp transcript. Like other flagellar protein genes in Chlamydomonas, this transcript was found to be up-regulated upon deflagellation, indicating that p58 is involved in flagellar structure or assembly (Figure 3A).
Figure 2.
Extraction and purification of the 58-kDa polypeptide. (A) KCl extraction of wild-type axonemes. Insoluble fractions after the extraction were subjected to a microtubule overlay assay. The 58-kDa polypeptide was not extracted with KCl of up to 0.6 M. (B) Urea extraction of wild-type axonemes. Supernatants (S) and precipitates (P) from the centrifuged samples were analyzed by the microtubule overlay assay. The 58-kDa polypeptide remained in the insoluble fraction after extraction with 2 M urea but was almost completely extracted with 8 M urea (asterisks). (C) Purification of the 58-kDa polypeptide. SDS gel patterns stained with CBB. Lane Ax, intact axonemes. Lane K, precipitate after 0.6 M KCl extraction. Lane U, precipitate after 2 M urea extraction of the KCl-extracted sample. Lane E, eluate from a cation-exchange chromatography column.
Figure 3.
Northern blots of total RNA isolated from wild-type cells after deflagellation. Numbers on the lanes indicate the time after deflagellation. (A) The 3′-UTR fragment of the p58 cDNA used as the probe hybridized with a 2.7-kb band, which was up-regulated after deflagellation. (B) Same blot probed with an α-tubulin sequence, which is known to be up-regulated after deflagellation. (C) Same blot probed with a 16S rRNA sequence for a loading control.
Sequence Analysis of p58
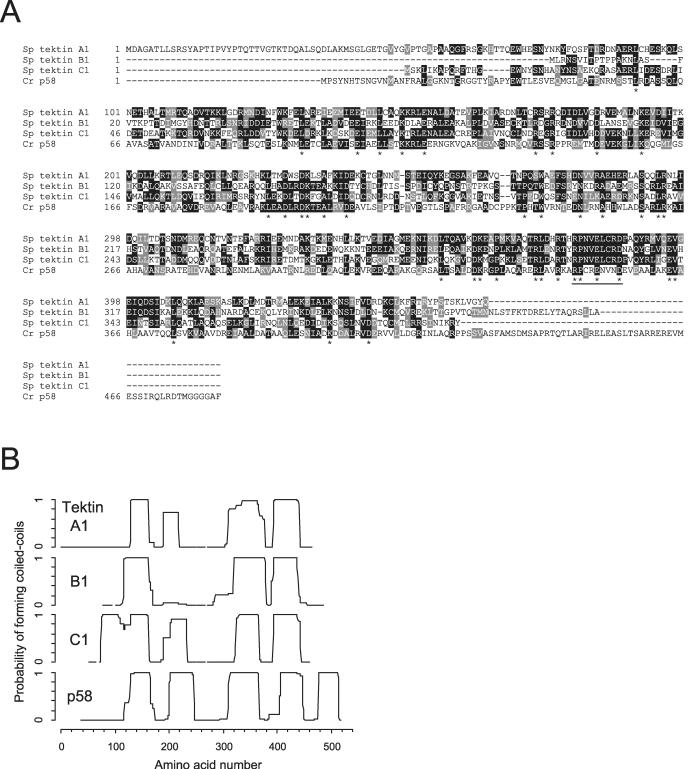
The sequence of p58 cDNA (GenBank accession no. AB111498) predicted that it encodes a 482-amino acid protein (Figure 4A). Unexpectedly, BLAST search revealed that p58 is homologous to tektins of sea urchin, with 23–25% identities and 39–41% similarities to each of the three isoforms. No other sequences currently registered in Chlamydomonas expressed sequence tag and genome databases show significant homologies to tektins. Although the homology is rather low, such low homology also has been found in other tektins; for example, putative tektins in C. elegans (NP_508689) and Drosophila melanogaster (NP_523577) show identities of 22–23% and 28–30%, respectively, to sea urchin tektins. Secondary structure analysis predicted that p58 has an ∼80-residue N-terminal sequence of nonhelical structure, followed by five coiled-coil regions separated by nonhelical linkers. Alignment of the amino-acid sequence with sea urchin tektins showed that p58 has a ∼60-residue extension in its C terminal (Figure 4A). This extension forms an additional helical segment (Figure 4B). Many residues, charged or hydrophobic, are conserved at the same positions of all four sequences (Figure 4A, asterisks). A sequence motif of unknown function, RPNVELCRD, conserved in several kinds of sea urchin and mammalian tektins (Norrander et al., 1996), is partially preserved in p58; amino acid residues are preserved at four of the nine positions (Figure 4A, underlined).
Figure 4.
