Abstract
The Cvt pathway is a biosynthetic transport route for a distinct subset of resident yeast vacuolar hydrolases, whereas macroautophagy is a nonspecific degradative mechanism that allows cell survival during starvation. Yet, these two vacuolar trafficking pathways share a number of identical molecular components and are morphologically very similar. For example, one of the hallmarks of both pathways is the formation of double-membrane cytosolic vesicles that sequester cargo before vacuolar delivery. The origin of the vesicle membrane has been controversial and various lines of evidence have implicated essentially all compartments of the endomembrane system. Despite the analogies between the Cvt pathway and autophagy, earlier work has suggested that the origin of the engulfing vesicle membranes is different; the endoplasmic reticulum is proposed to be required only for autophagy. In contrast, in this study we demonstrate that the endoplasmic reticulum and/or Golgi complex, but not endosomal compartments, play an important role for both yeast transport routes. Along these lines, we demonstrate that Berkeley bodies, a structure generated from the Golgi complex in sec7 cells, are immunolabeled with Atg8, a structural component of autophagosomes. Finally, we also show that none of the yeast t-SNAREs are located at the preautophagosomal structure, the presumed site of double-membrane vesicle formation. Based on our results, we propose two models for Cvt vesicle biogenesis.
INTRODUCTION
The lysosome/vacuole is the major cellular center for degradation, recycling, and storage of biological constituents. Several well-characterized transport routes are implicated in protein delivery to this organelle during normal growth conditions. These pathways are conserved among eukaryotic organisms, but work in the yeast Saccharomyces cerevisiae has had a major impact on their characterization and in the identification of the molecular machinery (Conibear and Stevens, 1998). The route called the alkaline phosphatase pathway provides a direct transport connection between the Golgi complex and the vacuole (Cowles et al., 1997; Piper et al., 1997). A second itinerary from the Golgi complex to the vacuole passes through the late endosome and is known as the carboxypeptidase Y pathway (Piper et al., 1995; Rieder et al., 1996; Babst et al., 1998; Conibear and Stevens, 1998). These two pathways are required for the biogenesis and maintenance of vacuoles, whereas a third route, endocytosis, has a catabolic function. Endocytosis mediates the down-regulation of plasma membrane proteins by delivering them via the late endosome to the vacuole for degradation (Hicke, 1999; Katzmann et al., 2002). Before reaching the late endosome, these proteins pass through the early endosome where some components are recycled back to the cell surface (Hicke et al., 1997; Prescianotto-Baschong and Riezman, 1998; Lewis et al., 2000). The carboxypeptidase Y pathway and endocytosis converge at the endosome where another route, the multivesicular body pathway, delivers both resident enzymes and substrates to the vacuole (Katzmann et al., 2002).
There is also an additional route to the vacuole called the cytoplasm to vacuole targeting (Cvt) pathway that mediates the transport of certain vacuolar hydrolases such as aminopeptidase I (Ape1) that form a large cytosolic oligomer after synthesis (Klionsky et al., 1992; Kim et al., 1997; Scott et al., 1997; Shintani et al., 2002). The existence of the Cvt pathway has only been demonstrated in the yeast Saccharomyces cerevisiae. However, this pathway has gained increased relevance with the discovery that it uses most of the same molecular machinery as autophagy, a catabolic process conserved among all eukaryotic cells (Harding et al., 1996; Scott et al., 1996). In yeast, autophagy is induced by starvation conditions, and it allows the cells to eliminate unnecessary proteins and organelles in the lysosome/vacuole to generate an internal pool of nutrients that can be exploited for survival (Klionsky and Emr, 2000; Reggiori and Klionsky, 2002). In addition, autophagy may play a protective role by removing damaged organelles, a function that may be associated with increased life span (reviewed in Klionsky and Emr, 2000; Reggiori and Klionsky, 2002; Ogier-Denis and Codogno, 2003). Conversely, defects in autophagy are associated with a number of diseases, including both cancer and neurodegeneration (Ogier-Denis and Codogno, 2003; Klionsky, 2004).
The basic mechanism of the Cvt pathway is the sequestration of biosynthetic cargo by a cytosolic double-membrane vesicle. This Cvt vesicle subsequently fuses with the vacuole liberating the inner vesicle into the organelle lumen where it is finally degraded by hydrolases, thus allowing the resident enzymes access to their final destination (Baba et al., 1997). Starvation conditions induce the expansion of these double-membrane vesicles and the bigger size of these structures, called autophagosomes, enables them to enwrap organelles and large fractions of the cytoplasm (Takeshige et al., 1992; Baba et al., 1994; Abeliovich et al., 2000). Most of the factors shared between these two pathways localize to a punctate perivacuolar site, called the pre-autophagosomal structure (PAS), that is believed to be the formation or organization site for the assembly of autophagosomes and Cvt vesicles (Suzuki et al., 2001; Kim et al., 2002; Noda et al., 2002). Some components such as Atg11 and Atg19 that are specific to the Cvt pathway are used to recruit cargo molecules to the PAS (Scott et al., 2001; Shintani et al., 2002). In contrast, the other components that are specific for the Cvt pathway or autophagy are mostly proteins involved in vesicular traffic. Autophagy, but not the Cvt pathway, requires Sec12 and some COPII components (Ishihara et al., 2001), whereas only the Cvt pathway uses the N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)–Sec1 complex composed of Tlg2, Tlg1, and Vps45, the Vps-fifty-three (VFT) tethering complex, and the two sorting nexins Atg24/Snx4 and Atg20/Snx42 (Abeliovich et al., 1999; Nice et al., 2002; Reggiori et al., 2003). Although the origin of the sequestering membrane has not been unequivocally established, these differences have led to the proposal that the membrane source differs for autophagy versus the Cvt pathway. In particular, it has been proposed that autophagy but not the Cvt pathway requires the endoplasmic reticulum (ER) (Ishihara et al., 2001), implying that the induction of autophagy involves a major shift in membrane flow that is under the direction of molecular changes in the basic Atg machinery.
The soluble SNARE proteins are the core of the machinery required for membrane fusion (Sollner et al., 1993; Burgoyne and Morgan, 2003). This event requires four SNAREs, typically three on the target membrane and one on the opposite, for example the surface of a vesicle. The trimeric SNARE complex on the target membrane always contains a syntaxin homologue also termed either target SNARE (t-SNARE) or Qa-syntaxin (Fasshauer et al., 1998; Pelham, 2001a; Burgoyne and Morgan, 2003). To gain clues as to the source of the sequestering membrane and how the PAS is generated, we took advantage of the fact that yeast has just seven t-SNAREs (Pelham, 2001a). None of these proteins was found in the PAS, suggesting that this structure may be created in a vesicle-independent manner. We also investigated which organelles of the yeast secretory pathway play an essential role in correct organization of the PAS. In particular, we show that early endosomes and the PAS are different compartments despite sharing some trafficking analogies. In addition, we also demonstrate that contrary to previous reports, the proper functioning of the ER and Golgi complex is essential for the normal progression of the Cvt pathway.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae knockout strains in the BY4742 background used in this study (atg1Δ, atg9Δ, atg14Δ, rcy1Δ, vps4Δ, vps5Δ, vps23Δ, vps27Δ, vps28Δ, vps29Δ, vps34Δ, vps35Δ, and vps38Δ) were purchased from ResGen (Invitrogen, Carlsbad, CA). The rest of the used strains are listed in Table 1. For the VPS38 and PHO13 gene disruptions, the entire coding regions were replaced with the Saccharomyces kluyveri HIS3 and the Kluveromyces lactis URA3 genes, respectively, flanked by coliphage loxP sites by using polymerase chain reaction (PCR) primers containing ∼40 bases of identity to the regions flanking the open reading frame.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | ResGen |
| FRY130 | BY4742 atg14Δ::kanMX4 vps38Δ::HIS3 S.k. | This study |
| SEY6210 | MATα his3-Δ200 leu2-3,112 lys2-801 ura3-52 trp1-Δ901 suc2-Δ9 | Robinson et al. (1988) |
| PSY82 | SEY6210 VPS51-YFP::TRP1 | This study |
| PSY83 | SEY6210 VPS54-YFP::TRP1 | This study |
| HAY456 | SEY6210 ATG9-PA::HIS5 S.p | This study |
| TDY2 | SEY6210 vam3Δ::LEU2 + pVAM3-6.414 | Darsow et al. (1997) |
| AFM69-1A | MATa sec7-4 his3,11-15 leu2-3,112 ura3-1 | A. Franzusoff |
| FBY217 | MATa sec12-4 his3 leu2 ura3 trp1 ade2 | A. Conzelmann |
| FRY208 | MATa sec7-4 his3,11-15 leu2-3,112 ura3-1 pho13Δ::URA3 K.l. pho8::ADH1-PHO8Δ60-HIS3 S.k. | This study |
| FRY209 | MATa sec12-4 his3 leu2 ura3 trp1 ade2 pho13Δ::URA3 K.l. pho8::ADH1-PHO8Δ60-HIS3 S.k | This study |
PCR-based integrations of yellow fluorescent protein (YFP) and protein A (PA) at the 3′ end of VPS51, VPS54, and ATG9 were used to generate strains expressing fusion proteins under the control of their native promoters. The templates for integration were pDH3 and pHAB102 (Drees et al., 2001; Abeliovich et al., 2002). Normal prApe1 processing and vacuolar morphology were used to confirm the functionality of all genomic fusions.
Strains were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base without amino acids, 2% glucose and auxotrophic amino acids as needed). Nitrogen starvation was carried out in SD-N medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose).
