Abstract
Atrial fibrillation (AF) is the most common arrhythmic disorder, and currently affects nearly 3 million Americans, 8.8 million Europeans, and an estimated 30 million individuals worldwide. The clinical risk factors for AF are numerous, with age, sex, hypertension, obesity, and ischemic heart disease among the most prevalent. Over the last ten years, a preponderance of evidence also suggests a large genetic contribution to AF. The earliest report of familial AF dates to the early 1940s1. Since then, it has become apparent that AF in referral populations2,3 and in the community is heritable4,5. Indeed, having a family member with AF is associated with a 40% increased risk for the arrhythmia6. Once the heritability was recognized, traditional genetics techniques for the discovery of rare, monogenic causes of AF were used to identify the initial AF genes. These studies in turn, informed candidate gene screening in AF cohorts. To identify additional sources of heritability for AF, large-scale analyses of common variation through genome wide association studies (GWAS) has recently yielded data identifying risk loci in many regions of the genome. In spite of these advances, the combination of these techniques has, as yet, failed to completely identify the heritability of AF in the population. It is the goal of this review to examine the previous studies on rare variants, address the findings of the recent GWAS studies, and describe future avenues towards defining the heritability of AF.
Keywords: Atrial fibrillation, genetics, arrhythmia
Mendelian and Candidate Gene Studies
Classic genetic techniques such as linkage analysis have been used with great success to identify the genetic basis of hypertrophic cardiomyopathy, long QT syndrome and many other heritable conditions. Although there had been sporadic reports of families with AF over the last 60 years, the first application of such methods for AF arose from work by Bob Roberts and colleagues published in the New England Journal of Medicine in 19977. In this manuscript, they identified a genetic locus for AF using a series of related families with early-onset AF. Although the specific causative gene at this locus remains unknown, this study helped to firmly establish a genetic basis for some patients with AF.
In a seminal manuscript published in Science in 2003, Yi-Han Chen and colleagues identified the first gene for familial AF8. Using a large Chinese kindred with autosomal dominant AF, they found a gain of function mutation in KCNQ1 or the gene encoding the alpha subunit of the slowly repolarizing potassium channel current, IKs. The identification of a well-known ion channel mutation for AF quickly led many groups to turn to candidate gene screening of a wide range of cardiac genes. Indeed, several additional gain of function variants have been identified in KCNQ19–14. A challenge with the interpretation of these candidate gene studies is that most lack convincing genetic support in the form of variant transmission in extended families. With this limitation in mind, we have provided an overview of the genes related to AF in the following section, and we have included a detailed compendium of known AF variants in Table 1.
Table 1.
Compendium of AF genetic variants identified in families and individuals.
| Gene | Gene Name | Function | Citation(s) |
|---|---|---|---|
| ABCC9 | ATP-binding cassette, subfamily C, member 9 |
IKATP current | 129 |
| GATA4 | Transcription factor GATA-4 | Cardiac development | 45–47,130 |
| GATA5 | Transcription factor GATA-5 | Cardiac development | 48,49,131 |
| GATA6 | Transcription factor GATA-6 | Cardiac development | 50,132,133 |
| GJA5 | Connexin 40 | Formation of atrial gap junctions | 38,134–138 |
| GREM2 | Gremlin-2 | BMP antagonist | 139 |
| HCN4 | Hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
If current | 103 |
| JPH2 | Junctophilin-2 | Ca2+ homeostasis | 140 |
| KCNA5 | Potassium voltage-gated channel, shaker-related subfamily, member 5 |
IKur current | 15,24–26 |
| KCND3 | Potassium voltage-gated channel, Shal- related subfamily, member 3 |
Ito1 current | 16 |
| KCNE1 | Potassium voltage-gated channel, Isk- related family, member 1 |
Kv channel activity modulation | 19 |
| KCNE2 | Potassium voltage-gated channel, Isk- related family, member 2 |
Kv channel activity modulation | 20 |
| KCNE3 | Potassium voltage-gated channel, Isk- related family, member 3 |
Kv channel activity modulation | 21 |
| KCNE5 | KCNE1-like | Kv channel activity modulation | 22 |
| KCNH2 | Potassium voltage-gated channel, subfamily H (eag-related), member 2 |
IKr current | 141,142 |
| KCNJ2 | Potassium inwardly-rectifying channel, subfamily J, member 2 |
IK1 current | 17,18 |
| KCNJ5 | Potassium inwardly-rectifying channel, subfamily J, member 5 |
IKACh current | 143 |
| KCNJ8 | Potassium inwardly-rectifying channel, subfamily J, member 8 |
IKATP current | 144 |
| KCNQ1 | Potassium voltage-gated channel, KQT- like subfamily, member 1 |
IKs current | 8 – 14 |
| LMNA | Lamin A/B | Nuclear envelope structure | 145,146 |
| NKX2.5 | Homeobox protein Nkx2.5 | Cardiac development | 43 |
| NPPA | Natriuretic Peptide Precursor A | Systemic sodium homeostasis | 39,147 |
| NUP155 | Nucleoporin 155 | Nuclear pore formation | 148 |
| PITX2c | Paired-like homeodomain 2c | Great vein development, left-right asymmetry |
44 |
| RYR2 | Ryanodine Receptor 2 | Ca2+ release from sarcoplasmic reticulum |
149 |
| SCN1B | Sodium channel, voltage-gated, type I, beta subunit |
INa current modulation | 34,37 |
| SCN2B | Sodium channel, voltage-gated, type II, beta subunit |
INa current modulation | 37 |
| SCN3B | Sodium channel, voltage-gated, type III, beta subunit |
INa current modulation | 35,36 |
| SCN4B | Sodium channel, voltage-gated, type IV, beta subunit |
INa current modulation | 33 |
| SCN5A | Sodium channel, voltage-gated, type V, alpha subunit |
INa current | 27 – 32 |
Ion channel variation in AF
In the broadest terms, the majority of functionally validated, AF-associated potassium channel variants have a gain of channel function, with an expected shortening of the atrial action potential duration and atrial refraction period. In addition to KCNQ1, mutations have been identified in potassium channels genes including KCNA5,15, KCND3,16 and KCNJ217,18 and accessory subunits KCNE1,19 KCNE2,20 KCNE3,21 and KCNE5.22 Alternatively, it has also been demonstrated that prolongation of atrial action potentials caused by loss of function potassium channel mutations can lead to early after-depolarizations and AF,23 After an initial description by the Olson laboratory,24 additional mutations in KCNA5 of the IKur current have been reported in subsequent years15,25,26.
Variation in sodium channel subunits has also been identified as an important factor in the development of familial AF. Voltage-gated sodium channels (NaV) are responsible for initiating the upstroke during phase 0 of cardiac action potential and for the coordinated propagation of the action potential throughout the atria. Cardiac sodium channels are composed of a pore-forming alpha subunit, and beta subunits, which can alter channel trafficking and inactivation kinetics. To date, AF-causing variants have been observed in both the major cardiac sodium channel, encoded by SCN5A27–32, and four of its associated beta subunits33–37. Similarly to reports of potassium channel variation, both loss and gain of function variation seem to be capable of creating a pro-arrhythmogenic substrate.
Other Genes Discovered in Individuals and Families with AF
Several variants have also been identified in genes which do not directly alter the atrial action potential, but instead would be expected to instigate the onset of AF through alternative mechanisms. Along these lines, Gollob et al discovered a series of somatic mutations in GJA538, which encodes the gap junctional protein, Connexin 40. Interestingly, while this mutation was observed in atrial biopsies, it was not found in DNA isolated from blood. The extent to which somatic mutation or mosaicism contribute to the AF is unclear, and further study is often limited by the difficulty in obtaining primary samples. Further lending support to GJA5 as an AF candidate gene, recently, several reports have identified additional GJA5 loss of function variants that associate with disease. Since gap junctions are responsible for propagation of action potentials between cardiomyocyes, disruption of these complexes can result in reduced conduction velocity throughout the atrium, conditions that would be predicted to promote reentry.
Another study identified a frameshift mutation which resulted in early truncation of NPPA in an extensive family with lone AF39. NPPA encodes the precursor for atrial naturetic peptide (ANP), an important factor in the regulation of sodium homeostasis and, by association, blood pressure. This mutation was shown to increase the resistance of ANP to degradation, in essence causing an increase in ANP-mediated signaling40. In this study, when the mutant, mature ANP was perfused in a rat, whole heart, Langendorff model there was significant shortening of the atrial action-potential duration. While the APD shortening may be the major phenotype observed following acute treatment, prolonged systemic exposure to the mutant ANP could also cause AF-inducing structural remodeling, as seen in canine models41 and supported by the recent identification of an autosomal recessive mutation in NPPA in a family with severe atrial dilated cardiomyopathy42.
Finally, genes broadly characterized under the umbrella of developmentally related cardiac transcription factors have also been identified as being associated with AF. Specifically, genetic variation in NKX2.543, PITX244, GATA445–47, GATA548,49, and GATA650 have been described, although the mechanisms whereby these lead to disease have remained unclear.