Sequence analysis of p58. (A) ClustalW (Thompson et al., 1994) alignment of the deduced amino acid sequence of p58 with the sequence of sea urchin tektins. The characters with black and gray backgrounds represent identical and conservatively substituted amino acids, respectively. Amino acid positions conserved in all four sequences are marked with asterisks. The identities/similarities (identities + conservative substitutions) between p58 and sea urchin tektins are as follows: tektin A1, 23%/39%; tektin B1, 25%/41%; and tektin C1, 23%/40%. The sequence motif RPNVELCRD (referenced to tektin A1 at residues 378–386, underlined) conserved in all sea urchin tektins is only partially preserved in p58. However, several blocks of amino acid residues are conserved between sea urchin tektins and p58 (e.g., ADLRDKTEA in tektin B1 and p58, referenced to tektin B1 at residues 147–155). The GenBank accession nos. for the sequences are as follows: Strongylocentrotus purpuratus (Sp) tektin A1, AAF14818; Sp tektin B1, Q26648; Sp tektin C1, AAB02680; and Chlamydomonas reinhardtii (Cr) p58, BAC77347. (B) Predicted coiled-coil structure of p58 according to the program COILS (Lupas et al., 1991). Probability of coiled-coil formation was calculated for a 28-residue window. Plots are aligned according to the ClustalW result. The horizontal axis shows the amino acid number referenced to tektin A1.
Characterization of the p58 Gene
To obtain the p58 genomic clone, a genomic library constructed from the wild-type strain was screened with the 3′-UTR probe. Two overlapping clones were obtained and partially sequenced. The structure of the gene is shown in Figure 5A. Sequence analysis identified eight exons within the coding region. When our cloning of the p58 gene was complete, the release 1 assembly of the C. reinhardtii genome sequence was announced from Department of Energy Joint Genome Institute (Walnut Creek, CA) (http://genome.jgipsf.org/chlre1/chlre1.home.html). The p58 gene was found within scaffold 22 of the assembly.
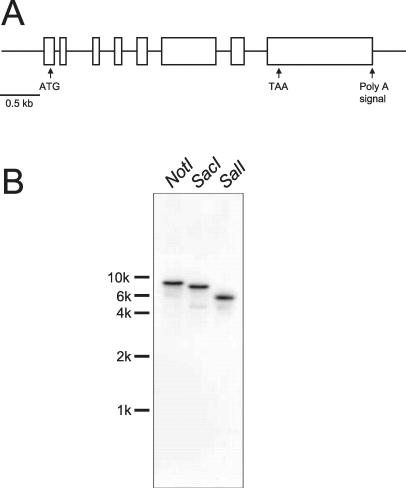
Figure 5.
(A) Structure of the p58 gene. Boxed areas indicate sequences contained in the p58 cDNA clone. The predicted translation start site (ATG), stop site (TAA), and the putative polyadenylation signal (Poly A) are indicated. (B) Southern blot of genomic DNA digested with the indicated restriction enzymes and hybridized with the 417-base pair HinfI fragment of p58 cDNA.
Southern blot analysis of genomic DNA was performed to determine the copy number of the p58 gene. A 417-base pair HinfI fragment containing a partial coding region was used as a probe. The hybridization patterns indicated the presence of only one copy of the p58 gene (Figure 5B).
The genetic locus of the p58 gene was determined by a PCR-based mapping method as described previously (Kathir et al., 2003). The p58 gene was mapped within 2 cM of S68, a molecular marker on linkage group XVIII. This result indicates that the p58 gene is different from the mutated genes in pf3 and ida6, which are mapped on linkage group VIII (Harris, 1989) and XIV (Kato et al., 1993), respectively. No mutant loci have been mapped near S68.
Antibody Production and Immunoblot Analysis
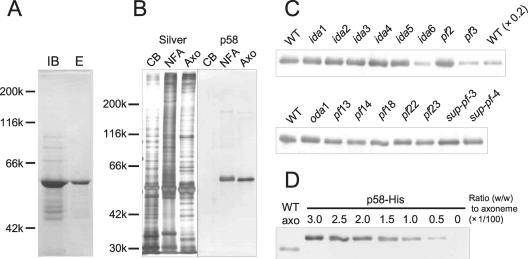
Polyclonal antibodies were raised against bacterially produced His-tagged p58 protein, which was solubilized from the inclusion body with 8 M urea and purified with a Ni2+ column (Figure 6A). Western blot analysis of wild-type samples demonstrated that the anti-p58 antiserum specifically detected a single band corresponding to p58 in the axoneme and the complex of nucleus and flagella (the nucleoflagellar apparatus; NFA), but not noticeably in the cell body (Figure 6B).
Figure 6.
