Abstract
The neural crest is a multipotent stem cell--like population that gives rise to a wide range of derivatives in vertebrate embryo including elements of the craniofacial skeleton and peripheral nervous system as well as melanocytes. The neural crest forms in a series of regulatory steps that include induction and specification of the prospective neural crest territory--neural plate border, specification of bona fide neural crest progenitors, and differentiation into diverse derivatives. These individual processes during neural crest ontogeny are controlled by regulatory circuits that can be assembled into a hierarchical gene regulatory network (GRN). Here we present an overview of the GRN that orchestrates the formation of cranial neural crest cells. Formulation of this network relies on information largely inferred from gene perturbation studies performed in several vertebrate model organisms. Our representation of the cranial neural crest GRN also includes information about direct regulatory interactions obtained from the cis-regulatory analyses performed to date, which increases the resolution of the architectural circuitry within the network.
Keywords: stem cell, transcription factors, cell migration, cis-regulation
INTRODUCTION
The neural crest, often referred to as the “fourth germ layer” (Hall 2000), is a multipotent stem cell--like population of highly migratory cells that contribute derivatives to a wide variety of tissues and organs in the vertebrate embryo. These include but are not limited to the sensory and autonomic ganglia, adrenal and thyroid glands, smooth muscle of major blood vessels, cartilage and bone of the face, and pigmentation of the skin. As a defining feature of vertebrates, neural crest formation has been extensively studied using vertebrate model organisms ranging from lampreys and fish to frogs, chicken, and mouse.
Neural crest cells form over a lengthy period of time during development that starts at gastrulation and extends into late organogenesis. This process is initiated by a combination of inductive signals emanating from environing tissues, such as the underlying mesoderm or adjacent neural and non-neural ectoderm, which set up the presumptive neural crest region. As a result, the territory between neural and non-neural ectoderm, termed the neural plate border, is competent to respond to signals specifying bona fide progenitors. These cells subsequently undergo an epithelial-to-mesenchymal transition (EMT), delaminate from the neuroepithelium, and migrate along stereotypical pathways. After settling in various and sometimes distant sites in the embryo, they differentiate into a multitude of derivatives.
For more than a century, the neural crest has provided a productive paradigm for addressing essential questions regarding cell interactions that underlie induction, specification, and differentiation events during development. As such, the neural crest is the subject of an extensive literature and descriptive database that, in combination with recent genomic cis-regulatory and gene knockdown data, provides a critical mass of information regarding the molecular underpinnings that guide neural crest formation. Such a compelling database calls for a systematic approach to integrate diverse information into a multistep gene regulatory network (GRN) that describes the process of neural crest formation.
The accrual of molecular information relevant to neural crest induction, specification, and migration has led to the formulation of a putative vertebrate GRN that orchestrates neural crest formation (Meulemans & Bronner-Fraser 2004, Sauka-Spengler & Bronner-Fraser 2008a, Steventon et al. 2005). Because of variation between species, the main challenge has been to incorporate the pertinent data, obtained from many vertebrate developmental models, into a single, pan-vertebrate network. In addition to discrepancies in the patterns of gene expression and differences in the deployment of paralogous genes among various vertebrates (Meulemans & Bronner-Fraser 2004), there are also remarkable differences between populations of neural crest cells originating from different axial levels within a given species. These include differences in mechanisms of delamination and developmental potential, such as the ability to generate skeletal structures (Graham et al. 2004). For example, although both cranial and trunk crest cells can generate the full repertoire of neural crest cell derivatives (McGonnell & Graham 2002), the skeletogenic potential of trunk crest cells is suppressed during normal development (Graham et al. 2004). Thus, different neural crest cell populations may well be exposed to at least a subset of unique regulatory interactions.
Finally, only a few cis-regulatory studies of neural crest genes have been reported thus far, which has made it difficult to discern direct regulatory interactions. Most known direct regulatory interactions have been elucidated in differentiating neural crest derivatives (Sauka-Spengler & Bronner-Fraser 2008a). Thus, the current formulation of the neural crest GRN is largely a consolidation of regulatory predictions. Nevertheless, many regulatory steps appear to be highly conserved even in basal vertebrate systems (Sauka-Spengler et al. 2007), which suggests that it should be possible to assemble a scaffold of regulatory interactions that may be common to all vertebrates and may function on all axial levels.
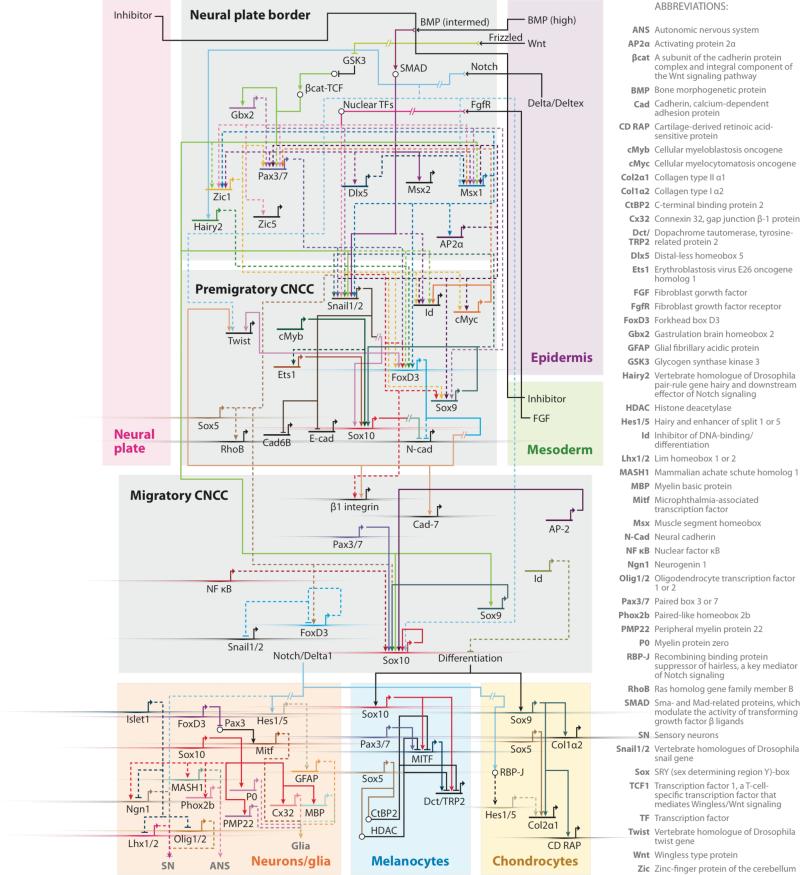
In this review, we attempt to integrate the most current neural crest regulatory information to generate an updated representation of the neural crest GRN. We present possible circuit connections inferred largely from loss-of-function analysis together with direct regulatory interactions, thus far documented mostly at later stages of differentiation. The goal is to build a model in which each link can be tested in several species. We also attempt to take into account separate spatial subpopulations of neural crest cells at different levels of the neural axis. As a starting point, we will focus on the regulatory state of cranial neural crest cells (Figures 1 and 2 and Table 1), which are the first crest population to form and initiate migration in the vertebrate embryo. These cells contribute derivatives mainly to the facial skeleton, peripheral nervous system, and pigmentation in the head.
Figure 1.
A gene regulatory network (GRN) model (a view from all nuclei) that maps vertebrate hierarchical gene regulatory interactions during cranial neural crest cell (CNCC) development. The model is built using the BioTapestry software (Longabaugh et al. 2009). The GRN is partitioned into subnetworks that regroup regulatory interactions during induction and specification at the neural plate border, in premigratory and migrating neural crest cells, and in differentiating neural crest derivatives. Most of the linkages in the GRN model are inferred from available gene perturbation data from Xenopus, chicken, mouse, zebrafish, and lamprey. Direct regulatory interactions, based on promoter and cis-regulatory analysis, are indicated with solid lines. Dashed lines show potential direct regulatory interactions inferred from gene perturbation studies. Broken lines represent potential indirect interactions. Bubble nodes indicate protein-protein interactions. Transcriptional orientation was not taken into consideration because it varies among different vertebrate models.