Genome-Wide Association Studies of AF
Until the mid-2000s, linkage and candidate gene sequencing methods were the predominant approaches used to identify AF genes. In 2005, a novel technique, termed a genome-wide association study or GWAS, was utilized to identify genetic loci associated with age-related macular degeneration51. A GWAS relies on the unbiased comparison of common single nucleotide polymorphisms or SNPs throughout the genome. SNPs that occur with different frequency in individuals with a disease versus controls can localize disease-related genetic loci. While a potentially powerful tool for identifying genetic variation associated with common diseases, careful correction for multiple testing is necessary. Since that initial publication, over 1,700 GWAS have been published listing associations at nearly 12,000 SNPs. Among cardiovascular diseases, this technique has successfully identified risk loci for premature myocardial infarction52, hypertension53, lipid levels54, and electrocardiographic intervals55–60, among others.
Initial GWAS Studies of AF
The first GWAS performed for AF was published in 2007 and identified a region on chromosome 4q25 (sentinel SNP rs2200733) which was associated with AF in those of European and Asian descent61. Subsequently, these findings were broadly replicated in individuals of European62,63, Asian64, and African-American65 descent. Further analysis also identified the same genomic region as being associated with an increased risk of cardioembolic stroke 64,66,67 and a prolonged PR interval 56,68. In a recent meta-analysis of AF GWAS data, carriers of a single copy of the 4q25 variant had a nearly 65% increased risk of AF (odds ratio (OR) of 1.64 for rs2634073 (p=1.8×10−74) 69. A follow up fine mapping study of the 4q25 locus identified at least three independent association signals within this region70. When these three signals are considered together, there is a subset of ~1% of the population that has all six risk alleles and a nearly six-fold risk of AF (OR of 6.02, p=1.2×10−36).
The 4q25 risk region lies in a relatively gene-sparse intergenic region approximately 150 kb upstream from the PITX2 gene. Although at present there is no data linking the SNPs in this region to the expression levels of Pitx2, our current understanding of Pitx2 function suggests a plausible link with AF. PITX2 encodes the paired-like homeodomain 2 protein, a transcription factor which is crucial during embryogenesis and, notably for AF, cardiogenesis71–75. Pitx2 expression is near the closing stages of the left/right asymmetry program in vertebrates, with 100 fold higher expression in the left versus the right atrium76. Critical roles for Pitx2 have also been identified for formation of the atrial septum, outflow tract, SA node, and the pulmonary vein myocardial sleeves77,78. The last of these is of particular note given the prevalence of ectopic electrical foci arising from the pulmonary vein in patients with AF, and the common approach of electrically isolating the pulmonary veins to treat recurrent AF.
Evaluation of Pitx2 knockout mice have also been informative for potential mechanisms whereby misregulation of Pitx2 could contribute to AF. Specifically, whereas homozygous knockout is embryonic lethal, haploinsufficiency of the predominant cardiac isoform, PITX2c, results in a shortened atrial action potential and an increased susceptibility to AF following burst pacing76. The same study also identified continued expression of Pitx2c in the left atrial myocardium, but whether altered adult expression in the myocardium contributes to the causation of AF in the absence of developmental differences is unclear. Atrial-specific conditional knockout of Pitx2c also results in perturbation of the action potential and resting membrane potential79. Further, deletion of PITX2c expression results in diminished expression of cardiac sodium and potassium channels80.
Following this initial study, the need for greater statistical power was recognized and led to the formation of the CHARGE-AF or AFGen Consortium. In 2009, two groups independently identified a second locus for AF at 16q22 in Europeans and Han Chinese81,82. These results were later replicated in individuals of African-American descent65. The AF risk SNP at this locus is intronic to the gene ZFHX3, alternatively known as ATBF1, which encodes a zinc finger homeobox transcription factor. ZFHX3 expression has been identified as a factor in the terminal differentiation of both neuronal and striated muscle tissues83,84, and also reported as a putative tumor suppressor gene85,86. Given these roles in other tissues, and its apparent expression within cardiac tissues87, a developmental role in the atria is possible. However, the lack of availability of model systems with altered ZFHX3 expression has limited the understanding of its potential role in AF. Development of these resources will undoubtedly aid in the discovery of the potential mechanisms whereby this gene, and this susceptibility locus, may be related to AF.
In a separate GWAS from the CHARGE-AF Consortium, patients with early-onset AF were used in hopes of minimizing any sample heterogeneity that may have been seen in previous analyses. In a meta-analysis of five GWAS studies with early-onset AF, a region intronic to the KCNN3 gene was identified88. Similar to the majority of targets identified in candidate gene studies in familial AF, KCNN3 encodes a potassium channel responsible for membrane repolarization. The encoded protein, the SK3 channel, is a calcium-activated, small conductance potassium channel which has largely been studied for its role in neuronal electrophysiology. In neurons, SK3 acts in late repolarization to reduce excitability of neurons following repeated stimulation, a phenomenon termed afterhyperpolarization89. The role of the KCNN3 in the heart is much less clear, but some evidence exists for a role of SK family members in AF pathogenesis. Among these, studies regarding the deletion of the SK2 channel in mice found a prolongation in cardiac action potentials and increased susceptibility to AF90. Further, blockade of the SK family-mediated IK,Ca current also confers an increased risk of atrial arrhythmias in rodents91 and canines92. Finally, recent reports utilizing a mouse model of altered SK3 expression demonstrated alterations in atrial myocyte repolarization93 and an increased incidence of inducible atrial arrhythmias94. Together, these data suggest a mechanism whereby altered expression of SK3 may have important implications on the electrical stability of the atrium.
Meta-analysis Identification of Novel AF Loci
In 2010, the AFGen Consortium published a meta-analysis of GWAS data from 16 different studies meta-analysis in which six novel AF loci were identified in individuals of European and Japanese descent69. The following section will detail the identified loci and possible mechanisms how they might contribute to AF.
Genetic variants at the 1q24 locus, approximately 46kb upstream of the PRRX1 gene, were associated with a modest, 14% increased risk of AF (p= 8.4×10−14). PRRX1 encodes a member of the paired related homeobox gene family, transcription factors which broadly contribute to differentiation and developmental patterning. In humans, mutations in PRRX1 lead to agnathia-otocephaly95,96, a generally fatal condition characterized by severely altered craniofacial development. In rodent models, homozygous deletion of PRRX1 results in early postnatal death, and abnormal development of craniofacial, limb and vertebral structures97. In addition, PRRX1 is highly expressed in the developing great vessels and is essential to the proper formation of the pulmonary vein98,99. As discussed for PITX2, ectopic depolarizations within the pulmonary venous regions are often responsible for the initiation of AF. It remains unclear whether PRRX1 regulatory variation is related to congenital alterations in the pulmonary vein structure or function during development, or is instead associated with altered activity later in life.
Another association signal was localized intronic to HCN4 on 15q24. Hcn4 is highly expressed in both sinoatrial and atrioventricular nodes and is responsible for the funny current (If) that controls cardiac pacemaking. Interestingly, mutations in HCN4 have been found in individuals and families with sick sinus syndrome100–102, tachy-brady syndrome and AF103. Whether the risk locus for AF alters overall HCN4 expression levels to a sufficient extent to confer a risk for AF, or if this region results in critical expression differences in a tissue-specific manner remains to be determined.
A novel locus was also located on 7q31 intronic to the CAV1 gene that encodes Caveolin-1, a protein essential for the formation and maintenance of caveolae. The caveolae are regions of the membrane with unique phospholipid composition that act as mediators of clathrin-independent endocytosis and as scaffolds for cellular, particularly integrin-mediated, signaling. In addition to these roles, caveolae also harbor many ion channels104, including those responsible for all phases of the cardiac action potential. Studies of cardiovascular function in CAV1-null mice reported aberrant calcium signaling, and an alteration in myogenic tone105. Dilated cardiomyopathy, right ventricular hypertrophy and pulmonary hypertension have also been observed106. Further evaluation of the risk locus may aid in determination of the tissue-localized effect of CAV1 which leads to an increased risk of AF.
SNPs significantly associated with AF were identified on chromosome 14q23 intronic to SYNE2. The SYNE2 gene encodes Nesprin2, a KASH protein family member that localizes to the nuclear outer membrane. Through its binding with the cytoskeleton, Nesprins are thought to provide a stable nuclear localization in the cell107,108 and also are crucial for microtubule-mediated migration of the nucleus during differentiation109. Missense mutations in SYNE2 have been reported to cause familial Emery-Dreifuss muscular dystrophy110, a disease which is also characterized by a spectrum of arrhythmic disorders, including AF.
On chromosome 9q22, an association signal with AF was identified within the gene C9ORF3 (rs10821415, OR=1.11, p=4.2×10-11). However, this region is relatively gene rich, with 3 additional genes and 3 identified MIRs within 300kb of the sentinel SNP. Since none of the genes in the region have an obvious relationship with cardiovascular function or development, further investigation of this locus will be necessary.