Antibody production and immunoblot analysis of mutant axonemes. (A) Purification of recombinant p58 produced in E. coli. The gel was stained with CBB. Lane IB, isolated inclusion bodies. Lane E, eluate from a Hitrap chelating column. (B) Western-blot analysis of the wild-type cell body (CB), NFA, and axoneme (Axo). Left, an SDS-PAGE gel stained with silver. Right, the blot immunostained with anti-p58 serum. A 58-kDa band was detected in both nucleoflagellar apparatus and axoneme. (C) Western blot analysis of wild-type and mutants axonemes. The blot was detected with the p58 antibody. Only an ∼58-kDa region is shown. Each lane contains 2.5 μg of axoneme, whereas lane WT (× 0.2) contains 0.5 μg. Densitometry indicated that p58 is decreased to <20% of the wild-type amount in both pf3 and ida6 (four independent experiments). (D) Quantification of p58 in the axoneme with Western blot by using known amounts of recombinant p58 (with a His tag) as a standard. Lane WT axo, wild-type axoneme (0.75 μg). The numbers on the lanes indicate the ratio (wt/wt) of recombinant p58 to the wild-type axoneme. The ratio was estimated to be 1:75.6 ± 7.9 (four independent experiments). Because the recombinant protein has a six-histidine tag and an additional linker, its mobility on the blot differs from that of p58. The p58 antibody did not react with other protein carrying the same tag and linker sequence (our unpublished data).
Immunoblot analysis indicated that p58 is greatly diminished, but not completely missing, in pf3 and ida6 axonemes (Figure 6C). Quantitative analysis, using a dilution series of wild-type axonemes as a standard, showed that the amount of p58 in these mutants is reduced to <20% (Figure 6C). Mutants sup-pf-3 and ida5, which also lack inner-arm species e, as well as mutants deficient in outer-arm dyneins, radial spokes, central pairs, or other inner-arm dyneins, had normal amounts of p58 (Figure 6C).
The amount of p58 in the wild-type axoneme was semiquantitatively estimated by Western blot analysis, with a dilution series of bacterially produced p58 (with a His-tag) as a standard. The estimate yielded a ratio of p58 to the total axonemal proteins of ∼1:76 (wt/wt) (Figure 6D). Because tubulin accounts for ∼60% of the total axonemal proteins, this means that the ratio of p58 to tubulin is ∼1:46 (wt/wt).
Localization of p58 by Immunofluorescence Microscopy
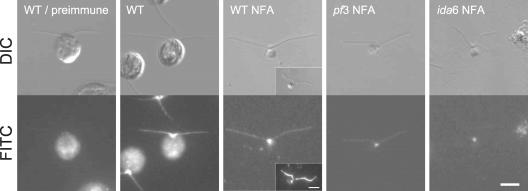
Immunofluorescence microscopy of whole cell samples demonstrated that p58 uniformly localized along the length of flagella and strongly on the basal body (Figure 7, WT). In isolated nucleoflagellar apparatuses, basal bodies were strongly stained (Figure 7, WT NFA). Staining of flagella was usually much weaker. However, flagellar staining became stronger when the flagella were apparently partially disrupted (Figure 7, WT NFA, inset). The staining intensity of the flagella in those samples was often comparable with that of the basal bodies. Thus, weak staining of flagella might be due to the presence of some structural barrier that limited antibody access. In the mutants pf3 and ida6, the staining on the flagella seemed to be uniformly weakened along the length (Figures 7, pf3 NFA and ida6 NFA)
Figure 7.
Immunofluorescence microscopy of whole cells and the NFA. Top, differential interference contrast images. Bottom, indirect immunofluorescence light microscopy. Cells and NFAs stained with preimmune serum or p58 antiserum, followed by FITC-conjugated goat anti-rabbit IgG. WT/preimmune, lack of staining with preimmune serum. WT, wild-type cell stained with the p58 antibody. Flagella and the basal body region are stained. WT NFA, wild-type NFA stained with the p58 antibody. Flagella and basal bodies are stained. Inset, intensive staining on flagella that seem partially disrupted. pf3 NFA, ida6 NFA, NFA of mutants stained with the p58 antibody. Staining of flagella and basal bodies is weak compared with wild-type cells. Bar, 5 μm.