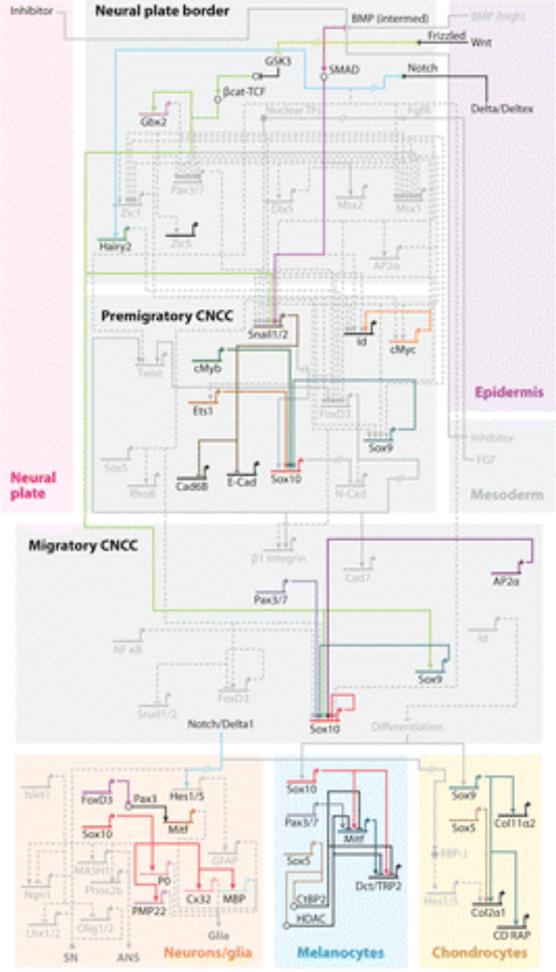
Figure 2.
A model of the GRN underlying CNCC formation that emphasizes only regulatory circuits with experimentally validated direct regulatory interactions.
Table 1.
Evidence for gene regulatory interactions in cranial neural crest cells
| Source | Interaction | Target | System | Evidence |
|---|---|---|---|---|
| BMP | Promotes* | Msx2 | Mouse | Brugger et al. 2004 |
| BMP | Promotes | Dlx5 | Xenopus | Luo et al. 2001 |
| BMP, FGF, Wnt, Notch, Gbx2, Dlx5, AP-2, Myc | Promotes | Msx1 | Xenopus, lamprey | Monsoro-Burq et al. 2005, Tribulo et al. 2003, Li et al. 2009, Woda et al. 2003, Nikitina et al. 2008 |
| BMP, Wnt, FGF, Gbx2, AP-2, Zic, Myc | Promotes | Pax3/7 | Xenopus, lamprey | Sato 2003, [**AU: This reference is not in the Lit Cited. Please add there or delete here.**] Hong & Saint-Jeannet 2007, Monsoro-Burq et al. 2005, Li et al. 2009, Nikitina et al. 2008 |
| BMP, Wnt, FGF, Msx | Promotes | Zic1 | Xenopus, lamprey | Sato 2003, [**AU: This reference is not in the Lit Cited. Please add there or delete here.**] Hong & Saint-Jeannet 2007, Nikitina et al. 2008 |
| FGF | Promotes | Zic5 | Xenopus | Monsoro-Burq et al. 2003 |
| Wnt | Promotes* | Gbx2 | Xenopus | Li et al. 2009 |
| Notch | Promotes* | Hairy2 | Xenopus | Glavic et al. 2004 |
| Msx | Promotes | AP-2 | Lamprey | Nikitina et al. 2008 |
| FGF, Hairy2, Zic1, Msx1, Pax3/7, Gbx2, AP-2, Sox9, Sox10, Sox5 | Promotes | Snail1/2 | Xenopus, chicken | Mayor et al. 1997, Villanueva et al. 2002, Glavic et al. 2004, Sato et al. 2005, Tribulo et al. 2003, Meulemans & Bronner-Fraser 2004, Li et al. 2009, Spokony et al. 2002, Aoki et al. 2003, Honore et al. 2003, Perez-Alcala et al. 2004 |
| BMP, Wnt, | Promotes* | Snail1/2 | Mouse, Xenopus | Sakai et al. 2005, Vallin et al. 2001 |
| FoxD3 | Represses | Snail1/2 | Zebrafish | Lister et al. 2006 |
| Notch | Promotes | Twist | Xenopus | Coffman et al. 1993, Cornell & Eisen 2005 |
| Snail1/2, FoxD3 | Promotes (Ind) | Twist | Xenopus | Aoki et al. 2003, Aybar et al. 2003, Sasai et al. 2001, Meulemans & Bronner-Fraser 2004 |
| cMyc | Promotes* | Id | Xenopus | Light et al. 2005 |
| BMP, Wnt, Zic, AP-2 | Promotes | Id | Xenopus, lamprey | Kee & Bronner-Fraser 2005, Nikitina et al. 2008 |
| Zic, AP-2 | Promotes | Myc | Lamprey | Nikitina et al. 2008 |
| Hairy2, Zic1, Pax3/7, Msx1, Sox10, Sox5 | Promotes | FoxD3 | Xenopus, chicken | Wettstein et al. 1997, Sato et al. 2005, Tribulo et al. 2003, Honore et al. 2003, Perez-Alcala et al. 2004 |
| Snail1/2 | Promotes (Ind) | FoxD3 | Xenopus | Aoki et al. 2003, Aybar et al. 2003 |
| FoxD3 | Represses | FoxD3 | Xenopus | Pohl & Knochel 2001 |
| cMyb | Promotes | Ets1 | Chicken | Betancur, unpublished data |
| Wnt | Promotes* | Sox9 | Mouse | Bagheri-Fam et al. 2006 |
| Ap-2, Gbx2, Zic1, Sox10 | Promotes | Sox9 | Xenopus, mouse | Lee et al. 2004, Luo et al. 2003, Saint-Germain et al. 2004, Bagheri-Fam et al. 2006, Li et al. 2009, Honore et al. 2003 |
| Id | Represses | Sox9 | Xenopus | Light et al. 2005 |
| cMyb, Ets1, Sox9, Wnt, Pax3/7, AP-2 | Promotes* | Sox10 | Chicken, mouse, zebrafish | Betancur et al. 2010, Werner et al. 2007, Dutton et al. 2008 |
| Sox5, Notch, NFKappaB | Promotes | Sox10 | Chicken | Perez-Alcala et al. 2004, Dutton et al. 2008 |
| Snail1/2 | Promotes (Ind) | Sox10 | Xenopus | Aoki et al. 2003, Aybar et al. 2003 |
| Id | Represses | Sox10 | Xenopus | Light et al. 2005 |
| Snail1/2 | Represses* | Cad6B, Ecad | Chicken, mouse, and human cell lines | Taneyhill et al. 2007, Cano et al. 2000 |
| Sox10 | Represses (Ind) | Ncad | Chicken, mouse | Cheung et al. 2005 |
| FoxD3 | Represses | Ncad | Chicken, mouse | Cheung et al. 2005 |
| RhoB | Modulates | Ncad | Groysman et al. 2008 | |
| FoxD3 | Promotes (Ind) | Cad7 | Chicken, mouse | Cheung et al. 2005 |
| FoxD3 | Promotes (Ind) | B1 Integrin | Chicken, mouse | Cheung et al. 2005 |
| Sox10 | Promotes | B1 integrin | Chicken, mouse | Cheung et al. 2005 |
| Sox5 | Promotes | RhoB | Chicken | Perez-Alcala et al. 2004 |
| Notch | Promotes* | Hes1/5 | HeLa cell line | Jarriault et al. 1995 |
| Hes1/5 | Promotes | GFAP, Col2a1 | Mouse | Ijuin et al. 2008 |
| Sox10 | Promotes* | Mitf | Cell lines | Verastegui et al. 2000 |
| Sox5 | Modulates* | Mitf | Mouse | Stolt et al. 2008 |
| Sox10, Mitf | Promotes* | Dct/TRP2 | Cell lines | Ludwig et al. 2004 |
| Sox5 | Modulates* | Dct/TRP2 | Mouse | Stolt et al. 2008 |
| Sox10 | Promotes* | P0, Cx32, MBP, PMP22 | Mouse, cell lines | Peirano et al. 2000, Bondurand et al. 2001 |
| Sox10 | Promotes | Phox2B, MASH1, Ngn1 | Rat cell culture, zebrafish | Kim et al. 2003, Carney et al. 2006 |
| Sox9, Sox5 | Promotes* | Col2a1 | Cell lines, mouse | Lefebvre et al. 1997, Hattori et al. 2008 |
| Sox9 | Promotes* | Col11a2, CD RAP | Cell lines, mouse | Bridgewater et al. 1998, Xie et al. 1999 |
| Islet1 | Represses | Ngn1, Lhx1/2, Olig1/2 | Mouse | Sun et al. 2008 |
Direct regulatory interaction (data available) (Ind) Possible indirect interaction.
We present this updated neural crest GRN, created using the generic drawing software BioTapestry (http://www.biotapestry.org/), which employs symbolic representation of genes to describe their regulatory interactions and to integrate experimentally derived network features (Figure 1; Longabaugh et al. 2009). Most data in the neural crest GRN relates to cells forming at cranial levels.