Genetic variants associated with AF were also localized to an intergenic region between two genes known to play crucial roles in striated muscle physiology, SYNPO2L and MYOZ1 (10q22, rs10824026, OR=0.87, p=4.0×10−9). This SYNPO2L/MYOZ1 locus illustrates the utility of expression quantitative trait loci (eQTL) data to identify a disease-associated gene. Many intronic and intergenic SNPs identified by GWAS are thought to mediate their effects by regulating the transcription of a gene in the region. Sometimes there can be many genes at a locus so it can be difficult to know which is related to disease. Therefore, in eQTL mapping, one examines the relation between a SNP genotype and transcript levels of all genes at the locus, ideally from a relevant tissue. If a disease-related SNP is associated with transcriptional differences in a gene, this cis-eQTL association provides compelling support for the role of this gene in disease.
Initially, a SNP in LD with the sentinel SNP at SYNPO2L/MYOZ1 locus was found to correlate with alterations in the expression of both genes. However, this data was derived from lymphoblastoid cell lines, a tissue type unlikely to truly reflect the transcriptional alterations associated with AF. Recently, an eQTL analysis from left atrial tissue found that the AF SNP was associated with transcriptional differences in MYOZ1 expression alone111. Therefore, it is likely that MYOZ1 is the AF related gene at this locus. The encoded protein, myozenin 1, is a cardiac-enriched, z-disk localized protein which aids in the binding of α-actinin and γ-filamin to confer stable sarcomeric organization112. Although no known disease causing variants of MYOZ1 have been identified, mutations in MYOZ2 result in familial hypertrophic cardiomyopathy113, and replication of these mutations or ablation of expression in a murine model114 resulted in hypertrophic program activation and disruption of z-disk structure in ventricular myocytes.
Integrating GWAS Data to Stratify AF Risk
In summary, genome wide studies have identified 9 genetic loci associated with AF. Although the odds ratios for any given region are modest, the potential risk in a given individual may be much higher when the AF SNPs are considered together. Ultimately, utilizing these combined data would be important in a clinical setting, where risk could be stratified based upon a combination of genetic and clinical risk factors. Along these lines, Dr. Albert and colleagues derived a clinical risk score for AF in women without previous cardiovascular disease. The addition of a genetic risk score, consisting of the top 9 GWAS variants, improved AF risk prediction, but it did alter the reclassification into ten year risk categories115.
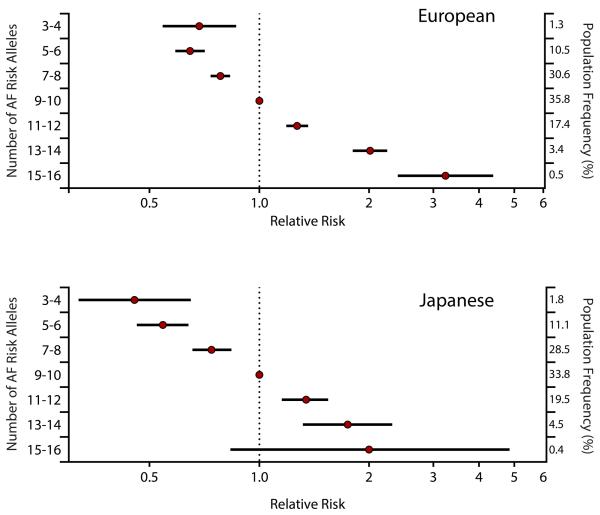
Interestingly, a recent large-scale conditional analysis in 17 cohorts from the AFGen Consortium, found that there are at least four different risk alleles at the 4q25/PITX2 locus for AF116. Consideration of these PITX2 SNPs plus the other 8 GWAS SNPs resulted in a nearly five-fold gradient in the risk of AF among individuals of European descent (Figure 1, European). The application of these same SNPs to a large Japanese population provided similar results (Figure 1, Japanese). As discussed below, with such a marked variation in AF risk in the population, it will be possible to identify individuals with both a marked increased and decreased risk for AF. Such genetic stratification of AF risk may ultimately enable an improved assessment of different treatment approaches or outcomes based on one’s risk.
Figure 1. Known genetic pathways for AF pathogenesis.
Schematic of known AF-related genes derived from previous studies. Genes listed include those where coding variation was identified in familial AF and candidate gene screens, as well as the genes suggested to be implicated in AF based upon GWAS. Names listed in red indicate those identified by familial studies and candidate gene screens. Those listed in gray are gene targets implicated by GWAS.
Future Directions for the Genetics of AF
Great strides have been made in determining the genetic risk for the development of AF; however, many challenges remain. In the following section, we outline a series of selected potential future directions for genetics studies of AF (Figure 1).
Identification of Additional AF Genetic Loci
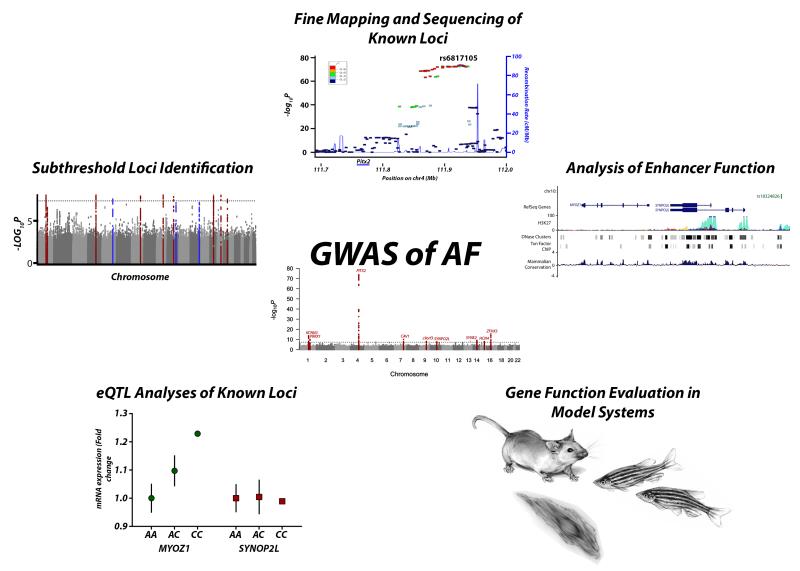
A qualitative viewing of the Manhattan plot from the latest publication by the AFGen Consortium reveals several association signals that rise well above the milieu of background noise, but do not exceed a genome-wide significance threshold (Figure 3). A logical extension of this work would then be to determine if these “subthreshold” loci are additional potential AF genetic risk loci. Genotyping AF associated SNPs from these subthreshold loci in a larger number of patient samples would likely lead a strengthening of an association signal for some loci. While genotyping these SNPs in additional cases is straightforward, the subthreshold loci that are found are likely to contribute to an ever decreasing fraction of AF risk. Thus, while newly identified loci are unlikely to have a large impact on clinical risk prediction, they could still be helpful to identifying more members of the molecular pathways that underlie AF.
Figure 3. Future directions for the study of GWAS risk loci.
Initial analyses of common variation have yielded 9 susceptibility loci for atrial fibrillation. Future pathways for confirming the causative variation include: Identification of subthreshold loci by increasing sample size or reduced sample heterogeneity in GWAS, fine mapping or direct sequencing of known risk loci for increased resolution of the causal region, in silico analyses of locus function to determine potential regulatory regions/causal variation, evaluation of AF candidate genes in model systems, and expression quantitative trait loci mapping to link common variation to altered gene expression in relevant tissues.
Future work should also focus on the identification of AF risk variants in different races and ethnicities. To date, the majority of discovery has been performed in populations of European descent, with limited work being done in individuals of Asian and African-American descent. Since AF prevalence varies greatly among races, it remains unclear whether the results from the studies of Europeans translate to other races, or if a different combination of risk loci are instead present. Studies in other races and ethnicities will be particularly important for the future application of genetic data to clinical care.
The Challenge of Causal Variant Identification at GWAS Loci
There are currently 9 identified genetic loci that are significantly associated with AF. However, despite the publication of the 4q25/PITX2 locus over 6 years ago, the causative variants at all of the AF loci remain unknown. One challenge is the sheer size of these genomic regions, as the PITX2 locus alone comprises a region of nearly 150 kilobases. Another challenge is that the top SNP identified by GWAS is rarely the causative variant, rather it is usually serving as a surrogate for a nearby causal variant. A final challenge is that the genetic mechanism for the association with AF is also unknown. We typically assume that AF risk is mediated by a SNP, but it is also quite possible that the association with AF could be due to a non-coding insertion or deletion, a genetic rearrangement, a variation in copy number, or an epigenetic modification.
To address these challenges a combination of techniques will be required (Figure 3). One approach could be to refine the genetic signal by fine mapping or increasing the density of SNPs within a target locus. This could be done directly genotyping more SNPs at a locus in a large population of cases and controls. Such an approach was used to identify multiple susceptibility signals at the 4q25 70,116. However, with the coverage of current genotyping platforms used for GWAS that consist of 1 to 5 million SNPs and the increasing resolution provided by the 1000 Genomes project, additional genotyping may have a limited incremental benefit.