Plasmids
DNA fragments encoding TLG1, TLG2, and SNC1 plus 350 base pairs of their terminator sequences were generated by PCR and used to replace the PHM5 gene as an AscI-BamHI fragment in the pYFPPHM52416 plasmid (Reggiori and Pelham, 2001). The new plasmids were called pYFPTLG1ter(416), pYFPTLG2ter(416), and pYFPSNC1ter(416), respectively. The YFP gene contained in these three plasmids was then replaced with cyan fluorescent protein (CFP) by excision and subsequent cloning by using HindIII and AscI. The promoter (TPI1) plus the new fusions were finally subcloned into a pRS414 vector (Sikorski and Hieter, 1989), generating pCFPTLG1ter(414), pCFPTLG2ter(414), and pCFPSNC1ter(414), respectively. All enzymes for manipulation of DNA were from New England Biolabs (Beverly, MA).
Plasmids expressing green fluorescent protein (GFP)-Snc1 (pGS416), GFP-Ape1 (pTS466), CFP-Ape1 (pTS470), GFP-Atg8 [pRS316GFP-Aut7 and pCuG-FPAUT7(416)], YFP-Atg8 (pRS414EYFP-Aut7), CFP-Atg8 (pRS316ECFP-Aut7), and Ape1 (pRC1) have been described previously (Klionsky et al., 1992; Lewis et al., 2000; Suzuki et al., 2001; Kim et al., 2002; Shintani et al., 2002). Vectors carrying the SEC7 and the SEC12 wild-type genes and complementing the corresponding thermosensitive mutations were a kind gift from Nava Segev (University of Illinois at Chicago, Chicago, IL) and Randy Schekman (University of California, Berkeley, CA), respectively. A plasmid to integrate pho8Δ60 at the PHO8 locus will be described elsewhere (Shintani and Klionsky, unpublished data). A plasmid expressing 3 × hemagglutinin (HA)-ATG8 was generated as follows. A BglII restriction site was introduced just after the initiation codon of the ATG8 gene on pAUT7(416) (Huang et al., 2000) by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the following primers: 5′-TAATTACTAGAGACATGAGATCTAAGTCTACATT-TAAGTC-3′ and 5′-GACTTAAATGTAGACTTAGATCTCATGTCTCTAGTAATTA-3′, to generate pATG8BglII(416). The DNA fragment encoding a triple hemagglutinin tag with BglII site on both sites was then ligated to the BglII site of pATG8BglII(306) to generate p3HA-ATG8(306).
Subcellular Fractionation
Overnight precultures in SMD medium were diluted in 500 ml of YPD medium and grown for 3–4 h to early log phase. Fractionation was carried out on a 22–60% sucrose step gradient as described by Black and Pelham (2000). After the final centrifugation, fourteen 0.86-ml fractions were collected from the top of the gradient and resolved by SDS-PAGE followed by Western blot by using antibodies or serum against PA (Sigma-Aldrich, St. Louis, MO), Atg8 (Huang et al., 2000), GFP (Covance, Princeton, NJ), Tlg1 (Holthuis et al., 1998), Sed5 (Banfield et al., 1994), Vam3 (Nichols et al., 1997), Sso2 (Aalto et al., 1993), and Pep12 (Molecular Probes, Eugene, OR).
Ape1 Pulse-Chase Radiolabeling Analysis
Wild-type and sec12 cells were grown overnight at 24°C to early log phase and then shifted to 37°C for 10, 55, and 85 min, respectively. Then 5 OD600 equivalents of cells were collected and resuspended in 75 μl of SMD medium without methionine and cysteine. After 5 min at the same temperature, cells were labeled with 10 μl of Tran35S-label (ICN Pharmaceuticals, Irvine, CA) for 10 min. Chase was initiated by addition of 5 ml of chase medium (SMD medium containing 1% casamino acids, 0.2% yeast extract, 4 mM methionine, and 2 mM cysteine) and continued for 2 h. As a control, cells were also pulse labeled and subjected to a nonradioactive chase at 24°C. At the beginning and end of the chase period, 2 ml of culture was removed, trichloroacetic acid (TCA) precipitated, and Ape1 immunoprecipitation carried out as described previously (Harding et al., 1995).
Immunoelectron Microscopy
Wild-type, sec7, and sec12 cells expressing HA-Atg8 under the control of the native ATG8 promoter were grown in rich medium at 24°C and then shifted to 37°C in either the same medium or in SD-N to induce autophagy for 2 h. Cells were then fixed with formaldehyde and glutaraldehyde, treated with sodium periodate, dehydrated with ethanol, and finally embedded with LR White resin (Ted Pella, Redding, CA) as described previously (Lybarger and Maddock, 2000). After resin polymerization, 70- to 80-nm sections were mounted on nickel grids, incubated first with anti-HA antibody (1:25 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) and successively with 12-nm colloidal gold-Affinipure goat anti-mouse IgG (1:30 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA). Sections were then stained with uranyl acetate to be finally imaged with a Philips CM-100 transmission electron microscope. Untransformed cells were also treated in the same way and used as background controls.
Miscellaneous Procedures
Fluorescence microscopy, FM 4-64 (Molecular Probes) staining, Pho8Δ60 activity assays, protein extraction, and Western blots were conducted as described previously (Reggiori et al., 2003).
RESULTS
The Late Endosome Is Not Required for Precursor Ape1 Sorting and Autophagy
The hallmark of the Cvt pathway and autophagy is the formation of sequestering double-membrane vesicles. A major unanswered question is the source of the membrane that forms these vesicles. To address this issue, we decided to analyze the biogenesis of the PAS that is thought to be central to the vesicle formation process. It has been shown in previous studies that mutations in the genes encoding the components of the VFT complex and two of the interacting SNAREs, Tlg1 and Tlg2, generate strains that are defective for Cvt vesicle formation (Abeliovich et al., 1999; Reggiori et al., 2003). We decided to investigate whether the observed block was characteristic for mutants affecting only early endosome/late Golgi function or whether it was common to all strains with a defect in the endosomal system. In particular, we decided to examine whether late endosomes play an essential role in precursor Ape1 (prApe1) trafficking as has been postulated for either yeast or mammalian cells (Shirahama et al., 1997; Nara et al., 2002).
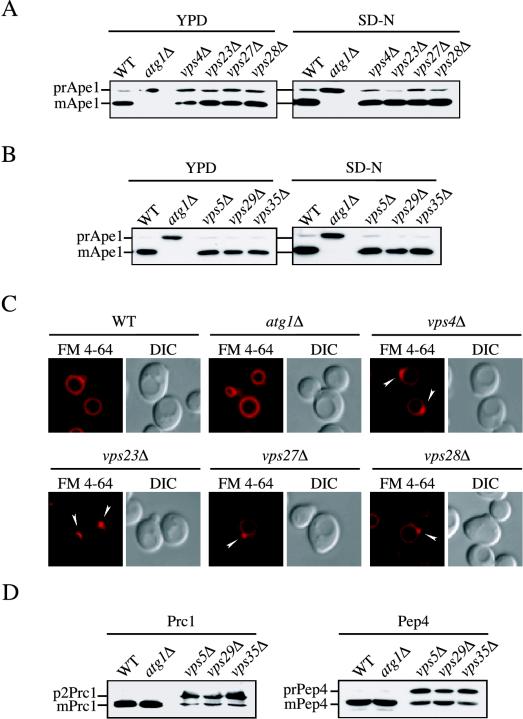
Class E VPS genes are required for the formation of multivesicular bodies in the late endosome (Odorizzi et al., 1998; Reggiori et al., 2000; Katzmann et al., 2001; Reggiori and Pelham, 2001), and the class E Vps proteins localize at least transiently to this compartment. In the corresponding mutant strains, the structure of this organelle is abnormally enlarged, severely affecting its functions, and both forward transport and retrieval from this compartment are compromised (Raymond et al., 1992; Piper et al., 1995; Rieder et al., 1996; Babst et al., 1997; Odorizzi et al., 1998). We examined whether prApe1 maturation was affected in the vps4Δ, vps23Δ, vps27Δ, and vps28Δ class E mutants. As shown in Figure 1A, prApe1 was essentially normally processed to the mature form either in rich medium (delivery to the vacuole via the Cvt pathway) or after nitrogen starvation (delivery to the vacuole by autophagy) in these mutant strains. As a control, we examined delivery of prApe1 to the vacuole in the atg1Δ strain. The atg1Δ mutant is defective in both the Cvt pathway and autophagy, and these cells accumulated only the precursor form of Ape1, indicating that processing to the mature form in the vps mutant strains did not occur during sample preparation but rather reflected normal vacuolar delivery. The class E compartment can become proteolytically active due to the accumulation and eventual maturation of vacuolar hydrolases that are blocked in forward transport to the vacuole (Raymond et al., 1992). We examined the localization of GFP-Ape1 in the class E mutants to verify that maturation correlated with vacuolar delivery. GFP-Ape1 was localized to the vacuolar lumen, not the class E compartment, in the class E vps mutants, indicating that normal transport had taken place (our unpublished data).
Figure 1.
The late endosome does not participate in the biogenesis of the PAS. (A) Wild-type (WT), atg1Δ, vps4Δ, vps23Δ, vps27Δ, and vps28Δ, and (B) WT, atg1Δ, vps5Δ, vps29Δ, and vps35Δ cells in the BY4742 background grown in YPD or nitrogen starved in SD-N medium for 4 h were TCA precipitated. Acetone washed proteins were then resolved by SDS-PAGE and prApe1 maturation analyzed by immunoblot with serum to Ape1. All strains showed normal prApe1 maturation by either the Cvt pathway (YPD) or autophagy (SD-N). (C) Morphological anomalies of class E vps mutants. Wild-type (WT), atg1Δ, vps4Δ, vps23Δ, vps27Δ, and vps28Δ cells were labeled with FM 4–64 and imaged as described previously (Reggiori et al., 2003). Arrowheads indicate enlarged endosomes. All four class E vps mutants accumulated an aberrant endosomal structure. DIC, differential interference contrast. (D) Vacuolar protease missorting in retromer mutants. Cell extracts analyzed in B were probed with anti-Prc1 and anti-Pep4 antisera. Retromer deletants accumulated unprocessed Prc1 (p2Prc1) and Pep4 (prPep4).