INITIAL SIGNALING INPUTS INTO THE NEURAL CREST GENE REGULATORY NETWORK: BMP, WNT, FGF, AND NOTCH
PATHWAYS IN INDUCTION AND SPECIFICATION
The classical view suggested that neural crest cell induction occurred during the process of neurulation, as the neural folds elevated. This was thought to occur as a consequence of interactions resulting from the juxtaposition of the epidermis and the elevating neural plate (Mancilla & Mayor 1996, Selleck & Bronner-Fraser 1995). However, recent findings in frog (Monsoro-Burq et al. 2005) and chicken demonstrate that neural crest induction is underway much earlier, during gastrulation (Basch et al. 2006). In chicken, for instance, the transcription factor Pax7 is expressed in the neural plate border domain, where neural crest cells originate, in the mid-gastrula as early as stage HH4+. When tissue explants from this Pax7-positive domain of the gastrula were cultured in the absence of exogenous inductive signals, they were able to generate neural crest cells (Basch et al. 2006) despite the lack of added factors or other tissue interactions. Recent fate map studies show that the neural plate border region is wider and overlaps partially with the bone morphogenetic protein (BMP) 4-expressing domain during gastrula stages (Ezin et al. 2009), which is consistent with the possibility that signaling cues are already in play at this place and time.
Evidence of early specification of the neural plate border in frog and chicken has been substantiated by studies in lamprey, where these events are conserved but occur at a much slower rate, which makes lamprey a suitable system for studying signaling inputs and neural plate border specifier readout with much better temporal resolution and therefore in much higher detail (Nikitina et al. 2008). Interestingly, the induction program and resulting expression of transcription factors specifying the neural plate border is shared by nonvertebrate chordates that do not possess a neural crest (Meulemans & Bronner-Fraser 2004) (Sauka-Spengler & Bronner-Fraser 2008b). Thus, all evidence suggests that neural crest cell induction in vertebrate embryos occurs during gastrulation. However, the early inductive events remain unexplored in some species, such as the mouse, which highlights the importance of performing comparative analysis in numerous vertebrates.
The induction of the prospective neural crest within the neural plate border is thought to occur in response to signaling molecules emanating from adjacent tissues. The response that sets future neural crest cells apart from other border cells requires the activation of a battery of transcription factors, which imbues them with multipotency, the characteristics of proliferating cells, and the competence to respond to later neural crest--specifying signals. Identifying the signaling inputs that initiate neural crest induction has been challenging because information obtained from different vertebrate systems is sometimes contradictory. Fate map studies suggest that presumptive neural crest cells are in proximity to three different regions: presumptive epidermis, neural plate, and mesoderm. These tissues are thought to secrete signaling ligands including BMPs, Wnts, [**AU: Spell out Wnt?**] and fibroblast growth factors (FGFs) that have all been demonstrated as essential for the early induction, maintenance, and differentiation of neural crest cells (Knecht & Bronner-Fraser 2002). Although there are differences between neural crest populations at various levels of the neural axis, the inductive signals appear similar regardless of axial level.
Bone Morphogenetic Proteins
In Xenopus embryos, high levels of BMP have been shown to be necessary for the acquisition of epidermal fate, whereas inhibition of BMPs is required for neural induction (LaBonne & Bronner-Fraser 1998). The neural plate border territory that lies between non-neural ectoderm (future epidermis) and neural ectoderm contains neural crest precursors, preplacodal ectoderm, dorsal neural tube, and epidermis, all of which are exposed to BMP signals. In chicken explant culture experiments, juxtaposition of non-neural ectoderm and intermediate neural plate tissue, which normally forms only neural tube, can generate neural crest cells. Addition of BMP4 and BMP7, which are endogenously expressed in the non-neural ectoderm, is able to substitute for non-neural ectoderm such that neural crest cells are induced from intermediate neural plate explants (Liem et al. 1995). It has been proposed that intermediate levels of BMP, obtained as a result of diffusion of secreted BMP molecules throughout the ectoderm (BMP gradient), are responsible for the induction of neural crest cells. In support of the gradient model, zebrafish BMP pathway mutants show either expansion or reduction of the neural crest cell domain depending on the alteration of BMP levels Knecht & Bronner-Fraser 2002, Nguyen et al. 1998). Alternatively, a gradient that would create the intermediate levels of BMP required for neural crest induction may be established by antagonistic interactions with Cerberus, noggin, chordin, and follistatins, ligands secreted by the forming neural plate cells (Sauka-Spengler & Bronner-Fraser 2008a, Tribulo et al. 2003, Wilson et al. 1997). Regardless of the way a BMP gradient is established, intermediate levels of BMP alone are not sufficient to induce expression of neural crest cell markers in Xenopus or any other vertebrate model organisms (Garcia- Castro et al. 2002, LaBonne & Bronner-Fraser 1998, Wilson et al. 1997). BMP signaling is therefore an important initial step, but additional signals are required for induction of the neural crest.
Fibroblast Growth Factors
The FGF family of growth factors represents another set of signaling cues implicated in neural crest induction. In Xenopus animal cap assays, FGF2 ligand, together with attenuated BMP signaling, upregulates expression of an early neural crest cell marker, Snail2, whereas overexpression of a dominant negative FGF receptor blocks Snail2 without affecting neural plate markers (Mayor et al. 1997, Villanueva et al. 2002). In Xenopus, overexpression of FGF8, normally expressed in the paraxial mesoderm, transiently induces neural crest cells (Monsoro-Burq et al. 2003). However, exogenous FGF8 alone is not sufficient to induce the full range of neural crest markers (Noden & Trainor 2005). Furthermore, the requirement for FGF signaling may vary between species, which makes it difficult to make definitive conclusions about its universality. For example, mouse null mutant embryos lacking either FGF or FGFR have no obvious defects in neural crest formation (Jones & Trainor 2005). This could be explained by functional redundancy of FGF signaling factors. Similarly, in zebrafish neural crest cells develop normally in the absence of mesoderm (Jones & Trainor 2005), and mutant embryos carrying mutations in FGF signaling components show no neural crest defects.
Wnt Signaling Pathway
Wnt family members are involved in many aspects of neural crest development. Numerous family members, e.g., Wnt6, Wnt7b, Wnt3a, Wnt1, and Wnt8, are expressed in the correct tissue and at the proper time to play a role in induction (Knecht & Bronner-Fraser 2002). Wnts are present in the paraxial mesoderm in frog (Christian et al. 1991, Knecht & Bronner-Fraser 2002) and in the non-neural ectoderm adjacent to the neural folds in chicken embryos (Garcia-Castro et al. 2002). Gain- and loss-of-function experiments in frog, chicken, and fish have shown that the activation of the Wnt pathway is essential for neural crest cell induction and specification (Garcia-Castro et al. 2002, LaBonne & Bronner-Fraser 1998, Lewis et al. 2004). For instance, in zebrafish, an inducible Wnt inhibitor activated during early neurulation specifically interferes with neural crest cell formation without altering the formation of neurons from the central nervous system (Lewis et al. 2004). In chicken, ectodermal cells express Wnt6 at the time of neural crest cell induction, and exposing neural plate explants to Wnt6 induces the formation of neural crest cells in culture (Garcia-Castro et al. 2002, Schubert et al. 2002) However, the role of Wnt signaling in induction of the neural crest during gastrulation has yet to be examined in the mouse embryo. Although Wnt1/Wnt3a double mutants exhibit defects in a wide range of neural crest derivatives (cranial skeleton, cranial and even dorsal root ganglia, and melanocytes), it is not yet clear if this results from early induction defects, as the analysis of a mutant phenotype in the neural plate border has yet to be performed (Ikeya et al. 1997). All other gene perturbation experiments used as evidence to suggest a role for Wnt signaling in mouse are confined to lineage specification and neural crest cell differentiation rather than early induction. These studies have targeted the Wnt signaling pathway components in the dorsal neural tube (Jones & Trainor 2005); this represents a relatively late time point by which bona fide neural crest progenitors reside within the dorsal aspects of the neural folds/tube. Thus, it is too late to address the role of Wnt signaling in induction events, which normally take place during gastrulation. Thus it remains unclear if Wnt signaling pathways play an inductive role at early stages.
Wnt/β-catenin in emigrating neural crest cells clearly promotes formation of sensory neurons at the expense of all other derivatives (Lee et al. 2004). Finally, due to gene duplications and the particularly large number of Wnt ligands in the mouse genome, it is possible that Wnts act redundantly during neural crest cell development in mouse. Their early inductive role may have been missed in single Wnt knockouts, but the effects of simultaneous inactivation of several Wnts have not been examined to date (Jones & Trainor 2005).