As the turnaround time from submission to results-in-hand is now measured in weeks and the cost continues to drop, a viable complementary approach would be to sequence an entire disease locus. Importantly, sequencing would provide nucleotide-level resolution of the genetic architecture within AF risk loci. Thus, it would be expected that sequencing of AF risk loci in a large number of cases and controls will aid the identification of the causative haplotypes and variants associated with AF. Since the genetic variants identified by GWAS are markers of an association rather than a causative variant, one would anticipate that a causative SNP identified by sequencing would have a greater effect size and significance than the original GWAS signal. Sequencing a locus could also identify insertions/deletions or copy number variants that associate with disease and may be poorly described in current public databases. Finally, it is possible that sequencing could reveal multiple causative variants within a given locus, something that may not be identifiable by fine mapping. While such large-scale sequencing is currently feasible, the overall benefits remain unclear particularly given the significant cost of such projects.
In addition to refinement of the loci with fine-mapping and sequencing, it will be essential to integrate the vast amount of emerging regulatory data. Although coding variation is a possibility at some loci, a large majority of GWAS loci reside in intergenic or non-coding areas of the genome. This observation led to the assumption that these associations may be due to alterations of regulatory elements such as enhancers or promoters which, in turn, alter the activity of distant genes. Indeed some GWAS loci have been shown to alter transcription factor binding sites that in turn lead to differential expression of an adjacent gene 117. For this purpose, in silico analyses of data provided by the ENCODE project can be incredibly useful for determining the causal mechanism of variation at a genetic locus. Genomic regions with high mammalian conservation, increased DNase hypersensivity, increased H3K27-acetylation, and identification of transcription factor binding sites through chromatin immunoprecipitation sequencing can prove useful for identifying altered functional elements within a risk locus.
Although both sequencing and in silico analyses can provide a higher resolution map of a genetic locus, there may still be many candidate regulatory regions across the locus. Studies that can identify the functional role of a regulatory region will be a critical next step. For example, one could postulate that, at the 4q25/PITX2 locus, sequencing would allow the identification of the critical haplotypes that are associated with AF. An in silico analysis would then identify a number of highly conserved regions with enhancer activity. One could then examine these potential enhancers for activity in a model system such as mice, zebrafish or in an atrial or cardiomyocyte cell line. The causative genetic variant would then likely be one that is both significantly associated with disease and results in an alteration in enhancer activity.
While methods currently exist for each of the steps outlined above, sequencing, in silico analyses and functional follow up is expensive, slow, and challenging. The limited number of causative variants that have been identified at GWAS loci is not a problem specific to AF. Indeed, thousands of GWAS loci have been described, but causative variants have only been identified at a handful. Ultimately, a larger scale effort to systematically identify causative variants at GWAS loci will be necessary to overcome the obstacles faced by any single laboratory.
Atrial and Pulmonary Vein Specific eQTL Maps
As detailed above for the MYOZ1 locus for AF, eQTL maps, which examine the changes in tissue specific expression of nearby genes when a given SNP genotype is present, can provide a useful link between GWAS loci and potential gene targets. Such analyses of gene expression have been useful in studies of atrial identity 111 and other cardiovascular traits 118,119. While these eQTL associations at genetic loci can be helpful if they are present, the tissue-specificity of an eQTL signal is critical. Current publically available datasets such as the eQTL browser or the GTEx repository 120 have a limited tissue composition that reflects the challenge in obtaining relevant human tissue samples, but they are quickly expanding. For AF, it would be ideal to have eQTL data from much more specific tissue sources that are more plausibly involved in the pathogenesis of the arrhythmia. One would expect that the generation of publically available left atrial, pulmonary venous, or AV nodal eQTL datasets would greatly aid in the discovery of the mechanism of causal variation in AF.
The Exome Chip will Enable Large-scale Assessment of Rare Coding Variation in AF
The evaluation of GWAS loci discussed above was focused on non-coding regions, but it is important to realize that many loci are in linkage disequilibrium, and thus effectively overlap, with coding region of one or more genes. In these cases, it is possible that the GWAS SNP is a marker or proxy for a coding SNP that actually underlies the association signal. SNPs within a gene could have many potential effects including non-synonymous variation that directly alters protein function, synonymous variation that alters splicing, affects transcript stability or influences codon efficiency, or untranslated region variation that affect translational efficiency or interactions with non-coding regulatory RNAs.
Once a locus is identified that overlaps with a gene, one could genotype every SNP within the gene in a large number of cases and controls to see if it has a stronger association with AF than that identified by GWAS. While straightforward in concept, in practice, GWAS loci are large, they may contain many genes each of which can have many rare and common variants and the cost of genotyping remains relatively expensive. One solution to address this issue has been the development of an exome genotyping array or exome chip.
In a GWAS genotyping array, SNPs are captured throughout the entire genome, while in an exome array, the focus is largely on coding SNPs. Current exome arrays include over a quarter of a million SNPs that essentially capture almost all of the common and rare coding variants for every gene in the genome. Within the past year, hundreds of thousands of individuals have been genotyped with these arrays. Much like a GWAS analysis, by comparing a large number of cases and controls, one can quickly identify any coding changes associated with AF. The exome chip analysis can be considered with the GWAS results to simultaneously identify the coding variants within all of the known AF loci.
Exome genotyping arrays will be incredibly powerful at systematically identifying any known coding variation for AF and we can expect to see the initial results of these studies within the next year. However, since these arrays are only genotyping known SNPs, they would not be useful for studying sporadic or novel genetic variation in an individual or family. Detection of such variation would require direct sequencing of individual genes, exomes, or genomes.
Candidate Gene Screening will be Replaced by Exome and Genome Sequencing
As described earlier, many mutations described for AF have been identified using a candidate gene approach. In brief, the coding region of a gene is sequenced in AF cases, a unique variant is identified, and that variant is then shown to alter the function of a protein. While such studies are straightforward, they are limited by 1) the time and cost restraints of sequencing that restrict the analysis to a small number of genes, 2) the inability to detect polygenic causative variation, 3) the focus on coding variation, and, perhaps most importantly, 4) the limited likelihood that a particular candidate gene or variant within a gene is pathologically related to AF.
In the upcoming years, the continually decreasing cost and improving quality of next generation sequencing will enable the widespread adoption of sequencing the exome or protein coding region of the genome. We can expect that exome sequencing of cohorts of individuals with early-onset AF will provide a more comprehensive initial approach for relating rare genetic variation to AF; however, several challenges remain. For every individual sequenced, one can expect to find hundreds of unique non-synonymous SNPs or insertion deletions that have never been described in publicly available resources such as the Exome Variant Server. Thus, determining which variants are truly related to AF and which are genetic noise can be difficult. The identification of multiple hits in a given gene or genetic pathway across individuals can provide compelling evidence for the role of the gene or pathway in AF, yet large, well-powered studies will be required to make definitive conclusions. Improvements in the yield of such efforts may come from sequencing extremes of a phenotype such as cases of early-onset AF.
As costs continue to further decrease, genome sequencing will also become more realistic in cohort studies, yet with an even greater number of variants identified, assigning causality to noncoding variants will prove even more difficult. Given the continued challenges with large-scale sequencing approaches, an important step forward would be the creation of a centralized repository of exome and genome sequencing-derived variants identified in patients with AF. Comparison of variation in larger datasets on the scale of thousands rather than tens or hundreds of patients will aid in determining variants and genes that are truly causative for the arrhythmia.
Families can Provide a Unique Window into the Mechanisms of AF
Although much of our discussion has focused on using genetics to identify risk markers for AF in populations, familial forms of AF remain an important investigational tool. While families with autosomal dominant AF are rare, even a single family can shed light on the underlying molecular mechanisms for AF. To date, convincing evidence from families has identified the role of KCNQ1 and ANP in AF. Challenges with using families to identify AF genes include the rarity of the families, the limited number of individuals with AF, and the difficulty in ensuring that all family members have a common genetic basis for the disease. The last point is particularly pertinent given that the background prevalence of AF can be as high as 10%. Traditional linkage analysis has become increasingly easy to perform by using SNP chips to genotype family members at a high density. Furthermore, exome and genome sequencing can be quickly performed in affected family members. Although with exome sequencing one will still identify hundreds of variants in each person, the familial transmission of disease enables a focus on those variants shared among all affected family members. A combination of using linkage analysis to identify a genetic locus and exome or genome sequencing, can further narrow the search for an underlying mutation. Ultimately, once identified, functional evaluation of a mutation on protein function will be necessary to provide convincing evidence of the role of gene in the pathogenesis of AF.
Given that only a handful of causative mutations have been identified in families with AF, the current HRS/EHRA consensus guideline states that there is no clinical utility for screening known AF-associated genes in patients with AF121. This includes utilization of any currently available commercial testing panels for AF genes and risk loci. As these gene panels are systematically tested in larger cohorts of individuals with familial AF, future evidence may emerge regarding the utility of commercial testing.
Other Forms of Genetic Variation
In addition to analyses of common and rare genetic variants described above, there are multiple other potential genetic analyses that could be considered to identify more of the heritability of AF. Variations in copy number have not been systematically examined in AF patients. High-resolution detection of deletions, insertions and duplications either in coding or non-coding regions has become increasingly straightforward using array-based or next generation sequencing methods. The major barriers to analyses of copy number variation at present are largely centered on cost and sample size necessary to ensure adequate statistical power.