The retromer complex is an essential part of the sorting machinery required for retrieval from the late endosome to the Golgi and is composed of the VPS35, VPS29, VPS26, VPS17, and VPS5 gene products, which fall into several of the VPS classes (Seaman et al., 1998). Similar to the class E mutants, defects in these proteins cause significant blocks in transport processes that depend on the late endosome. Analysis of prApe1 in vps35Δ, vps29Δ, and vps5Δ cells showed that the processing of this protease was unaffected (Figure 1B). Again, the atg1Δ strain served as a control and displayed a complete block in prApe1 transport.
To verify that the vps mutants were blocked in the expected pathways, we analyzed either sorting of vacuolar hydrolases or accumulation of the enlarged class E compartment. The four class E vps mutants were stained with FM 4-64, a lipophilic fluorescent dye that allows visualization of the yeast vacuole (Vida and Emr, 1995). Wild-type and atg1Δ cells had normal vacuole morphology (Figure 1C). In contrast, the vps4Δ, vps23Δ, vps27Δ, and vps28Δ strains showed the presence of an exaggerated perivacuolar punctate structure. This organelle corresponds to the enlarged late endosome typically accumulated in all class E vps deletants (Vida and Emr, 1995; Rieder et al., 1996; Babst et al., 1997). In retromer mutants, vacuolar proteases such as Prc1 (CPY) and Pep4 are partially missorted to the periplasmic space in the precursor form (Seaman et al., 1997; Seaman et al., 1998). To verify the phenotype of the vps5Δ, vps29Δ, and vps35Δ strains, the same cell extracts analyzed in Figure 1B were probed with antisera recognizing these two hydrolases. As shown in Figure 1D, unprocessed Prc1 (p2 or Golgi-modified form) and Pep4 (prPep4) were accumulated in retromer deletants but not in wild-type or atg1Δ cells, confirming that the former displayed the expected phenotypes of vps mutants. The essentially normal processing of prApe1 in these vps mutant strains led us to the conclusion that late endosome functions are not required for the formation of Cvt vesicles or autophagosomes.
The VFT Complex, Tlg1, and Tlg2 Catalyze a Membrane Fusion Event Required for Cvt Vesicle Formation That Does Not Occur at the PAS
The biogenesis of Cvt vesicles apparently requires a fusion step catalyzed by the VFT tethering factor and by the SNAREs Tlg1 and Tlg2 (Abeliovich et al., 1999; Reggiori et al., 2003). It remains unknown, however, where this event takes place. It has been proposed that Tlg1 and Tlg2 are required for the homotypic fusion that leads to the sealing of Cvt vesicles (Abeliovich et al., 1999). Because transmembrane proteins such as Atg9 are involved in Cvt vesicle biogenesis (Noda et al., 2000; Kim et al., 2002; Reggiori et al., 2004), a second possibility is that those proteins and the VFT complex are necessary for the transport of intermediate vesicles to the PAS that are required for Cvt vesicle formation, or for the retrieval of some components back to the donor membrane source. A third hypothesis is that in the absence of this fusion machinery, the proper homeostasis of either the early endosome or Golgi complex is severely compromised and that has an indirect effect on Cvt vesicle completion by altering the sorting of specific integral membrane proteins.
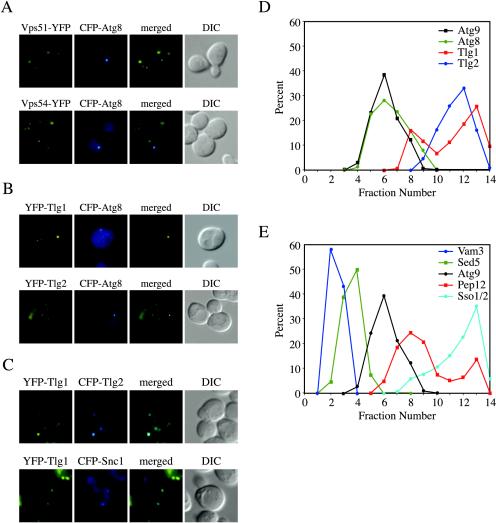
If Tlg1, Tlg2, and the VFT complex mediate Cvt vesicle completion or the fusion of the vesicles directed to the PAS, we would expect that those proteins would localize to the site of Cvt vesicle biogenesis. To distinguish among the above-mentioned possibilities, we examined the localization of the VFT complex. Accordingly, we integrated the gene encoding the yellow fluorescent protein (YFP) at the 3′ end of two genes, VPS51 and VPS54, coding for VFT complex subunits (Conibear and Stevens, 2000; Siniossoglou and Pelham, 2002; Conibear et al., 2003; Reggiori et al., 2003). The resulting strains were then transformed with a plasmid expressing the CFP-Atg8 fusion under the control of the authentic ATG8 promoter and analyzed by fluorescence microscopy. Atg8 is localized to the PAS where it is incorporated into nascent vesicles (Kirisako et al., 1999; Huang et al., 2000; Suzuki et al., 2001; Figure 2). The localization pattern of YFP-Vps51 and YFP-Vps54 was identical to that reported in the literature, a dispersed punctate staining (Conboy and Cyert, 2000; Siniossoglou and Pelham, 2001, 2002; Figure 2A). In all images, YFP chimeras and CFP-Atg8 were never observed to colocalize (Figure 2A). Tlg1 and Tlg2 were also tagged with YFP but on their N terminus and the plasmid carrying either functional fusion was used to transform wild-type cells together with the CFP-Atg8 chimera. As expected, Tlg1 and Tlg2 showed dispersed punctate staining that did not colocalize with CFP-Atg8 (Abeliovich et al., 1998; Holthuis et al., 1998; Seron et al., 1998; Coe et al., 1999; Siniossoglou and Pelham, 2001; Figure 2B). As a control, we examined the localization of YFP-Tlg1 with CFP-Tlg2 or CFP-Snc1, a Golgi-to-plasma membrane v-SNARE. It has been previously shown that these proteins colocalize to different extents (Holthuis et al., 1998; Lewis et al., 2000). As shown in Figure 2C, we could detect partial overlap among these proteins, indicating that the fluorescence approach we used allowed us to examine colocalization.
Figure 2.
The VFT complex, Tlg2, and all other t-SNAREs do not localize to the PAS. (A) The VFT complex does not localize to the PAS. Wild-type (SEY6210) cells expressing either Vps51-YFP (PSY82) or Vps54-YFP (PSY83) were transformed with the pRS316ECFP-Aut7 plasmid. The resulting strains were grown in SMD medium to an early log stage and analyzed with a fluorescence microscope. (B) Tlg1 and Tlg2 do not localize to the PAS. The wild-type (SEY6210) strain was cotransformed with plasmids encoding either YFP-Tlg1 or YFP-Tlg2 and CFP-Atg8. Transformed cells were analyzed as in A. (C) Tlg1 colocalizes with Tlg2 and partially with Snc1. The wild-type (SEY6210) strain was cotransformed with plasmids encoding YFP-Tlg1 and either CFP-Tlg2 or CFP-Snc1. Transformed cells were analyzed as in A. (D) Tlg1 and Tlg2 do not fractionate with the PAS. The wild-type (HAY456) strain expressing N-terminal PA-tagged Atg9 was transformed with the plasmid expressing YFP-Tlg2. Spheroplasts from the transformed cells were grown in rich medium until mid-log phase, osmotically lysed, and the membranes fractionated on a sucrose density gradient as described in MATERIALS AND METHODS. In total, 14 fractions were collected from the top of the gradient, resolved by SDS-PAGE, and membranes were probed with antisera to PA, Atg8, GFP, and Tlg1. (E) None of the yeast t-SNAREs localize to the PAS. The same gradient fractions analyzed in D were also probed for Vam3, Sed5, Sso2, and Pep12. None of these t-SNAREs colocalized with the PAS marker Atg9.
The absence of colocalization between Atg8 and either Vps51, Vps54, Tlg1, or Tlg2 established by fluorescence microscopy does not give the certitude that small populations of those proteins are not at the same place; this technique is not optimal for detecting small protein pools because they do not produce a signal sufficient to be visualized. To confirm the observations made with the fluorescence colocalization experiments, we decided to separate intracellular membranes on a sucrose density gradient. A strain carrying the ATG9 gene tagged on its 3′ end with PA was transformed with the plasmid expressing the YFP-Tlg2 fusion. Total cell extracts were centrifuged at 13,000 × g, and the supernatant, including the PAS, was resolved on density gradients as described in MATERIALS AND METHODS. Atg9 and Atg8 were used as PAS markers because they colocalize on gradients with the rest of the Atg proteins (Kim et al., 2002). As expected, these two proteins showed the same distribution and were concentrated in fractions 5–7 (Figure 2D). Two other PAS markers, Atg1 and Atg11, were also examined and found in the same fractions (our unpublished data). Because these two factors posses a cytosolic population, however, they had an additional peak corresponding to their soluble pool. In contrast, Tlg2 was mostly found in fractions 10–13 (Figure 2D). The same fractions were also probed with anti-Tlg1 antibodies. Tlg1 was mostly found in fraction 12–13, also well separated from the PAS marker (Figure 2D). We tried to localize Vps51 on the same sucrose gradient but the conditions used caused the complete dissociation of this protein from the membranes (our unpublished observations). These results confirmed the observation made with fluorescence microscopy; that is, Tlg1 and Tlg2 do not localize to the PAS under steady-state conditions.