Local cell-cell signals such as Notch/Delta are also found in the vicinity of and/or on developing neural crest cells (Endo et al. 2002, Glavic et al. 2004, Williams et al. 1995). In chick, Notch is confined to the neural folds together with Hairy2, its direct downstream effector, whereas Delta is expressed in the presumptive epidermis (Endo et al. 2002). It has been reported that Notch-Delta signaling acts upstream of BMP4 in chick and Xenopus embryos and can affect expression of Snail and other neural crest specifier genes (Endo et al. 2002, Glavic et al. 2004). However, the function of and requirement for Notch during neural crest cell development may vary among different vertebrates. In mouse, Delta1 null mutants have no apparent early neural crest defects even though cranial neural crest cells express several Notch genes (De Bellard et al. 2002, Williams et al. 1995); a different ligand may activate Notch signaling in those cells. In zebrafish, mutants in Notch pathway components appear to affect the trunk but not the cranial neural crest (Cornell & Eisen 2005), which is consistent with the possibility that this signaling pathway plays more of a role in the trunk than the cranial crest, where there may be functional redundancy with other signaling pathways.
Despite some species-specific differences, it is generally agreed that a combination of inductive signals activates a battery of immediately downstream genes in the neural plate border that give the cells the capacity to become neural crest cells. For instance, the combination of low levels of BMP plus Wnt family members can induce expression of Snail2 and other neural crest genes in Xenopus explants (LaBonne & Bronner-Fraser 1998).
NEURAL PLATE BORDER SPECIFIERS
Signaling inputs into the neural plate border territory activate a battery of transcription factors whose collective expression sets presumptive neural crest cells apart from other border progenitors by conferring on them the competence to respond to neural crest--specifying signals. These genes, termed neural plate border specifiers, appear early during neurulation and include homeobox transcription factors Mxs1/2, Dlx5, Pax3/7, and Gbx2, as well as zinc finger--containing Zic proteins. Although little is known about the direct inputs that regulate their expression or about the regulatory interactions that occur among them, gain- and loss-of-function experiments suggest possible hierarchical interrelationships. Understanding their regulatory interrelationships helps expand links within the GRN, adds several testable hypotheses, and can serve as an experimental guide.
In Xenopus, integration of inputs from the BMP, FGF, Wnt, and Notch signaling pathways activates expression of Msx1 (Monsoro-Burq et al. 2005, Tribulo et al. 2003). Zic1and Pax3 are also downstream of Wnt, BMP, and FGF signals (Sato et al. 2005), whereas FGF8 can experimentally induce Zic5 expression but is not required to do so endogenously (Monsoro-Burq et al. 2003). Although BMP and FGF signals can regulate individual expression of Zic1 and Pax3, both transcription factors need to be activated simultaneously to achieve neural crest specification. In Xenopus embryos, high levels of either transcription factor alone (Pax3 or Zic1) promote alternative neural plate border fates (hatching gland or preplacodal progenitors, respectively) (Hong & Saint-Jeannet 2007). Furthermore, FGF8 and Wnt signals act in parallel at the neural plate border and seem to converge independently onto Pax3 (Monsoro-Burq et al. 2005). Hairy2, a direct downstream effector of Delta/Notch input into the neural plate border territory, also participates in the regulation of neural crest specifier genes (Glavic et al. 2004). Dlx5, which is regulated by attenuated levels of BMP (Luo et al. 2001), expands the Msx1 expression domain upon ectopic activity (Woda et al. 2003).
Because neural plate border specifiers are the first transcription factors to appear at the border, it is not surprising that they may be directly activated by the simultaneous input of multiple signaling pathways. Although evidence for direct interactions is sparse, Brugger and colleagues show direct conversion of the intermediate levels of BMP signal onto the Msx2 promoter Msx2 (Brugger et al. 2004). Recently, Li and colleagues found that Gbx2, a gene essential for the anteroposterior partitioning of neural folds, is expressed in an ectodermal region that includes the future neural plate border from which crest cells will arise (Li et al. 2009). The authors demonstrated that Gbx2 is an immediate direct downstream target of Wnt signaling. Furthermore, epistatic rescue experiments reveal that Gbx2 is positioned upstream of the earliest previously reported neural plate border specifiers, Msx1 and Pax3. These results suggest Gbx2 as a candidate for mediating the earliest Wnt inductive signaling input into the neural crest GRN.
Studying the hierarchical interrelationships between newly activated neural plate border specifiers is challenging because of the inaccessibility and/or rapidity of the induction and border specification processes in most vertebrate models. Due to their slow development, however, lamprey embryos allow unprecedented temporal resolution of neural plate border specification. This has enabled chronological ordering of the onset of gene expression among neural plate border specifiers as well as gene perturbation assays to establish their hierarchical relationships. A study by Nikitina and colleagues establishes Msx, but also the neural crest specifier AP-2a, the top of the neural plate border cascade, with many of the factors present at the border (both known border specifiers such as Msx, Pax3/7, or Zic, as well as early crest specifiers such as AP-2a, n-Myc, or Id) feeding back and regulating each other's expression (Nikitina et al. 2008). It will be interesting to further investigate direct regulatory relationships at the border as well as to test similar interactions in higher vertebrates, such as chick embryo, which also have good temporal resolution of neural plate border specification.
NEURAL CREST SPECIFIER GENES
The regulatory state during neural crest specification is defined by the cumulative expression of a set of genes, termed neural crest specifiers, in the premigratory and early-migrating bona fide neural crest progenitors. Some neural crest specifiers persist in migrating and differentiating neural crest cells (such as Sox10), whereas others such as Snail2 are present only at the onset of the specification process and the EMT prior to their emigration. Some neural crest specifiers have a biphasic expression pattern in which they are present first in neural crest progenitors and again later in differentiating derivatives (e.g., Sox9). A subgroup of transcription factors such as AP-2a, Snail1/2, Id, c-Myc, and Twist are expressed even before neural crest progenitors become apparent, though the timing of their onset and presence within the neural plate border varies among different vertebrates. In a basal vertebrate, the lamprey, expression of this subgroup of early-expressing neural crest specifiers begins at the early neurula stage, preceding expression of canonical neural crest markers such as Sox10 and FoxD3 (Nikitina & Bronner-Fraser 2009, Sauka-Spengler et al. 2007). This raises the intriguing possibility these genes may function as a key regulatory link between the establishment of competence in the presumptive crest at the neural plate border and the specification of bona fide neural crest cells. During specification, neural plate border genes either directly or indirectly regulate neural crest specifier genes. They also receive signaling pathway inputs and undergo intricate cross-regulatory activity with other neural crest specifiers.
The regulatory control of Snail2 exemplifies how signaling pathways and regulatory factors merge to direct the expression of a key gene involved in the EMT of neural crest cells. Cis-regulatory analysis shows that Snail2 is directly regulated by intermediate levels of BMP, which are modulated by Wnt pathway input. Accordingly, the Snail2 regulatory region contains binding motifs for Smad1, a transcription factor that mediates BMP signaling input (Sakai et al. 2005), and Tcf/Lef1, which mediates the β-catenin-dependent Wnt signal (Vallin et al. 2001). Furthermore, in Xenopus animal cap explants, a combination of the BMP inhibitor chordin and Wnt8 is sufficient to induce the expression of Snail2 as well as Id3, a helix-loop-helix (HLH) transcriptional regulator involved in specification of the neural crest (Kee & Bronner-Fraser 2005). Overexpression of Hairy2, a direct downstream effector gene of Notch signaling, causes an expansion of Snail2 expression in Xenopus (Glavic et al. 2004) and has been proposed as a direct input into the Snail2 regulatory region. Finally, it has been demonstrated that the neural plate border specifiers Zic1, Msx1, and Pax3/7 are independently necessary and sufficient for the expression of a group of neural crest cell specifiers including Snail2 (Meulemans & Bronner-Fraser 2004, Sato et al. 2005, Tribulo et al. 2003). This suggests that regulatory signaling inputs activating Snail may be mediated by neural plate border specifiers such as Zic1, Msx1, and Pax3/7. Conversely, signaling inputs can act in parallel with upstream border specifiers to control neural crest specifier expression. For instance, in Xenopus embryos, β-catenin-dependent canonical Wnt signals cooperate with Zic1 and Pax3/7 to activate Snail2 expression (Sato et al. 2005).