Presence and Absence of p58 in Axonemes Extracted with Sarkosyl and Urea
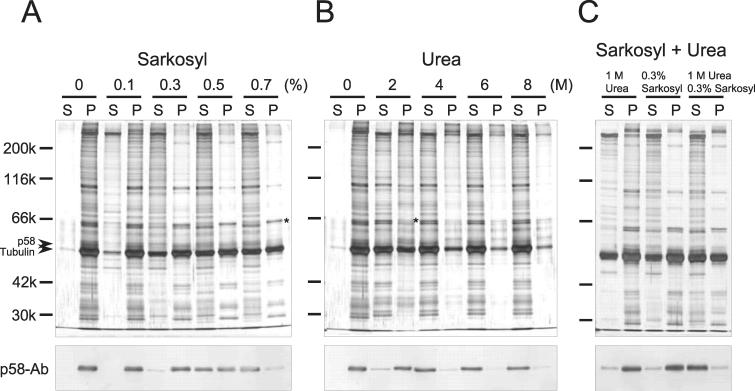
Extraction of axonemes with Sarkosyl has been reported to yield stable bundles of microtubule protofilaments, called the pf-ribbons (Witman et al., 1972; Linck, 1976). In sea urchin sperm, the pf-ribbons contain the three species of tektins as major components (Linck, 1976). To determine whether p58 is a component of a similar structure in the Chlamydomonas axoneme, axonemes were extracted with Sarkosyl in TED buffer and analyzed by Western blot with the p58 antibody (Figure 8A). We found that p58 was almost completely solubilized in 0.7% Sarkosyl, i.e., under the condition that yields pf-ribbons (Witman et al., 1972). Electron microscope observations confirmed the presence of pf-ribbons in the precipitate (Figure 9A). In addition, this fraction contained a prominent band of 66 kDa, the band of a Chlamydomonas pf-ribbon component, Rib72 (Figure 8A, top, asterisk) (Ikeda et al., 2003). It is therefore unlikely that p58 is a major component of the pf-ribbons.
Figure 8.
Presence and absence of p58 in axonemes extracted with Sarkosyl and urea. Top, silver-stained SDS-PAGE gels. Bottom, blots immunostained with p58 antibody. Lanes S and P indicate supernatants and precipitates from the centrifuged samples, respectively. Numbers on the lanes indicate the concentration of Sarkosyl or urea used. (A) Extraction of wild-type axonemes with Sarkosyl in TED buffer. p58 is insoluble in 0.3% Sarkosyl but completely soluble in 0.7% Sarkosyl, the condition wherein the protofilament ribbons remain in the precipitate (see also Figure 9). (B) Extraction of wild-type axonemes with urea in MESH buffer. The precipitate fraction obtained after 2 M urea extraction did not contain Rib72, a prominent 66-kDa band found in the precipitate after 0.7% Sarkosyl extraction (A and B, top, asterisk). (C) Extraction of wild-type axoneme with Sarkosyl and urea in TED buffer.
Figure 9.
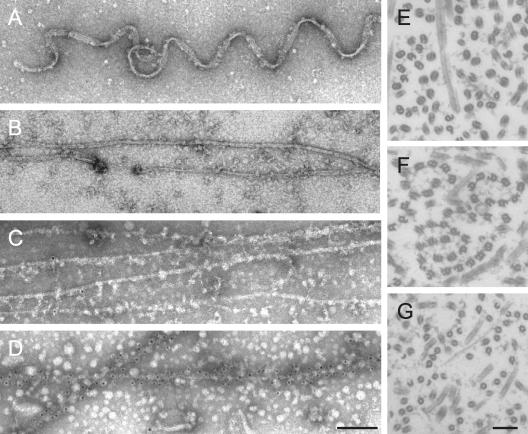
Electron microscopy of axonemes extracted with Sarkosyl or urea. (A) Negativestain image of wild-type axoneme treated with 0.7% Sarkosyl in TED buffer. A stable three-protofilament structure, previously described as the protofilament ribbons, remained. (B) Negative-stain image of wild-type axoneme treated with 2 M urea in MESH buffer. A filamentous structure remained. This filament is approximately two-protofilament wide. (C and D) Immunoelectron microscopy of the axonemes treated with 2 M urea. The specimens were stained with preimmune serum (C) or p58 antiserum (D), followed by 5-nm colloidal gold-conjugated secondary antibody. (E–G) Cross section images of detergent-treated axonemes. (E) Sarkosyl (0.3%) in TED. (F) Urea (1 M) in TED. (G) Sarkosyl (0.3%) and 1 M urea in TED. A-tubules remain intact in all three conditions. Bar, 100 nm.
Extraction of axonemes with 2 M urea in MESH buffer (pH 6.8) left p58 in the insoluble remnant while solubilizing the majority of tubulin and other axonemal proteins (Figure 8B), as stated above. Rib72 was not present in this remnant (Figure 8B, top, asterisk). We also found that p58 became partially solubilized in 2 M urea in TED buffer (pH 8.0) (our unpublished data). Negative-stain electron microscopy of the insoluble fraction obtained after 2 M urea/pH 6.8 extraction revealed filamentous structures with the width of 5–8 nm (Figure 9B). Immunoelectron microscopy with the p58 antiserum and a gold-conjugated second antibody indicated that these filaments contained p58 (Figure 9D). Because a large amount of tubulin was present in the insoluble fraction (Figure 8B), the thin filament may well be composed of microtubule protofilaments.