The Tlg2 protein is a t-SNARE (Abeliovich et al., 1998; Holthuis et al., 1998; Seron et al., 1998). The absence of Tlg2 at the PAS led to the question of which t-SNARE, if any, is localized to this compartment. Yeast offers an exceptional opportunity to investigate this issue because its genome contains only seven t-SNAREs (Pelham, 2001a). Thus, our gradient fractions were also probed with antibodies recognizing the t-SNAREs of the late endosome (Pep12), cis-Golgi compartment (Sed5), vacuole (Vam3), and plasma membrane (Sso1 and Sso2) (Aalto et al., 1993; Banfield et al., 1994; Becherer et al., 1996; Burd et al., 1997; Nichols et al., 1997). None of these proteins displayed complete cofractionation with the PAS marker Atg9 (Figure 2E). Although there was significant overlap between the Pep12 and Atg9 peaks, previous studies have shown that Pep12 is not required for prApe1 maturation (Abeliovich et al., 1999). Furthermore, the fractionation results are in agreement with previous studies reported in the literature. For example, it has been shown that Pep12 and Cvt vesicle markers are in organelles with a different density (Kim et al., 2001, 2002; Wang et al., 2001). PAS proteins are also in a different fraction than Anp1, a cis-Golgi protein that completely colocalizes with Sed5 (Jungmann and Munro, 1998; Wang et al., 2001). We did not localize the remaining t-SNARE, Ufe1 for two reasons. First, this protein is confined in the ER (Lewis and Pelham, 1996), an organelle that is clearly morphologically distinct from the perivacuolar punctate PAS. Second, the ER is eliminated by the centrifugation step performed before our gradient fractionation. Together, the data suggest that Tlg1, Tlg2, and the VFT complex are not involved in SNARE-dependent fusion events occurring at the PAS.
Early Endosomes and the PAS Are Different Structures
Late endosome function is not required for the Cvt pathway or autophagy, but the PAS possesses some characteristics typical of early endosomes. For example, the surface of both organelles is thought to be decorated with phosphatidylinositol-3-phosphate [PtdIns(3)P], and this lipid is required to recruit the two sorting nexins Atg24/Snx4 and Atg20/Snx42 (Nice et al., 2002; Hettema et al., 2003). In addition, proteins retrieved from these two compartments presumably transit in vesicles that undergo a fusion step, probably occurring at the Golgi complex, and catalyzed by Tlg1, Tlg2, and the VFT complex (Siniossoglou and Pelham, 2001, 2002; Reggiori et al., 2003). These analogies indicate the possibility that the PAS and early endosome are identical structures. However, although Tlg1, Tlg2, and the VFT complex are needed for prApe1 import, they do not seem to function at the PAS. This result suggests that the early endosome and PAS are separate structures. To resolve this issue we decided to investigate whether the PAS and early endosome are identical compartments through fractionation and microscopy analyses.
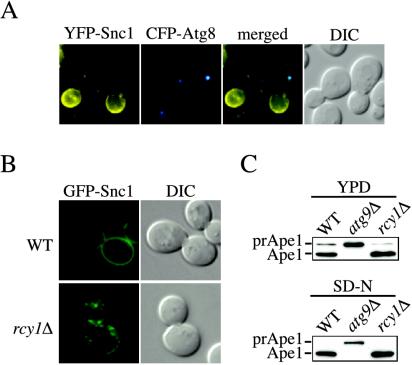
Tlg1 is spread all along the endosomal system, but it seems to be mostly concentrated at early endosomes (Holthuis et al., 1998; Lewis and Pelham, 2002). Our colocalization and fractionation studies showed that Tlg1 is not at the PAS (Figure 2, B and C). The vesicle SNARE Snc1 cycles through early endosomes and partially colocalizes with Tlg1 (Lewis et al., 2000; Figure 2C). We used this protein as a second early endosome marker and analyzed its localization relative to the PAS. For this reason, a YFP-Snc1 fusion was made and used to transform wild-type cells together with the plasmid expressing CFP-Atg8 to mark the PAS. Snc1 cycles between early endosomes and the plasma membrane. In wild-type cells, YFP-Snc1 displays a fluorescent signal predominantly at the cell surface along with some small punctate structures that presumably correspond to early endosomes. Fluorescence microscopy analysis did not show an overlap between YFP-Snc1 and CFP-Atg8/PAS signals, indicating the probable absence of Snc1 at the PAS (Figure 3A).
Figure 3.
The PAS and early endosomes are different structures. (A) Snc1 does not localize to the PAS. Wild-type (SEY6210) cells carrying plasmids expressing both YFP-Snc1 and CFP-Atg8 were grown in SMD medium to an early log stage and analyzed with a fluorescence microscope. (B) Snc1 recycling is blocked in the rcy1Δ strain. The wild-type (WT) and rcy1Δ cells in the BY4742 background were transformed with a plasmid expressing the GFP-Snc1 chimera. Transformed cells were grown to mid-log stage in SMD medium and imaged with a fluorescence microscope. In wild-type cells, Snc1 was concentrated on the plasma membrane, whereas it was accumulated in internal structures in the mutant. (C) Precursor Ape1 processing is normal in the rcy1Δ strain. Wild-type (WT), atg9Δ and rcy1Δ cells in the BY4742 background grown either in YPD or nitrogen starved in SD-N medium for 4 h were TCA precipitated. Acetone washed proteins were then resolved by SDS-PAGE and prApe1 maturation was analyzed by immunoblot with serum to Ape1. Both the wild-type and rcy1Δ strains showed normal prApe1 maturation either when grown in YPD medium or nitrogen starved for 4 h.
To assess independently from fluorescence data whether early endosomes play an essential role in the normal progression of the Cvt pathway, we decided to specifically disrupt the functions of this organelle and determine the effects on prApe1 transport. Rcy1 and Skp1 form a complex essential for sorting from early endosomes (Wiederkehr et al., 2000; Galan et al., 2001). In the absence of Rcy1, early endosomes become abnormally enlarged, severely compromising endocytosis and Snc1 recycling (Wiederkehr et al., 2000; Galan et al., 2001). We first transformed the rcy1Δ mutant with a plasmid expressing GFP-Snc1 to verify by fluorescence microscopy the early endosome-recycling defect of these cells. As previously reported, in wild-type cells most of the fluorescent staining corresponding to GFP-Snc1 was on the plasma membrane, whereas in the rcy1Δ mutant the same chimera was predominantly accumulated in internal punctate structures, probably early endosomes (Figure 3B; Lewis et al., 2000; Galan et al., 2001). We then examined whether prApe1 maturation was affected in the rcy1Δ mutant. As shown in Figure 3C, prApe1 was essentially normally processed either in rich medium or after nitrogen starvation, indicating that early endosome function does not seem to be required for prApe1 import through either the Cvt pathway or autophagy. We concluded that the PAS and early endosomes are different structures and that the early endosome does not play a direct role in Cvt pathway and autophagy progression.
The sole yeast PtdIns 3-kinase, Vps34, is found in two distinct tetrameric complexes that contain two other common components: Vps15 and Vps30/Atg6 (Schu et al., 1993; Kihara et al., 2001). These two complexes have been named complex I and II, and each one has a fourth subunit that is specific, Atg14 and Vps38, respectively (Kihara et al., 2001; Burda et al., 2002). Complex I generates PtdIns(3)P at the PAS, and it is essential for autophagy and the Cvt pathway (Kihara et al., 2001; Kim et al., 2002; Nice et al., 2002). Complex II produces the PtdIns(3)P population found on endosomes, and its presence is required to ensure the transport of carboxypeptidase Y from the Golgi complex to the vacuole lumen (Schu et al., 1993; Kihara et al., 2001; Burda et al., 2002). It seems that this complex is also involved in PtdIns(3)P synthesis at the early endosomes because its function is essential to recruit sorting nexins to those compartments (Hettema et al., 2003). We decided to analyze prApe1 and Snc1 trafficking in mutant strains that specifically affect the function of one of the two PtdIns 3-kinase complexes to see whether it was possible to distinguish the PAS from early endosomes by the fact that their PtdIns(3)P pools are generated in different ways.
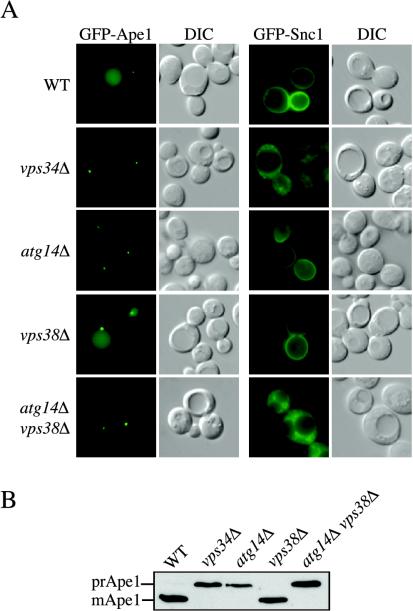
Wild-type, atg14Δ, and vps38Δ cells were transformed with a plasmid expressing either GFP-Snc1 or GFP-Ape1 (Lewis et al., 2000; Shintani et al., 2002). We also checked the localization of these two chimeras in the vps34Δ strain; the vps34Δ cells are unable to synthesize PtdIns(3)P, and all pathways requiring this lipid are blocked. When expressed in wild-type cells, the two fluorescent constructs showed the localization pattern that has been described in the literature. GFP-Ape1 was localized to the vacuole lumen and PAS, whereas GFP-Snc1 was on the plasma membrane and in small punctate structures (Lewis et al., 2000; Shintani et al., 2002; Reggiori et al., 2003; Figure 4A). The VPS34 deletion affected the trafficking of both constructs. In the vps34Δ strain GFP-Ape1 was exclusively concentrated at the PAS in a manner characteristic of mutants with a defect in the formation of Cvt vesicles, whereas GFP-Snc1 was completely accumulated in small spherical cytosolic structures representative of a recycling defect (Lewis et al., 2000; Panek et al., 2000; Siniossoglou et al., 2000; Galan et al., 2001; Siniossoglou and Pelham, 2001; Figure 4A). Identical results were also obtained when the two other subunits common to the PtdIns 3-kinase complex, Vps15 and Vps30/Atg6, were absent (our unpublished data).