Far less is known about the regulation of other neural crest specifiers. Twist, for instance, is ectopically activated upon Snail2 and FoxD3 misexpression in Xenopus embryos and ectodermal explants, perhaps indirectly via Zic (Meulemans & Bronner-Fraser 2004, Sasai et al. 2001). In contrast, expression of a constitutively activated truncated version of a Notch receptor in Xenopus embryos downregulates Twist expression, simultaneously causing the neural plate to expand and the epidermis to regress. Thus, it is not clear if the loss of Twist expression is a result of regulatory changes caused by a shift in signaling or a secondary effect owing to neural plate expansion at the expense of the neural crest (Coffman et al. 1993, Cornell & Eisen 2005). Although it is intriguing to speculate that Zic1 mediates Notch-Twist regulation, currently no data either support or refute this possibility. Some early neural crest cell specifiers, such as Id and cMyc, appear to function within the neural crest GRN to maintain the neural crest cells in a multipotent state, mediating critical cell cycle and/or cell fate (Bellmeyer et al. 2003, Kee & Bronner-Fraser 2005, Light et al. 2005). Id is a transcriptional repressor that possesses a HLH domain for dimerization but lacks a basic domain for DNA binding. Id proteins interfere with gene expression by binding to transcriptional activators from bHLH families and preventing them from activating their direct targets. In lamprey, initial expression of Id at the neural plate border precedes that of cMyc (Nikitina & Bronner-Fraser 2009). However, in Xenopus embryos cMyc can directly regulate Id expression (Light et al. 2005), which indicates that other factors, such as AP-2a or Zic1, may be responsible for the initial expression of Id (Nikitina et al. 2008). Therefore, cMyc functions directly upstream of Id, via the identified cis-regulatory region, to maintain its expression in premigratory neural crest cells.
By the time premigratory and delaminating neural crest cells express transcription factors such as FoxD3, Sox9, Snail2, or Sox10, they are specified to a neural crest fate. The winged-helix transcription factor FoxD3 appears to play a role in maintaining neural crest multipotency by preventing early differentiation (Lister et al. 2006). Direct regulatory inputs responsible for FoxD3 activation and maintenance in premigratory and migrating neural crest cells have yet to be described. Similar to Snail2 activation, evidence from studies in Xenopus embryos suggests that a Hairy2-mediated Notch signal regulates FoxD3 expression (Wettstein et al. 1997). In addition, the collective activity of Zic1 and Pax3/7 complemented with Wnt input induces FoxD3 expression (Sato et al. 2005). Gain- and loss-of-function experiments in Xenopus have also shown that Msx1 regulates FoxD3 expression (Tribulo et al. 2003).
The SoxE family of transcription factors, most notably Sox9 and Sox10, have well-established roles in neural crest development. In Xenopus, Sox9 expression is dependent on the activity of AP-2a (Lee et al. 2004, Luo et al. 2003, Saint-Germain et al. 2004). Moreover, in silico database searches have identified AP-2a binding motifs within the early-acting Sox9 cis-regulatory region in mouse (Bagheri-Fam et al. 2006). In Xenopus it has been shown that Gbx2 together with Zic1 can induce the expression of neural crest specifier genes including Sox9 and Snail2 while inhibiting preplacodal fate (Li et al. 2009). However, the direct regulatory inputs into Sox9 have yet to be experimentally demonstrated.
Recently Ets1 and cMyb have been added to the neural crest GRN as new neural crest specifier genes directly regulating the onset of Sox10 expression. Extensive characterization of the initial Sox10-activating cis-regulatory element in chicken embryo (Betancur et al. 2010) reveals that the synergistic activity of Ets1, cMyb, and Sox9 directly regulates the onset of Sox10 in the cranial neural crest via an early cranial Sox10 enhancer. The possible role of the proto-oncogene cMyb in neural crest cell development was first suggested in migrating trunk neural crest cells, where the knockdown of cMyb reduced Snail2 expression (Karafiat et al. 2005). However, cMyb expression in chicken begins much earlier, at the gastrula stage. It becomes confined to the neural folds as the neural plate begins to invaginate and later continues to be expressed in migrating crest cells (Betancur et al. 2010). Knockdown of cMyb in the cranial neural crest causes a diminution of Sox10 expression, which confirms that this factor acts upstream of Sox10. Ets1 expression is specific to the cranial crest population and first appears in neural crest progenitors in chicken embryos as the neural folds are closing (Theveneau et al. 2007). Trunk neural crest cells, which normally do not express Ets1, arrest in the G1 phase of the cell cycle prior to separating from the neuroepithelium and synchronously enter the S phase upon delamination. Interfering with the G1/S transition prevents the delamination process from occuring (Burstyn-Cohen & Kalcheim 2002). Ectopic expression of Ets-1 in the trunk region suggests that it promotes massive migration independent of the cell cycle (Sauka-Spengler & Bronner-Fraser 2008a, Theveneau et al. 2007), which is more like migration in the cranial region. These data, together with the finding that Ets1 directly regulates Sox10 specifically in cranial crest cells, raise the intriguing possibility that in the cranial neural crest Ets1 may have the unique function of establishing a regulatory state that activates cranial crest--specific effector genes responsible for the transition from the premigratory to migratory state. The differential expression of Ets1 and its regulatory relationship to other neural crest genes highlights interesting differences between neural crest populations at different levels of the neural axis.
Neural crest cell specifiers, in general, represent a node point onto which inductive inputs mediated by or acting in parallel with neural plate border specifiers converge. Those specifying transcription factors in turn control the expression of effector genes that will give neural crest cells their unique migratory and multipotent characteristics. Therefore, in the life cycle of a neural crest cell, it is critical to keep the specifier genes running as a unit in the network. For this purpose in frog, high interdependence among neural crest cell specifiers seems to exist. Gain- and loss-of-function experiments suggest that Snail2 regulates FoxD3, Twist, and Sox10 expression, probably in an indirect fashion (Aoki et al. 2003, Aybar et al. 2003). Ectopic expression of AP-2a in the neural plate activates the ectopic expression of Snail2 (Spokony et al. 2002), whereas Sox10 feeds back to maintain Snail2, Sox9, and FoxD3 expression (Honore et al. 2003). However, in mouse and zebrafish, cross-regulation among neural crest cell specifiers is less tight because knockouts of Snail1 and 2, Sox10, and AP-2a have effects later, during differentiation in selective neural crest derivatives, rather than at this state of specification (Meulemans & Bronner-Fraser 2004). Perhaps in other organisms, neural crest specifier genes have a more redundant function during specification, and then their function becomes more restricted as the neural crest advances to the differentiating state. Conversely, this discrepancy may be due to the higher rate of gene duplication and functional compensation by redundant paralogs (Lister et al. 1999, Luo et al. 2001, Yan et al. 2005). Only through characterization of cis-regulatory modules will we be able to understand the degree of importance of these neural crest cell specifier cross-regulatory events.
GENES REGULATING NEURAL CREST EMIGRATION AND MIGRATION
To initiate migration, premigratory neural crest cells must delaminate from the neuroepithelium. Thus, transcription factors acting on the neural crest precursor pool must not only maintain the precursors in a multipotent and proliferating state, but also activate or repress effector genes involved in their EMT. To allow cells to become less compact and acquire motility, the EMT induces changes at the cellular level that include switches in cell junctions and adhesion properties as well as major cytoskeletal rearrangements. One characteristic of the EMT process is a switch in cadherin expression that involves upregulation of type II cadherins that allow for less adhesiveness and concomitant downregulation of type I cadherins and other factors characteristic of epithelial cell types. For example, in trunk neural crest cells in the chick, forced expression of FoxD3 downregulates N-cadherin (N-Cad, a type I cadherin) while concomitantly upregulating expression of Cad-7, a type II cadherin, and β1 integrin (Cheung et al. 2005). Because FoxD3 is a repressor, the upregulation is likely to be indirect. Confirming a role for FoxD3 during delamination, misexpression of FoxD3 along the entire dorsoventral axis of the chicken neural tube caused an increase in expression of neural crest cell markers, including Cad-7, and promoted delamination and migration from more ventral regions of the neural tube while simultaneously repressing interneuron differentiation (Dottori et al. 2001). Normally, Cad-7 is only expressed in migrating crest cells and excluded from the neural tube (Nakagawa & Takeichi 1995). Similar to FoxD3, Sox10 overexpression induces β1 integrin expression while inhibiting N-Cad expression (Cheung et al. 2005). Although it is difficult to ascribe direct gene regulatory interactions, it is clear that both FoxD3 and Sox10 affect expression of EMT effector genes, such as cadherins, whose orchestrated regulation is crucial for EMT to occur.