The detection of epigenetic DNA methylation patterns in a tissue is also a straightforward technique; however, as DNA methylation is a highly tissue-specific process, multiple challenges exist with respect to AF. Ideally one would want to analyze left atrial or pulmonary venous tissue from both patients with and without lone AF, yet it is not practical to obtain these samples. Rather, most samples are obtained at the time of cardiac surgery for coronary disease, valvular heart disease, or transplant, and as such the analyses are limited by the inherent co-morbidities present with each type of patient population.
Given that AF increases in prevalence with age, it is possible that somatic or acquired mutations underlie some portion of the heritability of AF. In an intriguing paper, Dr. Gollob and colleagues found somatic mutations in GJA5 among patients with lone atrial fibrillation 38. Presently, one could identify total somatic variation by whole genome or whole exome sequencing rather than on a gene-by-gene basis; however, the same challenges mentioned above regarding the need for left atrial tissue from healthy individuals will limit these analyses.
Finally, it will be interesting to determine whether de novo genetic variation could be responsible for AF. With each successive generation, there is a background rate of spontaneous genetic variation that occurs. By performing exome or genome sequencing in an affected child and unaffected parents, it is possible to identify the handful of novel coding variants present in the child, but not in the parents that may be associated with a disease. Recently, such an approach has identified a novel pathway for autism spectrum disorders122.
Integration of Genetic Data to Predict AF and Outcomes
One ultimate goal of research into the genetic basis of AF is the potential return of this data to clinical arena. It is hoped that with the current trajectory of novel findings and the integration of the additional studies outlined above, that SNP data could be clinically useful in the near future. However, it is important to note that, in addition to not recommending the testing of known AF genes, the current HRS/EHRA guidelines recommend against the testing of individual GWAS-associated SNPs in AF patients. This decision was likely based on the small number of AF SNPs that had been identified at that time, and the limited data on the clinical utility of these variants.
Since the publication of these guidelines, there have been a number of studies examining the relation between AF SNPs and treatment outcomes. Specifically, the risk of AF recurrence after cardioversion 123, pulmonary vein isolation 124,125, or the intiation of antiarrhythmic medication 126 has been studied; however, the observed sample and effect sizes have limited the applicability of these results to the broader population. More compelling results have been seen in stroke patients. Interestingly, the top two genetic variants identified in a large GWAS for cardioembolic stroke are also the top two regions (PITX2 and ZFHX3) associated with AF 64,66,67,82.
Over the last five years, it has become clear that clinical risk factors127, biomarkers128 and now genetic variants can all help to identify individuals at risk for AF. Rather than using any single one of these approaches alone, we should seek to combine each of these risk factors to enhance the detection of AF. One could imagine that in high-risk populations, such as cryptogenic stroke, that we will be able to stratify patients into varying degrees of AF risk, and in turn consider alternative strategies to AF monitoring or anticoagulation.
Conclusions
Recent studies have identified a number of rare and common genetic variants associated with AF. However, the present data only account for a limited percentage of the heritability of AF. Integration of next generation sequencing technologies, improved gene expression data repositories, the identification of additional AF risk loci and a more complete understanding of causative mechanisms behind AF risk loci will be required. Ultimately, with a more complete picture of the genetic risk for AF, we can seek to develop genetically-driven clinical interventions and treatment strategies.
Supplementary Material
Figure 2. Graded relative risk of atrial fibrillation in European and Japanese populations.
The risk of atrial fibrillation is plotted according to the estimated atrial fibrillation risk alleles, relative to that among individuals with the most common number of estimated risk alleles for all genome-wide significant atrial fibrillation susceptibility loci. Data is plotted from individuals of European ancestry from AFGen and Japanese ancestry from BioBank Japan. Right axis denotes the population frequency of each category. Error bars represent the 95% confidence intervals. Adapted from Lubitz et. al, JACC 2014116.
Table 2.
GWAS-derived risk loci for AF
| Locus | Sentinel SNP | RR | P-value | Nearest gene symbol |
Relative location |
Citations |
|---|---|---|---|---|---|---|
| 4q25 | rs6817105 | 1.64 | 1.8×10-74 | PITX2 | 150kb upstream |
61–65,69,70,81,82,88 |
| 16q22 | rs2106261 | 1.24 | 3.2×10-16 | ZFHX3 | Intronic | 65,69,82 |
| 1q21 | rs6666258 | 1.18 | 2.0×10-14 | KCNN3 | Intronic | 65,69,88 |
| 1q24 | rs3903239 | 1.14 | 8.4×10-14 | PRRX1 | 46kb upstream |
69 |
| 7q31 | rs3807989 | 0.90 | 3.6×10-12 | CAV1 | Intronic | 69 |
| 14q23 | rs1152591 | 1.13 | 5.8×10-13 | SYNE2 | Intronic | 69 |
| 9q22 | rs10821415 | 1.11 | 4.2×10-11 | C9orf3 | Intronic | 69 |
| 15q24 | rs7164883 | 1.19 | 2.8×10-17 | HCN4 | Intronic | 69 |
| 10q22 | rs10824026 | 0.87 | 4.0×10-9 | MYOZ1 | 20kb upstream |
69 |
ACKNOWLEDGMENT
The authors thank Julie Herndon for the creation of original artwork utilized in accompanying figures.
SOURCES OF FUNDING Dr. Tucker is supported by a T32 training grant from the National Heart, Lung, and Blood Institute (T32HL007208). Dr. Ellinor is supported by grants from the National Institutes of Health (R01HL092577, R01HL104156, 1K24HL105780) and the American Heart Association to (13EIA14220013).
Footnotes
Disclosures The authors have no conflicts of interest to report.
References
- 1.Wolff L. Familial Auricular Fibrillation. N Engl J Med. 1943;229:396–398. [Google Scholar]
- 2.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–84. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 3.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen W-K, Hammill SC, Packer DL, Olson TM. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 4.Fox CS, Parise H, D’Agostino RB, Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–5. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 5.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–12. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 6.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304:2263–9. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, Brugada J, Girona J, Domingo A, Bachinski LL, Roberts R. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–11. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y-H, Xu S-J, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–4. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa K, Ohno S, Ashihara T, Itoh H, Ding W-G, Toyoda F, Makiyama T, Aoki H, Nakamura Y, Delisle BP, Matsuura H, Horie M. A novel KCNQ1 missense mutation identified in a patient with juvenile-onset atrial fibrillation causes constitutively open IKs channels. Heart Rhythm. 2013 doi: 10.1016/j.hrthm.2013.09.073. [DOI] [PubMed] [Google Scholar]
- 10.Ki C-S, Jung CL, Kim H-J, Baek K-H, Park SJ, On YK, Kim K-S, Noh SJ, Youm JB, Kim JS, Cho H. A KCNQ1 mutation causes age-dependant bradycardia and persistent atrial fibrillation. Pflugers Arch. 2013 doi: 10.1007/s00424-013-1337-6. [DOI] [PubMed] [Google Scholar]
- 11.Bartos DC, Anderson JB, Bastiaenen R, Johnson JN, Gollob MH, Tester DJ, Burgess DE, Homfray T, Behr ER, Ackerman MJ, Guicheney P, Delisle BP. A KCNQ1 mutation causes a high penetrance for familial atrial fibrillation. J Cardiovasc Electrophysiol. 2013;24:562–9. doi: 10.1111/jce.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartos DC, Duchatelet S, Burgess DE, Klug D, Denjoy I, Peat R, Lupoglazoff J-M, Fressart V, Berthet M, Ackerman MJ, January CT, Guicheney P, Delisle BP. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Heart Rhythm. 2011;8:48–55. doi: 10.1016/j.hrthm.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, Makino S, Melman YF, Shea MA, Goyal SB, Rosenzweig A, Macrae CA, Ellinor PT. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm. 2009;6:1146–53. doi: 10.1016/j.hrthm.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundby A, Ravn LS, Svendsen JH, Olesen S-P, Schmitt N. KCNQ1 mutation Q147R is associated with atrial fibrillation and prolonged QT interval. Heart Rhythm. 2007;4:1532–41. doi: 10.1016/j.hrthm.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Christophersen IE, Olesen MS, Liang B, Andersen MN, Larsen AP, Nielsen JB, Haunsø S, Olesen S-P, Tveit A, Svendsen JH, Schmitt N. Genetic variation in KCNA5: impact on the atrial-specific potassium current IKur in patients with lone atrial fibrillation. Eur Heart J. 2013;34:1517–25. doi: 10.1093/eurheartj/ehs442. [DOI] [PubMed] [Google Scholar]