Figure 4.
Both PtdIns 3-kinase complexes generate the PtdIns(3)P on the early endosome surface. (A) Wild-type (WT), vps34Δ, vps38Δ, atg14Δ, and atg14Δ vps38Δ cells in the BY4742 background were transformed with a plasmid expressing either GFP-Ape1 or GFP-Snc1. Transformed cells were grown to a mid-log stage in SMD medium and imaged with a fluorescence microscope. GFP-Ape1 was delivered to the vacuole lumen in wild-type and vps38Δ cells but not in vps34Δ, atg14Δ and atg14Δ vps38Δ mutants where it was accumulated at the PAS. GFP-Snc1 displayed normal cycling in wild-type, vps38Δ, and atg14Δ cells but not in vps34Δ and atg14Δ vps38Δ mutants, indicating that its transport between the early endosome and Golgi complex depends on the PtdIns(3)P produced by both PtdIns 3-kinase complexes. (B) The maturation of endogenous prApe1 is blocked in vps34Δ, atg14Δ, and atg14Δ vps38Δ strains.
The specific disruption of the PtdIns 3-kinase complex I resulting from the ATG14 disruption blocked GFP-Ape1 transport but not GFP-Snc1 recycling (Figure 4A). In contrast, altering the function of PtdIns 3-kinase complex II by removal of Vps38 did not completely interfere with either GFP-Snc1 or proper prApe1 trafficking (Figure 4A). The latter result led us to propose that maybe both PtdIns 3-kinase complexes were involved in the synthesis of PtdIns(3)P on early endosomes. To test this hypothesis, a vps38Δ atg14Δ double deletion strain was generated and transformed with our two reporter constructs. The simultaneous elimination of these two PtdIns 3-kinase complex subunits had the same effect as the VPS34 deletion; the transport of both GFP-Ape1 and GFP-Snc1 was severely compromised (Figure 4A). This result is surprising because it has been published that the PtdIns 3-kinase complex II (Vps38-Vps30/Atg6-Vps15-Vps34) synthesizes the majority of the PtdIns(3)P required to recruit Atg24/Snx4 and Atg20/Snx42 to early endosome membranes (Hettema et al., 2003). These two sorting nexins are essential for Snc1 trafficking through the early endosome (Hettema et al., 2003). Accordingly, it would be expected that Snc1 recycling is severely affected in vps38Δ cells, but our data showed that the low amounts of PtdIns(3)P generated by the PtdIns 3-kinase complex I (Atg14-Vps30/Atg6-Vps15-Vps34) were sufficient to allow correct Snc1 transport (Figure 4A). We extended our analysis by examining the processing of prApe1 in the different PtdIns 3-kinase complex mutants. In agreement with the fluorescent data obtained with the GFP-Ape1 fusion, we found that only the vps34Δ, atg14Δ, and atg14Δ vps38Δ mutant strains displayed a defect in the processing of endogenous prApe1 (Figure 4B). Based on these results, we concluded that both PtdIns 3-kinase complexes produce the PtdIns(3)P pool on early endosomes. Alternatively, complex I is able to compensate for complex II in the absence of the latter. Because the PtdIns(3)P on the PAS surface is generated exclusively by the PtdIns 3-kinase complex I, we confirmed that the PAS and early endosomes are different compartments.
Both the ER and the Golgi Complex Are Involved in PAS Formation and Autophagy
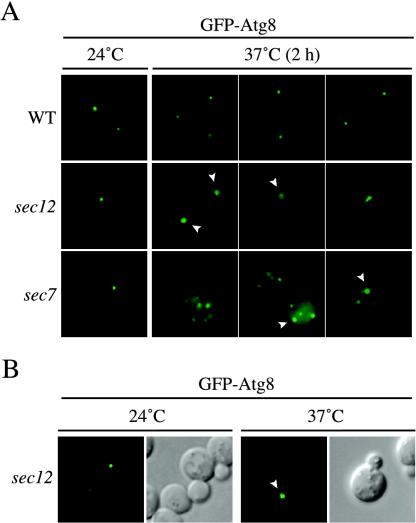
Having established that both early and late endosomes are not participating in the biogenesis of the PAS, we decided to focus on the remaining intracellular organelles that generate vesicular structures, the ER and the Golgi complex. To further test the requirement of secretory pathway components on PAS biogenesis, we examined various sec mutants under steady-state conditions. Wild-type and two thermosensitive mutants that block the secretory pathway at different stages, sec12 and sec7, were transformed with a construct expressing GFP-Atg8 to monitor PAS alterations after long incubation time at restrictive temperatures. The SEC12 gene codes for a GDP/GTP exchange factor that is required for vesicle budding and exit from the ER (Barlowe and Schekman, 1993). Sec7 is a GDP/GTP exchange factor required for trafficking through the Golgi (Franzusoff and Schekman, 1989; Jackson and Casanova, 2000). Wild-type and mutant cells expressing GFP-Atg8 were grown at 24°C and then analyzed with a fluorescence microscope and successively transferred to a 37°C incubator. Cells were imaged again after 30, 60, 90, and 120 min at restrictive temperature.
When the cell culture was maintained at permissive temperature, the localization pattern of the chimera in the secretion mutants was indistinguishable from that in wild-type cells; GFP-Atg8 was present at a perivacuolar punctate structure (Figure 5A). The temperature shift did not alter GFP-Atg8 localization in wild-type cells during the 2-h incubation period at 37°C (Figure 5A). This was also true for sec12 and sec7 cells during the first hour at restrictive temperature (our unpublished data). However, different fluorescent structures started to occur in these two mutant strains after 90 min at 37°C becoming more evident after 2 h. In several sec12 cells (∼22% of the cells at the 2-h time point; 100 cells scored), the PAS size was larger and/or its shape assumed toroid and ellipsoid forms that sometimes resembled a cluster of punctate vesicles (Figure 5A). Identical results were also obtained with sec23, another mutant strain that affects ER-to-Golgi transport (our unpublished data; Kaiser and Schekman, 1990). The sec7 cells showed the presence of several punctate structures (∼82% of the cells at the 2-h time point; 200 cells scored), often larger than the normal PAS, that in some cases also assumed toroid or ellipsoid forms. The transfer of both strains back to 24°C for 4 h caused the dissolution of most of these abnormal formations, indicating that they were not terminal structures reflecting cell death (our unpublished data).
Figure 5.
Early secretory mutants accumulate an aberrant PAS. (A) The wild-type (WT, SEY6210), sec12 (FBY217), and sec7 (AFM69–1A) strains were transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter. Transformed cells were grown at 24°C to early log stage in SMD medium and imaged with a fluorescence microscope. Cultures were then transferred to 37°C, and cells were imaged every 30 min. Between 90 min and 2 h at restrictive temperature, sec12 and sec7 cells started to display the presence of aberrant PAS some of which assumed toroid and ellipsoid forms, suggesting that early secretory mutations affect proper PAS organization. Examples of images taken after 2 h are shown and toroid structures indicated with a white arrowhead. (B) Overexpression of Atg8 does not promote the formation of aberrant PAS structures in early secretory mutants. The sec12 strain (FBY217) was transformed with a plasmid carrying a GFP-Atg8 fusion driven by the authentic ATG8 promoter. Transformed cells were analyzed as in A. GFP-Atg8–marked toroid and ellipsoid structures were observed in sec12 cells incubated at 37°C for 2 h.
The temperature shift experiments described above were performed using a GFP-Atg8 chimera under the control of the strong CUP1 promoter to facilitate cell imaging. To verify that our observations were not an artifact caused by higher Atg8 expression levels, sec12 cells were transformed with a plasmid expressing the GFP-Atg8 fusion under the control of its authentic promoter. Transformed cells were successively grown at 24°C or shifted to 37°C for 2 h. Fluorescence images showed that GFP-Atg8 was rearranged in the same abnormal structures at the nonpermissive temperature (Figure 5B). These results suggest that early sec mutants affect localization of Atg8, a component required for the Cvt pathway in rich media conditions.
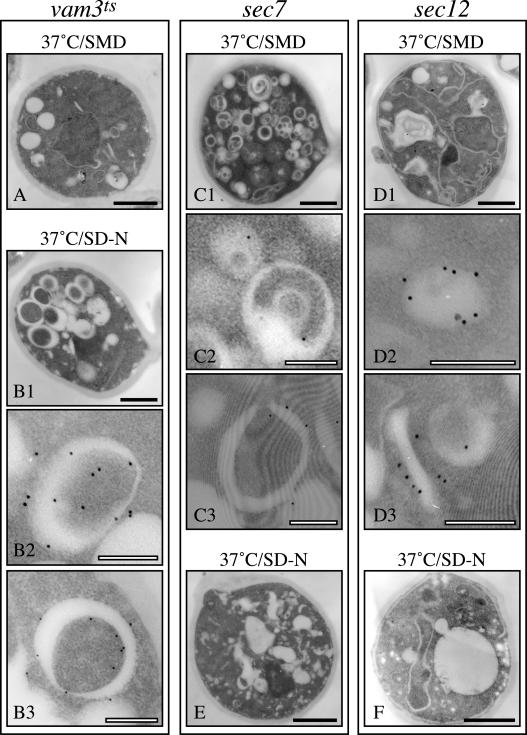
The size of the observed GFP-Atg8–containing structures was comparable with that of autophagosomes; thus, we decided to try to analyze them by immunoelectron microscopy. Autophagosomes are a transient structure and in order to stabilize and visualize them, we took advantage of a vam3Δ strain carrying a conditional allele of VAM3 (vam3ts), the gene coding for the vacuolar t-SNARE. When these cells are nitrogen starved at restrictive temperatures, autophagosomes are accumulated in the cytoplasm (Darsow et al., 1997; Reggiori et al., 2003). A triple hemagglutinin-ATG8 fusion under the control of the authentic promoter was integrated in vam3ts, sec7, and sec12 strains. Transformed cells were grown at 24°C in YPD medium and then shifted to 37°C for 2 h in either the same medium or in SD-N. Samples were collected before and after the temperature and growth media shifts.