Finally, combined extraction with Sarkosyl and urea yielded important information about the p58 localization in doublet microtubules. Extraction with 0.3% Sarkosyl in TED alone yielded apparently intact A-tubules with remnants of B-tubules attached, as reported previously (Witman et al., 1972) (Figure 9E). SDS-PAGE/Western blot indicated that the remaining structure still contained p58 (Figure 8C). Extraction with 1 M urea in TED alone yielded a similar structure containing p58 (Figures 8C and 9F). Extraction with 0.3% Sarkosyl and 1 M urea in TED also yielded apparently intact A-tubules (Figure 9G). However, this structure lacked B-tubules more thoroughly and did not contain p58 (Figure 8C). Thus, p58 may be somehow involved in the attachment of B-tubule to the A-tubule.
Association between p58 and Other Axonemal Proteins
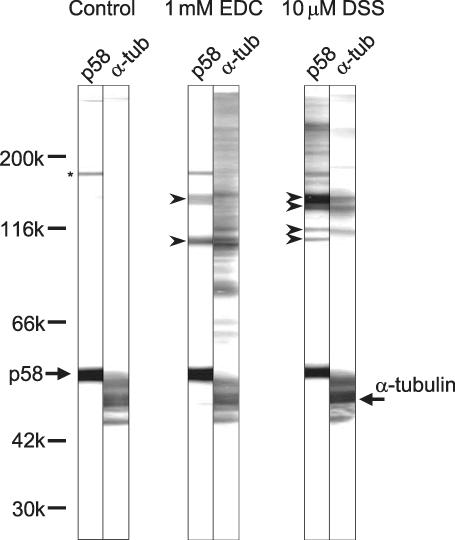
Chemical cross-linking of axonemes was performed to examine possible association between p58 and other axonemal proteins. Wild-type axonemes were treated with EDC or DSS and analyzed by immunoblot with the p58 antibody and α-tubulin antibody (Figure 10). With EDC, ∼107- and 150-kDa products were detected with the p58 antibody. With DSS, ∼112-kDa, 117-kDa, 143-kDa, and 151-kDa products were detected. Some of these bands might have been derived from cross-linking with p58 itself or with other ∼50-kDa proteins such as tubulin. With α-tubulin antibody, a few intense bands were detected overlapping with those detected with the p58 antibody. This may be indicating p58-tubulin association. However, because the molecular weight of p58 is similar to that of tubulin and p58 is present in only 1/46 amount of tubulin, it is also possible that these bands were derived from cross-linking between tubulin and other protein(s) or between α- and β-tubulins.
Figure 10.
Chemical cross-linking analysis of axonemes. Wild-type axonemes were treated with either EDC or DSS, followed by Western blot. The concentration of cross-linkers used is noted above lanes. Blots were immunostained with p58 antibody (p58) and α-tubulin antibody (α-tub). In the control sample without chemical cross-linking, p58 antibody produced weak bands at 168 kDa (asterisks) in addition to p58 itself (arrow). These bands were probably due to nonspecific staining of the antibody. In the EDC-treated sample, 107- and 150-kDa bands were detected with p58 antibody (arrowheads). In the DSS-treated sample, 112-, 117-, 143-, and 151-kDa bands were detected with p58 antibody (arrowheads).
DISCUSSION
In this study, we found that a novel Chlamydomonas axonemal protein, p58, is greatly diminished in mutants that lack inner-arm dynein e, and identified it as a homologue of tektin. Until recent release of an early version of Chlamydomonas genome database, presence of tektin had not been known in this organism. Tektin has been thought to play important roles in the formation of doublet and triplet microtubules (Linck, 1976; Linck and Langevin, 1982; Linck and Stephens, 1987; Stephens and Lemieux, 1998). Current models postulate that tektin forms a filament that is incorporated as an integral part of the outer doublet (Pirner and Linck, 1994); an interesting possibility is that it forms a protofilament of the outer-doublet microtubule (Nojima et al., 1995). It is therefore surprising that the mutants pf3 and ida6 with a greatly reduced amount of p58 can produce flagella. Tektin may function in a significantly different manner in Chlamydomonas than in other organisms.
Localization and Structural Organization of p58 in Axonemes
Immunofluorescence microscopy showed that p58 was uniformly distributed along the axoneme, as well as on the basal bodies. This localization pattern is consistent with previous observations with other species (Linck et al., 1985; Steffen and Linck, 1988; Steffen and Linck, 1992; Steffen et al., 1994). In sea urchin sperm, the Sarkosyl-resistant protofilament ribbon contains tektin as a major component (Linck and Langevin, 1982; Linck and Stephens, 1987). In contrast, our result suggested that no p58 is contained in the protofilament ribbons. Possibly, protofilament ribbons of sea urchin sperm and Chlamydomonas are composed of different sets of protein subunits.