- 16.Olesen MS, Refsgaard L, Holst AG, Larsen AP, Grubb S, Haunsø S, Svendsen JH, Olesen S-P, Schmitt N, Calloe K. A novel KCND3 gain-of-function mutation associated with early-onset of persistent lone atrial fibrillation. Cardiovasc Res. 2013;98:488–95. doi: 10.1093/cvr/cvt028. [DOI] [PubMed] [Google Scholar]
- 17.Deo M, Ruan Y, Pandit SV, Shah K, Berenfeld O, Blaufox A, Cerrone M, Noujaim SF, Denegri M, Jalife J, Priori SG. KCNJ2 mutation in short QT syndrome 3 results in atrial fibrillation and ventricular proarrhythmia. Proc Natl Acad Sci U S A. 2013;110:4291–6. doi: 10.1073/pnas.1218154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia M, Jin Q, Bendahhou S, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 19.Olesen MS, Bentzen BH, Nielsen JB, Steffensen AB, David J-P, Jabbari J, Jensen HK, Haunsø S, Svendsen JH, Schmitt N. Mutations in the potassium channel subunit KCNE1 are associated with early-onset familial atrial fibrillation. BMC Med Genet. 2012;13:24. doi: 10.1186/1471-2350-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Xia M, Jin Q, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundby A, Ravn LS, Svendsen JH, Hauns S, Olesen S-P, Schmitt N. KCNE3 mutation V17M identified in a patient with lone atrial fibrillation. Cell Physiol Biochem. 2008;21:47–54. doi: 10.1159/000113746. [DOI] [PubMed] [Google Scholar]
- 22.Ravn LS, Aizawa Y, Pollevick GD, et al. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart Rhythm. 2008;5:427–35. doi: 10.1016/j.hrthm.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemoine MD, Duverger JE, Naud P, Chartier D, Qi XY, Comtois P, Fabritz L, Kirchhof P, Nattel S. Arrhythmogenic left atrial cellular electrophysiology in a murine genetic long QT syndrome model. Cardiovasc Res. 2011;92:67–74. doi: 10.1093/cvr/cvr166. [DOI] [PubMed] [Google Scholar]
- 24.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–91. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 25.Yang YY, Li J, Lin X, et al. Novel KCNA5 loss-of-function mutations responsible for atrial fibrillation. J Hum Genet. 2009;54:277–83. doi: 10.1038/jhg.2009.26. [DOI] [PubMed] [Google Scholar]
- 26.Yang T, Yang P, Roden DM, Darbar D. Novel KCNA5 mutation implicates tyrosine kinase signaling in human atrial fibrillation. Heart Rhythm. 2010;7:1246–52. doi: 10.1016/j.hrthm.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellinor PT, Nam EG, Shea MA, Milan DJ, Ruskin JN, MacRae CA. Cardiac sodium channel mutation in atrial fibrillation. Heart Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 29.Laitinen-Forsblom PJ, Mäkynen P, Mäkynen H, Yli-Mäyry S, Virtanen V, Kontula K, Aalto-Setälä K. SCN5A mutation associated with cardiac conduction defect and atrial arrhythmias. J Cardiovasc Electrophysiol. 2006;17:480–5. doi: 10.1111/j.1540-8167.2006.00411.x. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Huang H, Liu G, Lam K, Rutberg J, Green MS, Birnie DH, Lemery R, Chahine M, Gollob MH. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem Biophys Res Commun. 2009;380:132–7. doi: 10.1016/j.bbrc.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 31.Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, Ohno S, Nishio Y, Tsuji K, Itoh H, Kimura T, Kita T, Horie M. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326–34. doi: 10.1016/j.jacc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Olesen MS, Yuan L, Liang B, Holst AG, Nielsen N, Nielsen JB, Hedley PL, Christiansen M, Olesen S-P, Haunsø S, Schmitt N, Jespersen T, Svendsen JH. High prevalence of long QT syndrome-associated SCN5A variants in patients with early-onset lone atrial fibrillation. Circ Cardiovasc Genet. 2012;5:450–9. doi: 10.1161/CIRCGENETICS.111.962597. [DOI] [PubMed] [Google Scholar]
- 33.Li R-G, Wang Q, Xu Y-J, Zhang M, Qu X-K, Liu X, Fang W-Y, Yang Y-Q. Mutations of the SCN4B-encoded sodium channel β4 subunit in familial atrial fibrillation. Int J Mol Med. 2013;32:144–50. doi: 10.3892/ijmm.2013.1355. [DOI] [PubMed] [Google Scholar]
- 34.Olesen MS, Holst AG, Svendsen JH, Haunsø S, Tfelt-Hansen J. SCN1Bb R214Q found in 3 patients: 1 with Brugada syndrome and 2 with lone atrial fibrillation. Heart Rhythm. 2012;9:770–3. doi: 10.1016/j.hrthm.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Olesen MS, Jespersen T, Nielsen JB, Liang B, Møller DV, Hedley P, Christiansen M, Varró A, Olesen S-P, Haunsø S, Schmitt N, Svendsen JH. Mutations in sodium channel β-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786–93. doi: 10.1093/cvr/cvq348. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Yang Q, Wu X, et al. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98–104. doi: 10.1016/j.bbrc.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe H, Darbar D, Kaiser DW, Jiramongkolchai K, Chopra S, Donahue BS, Kannankeril PJ, Roden DM. Mutations in sodium channel β1- and β2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268–75. doi: 10.1161/CIRCEP.108.779181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gollob MH, Jones DL, Krahn AD, et al. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677–88. doi: 10.1056/NEJMoa052800. [DOI] [PubMed] [Google Scholar]
- 39.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, Ballew JD, de Andrade M, Burnett JC, Olson TM. Atrial natriuretic peptide frameshift mutation in familial atrial fibrillation. N Engl J Med. 2008;359:158–65. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickey DM, Yoder AR, Potter LR. A familial mutation renders atrial natriuretic Peptide resistant to proteolytic degradation. J Biol Chem. 2009;284:19196–202. doi: 10.1074/jbc.M109.010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Fareh S, Leung TK, Nattel S. Promotion of atrial fibrillation by heart failure in dogs: atrial remodeling of a different sort. Circulation. 1999;100:87–95. doi: 10.1161/01.cir.100.1.87. [DOI] [PubMed] [Google Scholar]
- 42.Disertori M, Quintarelli S, Grasso M, et al. Autosomal recessive atrial dilated cardiomyopathy with standstill evolution associated with mutation of Natriuretic Peptide Precursor A. Circ Cardiovasc Genet. 2013;6:27–36. doi: 10.1161/CIRCGENETICS.112.963520. [DOI] [PubMed] [Google Scholar]
- 43.Huang R-T, Xue S, Xu Y-J, Zhou M, Yang Y-Q. A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2013;31:1119–26. doi: 10.3892/ijmm.2013.1316. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y-M, Zheng P-X, Yang Y-Q, Ge Z-M, Kang W-Q. A novel PITX2c loss-of-function mutation underlies lone atrial fibrillation. Int J Mol Med. 2013;32:827–34. doi: 10.3892/ijmm.2013.1463. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y-Q, Wang M-Y, Zhang X-L, Tan H-W, Shi H-F, Jiang W-F, Wang X-H, Fang W-Y, Liu X. GATA4 loss-of-function mutations in familial atrial fibrillation. Clin Chim Acta. 2011;412:1825–30. doi: 10.1016/j.cca.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 46.Jiang J-Q, Shen F-F, Fang W-Y, Liu X, Yang Y-Q. Novel GATA4 mutations in lone atrial fibrillation. Int J Mol Med. 2011;28:1025–32. doi: 10.3892/ijmm.2011.783. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Sun Y-M, Yang Y-Q. Mutation spectrum of the GATA4 gene in patients with idiopathic atrial fibrillation. Mol Biol Rep. 2012;39:8127–35. doi: 10.1007/s11033-012-1660-6. [DOI] [PubMed] [Google Scholar]
- 48.Gu J-Y, Xu J-H, Yu H, Yang Y-Q. Novel GATA5 loss-of-function mutations underlie familial atrial fibrillation. Clinics (Sao Paulo) 2012;67:1393–9. doi: 10.6061/clinics/2012(12)08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X-H, Huang C-X, Wang Q, Li R-G, Xu Y-J, Liu X, Fang W-Y, Yang Y-Q. A novel GATA5 loss-of-function mutation underlies lone atrial fibrillation. Int J Mol Med. 2013;31:43–50. doi: 10.3892/ijmm.2012.1189. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y-Q, Li L, Wang J, Zhang X-L, Li R-G, Xu Y-J, Tan H-W, Wang X-H, Jiang J-Q, Fang W-Y, Liu X. GATA6 loss-of-function mutation in atrial fibrillation. Eur J Med Genet. 2012;55:520–6. doi: 10.1016/j.ejmg.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kathiresan S, Voight BF, Purcell S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–22. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 56.Kolek MJ, Parvez B, Muhammad R, Shoemaker MB, Blair MA, Stubblefield T, Kucera GA, Denny JC, Roden DM, Darbar D. A Common Variant on Chromosome 4q25 is Associated With Prolonged PR Interval in Subjects With and Without Atrial Fibrillation. Am J Cardiol. 2013 doi: 10.1016/j.amjcard.2013.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newton-Cheh C, Eijgelsheim M, Rice KM, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–14. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith JG, Magnani JW, Palmer C, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304. doi: 10.1371/journal.pgen.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 62.Kääb S, Darbar D, van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–9. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viviani Anselmi C, Novelli V, Roncarati R, Malovini A, Bellazzi R, Bronzini R, Marchese G, Condorelli G, Montenero AS, Puca AA. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart. 2008;94:1394–6. doi: 10.1136/hrt.2008.148544. [DOI] [PubMed] [Google Scholar]