All three strains showed normal structures when grown in rich medium at permissive temperature (our unpublished data). Shifting to nonpermissive temperature in rich medium, a condition that does not induce autophagy, did not cause any morphological alteration in vam3ts cells; gold particles were not associated with a defined membrane (Figure 6A). In contrast, when vam3ts cells were shifted to nitrogen starvation conditions to induce autophagy at 37°C, several autophagosomes accumulated in the cytoplasm (Figure 6B1). The external and internal surfaces of these double-membrane vesicles were surrounded with gold particles consistent with Atg8 function and previous reports on the localization of this protein (Figure 6, B2 and B3; Kirisako et al., 1999). The incubation of the sec7 strain in rich medium at 37°C provoked the accumulation of numerous aberrant structures (Figure 6C1). These membranous arrangements in sec7 cells have been previously characterized and named Berkeley bodies (Novick et al., 1980; Esmon et al., 1981). Interestingly, Berkeley bodies were marked with gold particles, allowing us to identify the circular GFP-Atg8–containing structures (Figure 6, C2 and C3). The decoration of these bodies with gold particles was less intense compared with that of autophagosomes that were accumulated in the vam3ts strain in SD-N but that may reflect the fact that nitrogen starvation induces Atg8 expression (Kirisako et al., 1999; Huang et al., 2000). When incubated at 37°C in rich medium, sec12 cells showed the typical ER expansion and in some cases, consistent with the fluorescent data, there was also the presence of a Berkeley body decorated with gold particles (Figure 6, D2 and D3). These cells also often displayed one cluster of gold particles in the proximity of an unidentified membrane (Figure 6D3).
Figure 6.
Atg8 is targeted to the Berkeley bodies in sec7 and sec12 cells. The vam3ts (A and B), sec7 (C and E), and sec12 (D and F) strains expressing a triple hemagglutinin-ATG8 fusion under the control of the authentic ATG8 promoter were grown at 24°C in YPD medium and then shifted to 37°C for 3 h in either the same medium (A, C, and D) or in SD-N (B, E, and F). Cells were collected before and after the temperature and the media shifts. All samples were prepared for immunoelectron microscopy as described in MATERIALS AND METHODS. Black bar, 1 μm; white bar, 0.2 μm.
When both sec7 and sec12 strains were starved for nitrogen at nonpermissive temperatures, the cell morphology was severely affected and autophagosomes or autophagosome-like structures were not formed (Figure 6, E and F). The results obtained with the sec12 mutant correlate with previous data reporting that autophagy is blocked in this strain (Ishihara et al., 2001). Our data illustrating that this pathway is also affected by the sec7 mutation emphasize the relevance of the Golgi complex in autophagy.
To further demonstrate the importance of the Golgi complex for the normal progression of this degradative pathway, two different biochemical approaches were used to explore the effects of the sec7 mutation on autophagy. The same experiments were performed in parallel in the sec12 mutant as a control because, as noted above, it has been previously shown that autophagy is severely impaired when these cells are grown at restrictive temperatures (Ishihara et al., 2001). Overexpression of Ape1 causes the saturation of the Cvt pathway, leading to the accumulation of its precursor form in the cytoplasm (Klionsky et al., 1992). Starvation conditions induce the formation of the larger autophagic double-membrane vesicles, and these structures are able to sequester all this amassed material and deliver it to the vacuole interior where it is processed to the mature form (Scott et al., 1996). Thus, starvation-induced maturation of overexpressed prApe1 can be used to monitor autophagy.
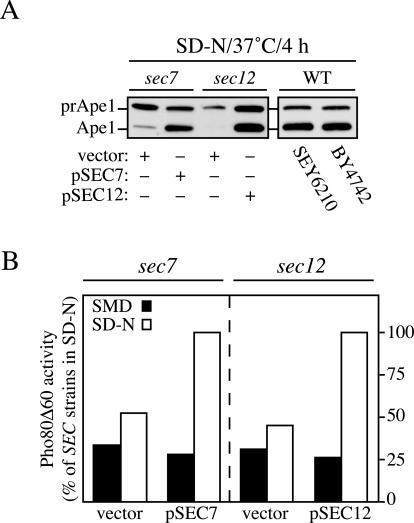
The sec7 and sec12 strains were transformed with both a multi-copy plasmid expressing Ape1 and either an empty vector or one carrying the SEC wild-type gene. The complementation of the mutated gene was carried out to generate an isogenic wild-type strain. Transformed cells and two different wild-type control strains revealed a clear prApe1 accumulation phenotype similar to that reported in the literature when cells were grown in rich medium at permissive temperature (Klionsky et al., 1992; Scott et al., 1996; our unpublished data). Only wild-type cells and complemented sec mutants showed an increase in prApe1 processing when nitrogen starved for 4 h at 37°C (Figure 7A). The level of prApe1 processing seen in the complemented sec mutant strains was lower than that described in the literature (Scott et al., 1996); however, a similar result was seen with the two different wild-type strains analyzed in this experiment. This finding suggests that the heat shock caused by the culture passage from 24 to 37°C resulted in reduced autophagic activity. In comparison, the noncomplemented sec mutants showed a clear reduction in autophagic activity (Figure 7A); there was minimal or no processing of the accumulated prApe1 after a shift to starvation conditions at the nonpermissive temperature, suggesting that autophagy was almost completely blocked.
Figure 7.
A functional Golgi complex is essential for autophagy. (A) The sec12 (FBY217) and sec7 (AFM69-1A) strains were transformed with both a multi-copy plasmid expressing Ape1 and either an empty vector (pRS415) or one carrying the wild-type allele of the mutated gene (pSEC12 or pSEC7, respectively). Transformed cells and two wild-type strains (WT, SEY6210, and BY4742) were grown to 1 OD600 in SMD medium at 24°C. After washing twice with water, cells were nitrogen starved in SD-N medium for 4 h at 37°C before collecting the proteins by TCA precipitation. Polypeptides were finally resolved by SDS-PAGE and prApe1 maturation analyzed by immunoblot with serum to Ape1. (B) The sec12 (FRY208) and sec7 (FRY209) strains expressing Pho8Δ60 and carrying either an empty vector (pRS415) or a plasmid complementing the corresponding sec mutation (pSEC12 or pSEC7, respectively) were shifted from SMD medium at 24°C (black bars) to SD-N medium at 34°C (white bars) for 4 h. Autophagy induction was determined by a Pho8 activity assay. Results were expressed as percentage of the activity measured for the complemented strains starved for nitrogen.
To extend our analysis, we decided to measure the vacuolar processing of the cytosolic marker protein Pho8Δ60. This truncated form of the vacuolar alkaline phosphatase (Pho8) lacks the transmembrane domain and consequently localizes to the cytosol (Noda et al., 1995). This protein is delivered to the vacuole exclusively by autophagy. Proteolytic cleavage of the Pho8Δ60 propeptide in the vacuole lumen generates the active form of the enzyme, which can be detected by an activity assay (Noda et al., 1995). To measure autophagy in the sec7 and sec12 cells, the chromosomal PHO8 gene was replaced with pho8Δ60, and the PHO13 open reading frame (encoding cytosolic alkaline phosphatase) was knocked out. The resulting strains were transformed with either an empty vector or a plasmid encoding the wild-type allele of the mutated sec gene. Phosphatase activity was determined either before or after nitrogen starvation for 4 h at 34°C. In all strains, there was low alkaline phosphatase activity when cells were grown in rich medium (Figure 7B). There was an ∼3.5-fold increase in activity after the induction of autophagy when complemented mutant cells were shifted to SD-N. In contrast, nitrogen starvation resulted in a minimal increase in alkaline phosphatase activity from Pho8Δ60 in the noncomplemented mutant strains.
Together with the electron microscopy analysis, these data suggest that the Golgi complex plays an essential role during autophagy that is comparable with that of the endoplasmic reticulum.
The Cvt Pathway Is Blocked in Early Secretory Mutants
The ER and the secretory flux from this compartment have been shown to be essential for autophagy but unnecessary for the Cvt pathway (Klionsky et al., 1992; Ishihara et al., 2001; Hamasaki et al., 2003). In particular, those experiments analyzed the Cvt pathway functionality by pulse-chase analyses of radiolabeled prApe1, after the kinetics of precursor processing. For example, it was shown that a strain carrying a thermosensitive allele of SEC12 was able to transport 50–60% of prApe1 into the vacuole at the nonpermissive temperature (Klionsky et al., 1992). The preincubation times at restrictive temperature in those prApe1 processing analyses were between 5 and 60 min. The maturation time of prApe1 is 90 min, and this time presumably reflects the transport rate of the Cvt pathway rather than the processing time once the zymogen has reached the vacuole (Klionsky et al., 1992). For this reason, it is possible that prApe1 processing in the conditional sec mutants would not be affected during the first 90 min after the induction of the transport block. That is, even ER-derived membrane that was involved in the Cvt pathway might not reveal a defect in prApe1 processing for up to 90 min if that membrane had already been directed into the pathway before the induction of the sec phenotype.