Extraction of axonemes with 2 M urea yielded a filamentous structure that contained p58, tubulin, and small amounts of other polypeptides. Immunoelectron microscopy showed that p58 is present along the filaments. It is possible that p58, like tektin in other organisms (Pirner and Linck, 1994), forms filaments itself, although we failed to isolate pure p58 filaments. However, because extraction of axonemes with 0.3% Sarkosyl and 1 M urea almost completely solubilized p58 while leaving A-tubules apparently intact, none of the protofilaments in A-microtubules should be composed of p58.
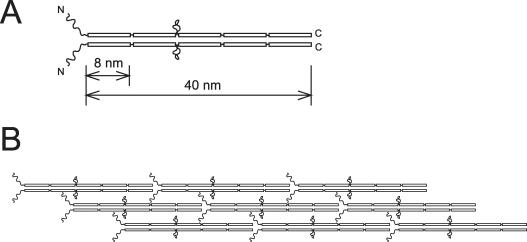
Secondary structure prediction suggested that p58 has a similar molecular organization to that of other tektins (Norrander et al., 1992; Chen et al., 1993; Norrander et al., 1996), except that it has an extended C terminal sequence that forms an additional helical segment (Figure 4, A and B). Each one of the five α-helical segments in p58 can be ∼8 nm in length. If we adopt the model for sea urchin tektins (Norrander et al., 1996), p58 may possibly form ∼40-nm-long homodimers that are concatenated to form filaments along the length of the outer doublet (Figure 11, A and B). This structural organization is supported by the cross-link patterns (Figure 10), although more experiments are necessary to establish whether p58 can associate with itself. Our estimate of the ratio of p58 to tubulin in the wild-type axoneme was ∼1:46 (wt/wt). Considering the molecular weight of the proteins and the number of tubulin protofilaments in the axoneme, we hence estimate the number of tektin molecules to be ∼1.1 per 8 nm of an outer doublet, or ∼5.5 per 40 nm. Because a p58 filament in the above-mentioned model contains two p58 molecules per 40 nm, approximately three filaments may be present along one outer doublet. In pf3 and ida6, however, p58 is diminished to <20% of the wild-type amount. Because the structure of the outer doublet microtubules in these mutants does not seem to significantly differ from the wild-type outer doublets (Kato et al., 1993; Gardner et al., 1994), as observed by light and electron microscopy, we are inclined to think that only <20% tektin present in the wild-type axoneme is necessary and sufficient for the formation of outer-doublet microtubules. In other words, the amount of essential p58 may be as small as 1/230 (wt/wt) of that of tubulin. This value is significantly smaller than the ratio, ∼1:20 (wt/wt), of tektin A+B+C to tubulin estimated in surf clam sperm flagella (Stephens and Lemieux, 1998). The origin of such a big difference is not clear. A possible explanation would be that p58 in the Chlamydomonas axoneme assumes a structure significantly different from the tektin filament model suggested for other organisms. An alternative possibility is that another protein is present in Chlamydomonas and forms heteropolymers with p58. Because neither immunoblot analysis nor expressed sequence tag and genome database search revealed presence of other tektin-like proteins, such a protein, if any, may be very distantly related to tektin.
Figure 11.
Model of the p58 structure. (A) Model of a p58 homodimer. Boxes indicate helical domains. Following the previous structural model of sea urchin tektins (Norrander et al., 1996), two p58 polypeptides are assumed to form a homodimer. Because p58 has five ∼8-nm-long helical domains, the total length of the homodimer is ∼40 nm. The homodimer could be concatenated to form a filament. (B) Model of p58 filaments on A-tubule. The ratio of p58 to the total axoneme is ∼1:76 (wt/wt) (Figure 6C). From this ratio, the number of p58 molecules within a 40-nm period of one outer doublet is calculated to be ∼5.5. This means that approximately three p58 filaments exist per one outer doublet.
Possible Functions of p58 in the Assembly of Inner-Arm Dyneins
p58 is greatly diminished in the axonemes of the mutants pf3 and ida6, which completely lack inner-arm dynein e, but not diminished in pf2 and sup-pf-3, which partially lack it (Gardner et al., 1994). Biochemical analyses indicated that pf3 axonemes lack, besides dynein e, four of the seven polypeptides that make up a protein complex called the dynein regulatory complex (DRC), whereas pf2 and sup-pf-3 lack five or one of the DRC polypeptides, respectively (Huang et al., 1982; Piperno et al., 1994). Whether ida6 lacks any DRC component is unknown. p58 is present in a normal amount in ida5, which lacks dynein a, c, d, and e due to the absence of actin, a subunit of these dyneins (Kato et al., 1993). Naturally, the docking of dynein e on the outer doublet must involve multiple factors. The above-mentioned findings suggest that some DRC components and tektin are intimately related to the docking of dynein e. It is likely that the pf3 and ida6 axonemes lack some components that are important for docking dynein e, some DRC components, and a large fraction of p58 molecules to specific sites on the outer doublet. p58 does not seem to be one of the known seven DRC subunits, because DRC3 and DRC4, the two DRC subunits with molecular weights similar to that of p58, are absent from the pf2 axonemes in which p58 is present in a normal amount. However, it is possible that some DRC components cooperate with p58 in the docking of dynein e.