- 64.Shi L, Li C, Wang C, et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum Genet. 2009;126:843–9. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- 65.Delaney JT, Jeff JM, Brown NJ, Pretorius M, Okafor HE, Darbar D, Roden DM, Crawford DC. Characterization of genome-wide association-identified variants for atrial fibrillation in African Americans. PLoS One. 2012;7:e32338. doi: 10.1371/journal.pone.0032338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–9. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 67.Lemmens R, Buysschaert I, Geelen V, et al. The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke. 2010;41:1850–7. doi: 10.1161/STROKEAHA.110.587980. [DOI] [PubMed] [Google Scholar]
- 68.Goodloe AH, Herron KJ, Olson TM. Uncovering an intermediate phenotype associated with rs2200733 at 4q25 in lone atrial fibrillation. Am J Cardiol. 2011;107:1802–5. doi: 10.1016/j.amjcard.2011.02.326. [DOI] [PubMed] [Google Scholar]
- 69.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–5. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lubitz SA, Sinner MF, Lunetta KL, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–84. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–24. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 72.Liu C, Liu W, Lu MF, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–48. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- 73.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–8. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 74.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisúa-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–82. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 75.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–58. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 76.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld H-H, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–33. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 77.Mommersteeg MTM, Brown NA, Prall OWJ, de Gier-de Vries C, Harvey RP, Moorman AFM, Christoffels VM. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–9. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 78.Mommersteeg MTM, Hoogaars WMH, Prall OWJ, de Gier-de Vries C, Wiese C, Clout DEW, Papaioannou VE, Brown NA, Harvey RP, Moorman AFM, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–62. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 79.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpón E, Tamargo J, Cinca J, Hove-Madsen L, Aranega AE, Franco D. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–79. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 80.Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, Kaminski HJ, Martin JF. Pitx2, an Atrial Fibrillation Predisposition Gene, Directly Regulates Ion Transport and Intercalated Disc Genes. Circ Cardiovasc Genet. 2014 doi: 10.1161/CIRCGENETICS.113.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jung C-G, Kim H-J, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–45. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 84.Berry FB, Miura Y, Mihara K, Kaspar P, Sakata N, Hashimoto-Tamaoki T, Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J Biol Chem. 2001;276:25057–65. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- 85.Sun X, Frierson HF, Chen C, Li C, Ran Q, Otto KB, Cantarel BL, Cantarel BM, Vessella RL, Gao AC, Petros J, Miura Y, Simons JW, Dong J-T. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407–12. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 86.Kim CJ, Song JH, Cho YG, Cao Z, Lee YS, Nam SW, Lee JY, Park WS. Down-regulation of ATBF1 is a major inactivating mechanism in hepatocellular carcinoma. Histopathology. 2008;52:552–9. doi: 10.1111/j.1365-2559.2008.02980.x. [DOI] [PubMed] [Google Scholar]
- 87.Ido A, Miura Y, Watanabe M, Sakai M, Inoue Y, Miki T, Hashimoto T, Morinaga T, Nishi S, Tamaoki T. Cloning of the cDNA encoding the mouse ATBF1 transcription factor. Gene. 1996;168:227–231. doi: 10.1016/0378-1119(95)00740-7. [DOI] [PubMed] [Google Scholar]
- 88.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-Conductance, Calcium-Activated Potassium Channels from Mammalian Brain. Science (80- ) 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 90.Li N, Timofeyev V, Tuteja D, Xu D, Lu L, Zhang Q, Zhang Z, Singapuri A, Albert TR, Rajagopal AV, Bond CT, Periasamy M, Adelman J, Chiamvimonvat N. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J Physiol. 2009;587:1087–100. doi: 10.1113/jphysiol.2008.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diness JG, Sørensen US, Nissen JD, Al-Shahib B, Jespersen T, Grunnet M, Hansen RS. Inhibition of small-conductance Ca2+-activated K+ channels terminates and protects against atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3:380–90. doi: 10.1161/CIRCEP.110.957407. [DOI] [PubMed] [Google Scholar]
- 92.Qi X-Y, Diness JG, Brundel B, Zhou X-B, Naud P, Wu C-T, Huang H, Harada M, Aflaki M, Dobrev D, Grunnet M, Nattel S. Role of Small Conductance Calcium-Activated Potassium Channels in Atrial Electrophysiology and Fibrillation in the Dog. Circulation. 2013 doi: 10.1161/CIRCULATIONAHA.113.003019. [DOI] [PubMed] [Google Scholar]
- 93.Zhang X-D, Timofeyev V, Li N, Myers RE, Zhang D, Singapuri A, Lau VC, Bond CT, Adelman J, Lieu DK, Chiamvimonvat N. Critical Roles of a Small Conductance Ca2+-Activated K+ Channel (SK3) in the Repolarization Process of Atrial Myocytes. Cardiovasc Res. 2013 doi: 10.1093/cvr/cvt262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mahida S, Mills R, Tucker NR, Simonson B, Macri V, Lemoine MD, Das S, Milan DJ, Ellinor PT. Overexpression of KCNN3 Results in Sudden Cardiac Death. Cardiovasc Res. 2013 doi: 10.1093/cvr/cvt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schiffer C, Tariverdian G, Schiesser M, Thomas MC, Sergi C. Agnathia-otocephaly complex: report of three cases with involvement of two different Carnegie stages. Am J Med Genet. 2002;112:203–8. doi: 10.1002/ajmg.10672. [DOI] [PubMed] [Google Scholar]
- 96.Dasouki M, Andrews B, Parimi P, Kamnasaran D. Recurrent agnathia-otocephaly caused by DNA replication slippage in PRRX1. Am J Med Genet A. 2013;161A:803–8. doi: 10.1002/ajmg.a.35879. [DOI] [PubMed] [Google Scholar]
- 97.Martin JF, Bradley A, Olson EN. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–49. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- 98.Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, Olson EN, Kern MJ. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–9. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- 99.Ihida-Stansbury K, McKean DM, Gebb SA, Martin JF, Stevens T, Nemenoff R, Akeson A, Vaughn J, Jones PL. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ Res. 2004;94:1507–14. doi: 10.1161/01.RES.0000130656.72424.20. [DOI] [PubMed] [Google Scholar]
- 100.Schulze-Bahr E, Neu A, Friederich P, Kaupp UB, Breithardt G, Pongs O, Isbrandt D. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest. 2003;111:1537–45. doi: 10.1172/JCI16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ueda K, Nakamura K, Hayashi T, et al. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem. 2004;279:27194–8. doi: 10.1074/jbc.M311953200. [DOI] [PubMed] [Google Scholar]
- 102.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–7. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 103.Duhme N, Schweizer PA, Thomas D, Becker R, Schröter J, Barends TRM, Schlichting I, Draguhn A, Bruehl C, Katus HA, Koenen M. Altered HCN4 channel C-linker interaction is associated with familial tachycardia-bradycardia syndrome and atrial fibrillation. Eur Heart J. 2013;34:2768–75. doi: 10.1093/eurheartj/ehs391. [DOI] [PubMed] [Google Scholar]
- 104.Maguy A, Hebert TE, Nattel S. Involvement of lipid rafts and caveolae in cardiac ion channel function. Cardiovasc Res. 2006;69:798–807. doi: 10.1016/j.cardiores.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 105.Adebiyi A, Zhao G, Cheranov SY, Ahmed A, Jaggar JH. Caveolin-1 abolishment attenuates the myogenic response in murine cerebral arteries. Am J Physiol Heart Circ Physiol. 2007;292:H1584–92. doi: 10.1152/ajpheart.00584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y-Y, Liu Y, Stan R-V, Fan L, Gu Y, Dalton N, Chu P-H, Peterson K, Ross J, Chien KR. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc Natl Acad Sci U S A. 2002;99:11375–80. doi: 10.1073/pnas.172360799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106:10207–12. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lüke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, Padmakumar VC, Gloy J, Walz G, Noegel AA. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci. 2008;121:1887–98. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- 109.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–87. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Q, Bethmann C, Worth NF, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007;16:2816–33. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 111.Lin H, Dolmatova EV, Morley MP, Lunetta KL, McManus DD, Magnani JW, Margulies KB, Hakonarson H, Del Monte F, Benjamin EJ, Cappola TP, Ellinor PT. Gene Expression and Genetic Variation in Human Atria. Heart Rhythm. 2013 doi: 10.1016/j.hrthm.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Takada F, Vander Woude DL, Tong HQ, Thompson TG, Watkins SC, Kunkel LM, Beggs AH. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci U S A. 2001;98:1595–600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Osio A, Tan L, Chen SN, Lombardi R, Nagueh SF, Shete S, Roberts R, Willerson JT, Marian AJ. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ Res. 2007;100:766–8. doi: 10.1161/01.RES.0000263008.66799.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frey N, Barrientos T, Shelton JM, Frank D, Rütten H, Gehring D, Kuhn C, Lutz M, Rothermel B, Bassel-Duby R, Richardson JA, Katus HA, Hill JA, Olson EN. Mice lacking calsarcin-1 are sensitized to calcineurin signaling and show accelerated cardiomyopathy in response to pathological biomechanical stress. Nat Med. 2004;10:1336–43. doi: 10.1038/nm1132. [DOI] [PubMed] [Google Scholar]