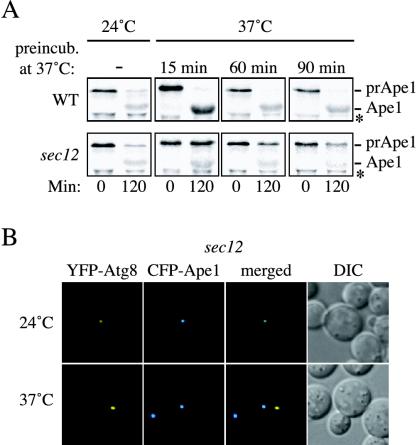
Because the previously published pulse-chase analyses of radiolabeled prApe1 in early secretory mutants suggested that the ER was not required for prApe1 import into the vacuole (Klionsky et al., 1992; Ishihara et al., 2001), we decided to extend our analysis. We chose different preincubation times at restrictive temperature to explore the effects on the Cvt pathway of longer ER-to-Golgi transport blocks. Wild-type and sec12 strains were grown at 24°C and then shifted to 37°C for different times before being labeled for 10 min and chased for 2 h. Cells were collected at the beginning and end of the chase period and Ape1 immunoprecipitated. Incubation at the restrictive temperature did not interfere with prApe1 transport in wild-type cells (Figure 8A). In contrast, a shift of sec12 cells for 15 min at 37°C before the labeling was enough to severely affect the transport of prApe1 into the vacuole (Figure 8A). Longer preincubation times had an increasing inhibitory effect without completely blocking the Cvt pathway. This observation suggested that the components required for vesicular exit from the ER did not affect the function of the existing PAS, but their inactivation may affect biogenesis of new PAS that are required for additional rounds of prApe1 import through the Cvt pathway.
Figure 8.
Early stages of the secretory pathway are essential for the Cvt pathway. (A) Precursor Ape1 transport is affected in sec12 cells. Wild-type (WT and SEY6210) and sec12 (FBY217) cells were grown at 24°C and then shifted to 37°C for 15, 60, and 90 min, respectively. Both strains were pulse-labeled and subjected to a nonradioactive chase as described in MATERIALS AND METHODS. Samples were removed at the beginning (0 min) and at the end (120 min) of the chase period. Proteins were TCA precipitated and then immunoprecipitated with antiserum to Ape1. Precursor Ape1 transport was perturbed in sec12 cells at prolonged restrictive temperatures but not in wild-type cells. The asterisk marks a cross-reacting band. (B) Precursor Ape1 is not correctly targeted to the PAS when ER exit is blocked. The sec12 strain (FBY217) was transformed with plasmids expressing CFP-Ape1 and YFP-Atg8. Transformed cells were grown at 24°C in SMD and imaged. The temperature was then shifted to 37°C and cells imaged again after 2 h. CFP-Ape1 and YFP-Atg8 colocalized to the PAS when the cells were grown at 24°C. Incubation at restrictive temperature for 2 h disrupted the correct CFP-Ape1 recruitment to the PAS marked with YFP-Atg8 in 50–60% of the imaged cells.
We next decided to investigate if prApe1 was recruited to the abnormal structures that formed in the sec mutants at the nonpermissive temperature. In mutants that are defective in the Cvt pathway such as atg1Δ, CFP-Ape1 can be observed to colocalize with the PAS (Shintani et al., 2002). To examine colocalization, sec12 cells were cotransformed with plasmids expressing CFP-Ape1 and YFP-Atg8 (to mark the PAS) under the control of their own promoters. Transformants were then grown at 24°C to early log phase and subsequently shifted to 37°C for 90 min. The two fluorescent signals were separated in 50–60% of the cells incubated at the nonpermissive temperature, indicating that the normal recruitment of prApe1 to the PAS was severely affected (Figure 8B). The PAS is defined as the location where Atg proteins are assembled to catalyze double-membrane vesicle formation (Suzuki et al., 2001; Kim et al., 2002). Because prApe1 in not correctly targeted to the Atg8-containing structures in sec12 cells, there is a possibility that, in addition to being required to form a normal PAS, the ER and Golgi complex are the source for the lipids used to make the double-membrane–sequestering vesicles used in the Cvt and autophagy pathways. Together, these results suggest that the normal progression of the Cvt pathway, possibly the biogenesis of the PAS, requires the proper functioning of ER and Golgi complex cisternae, similar to autophagy (Ishihara et al., 2001; Hamasaki et al., 2003).
DISCUSSION
The Early and Late Endosome Are Not Required for Cvt Vesicle and Autophagosome Biogenesis
The origin of the sequestering membrane in autophagy has been the focus of a substantial body of research (Ogier-Denis and Codogno, 2003; reviewed in Fengsrud et al., 2004) but the resulting data have been inconclusive. Previous studies have suggested that the formation site of Cvt vesicles and autophagosomes is the PAS (Suzuki et al., 2001; Kim et al., 2002). The PAS is a punctate, perivacuolar structure similar to the late endosome. Analyses of prApe1 transport in mutants defective in endosomal function, however, revealed that the late endosome and PAS are physically and functionally separate structures (Figure 1).
Tlg1, Tlg2, and Vps45 are required for protein sorting between the late Golgi and early endosome. It has been proposed that Tlg1, Tlg2, and Vps45 are required for the homotypic fusion that leads to Cvt vesicle completion (Abeliovich et al., 1999). Our fluorescence and fractionation experiments, however, indicated instead that Tlg1, Tlg2, and the VFT complex carry out a function required for Cvt vesicle formation in a location different from that of the PAS (Figure 2, A, B, and D). Previous data made evident the fact that Cvt vesicle biogenesis needs at least some of the same factors used for the retrieval of the v-SNARE Snc1 from early endosomes to the late Golgi compartments. In both processes, PtdIns(3)P is generated on the organelle surface catalyzing the recruitment of the two sorting nexins Atg24/Snx4 and Atg20/Snx42 (Nice et al., 2002; Hettema et al., 2003). In addition, a fusion step probably occurring at the Golgi complex and catalyzed by Tlg1, Tlg2, and the VFT tethering complex is necessary for the normal progression of the two pathways (Abeliovich et al., 1999; Siniossoglou and Pelham, 2001, 2002; Reggiori et al., 2003). Despite sharing this analogy, three diverse results suggest that the PAS and early endosomes are different structures. First, fluorescence and fractionation studies show that the early endosome markers Tlg1 and Snc1 do not colocalize with the PAS (Figures 2, A and D, and 3A). Second, deletion of RCY1 specifically affects early endosome functions but it does not perturb the Cvt pathway and autophagy (Figure 3B). Third, the PtdIns(3)P pool required for the Cvt pathway and Snc1 recycling is generated in different ways (Figure 4).
The PAS and Double-Membrane Vesicle Biogenesis Depends on Early Secretory Compartment Functions
The identification of the SNAREs that are required for autophagy and the Cvt pathway is one approach that might provide insight into the origin of the sequestering membrane. Three membrane fusion events have been postulated to be required for the formation and consumption of Cvt vesicles and autophagosomes: condensation of transient vesicles with the PAS, sealing of the cytosolic double-membrane vesicles, and fusion of the completed vesicle with the vacuole. The last reaction is the best characterized. It requires the same factors used for all fusion events with the vacuole such as the class C Vps tethering complex, the small rab-GTPase Ypt7, and a set of four SNAREs: Vam7, Vam3, Vti1, and Ykt6 (reviewed in Huang and Klionsky, 2002; Reggiori and Klionsky, 2002).
As with Tlg2, we could not find any other t-SNAREs at the PAS, suggesting that conventional SNARE-mediated fusion events do not take place at that location (Figure 2E). We cannot rule out the possibility, however, that a small population of SNARE proteins, below the level that we can detect, functions at the PAS. Sec18 is the yeast homologue of the mammalian N-ethylmaleimide-sensitive fusion protein, and it is an essential component of the SNARE-mediated fusion machinery (Wilson et al., 1989). When Sec18 activity is blocked, sealed autophagosomes accumulate, indicating that the closure step of these vesicles does not require a SNARE-mediated fusion event (Ishihara et al., 2001). That observation supports our analysis that fails to detect any t-SNARE at the PAS (Figure 2, D and E), and it suggests a different mechanism takes place during double-membrane vesicle sealing. This suggestion would be in agreement with the finding that microautophagy, another process that requires the fusion of double membranes, does not use the SNARE proteins involved in homotypic vacuole fusion (Sattler and Mayer, 2000); that is, a different type of fusion event is taking place that is independent of SNAREs.
Previous studies have shown that part of the machinery involved in COPII vesicle formation is also required for autophagy but not the Cvt pathway (Klionsky et al., 1992; Ishihara et al., 2001). One problem is that those studies used conditional alleles, and it is not clear how long it takes to interfere with either the preexisting or the forming PAS after the block is induced by the temperature shift. We analyzed the effects of long temperature shifts on the PAS morphology in secretory mutants grown in rich medium by monitoring the GFP-Atg8 fusion. We discovered that blocking exit from the ER or cycling through the Golgi complex caused severe phenotypic changes of those structures that were accompanied by the loss of the ability to correctly recruit and transport prApe1 (Figures 5 and 7). This result indicates that those two organelles, which ultimately depend on each other for their proper function, play a relevant role in Cvt pathway organization. The ER has already been shown to be essential for autophagosome biogenesis in both yeast and mammalian cells (Dunn, 1990; Ishihara et al., 2001; Munafo and Colombo, 2002). Our result with sec7 cells showing that autophagosome formation is blocked in this mutant also agrees with this finding (Figure 6E). The difficulty in demonstrating a correlation between the Cvt pathway and early compartments of the secretory pathway is probably due to the fact that this transport route requires a minor membrane supply. The membrane demand is higher during autophagy because multiple large double-membrane vesicles are formed, and this pathway is consequently more sensitive to structural alterations of those compartments— autophagosomes have between 4–36 times the membrane surface area of Cvt vesicles. Our results cannot discern whether the PAS is derived from the ER or Golgi complex because the sec mutants analyzed do not completely block prApe1 transport (Figure 8A).