Our present study has thus suggested possible involvement of tektin in the docking of dynein e and DRC, but the true causality between tektin and dynein/DRC docking awaits further studies. Cross-linking experiments suggested that p58 is associated with multiple proteins of 50–80 kDa (Figure 10). Identification of these proteins will be important for obtaining further information. A most puzzling finding is that the pf3 and ida6 axonemes are formed with only <20% of the normal amount of tektin. The organization of the extra >80% tektin, as well as that of essential tektin, in the wild-type axoneme is an intriguing subject of future studies.
Acknowledgments
We thank Drs. Masafumi Hirono (University of Tokyo) and Pete Lefebvre (University of Minnesota) for carrying out genetic mapping. This study has been supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–11–0854. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–11–0854.
References
- Casey, D.M., Inaba, K., Pazour, G.J., Takada, S., Wakabayashi, K., Wilkerson, C.G., Kamiya, R., and Witman, G.B. (2003). DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol. Biol. Cell 14, 3650–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Perrone, C.A., Amos, L.A., and Linck, R.W. (1993). Tektin B1 from ciliary microtubules: primary structure as deduced from the cDNA sequence and comparison with tektin A1. J. Cell Sci. 106, 909–918. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. (1995). Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 11, 398–404. [DOI] [PubMed] [Google Scholar]
- Gardner, L.C., O'Toole, E., Perrone, C.A., Giddings, T., and Porter, M.E. (1994). Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J. Cell Biol. 127, 1311–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons, I.R. (1981). Cilia and flagella of eukaryotes. J. Cell Biol. 91, 107s–124s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1985). Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E. (1989). The Chlamydomonas Sourcebook, San Diego: Academic Press.
- Huang, B., Piperno, G., Ramanis, Z., and Luck, D.J. (1981). Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell biol. 88, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Ramanis, Z., and Luck, D.J. (1982). Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell 28, 115–124. [DOI] [PubMed] [Google Scholar]
- Hyman, A., Drechsel, D., Kellogg, D., Salser, S., Sawin, K., Steffen, P., Wordeman, L., and Mitchison, T. (1991). Preparation of modified tubulins. Methods Enzymol. 196, 478–485. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., Brown, J.A., Yagi, T., Norrander, J.M., Hirono, M., Eccleston, E., Kamiya, R., and Linck, R.W. (2003). Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J. Biol. Chem. 278, 7725–7734. [DOI] [PubMed] [Google Scholar]
- Kamiya, R. (2002). Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219, 115–155. [DOI] [PubMed] [Google Scholar]
- Kamiya, R., Kurimoto, E., and Mutto, E. (1991). Two types of Chlamydomonas flagellar mutants missing different components of inner-arm denein. J. Cell Biol. 112, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir, P., LaVoie, M., Brazelton, W.J., Haas, N.A., Lefebvre, P.A., and Silflow, C.D. (2003). Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2, 362–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Kagami, O., Yagi, T., and Kamiya, R. (1993). Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct. Funct. 18, 371–377. [DOI] [PubMed] [Google Scholar]
- King, S.M., Wilkerson, C.G., and Witman, G.B. (1991). The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with alphatubulin in situ. J. Biol. Chem. 266, 8401–8407. [PubMed] [Google Scholar]
- Koutoulis, A., Pazour, G.J., Wilkerson, C.G., Inaba, K., Sheng, H., Takada, S., and Witman, G.B. (1997). The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Linck, R.W. (1976). Flagellar doublet microtubules: fractionation of minor components and alpha-tubulin from specific regions of the A-tubule. J. Cell Sci. 20, 405–439. [DOI] [PubMed] [Google Scholar]
- Linck, R.W., and Langevin, G.L. (1982). Structure and chemical composition of insoluble filamentous components of sperm flagellar microtubules. J. Cell Sci. 58, 1–22. [DOI] [PubMed] [Google Scholar]
- Linck, R.W., Amos, L.A., and Amos, W.B. (1985). Localization of tektin filaments in microtubules of sea urchin sperm flagella by immunoelectron microscopy. J. Cell Biol. 100, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck, R.W., and Stephens, R.E. (1987). Biochemical characterization of tektins from sperm flagellar doublet microtubules. J. Cell Biol. 104, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., O'Toole, E.T., McDonald, K.L., McIntosh, J.R., and Porter, M.E. (1992). Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J. Cell Biol. 118, 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko, Y., Nabeshima, K., Kinoshita, K., and Yanagida, M. (1996). Dissection of fission yeast microtubule associating protein p93Dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Nojima, D., Linck, R.W., and Egelman, E.H. (1995). At least one of the protofilaments in flagellar microtubules is not composed of tubulin. Curr. Biol. 5, 158–167. [DOI] [PubMed] [Google Scholar]