Snail1 and Snail2 genes have a clear role in controlling cell adhesiveness and many other aspects of EMTs in embryonic and metastatic cells (Thiery & Sleeman 2006). Snail1 is directly responsible for the negative regulation of E-cadherin, a cell adhesion molecule characteristic of epithelial cells (Cano et al. 2000). Similarly, Snail2 acts directly to negatively regulate the expression of Cad-6B, a molecule that characterizes cell-cell adhesion among dorsal neural tube cells, most of which are premigratory neural crest progenitors (Taneyhill et al. 2007). Sox5, a member of the SoxD family, is another transcription factor proposed to have a regulatory role during neural crest cell delamination. Sox5 misexpression causes an increase in the number of cranial neural crest cells generated. Sox5 upregulates Snail2, FoxD3, and Sox10 in migrating crest cells and cell autonomously upregulates RhoB, a member of the Rho family of small GTPases that controls a variety of signal transduction pathways (Perez-Alcala et al. 2004). RhoB is a well-known regulator of events that change cell morphology such as actin cytoskeleton rearrangements as well as the formation of focal adhesions and stress fibers (Liu & Jessell 1998). All these cellular changes are necessary for neural crest delamination (Nobes & Hall 1995). The function of RhoB in cranial crest cells appears to be distinct from that in the trunk, where it acts as a negative modulator, downregulating N-Cad and preparing cells for delamination (Groysman et al. 2008). Again, cis-regulatory profiling will confirm if the subcircuit initiated by Sox5 consists of direct feed-onto Snail, FoxD3, Sox10, and RhoB regulatory modules in delaminating cranial crest cells. Other studies have demonstrated that Sox5 can bind to cis-regulatory modules via known motifs, previously identified as Sox9 and Sox10 binding sites, and can modulate expression of downstream target genes by recruiting specific cofactors during neural crest cell differentiation (Hattori et al. 2008, Stolt et al. 2008). The same regulatory mechanism likely is used during cranial crest delamination. Because Sox5 appears early in the premigratory neural crest, it may be also be involved in the regulatory interactions that take place during neural crest specification. However, this possibility remains to be explored.
Most of the transcription factors that are involved in neural crest cell specification continue to be expressed in neural crest cells as they migrate. However, other unidentified upstream inputs, different from those that initiate expression of neural crest specifiers in the premigratory state, may be responsible for maintaining their expression during migration. Moreover, different upstream regulators may be characteristic of neural crest cells with various differentiation potentials, correlated with their future fate. For example, inactivation of Wnt signaling input sites within the Sox9 enhancer decreased reporter expression exclusively in neural crest cells migrating into the first but not the second or third branchial arches (Bagheri-Fam et al. 2006). Cis-regulatory analysis in mouse has shown that during neural crest migration, Sox10 is directly regulated by Pax3, AP-2, and Sox9 but also receives Wnt signaling input (Werner et al. 2007). Analysis in zebrafish confirms that a Wnt signal feeds directly to the Sox10 regulatory element during migration but also strongly suggests SoxE, NFkB, and Notch signals as potential direct Sox10 regulatory inputs (Dutton et al. 2008). These studies also demonstrated that there is no direct regulatory interaction between FoxD3 and Sox10 despite the presence of FoxD3 binding motifs in Sox10 cis-regulatory regions. However, FoxD3 has been reported as a negative regulator of Sox10 (Pohl & Knochel 2001, Sasai et al. 2001). It is plausible that the negative feed of a FoxD3 repressor on the Sox10 regulatory module may have been missed because assays employed to identify direct regulators are more targeted to isolation of positive regulatory influences (activators). Conversely, FoxD3 may function as a regulator of Sox10 activity via currently unidentified enhancers. Alternatively, the loss of Sox10 expression after FoxD3 inactivation may suggest that their functional interactions are not direct and perhaps involve an intermediary inhibitor.
Prior to and during neural crest migration, cells acquire signaling receptors that allow them to interact with their environment and help guide them along specific pathways. In the cranial region such molecules include Neuropilin-1/2, Robo-1/2, and Ephrin receptors (Sauka-Spengler & Bronner-Fraser 2008a). However, the transcription factors that regulate expression of these signaling molecules remain elusive. Similarly, not much information is available regarding the upstream regulators of genes that are involved in cell cycle decisions prior to cranial neural crest cell delamination; only a few regulatory interactions that prevent cells from undergoing apoptosis have been described. In Sox9 null mice, neural crest cells undergo massive apoptosis (Cheung et al. 2005). Similarly, zebrafish neural crest cells lacking Sox9 within the branchial arches show a predominant cell death phenotype (Yan et al. 2005). Gain- and loss-of-function experiments in Xenopus suggest a direct regulatory connection between Sox9 and another antiapoptotic factor, Snail1 (Aoki et al. 2003).
In summary, the combined regulatory function of neural crest specifier genes and their downstream effectors endows neural crest cells with the characteristics that renderi them mesenchymal, proliferative, and motile. However, out of the many neural crest downstream effector genes, the direct regulatory inputs and links to upstream neural crest specifiers are known for only a few, which makes it difficult to assign their precise positions within the neural crest GRN.
THE TRANSITION FROM MIGRATION TO DIFFERENTIATION
How neural crest cells lose their migratory and multipotent characteristics as they prepare to differentiate remains an open question. It is logical to postulate that a separate set of gene batteries is deployed in each neural crest lineage. Cis-regulatory analysis combined with functional and binding affinity assays have revealed several subcircuits of direct gene regulatory interactions for each lineage. After neural crest cells have migrated and reached their final destinations, typically expression of most early neural crest cell specifiers, including Snail/Snail2, FoxD3, Id, and AP-2, is downregulated, although the direct regulatory interactions triggering or mediating this downregulation are elusive (Meulemans & Bronner-Fraser 2004). Nevertheless, some evidence suggests that FoxD3 participates in repression of Snail1b (previously Snail2) in zebrafish. Its absence causes prolonged expression of Snail1b when it would normally be turned off (Lister et al. 2006). Exogenous expression of FoxD3 in Xenopus causes repression of endogenous FoxD3, indicating that FoxD3 can directly downregulate its own expression in a negative autoregulatory loop (Pohl & Knochel 2001). Downregulation of FoxD3 in migrating cells prior to differentiation does not take place in all neural crest--derived lineages. Although absent from melanoblasts, FoxD3 expression persists in neural/glial precursors, where it prevents Pax3 from binding to the promoter of Microphtalmia-associated transcription factor (MITF) and thus prevents sensory precursors from assuming a pigment cell fate (Thomas & Erickson 2009). These data demonstrate the importance of negative regulation in cell fate acquisition in cell types with multiple developmental potentials such as the neural crest. It will be essential to study the differential upstream inputs that confine FoxD3 or other repressive circuits that could act as regulatory switches between different lineages.
Notable exceptions are SoxE transcription factor family members Sox9 and Sox10, which persist in specific subpopulations of neural crest cell derivatives and appear to be master regulators of terminal differentiation in the majority of neural crest derivatives (Kelsh 2006, Sauka-Spengler & Bronner-Fraser 2008b). The necessity of different SoxE genes for specification of distinct neural crest sublineages has recently been demonstrated in zebrafish (Arduini et al. 2009). Sox9 and Sox10 are maintained in cartilage and neuron/glial/melanocyte lineages, respectively, such that Sox10 persists in melanoblasts and elements of the peripheral nervous system, whereas Sox9 is characteristic of neural crest--derived chondrocytes. Experiments in Xenopus suggest that the HLH transcriptional repressor Id prevents premature neural crest cell differentiation during neural crest migration. Constitutive expression of Id family members in migrating neural crest cells populating the pharyngeal arches, most of which would normally adopt a cartilage fate, extends Sox10 expression, which is normally downregulated in this population when the cells enter the arches (Light et al. 2005). Furthermore, overexpression of Id3 in Sox10-expressing melanoblasts or Sox9-expressing neural crest--derived cartilage cells inhibits SoxE expression, which affects melanocyte and chondrocyte differentiation (Light et al. 2005). Thus, downregulation of Id is necessary for the initial steps of neural crest cell differentiation to occur. It is plausible that endogenous downregulation of Id indirectly releases inhibitors that feed into the neural crest specifier module. Maintaining expression of Sox9 and Sox10 until the time of differentiation, however, may be independent of the Id regulatory cascade. Strong evidence indicates that Id helps establish the time window during which cells respond to differentiating signals (Light et al. 2005). At the proper time, it may release activator genes involved in differentiation and maintenance of Sox9 and Sox10 expression in their respective differentiated lineages.
Another possibility is that the inhibitory activity of Id maintains Sox10 and perhaps Sox9 expression at low levels. It has been suggested that low concentrations of Sox10 sustain the multipotency of neural crest cells and at higher levels inhibit neuronal differentiation and promote glia and melanoblast formation (Kim et al. 2003, Paratore et al. 2001). Resolving regulatory interactions to the detail that would allow unraveling of these complex events remains a challenge. The battery of genes involved in maintaining neural crest cells may change such that new regulatory interactions emerge, some of which may involve redeployment of transcription factors involved in early neural crest cell specification to perform a later function in cell differentiation. For example, the way that Sox9 and Sox10 acquire new, instructive roles in directing the fate of certain neural crest derivatives may involve acquisition of new cofactors.