- 115.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J. 2013;34:2243–51. doi: 10.1093/eurheartj/eht033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lubitz SA, Lunetta KL, Lin H, et al. Novel Genetic Markers Associate with Atrial Fibrillation Risk in Europeans and Japanese. JAMA J Am Med Assoc. doi: 10.1016/j.jacc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Petretto E, Sarwar R, Grieve I, et al. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet. 2008;40:546–52. doi: 10.1038/ng.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Monti J, Fischer J, Paskas S, et al. Soluble epoxide hydrolase is a susceptibility factor for heart failure in a rat model of human disease. Nat Genet. 2008;40:529–37. doi: 10.1038/ng.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 122.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parvez B, Shoemaker MB, Muhammad R, Richardson R, Jiang L, Blair MA, Roden DM, Darbar D. Common genetic polymorphism at 4q25 locus predicts atrial fibrillation recurrence after successful cardioversion. Heart Rhythm. 2013;10:849–55. doi: 10.1016/j.hrthm.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–53. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 125.Benjamin Shoemaker M, Muhammad R, Parvez B, et al. Common atrial fibrillation risk alleles at 4q25 predict recurrence after catheter-based atrial fibrillation ablation. Heart Rhythm. 2013;10:394–400. doi: 10.1016/j.hrthm.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parvez B, Vaglio J, Rowan S, Muhammad R, Kucera G, Stubblefield T, Carter S, Roden D, Darbar D. Symptomatic response to antiarrhythmic drug therapy is modulated by a common single nucleotide polymorphism in atrial fibrillation. J Am Coll Cardiol. 2012;60:539–45. doi: 10.1016/j.jacc.2012.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schnabel RB, Sullivan LM, Levy D, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–45. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patton KK, Ellinor PT, Heckbert SR, Christenson RH, DeFilippi C, Gottdiener JS, Kronmal RA. N-terminal pro-B-type natriuretic peptide is a major predictor of the development of atrial fibrillation: the Cardiovascular Health Study. Circulation. 2009;120:1768–74. doi: 10.1161/CIRCULATIONAHA.109.873265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Olson TM, Alekseev AE, Moreau C, Liu XK, Zingman LV, Miki T, Seino S, Asirvatham SJ, Jahangir A, Terzic A. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med. 2007;4:110–6. doi: 10.1038/ncpcardio0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Posch MG, Boldt L-H, Polotzki M, Richter S, Rolf S, Perrot A, Dietz R, Ozcelik C, Haverkamp W. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur J Med Genet. 53:201–3. doi: 10.1016/j.ejmg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 131.Yang Y-Q, Wang J, Wang X-H, Wang Q, Tan H-W, Zhang M, Shen F-F, Jiang J-Q, Fang W-Y, Liu X. Mutational spectrum of the GATA5 gene associated with familial atrial fibrillation. Int J Cardiol. 2012;157:305–7. doi: 10.1016/j.ijcard.2012.03.132. [DOI] [PubMed] [Google Scholar]
- 132.Yang Y-Q, Wang X-H, Tan H-W, Jiang W-F, Fang W-Y, Liu X. Prevalence and spectrum of GATA6 mutations associated with familial atrial fibrillation. Int J Cardiol. 2012;155:494–6. doi: 10.1016/j.ijcard.2011.12.091. [DOI] [PubMed] [Google Scholar]
- 133.Li J, Liu W-D, Yang Z-L, Yang Y-Q. Novel GATA6 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2012;30:783–90. doi: 10.3892/ijmm.2012.1068. [DOI] [PubMed] [Google Scholar]
- 134.Christophersen IE, Holmegard HN, Jabbari J, Sajadieh A, Haunsø S, Tveit A, Svendsen JH, Olesen MS. Rare variants in GJA5 are associated with early-onset lone atrial fibrillation. Can J Cardiol. 2013;29:111–6. doi: 10.1016/j.cjca.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 135.Shi H-F, Yang J-F, Wang Q, Li R-G, Xu Y-J, Qu X-K, Fang W-Y, Liu X, Yang Y-Q. Prevalence and spectrum of GJA5 mutations associated with lone atrial fibrillation. Mol Med Rep. 2013;7:767–74. doi: 10.3892/mmr.2012.1252. [DOI] [PubMed] [Google Scholar]
- 136.Sun Y, Yang Y-Q, Gong X-Q, Wang X-H, Li R-G, Tan H-W, Liu X, Fang W-Y, Bai D. Novel germline GJA5/connexin40 mutations associated with lone atrial fibrillation impair gap junctional intercellular communication. Hum Mutat. 2013;34:603–9. doi: 10.1002/humu.22278. [DOI] [PubMed] [Google Scholar]
- 137.Yang Y-Q, Liu X, Zhang X-L, Wang X-H, Tan H-W, Shi H-F, Jiang W-F, Fang W-Y. Novel connexin40 missense mutations in patients with familial atrial fibrillation. Europace. 2010;12:1421–7. doi: 10.1093/europace/euq274. [DOI] [PubMed] [Google Scholar]
- 138.Yang Y-Q, Zhang X-L, Wang X-H, Tan H-W, Shi H-F, Jiang W-F, Fang W-Y, Liu X. Connexin40 nonsense mutation in familial atrial fibrillation. Int J Mol Med. 2010;26:605–10. doi: 10.3892/ijmm_00000505. [DOI] [PubMed] [Google Scholar]
- 139.Müller II, Melville DB, Tanwar V, Rybski WM, Mukherjee A, Shoemaker MB, Wang W-D, Schoenhard JA, Roden DM, Darbar D, Knapik EW, Hatzopoulos AK. Functional modeling in zebrafish demonstrates that the atrial-fibrillation-associated gene GREM2 regulates cardiac laterality, cardiomyocyte differentiation and atrial rhythm. Dis Model Mech. 2013;6:332–41. doi: 10.1242/dmm.010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Beavers DL, Wang W, Ather S, Voigt N, Garbino A, Dixit SS, Landstrom AP, Li N, Wang Q, Olivotto I, Dobrev D, Ackerman MJ, Wehrens XHT. Mutation E169K in Junctophilin-2 Causes Atrial Fibrillation Due to Impaired RyR2 Stabilization. J Am Coll Cardiol. 2013;62:2010–9. doi: 10.1016/j.jacc.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hong K, Bjerregaard P, Gussak I, Brugada R. Short QT syndrome and atrial fibrillation caused by mutation in KCNH2. J Cardiovasc Electrophysiol. 2005;16:394–6. doi: 10.1046/j.1540-8167.2005.40621.x. [DOI] [PubMed] [Google Scholar]
- 142.Sinner MF, Pfeufer A, Akyol M, et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG) Eur Heart J. 2008;29:907–14. doi: 10.1093/eurheartj/ehm619. [DOI] [PubMed] [Google Scholar]
- 143.Jabbari J, Olesen MS, Holst AG, Nielsen JB, Haunso S, Svendsen JH. Common polymorphisms in KCNJ5 [corrected] are associated with early-onset lone atrial fibrillation in Caucasians. Cardiology. 2011;118:116–20. doi: 10.1159/000323840. [DOI] [PubMed] [Google Scholar]
- 144.Delaney JT, Muhammad R, Blair MA, Kor K, Fish FA, Roden DM, Darbar D. A KCNJ8 mutation associated with early repolarization and atrial fibrillation. Europace. 2012;14:1428–32. doi: 10.1093/europace/eus150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Brauch KM, Chen LY, Olson TM. Comprehensive mutation scanning of LMNA in 268 patients with lone atrial fibrillation. Am J Cardiol. 2009;103:1426–8. doi: 10.1016/j.amjcard.2009.01.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Saj M, Dabrowski R, Labib S, Jankowska A, Szperl M, Broda G, Szwed H, Tesson F, Bilinska ZT, Ploski R. Variants of the lamin A/C (LMNA) gene in non-valvular atrial fibrillation patients: a possible pathogenic role of the Thr528Met mutation. Mol Diagn Ther. 2012;16:99–107. doi: 10.1007/BF03256434. [DOI] [PubMed] [Google Scholar]
- 147.Ren X, Xu C, Zhan C, et al. Identification of NPPA variants associated with atrial fibrillation in a Chinese GeneID population. Clin Chim Acta. 2010;411:481–5. doi: 10.1016/j.cca.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 148.Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, de la Fuente R, Wang L, Chen Q, Wang QK. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–27. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 149.Zhabyeyev P, Hiess F, Wang R, Liu Y, Wayne Chen SR, Oudit GY. S4153R is a gain-of-function mutation in the cardiac Ca(2+) release channel ryanodine receptor associated with catecholaminergic polymorphic ventricular tachycardia and paroxysmal atrial fibrillation. Can J Cardiol. 2013;29:993–6. doi: 10.1016/j.cjca.2012.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.