- Norrander, J.M., Amos, L.A., and Linck, R.W. (1992). Primary structure of tektin A 1, comparison with intermediate-filament proteins and a model for its association with tubulin. Proc. Natl. Acad. Sci. USA 89, 8567–8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander, J.M., Linck, R.W., and Stephens, R.E. (1995). Transcriptional control of tektin A mRNA correlates with cilia development and length determination during sea urchin embryogenesis. Development 121, 1615–1623. [DOI] [PubMed] [Google Scholar]
- Norrander, J.M., Perrone, C.A., Amos, L.A., and Linck, R.W. (1996). Structural comparison of tektins and evidence for their determination of complex spacings in flagellar microtubules. J. Mol. Biol. 257, 385–397. [DOI] [PubMed] [Google Scholar]
- Piperno, G., Mead, K., LeDizet, M., and Moscatelli, A. (1994). Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 125, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Mead, K., and Shestak, W. (1992). The inner dynein arms 12 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J. Cell Biol. 118, 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirner, M.A., and Linck, R.W. (1994). Tektins are heterodimeric polymers in flagellar microtubules with axial periodicities matching the tubulin lattice. J. Biol. Chem. 269, 31800–31806. [PubMed] [Google Scholar]
- Rosenfeld, J., Capdevielle, J., Guillemot, J.C., and Ferrara, P. (1992). In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203, 173–179. Chlamydomonas is a homologue of a growth arrest-specific gene product. J. Cell Biol. 162, 47–57. [DOI] [PubMed] [Google Scholar]
- Rupp, G., and Porter, M.E. (2003). A subunit of the dynein regulatory complex in Chlamydomonas is a homologue of a growth arrest-specific gene product. J. Biol. 162, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M.A., and Salisbury, J.L. (1995). Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 47, 163–169. [DOI] [PubMed] [Google Scholar]
- Shelanski, M.L., Gaskin, F., and Cantor, C.R. (1973). Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. USA 70, 765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, W., and Linck, R.W. (1988). Evidence for tektins in centrioles and axonemal microtubules. Proc. Natl. Acad. Sci. USA 85, 2643–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen, W., and Linck, R.W. (1992). Evidence for a non-tubulin spindle matrix and for spindle components immunologically related to tektin filaments. J. Cell Sci. 101, 809–822. [DOI] [PubMed] [Google Scholar]
- Steffen, W., Fajer, E.A., and Linck, R.W. (1994). Centrosomal components immunologically related to tektins from ciliary and flagellar microtubules. J. Cell Sci. 107, 2095–2105. [DOI] [PubMed] [Google Scholar]
- Stephens, R.E. (1989). Quantal tektin synthesis and ciliary length in sea-urchin embryos. J. Cell Sci. 92, 403–413. [DOI] [PubMed] [Google Scholar]
- Stephens, R.E., Oleszko Szuts, S., and Linck, R.W. (1989). Retention of ciliary ninefold structure after removal of microtubules. J. Cell Sci. 92, 391–402. [DOI] [PubMed] [Google Scholar]
- Stephens, R.E., and Lemieux, N.A. (1998). Tektins as structural determinants in basal bodies. Cell Motil. Cytoskeleton 40, 379–392. [DOI] [PubMed] [Google Scholar]
- Taillon, B.E., and Jarvik, J.W. (1995). Release of the cytoskeleton and flagellar apparatus from Chlamydomonas. Methods Cell Biol. 47, 307–313. [DOI] [PubMed] [Google Scholar]
- Takada, S., and Kamiya, R. (1994). Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J. Cell Biol. 126, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Wilkerson, C.G., Wakabayashi, K., Kamiya, R., and Witman, G.B. (2002). The outer dynein arm-docking complex: composition and characterization of a subunit (Oda1) necessary for outer arm assembly. Mol. Biol. Cell 13, 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi, K., Takada, S., Witman, G.B., and Kamiya, R. (2001). Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil. Cytoskeleton 48, 277–286. [DOI] [PubMed] [Google Scholar]
- Witman, G.B., Carlson, K., Berliner, J., and Rosenbaum, J.L. (1972). Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 54, 507–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G.B., Plummer, J., and Sander, G. (1978). Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76, 729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]