DIFFERENTIATION OF THE CRANIAL NEURAL CREST
Neural crest cells give rise to a wide variety of derivatives ranging from melanocytes, glia, and neurons to skeletal components of the head. In general, the type of derivative depends upon the axial level from which the neural crest cells originate and the time of their emigration from the neuroepithelium. For example, midbrain and rhombomere (r) 1 and r2 neural crest cells contribute to the neurons and glia of the trigeminal ganglion as well as to the skeleton of the upper and lower jaw. Neural crest cells from r4 give rise to neurons of the proximal facial ganglion and the hyoid bone. Neurons of the proximal and jugular ganglia and skeletal components of the postpharyngeal arches are derived from post-otic neural crest streams r6 and r7 (Graham et al. 2004, Lumsden et al. 1991, Schilling & Kimmel 1994). The vagal neural crest forms the enteric nervous system as well as cardiac and aortic arch components. The trunk neural crest forms sensory and autonomic ganglia and the adrenal medulla.
The time of migration also influences the types of derivatives that neural crest cells form. Early migrating cranial neural crest cells populate the pharyngeal arches to generate bone, cartilage, and connective tissue (skeletal structures), whereas the later wave stays close to the central nervous system and generates the neurons and glia of the cranial ganglia (Graham et al. 2004). Melanocytes are derived from neural crest cells from all axial levels. In mouse, a subpopulation of neural crest cells within a dorsomedial domain of the neural tube at the midbrain-hindbrain junction migrates exclusively into the developing dermis and expresses melanocyte lineage markers (Trainor 2005).
Of all the cranial neural crest derivatives, melanocytes and chondrocytes are the two lineages in which the most cis-regulatory work has been performed, and this research allows predictions regarding regulatory subcircuits. In melanocytes, Sox10, in synergy with Pax3, directly regulates Mitf by binding to a proximal region of its promoter (Bondurand et al. 2001, Verastegui et al. 2000). Then, in collaboration with Mitf, Sox10 directly regulates expression of an enzyme necessary for melanin synthesis, dopachrome tautomerase (Dct/TRP2; Ludwig et al. 2004).
Sox5 also plays a direct modulatory role in melanocyte differentiation. Sox5 and members of the SoxD family of transcription factors are characterized by their lack of a transactivation domain (Lefebvre et al. 1998). It has been speculated that they regulate transcription by recruiting other coactivators or corepressors to regulatory regions. On one hand, in melanocytes, Sox5 binds to the Mitf and Dct/TRP2 promoter regions through Sox10-identified binding elements. It recruits the corepressors CtBP2 and HDAC to compete with Sox10 for binding of these regulatory regions and therefore modulates Sox10-inducing activity (Stolt et al. 2008). During chondrocyte development, on the other hand, Sox9 directly regulates expression of important cartilage markers such as Collagen type II a (Col2a1; Lefebvre et al. 1997), Col1a2 (Bridgewater et al. 1998), and CD RAP (Xie et al. 1999) by binding to sites in identified enhancer regions. Interestingly, Sox5 null mice have skeletal defects and particularly craniofacial defects (Smits et al. 2001). This suggests another role for Sox5 in chondrocyte development. Consistent with this possibility, Sox5 was recently found to cooperate with Sox9 and other cofactors in chondrocytes to regulate expression of Col2a1 by binding to Sox9 target sites (Hattori et al. 2008). These inputs at the effector level of the neural crest GRN are a few notable examples of how precise gene regulatory subcircuits can guide a neural crest subpopulation to differentiate into specific derivatives.
Little is known about direct regulatory interactions in the specification and differentiation of cranial neural crest cells into glia and neurons. Most knowledge about direct regulatory interactions in neurogenic neural crest derivatives comes from experiments performed in trunk neural crest cells. These studies show that differentiation into neural crest--derived neurons and glia requires redeployment of factors utilized earlier during neural crest induction and specification. As an example, Notch and Delta proteins are expressed in neural crest cells that populate the presumptive trigeminal ganglion region, where they undergo gliogenesis and neurogenesis. The Notch signaling pathway promotes gliogenic differentiation while inhibiting neuronal differentiation (Nakamura et al. 2000, Ohtsuka et al. 1999). Furthermore, different mediators of Notch signaling appear to control, in part, the cell fate decision between gliogenic and skeletogenic differentiation. Whereas the Deltex-mediated Notch pathway controls gliogenesis, simultaneous activation of the RBP-J and the Deltex-dependent Notch pathways leads to chondrogenic specification (Ijuin et al. 2008), which is mediated by the previously characterized Notch downstream effectors Hes1 and Hes5 (Jarriault et al. 1995). The downstream readouts used to differentiate the gliogenic and chondrogenic lineages were glial fibrillary acidic protein (GFAP) and Col2a1, respectively. Thus, the above-mentioned studies place these specific markers as potential effector genes that act directly downstream of Notch signaling inputs mediated by Hes1 and Hes5 (Ijuin et al. 2008).
In addition to its role in melanocyte differentiation, Sox10 also controls specification of glial and neuronal fates in neural crest derivative specification. Sox10 appears to further participate in the differentiation of glia, as its expression within this lineage persists into terminal differentiation stages (Kelsh 2006). During glial differentiation, Sox10 directly regulates the expression of protein zero (P0) (Peirano et al. 2000), myelin basic protein (MBP), peripheral myelin protein 22 (PMP22) and the gap junction protein connexin 32 (Cx32), thus affecting all major components of the myelination process (Bondurand et al. 2001). Finally, evidence concerning the direct regulatory role of Sox10 during differentiation of neural crest--derived neurons comes from studies of sensory and autonomic lineages in the trunk. In mouse neural crest cell cultures, Sox10 regulates the expression of mouse achaete-scute homolog 1 (MASH1) and the paired homeodomain (Phox2b), transcription factors that are essential for autonomic neurogenesis (Kim et al. 2003). Sensory neurons derived from the dorsal root ganglia transiently express Sox10, which has been shown to regulate the expression of proneural gene neurogenin 1 in zebrafish (Carney et al. 2006). Similar interactions involving direct Sox10 regulatory inputs and expression of the sensory neuronal marker Neurogenin1 may take place during cranial neurogenesis.
Finally, it is important to stress the role of negative regulation during the steps of terminal differentiation into neural crest derivatives. A recent study by Sun and colleagues shows that LIM-homeodomain factor Islet1 specifically regulates subprograms within different sensory neuron lineages (Sun et al. 2008). At the end of the neurogenic phase of development, Islet1 is specifically required to repress/terminate the expression of genes such as Neurogenin1 or NeuroD family members. Interestingly, Islet1 is also required to repress several transcription factors not normally expressed in the sensory ganglia but found in the spinal cord and hindbrain, such as LIM-homeobox factors Lhx1 and Lhx2 and oligodendrocyte markers Olig1 and Olig2. This suggests that Islet1 inhibition also serves as a control switch that keeps cells within the sensory lineage (Sun et al. 2008).
CONCLUSION AND FUTURE PROSPECTS
In this review, we present an overview of the GRN orchestrating the formation of neural crest cells, with a focus on the cranial level. Formulation of this network relies on information largely inferred from studies of the molecular mechanisms underlying neural crest formation in several vertebrate model organisms. It also includes all known cis-regulatory information obtained to date, which provides evidence for direct regulatory interactions and architectural circuitry between the molecular factors involved. The neural crest GRN presented here can be used as a guide for future experiments to test if predicted direct regulatory connections hold true for different vertebrate model organisms. The future promise of high throughput cis-regulatory and transcriptional profiling of neural crest cells at each regulatory step will provide further information that can be assembled into a high resolution GRN.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, et al. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 2003;259:19–-33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Arduini BL, Bosse KM, Henion PD. Genetic ablation of neural crest cell diversification. Development. 2009;136:1987–-94. doi: 10.1242/dev.033209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–-94. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, et al. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 2006;291:382–-97. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–-22. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in Xenopus. Dev. Cell. 2003;4:827–-39. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Betancur PA, Sauka-Spengler T, Bronner-Fraser M. A key regulatory enhancer for cranial neural crest: genomic code for Sox10. Proc. Natl. Acad. Sci. USA. 2010 doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N, Girard M, Pingault V, Lemort N, Dubourg O, Goossens M. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum. Mol. Genet. 2001;10:2783–-95. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J. Biol. Chem. 1998;273:14998–-5006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, et al. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development. 2004;131:5153–-65. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Kalcheim C. Association between the cell cycle and neural crest delamination through specific regulation of G1/S transition. Dev. Cell. 2002;3:383–-95. doi: 10.1016/s1534-5807(02)00221-6. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–-83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–-30. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005;8:179–-92. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Christian JL, McMahon JA, McMahon AP, Moon RT. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development. 1991;111:1045–-55. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- Coffman CR, Skoglund P, Harris WA, Kintner CR. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–-71. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 2005;16:663–-72. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- De Bellard ME, Ching W, Gossler A, Bronner-Fraser M. Disruption of segmental neural crest migration and ephrin expression in delta-1 null mice. Dev. Biol. 2002;249:121–-30. doi: 10.1006/dbio.2002.0756. [DOI] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–-38. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 2008;8:105. doi: 10.1186/1471-213X-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Osumi N, Wakamatsu Y. Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development. 2002;129:863–-73. doi: 10.1242/dev.129.4.863. [DOI] [PubMed] [Google Scholar]
- Ezin AM, Fraser SE, Bronner-Fraser M. Fate map and morphogenesis of presumptive neural crest and dorsal neural tube. Dev. Biol. 2009;330:221–-36. doi: 10.1016/j.ydbio.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–-51. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004;131:347–-59. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Graham A, Begbie J, McGonnell I. Significance of the cranial neural crest. Dev. Dyn. 2004;229:5–-13. doi: 10.1002/dvdy.10442. [DOI] [PubMed] [Google Scholar]
- Groysman M, Shoval I, Kalcheim C. A negative modulatory role for rho and rho-associated kinase signaling in delamination of neural crest cells. Neural. Dev. 2008;3:27. doi: 10.1186/1749-8104-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol. Dev. 2000;2:3–-5. doi: 10.1046/j.1525-142x.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- Hattori T, Coustry F, Stephens S, Eberspaecher H, Takigawa M, et al. Transcriptional regulation of chondrogenesis by coactivator Tip60 via chromatin association with Sox9 and Sox5. Nucleic Acids Res. 2008;36:3011–-24. doi: 10.1093/nar/gkn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 2007;18:2192–-202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore SM, Aybar MJ, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev. Biol. 2003;260:79–-96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Ijuin K, Nakanishi K, Ito K. Different downstream pathways for Notch signaling are required for gliogenic and chondrogenic specification of mouse mesencephalic neural crest cells. Mech. Dev. 2008;125:462–-74. doi: 10.1016/j.mod.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–-70. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–-58. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jones NC, Trainor PA. Role of morphogens in neural crest cell determination. J. Neurobiol. 2005;64:388–-404. doi: 10.1002/neu.20162. [DOI] [PubMed] [Google Scholar]
- Karafiat V, Dvorakova M, Krejci E, Kralova J, Pajer P, et al. Transcription factor c-Myb is involved in the regulation of the epithelial-mesenchymal transition in the avian neural crest. Cell Mol. Life Sci. 2005;62:2516–-25. doi: 10.1007/s00018-005-5297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Bronner-Fraser M. To proliferate or to die: role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes Dev. 2005;19:744–-55. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN. Sorting out Sox10 functions in neural crest development. Bioessays. 2006;28:788–-98. doi: 10.1002/bies.20445. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–-31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat. Rev. Genet. 2002;3:453–-61. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–-14. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Lee HY, Kleber M, Hari L, Brault V, Suter U, et al. Instructive role of Wnt/β-catenin in sensory fate specification in neural crest stem cells. Science. 2004;303:1020–-23. doi: 10.1126/science.1091611. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the Proα1(II) collagen gene. Mol. Cell Biol. 1997;17:2336–-46. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–-33. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JL, Bonner J, Modrell M, Ragland JW, Moon RT, et al. Reiterated Wnt signaling during zebrafish neural crest development. Development. 2004;131:1299–-308. doi: 10.1242/dev.01007. [DOI] [PubMed] [Google Scholar]
- Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–-78. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82:969–-79. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Light W, Vernon AE, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005;132:1831–-41. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev. Biol. 2006;290:92–-104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–-67. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–-67. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim. Biophys. Acta. 2009;1789:363–-74. doi: 10.1016/j.bbagrm.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 2004;556:236–-44. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–-91. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2α. Proc. Natl. Acad. Sci. USA. 2003;100:532–-37. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Matsuo-Takasaki M, Lim JH, Sargent TD. Differential regulation of Dlx gene expression by a BMP morphogenetic gradient. Int. J. Dev. Biol. 2001;45:681–-84. [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev. Biol. 1996;177:580–-89. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Mayor R, Guerrero N, Martinez C. Role of FGF and noggin in neural crest induction. Dev. Biol. 1997;189:1–-12. doi: 10.1006/dbio.1997.8634. [DOI] [PubMed] [Google Scholar]
- McGonnell IM, Graham A. Trunk neural crest has skeletogenic potential. Curr. Biol. 2002;12:767–-71. doi: 10.1016/s0960-9822(02)00818-7. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev. Cell. 2004;7:291–-99. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Fletcher RB, Harland RM. Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development. 2003;130:3111–-24. doi: 10.1242/dev.00531. [DOI] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell. 2005;8:167–-78. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–-32. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, et al. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J. Neurosci. 2000;20:283–-93. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VH, Schmid B, Trout J, Connors SA, Ekker M, Mullins MC. Ventral and lateral regions of the zebrafish gastrula, including the neural crest progenitors, are established by a bmp2b/swirl pathway of genes. Dev. Biol. 1998;199:93–-110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc. Natl. Acad. Sci. USA. 2008;105:20083–-88. doi: 10.1073/pnas.0806009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitina NV, Bronner-Fraser M. Gene regulatory networks that control the specification of neural-crest cells in the lamprey. Biochim. Biophys. Acta. 2009;1789:274–-78. doi: 10.1016/j.bbagrm.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem. Soc. Trans. 1995;23:456–-59. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat. 2005;207:575–-601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–-207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratore C, Goerich DE, Suter U, Wegner M, Sommer L. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 2001;128:3949–-61. doi: 10.1242/dev.128.20.3949. [DOI] [PubMed] [Google Scholar]
- Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol. Cell Biol. 2000;20:3198–-209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–-65. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- Pohl BS, Knochel W. Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech. Dev. 2001;103:93–-106. doi: 10.1016/s0925-4773(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Saint-Germain N, Lee YH, Zhang Y, Sargent TD, Saint-Jeannet JP. Specification of the otic placode depends on Sox9 function in Xenopus. Development. 2004;131:1755–-63. doi: 10.1242/dev.01066. [DOI] [PubMed] [Google Scholar]
- Sakai D, Tanaka Y, Endo Y, Osumi N, Okamoto H, Wakamatsu Y. Regulation of Slug transcription in embryonic ectoderm by β-catenin-Lef/Tcf and BMP-Smad signaling. Dev. Growth Differ. 2005;47:471–-82. doi: 10.1111/j.1440-169X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–-36. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–-63. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 2008a;9:557–-68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol. Bull. 2008b;214:303–-14. doi: 10.2307/25470671. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell. 2007;13:405–-20. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–-94. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Schubert FR, Mootoosamy RC, Walters EH, Graham A, Tumiotto L, et al. Wnt6 marks sites of epithelial transformations in the chick embryo. Mech. Dev. 2002;114:143–-48. doi: 10.1016/s0925-4773(02)00039-4. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Origins of the avian neural crest: the role of neural plate-epidermal interactions. Development. 1995;121:525–-38. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell. 2001;1:277–-90. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–-32. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol. 2005;16:647–-54. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Hillgartner S, Wegner M. The transcription factor Sox5 modulates Sox10 function during melanocyte development. Nucleic Acids Res. 2008;36:5427–-40. doi: 10.1093/nar/gkn527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 2008;11:1283–-93. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–-90. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006;7:131–-42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–-58. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA. Specification and patterning of neural crest cells during craniofacial development. Brain Behav. Evol. 2005;66:266–-80. doi: 10.1159/000088130. [DOI] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Nguyen VH, Mullins MC, Mayor R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development. 2003;130:6441–-52. doi: 10.1242/dev.00878. [DOI] [PubMed] [Google Scholar]
- Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/β-catenin signaling. J. Biol. Chem. 2001;276:30350–-8. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J. Biol. Chem. 2000;275:30757–-60. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- Villanueva S, Glavic A, Ruiz P, Mayor R. Posteriorization by FGF, Wnt, and retinoic acid is required for neural crest induction. Dev. Biol. 2002;241:289–-301. doi: 10.1006/dbio.2001.0485. [DOI] [PubMed] [Google Scholar]
- Werner T, Hammer A, Wahlbuhl M, Bosl MR, Wegner M. Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 2007;35:6526–-38. doi: 10.1093/nar/gkm727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–-702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech. Dev. 1995;53:357–-68. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Wilson PA, Lagna G, Suzuki A, Hemmati-Brivanlou A. Concentration-dependent patterning of the Xenopus ectoderm by BMP4 and its signal transducer Smad1. Development. 1997;124:3177–-84. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- Woda JM, Pastagia J, Mercola M, Artinger KB. Dlx proteins position the neural plate border and determine adjacent cell fates. Development. 2003;130:331–-42. doi: 10.1242/dev.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J. Bone Miner. Res. 1999;14:757–-63. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–-83. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]