Our study reveals the striking structural similarities between Berkeley bodies and autophagosomes (Figure 6). Both are large double-membrane vesicles enwrapping ribosomes and cytosol (Novick et al., 1980; Baba et al., 1994; Figure 6). We have now shown that Atg8 localizes to membranes derived from the Golgi complex and/or ER that give rise to Berkeley bodies (Novick et al., 1980; Esmon et al., 1981; Rambourg et al., 1993; Rambourg et al., 1996; Figure 6).
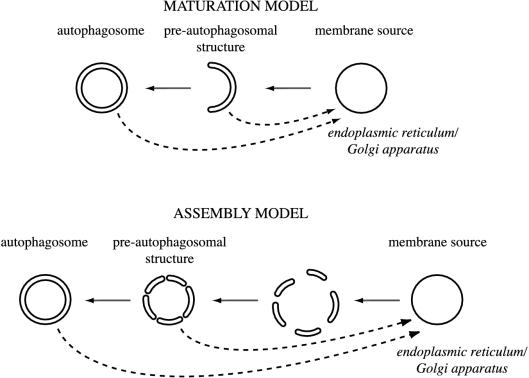
A Maturation Model for Formation of the PAS
The absence of t-SNAREs at the PAS raises the question of how membranes can be delivered to this location without the use of transport vesicles. An organelle maturation model similar to that established for the Golgi complex would explain our data. Proteins reaching their final destinations via the Golgi complex progress through this organelle stack by a process of cisternal maturation, balanced by a return flow of Golgi resident proteins in COPI-coated vesicles (Pelham, 2001b). Similarly, in our model, integral membrane proteins such as Atg9 (Noda et al., 2000) are segregated in a compartment possibly derived from either the ER or Golgi complex (Figure 9, maturation model). This organelle then matures into the PAS in a process balanced by the retrieval of specific proteins back to the donor compartment. The block of this retrograde transport stops the maturation process, creating an organelle unable to fulfill all its functions such as the correct recruitment of the prApe1–Atg19–Atg11 complex (Reggiori et al., 2003). This general sorting mechanism also resembles that taking place at the late endosome, where PtdIns(3)P is needed to recruit the sorting nexin-containing retromer complex and consequently initiate the retrograde transport from this organelle that permits, for example, the recycling of the Vps10 receptor back to the late Golgi (Burda et al., 2002). According to current models, a single compartment, possibly the PAS, vesiculates and forms the double-membrane vesicles in the Cvt and autophagy pathways. However, it cannot be excluded that several discrete sets of membrane are generated and successively assembled at the PAS in a SNARE-independent manner (Figure 9, assembly model). In either case, some proteins would need to be recycled from the forming vesicle. We have recently shown that indeed, the transmembrane protein Atg9 cycles between the PAS and small punctate structures dispersed in the cytosol (Reggiori et al., 2004).
Figure 9.
Models for the PAS and double-membrane vesicle biogenesis. In the maturation model, integral membrane proteins involved in Cvt vesicle and autophagosome formation are segregated in a compartment derived from the membrane source. This organelle then matures into the PAS and finally becomes the double-membrane vesicle in a process balanced by the retrieval of specific proteins back to the donor compartment. In the assembly model, several sets of lipid bilayers are derived from the membrane source by a maturation process similar to the one described above. Those structures are then assembled into double-membrane vesicles at the PAS. Both models suggest a SNARE-independent fusion event required for the sealing of double-membrane vesicles and a retrieval transport route (dashed lines). During Cvt vesicle formation, this route involves sorting nexins and possibly Tlg1, Tlg2, Vps45, and the VFT complex.
It is tempting to speculate that the Tlg1–Tlg2–VFT and Atg20–Atg24 complexes, in analogy to Snc1 transport, are carrying out a similar recycling function during Cvt vesicle biogenesis; however, Atg9 retrieval from the PAS/autophagosomes does not require these factors (Reggiori et al., 2004; our unpublished data). Another possibility is that some Golgi complex functions are severely altered in the absence of Tlg1, Tlg2, and the VFT complex and that interferes with biogenesis of the PAS in a way similar to that observed in sec7 mutants (Figures 5A and 6). Both VFT complex deletants and sec mutants are unable to correctly target the prApe1–Atg19–Atg11 complex to the PAS, suggesting that they are essential for the organization of this structure (Figure 8B; Reggiori et al., 2003). This idea is also supported by the inability of VFT complex mutants to properly deliver Atg9 to the PAS (our unpublished data).
Several vesicular trafficking components have been implicated in either the Cvt pathway or autophagy, creating a complex view of the double-membrane vesicle formation process. In this study, we have investigated the direct involvement of several of these factors. Our data indicate that the early stages of the secretory pathway play an essential role in the Cvt pathway, whereas early and late endosomes do not directly participate. The contribution of similar membranes to both the Cvt pathway and autophagy makes sense because these processes share most of the same molecular machinery. Neither Cvt vesicle nor autophagosome formation requires SNARE-mediated fusion events at the PAS. To understand the mechanism of these unique transport routes, future work will be needed to determine how double-membrane vesicles are generated from early compartments of the secretory pathway and the role of Atg proteins in this event.
Acknowledgments
We thank Drs. Andreas Conzelmann, Alex Franzusoff, Serkka Keranen, Yoshinori Ohsumi, Hugh Pelham, Randy Schekman, Nava Segev, Per Stromhaug, and Bill Wickner for yeast strains, plasmids, and antibodies. This work was supported by the National Institutes of Health Public Health Service grant GM-53396 (to D.J.K.) and by a European Molecular Biology Organization long-term fellowship and a Swiss National Science Foundation fellowship for advanced researchers (to F.R.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–07–0479. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–07–0479.
Abbreviations used: Ape1, aminopeptidase I; CFP, cyan fluorescent protein; Cvt, cytoplasm to vacuole targeting; ER, endoplasmic reticulum; GFP, green fluorescent protein; PA, protein A; PAS, pre-autophagosomal structure; prApe1, precursor Ape1; PtdIns(3)P, phosphatidylinositol-3-phosphate; SNARE, N-ethylmaleimide-sensitive factor attachment protein receptor; t-SNARE, target-N-ethylmaleimide-sensitive factor attachment protein receptor; ts, temperature-sensitive; VFT, Vps-fifty-three; vps, vacuolar protein sorting; YFP, yellow fluorescent protein.
References
- Aalto, M.K., Ronne, H., and Keranen, S. (1993). Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12, 4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich, H., Darsow, T., and Emr, S.D. (1999). Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 18, 6005–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich, H., Dunn, W.A., Jr., Kim, J., and Klionsky, D.J. (2000). Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich, H., Grote, E., Novick, P., and Ferro-Novick, S. (1998). Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273, 11719–11727. [DOI] [PubMed] [Google Scholar]
- Abeliovich, H., Zhang, C., Dunn, W.A., Jr., Shokat, K.M., and Klionsky, D.J. (2002). Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Osumi, M., Scott, S.V., Klionsky, D.J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Takeshige, K., Baba, N., and Ohsumi, Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Sato, T.K., Banta, L.M., and Emr, S.D. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Wendland, B., Estepa, E.J., and Emr, S.D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield, D.K., Lewis, M.J., Rabouille, C., Warren, G., and Pelham, H.R.B. (1994). Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J. Cell Biol. 127, 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., and Schekman, R. (1993). SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349. [DOI] [PubMed] [Google Scholar]
- Becherer, K.A., Rieder, S.E., Emr, S.D., and Jones, E.W. (1996). Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7, 579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, M.W., and Pelham, H.R.B. (2000). A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 151, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., Peterson, M., Cowles, C.R., and Emr, S.D. (1997). A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell 8, 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, P., Padilla, S.M., Sarkar, S., and Emr, S.D. (2002). Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115, 3889–3900. [DOI] [PubMed] [Google Scholar]
- Burgoyne, R.D., and Morgan, A. (2003). Secretory granule exocytosis. Physiol. Rev. 83, 581–632. [DOI] [PubMed] [Google Scholar]
- Coe, J.G., Lim, A.C., Xu, J., and Hong, W. (1999). A role for Tlg1p in the transport of proteins within the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2407–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy, M.J., and Cyert, M.S. (2000). Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol. Biol. Cell 11, 2429–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear, E., Cleck, J.N., and Stevens, T.H. (2003). Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 14, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear, E., and Stevens, T.H. (1998). Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim. Biophys. Acta 1404, 211–230. [DOI] [PubMed] [Google Scholar]
- Conibear, E., and Stevens, T.H. (2000). Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11, 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C.R., Snyder, W.B., Burd, C.G., and Emr, S.D. (1997). Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 16, 2769–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow, T., Rieder, S.E., and Emr, S.D. (1997). A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L., et al. (2001). A protein interaction map for cell polarity development. J. Cell Biol. 154, 549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, W.A., Jr. (1990). Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 110, 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon, B., Novick, P., and Schekman, R. (1981). Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell 25, 451–60. [DOI] [PubMed] [Google Scholar]
- Fasshauer, D., Sutton, R.B., Brunger, A.T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengsrud, M., Sneve, L., Øverbye, A., and Seglen, P. O. (2004). Structural aspects of mammalian autophagy. In: Autophagy, ed. D.J. Klionsky, Georgetown, TX: Landes Bioscience.
- Franzusoff, A., and Schekman, R. (1989). Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 8, 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan, J.M., Wiederkehr, A., Seol, J.H., Haguenauer-Tsapis, R., Deshaies, R.J., Riezman, H., and Peter, M. (2001). Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21, 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki, M., Noda, T., and Ohsumi, Y. (2003). The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28, 49–54. [DOI] [PubMed] [Google Scholar]
- Harding, T.M., Hefner-Gravink, A., Thumm, M., and Klionsky, D.J. (1996). Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 271, 17621–17624. [DOI] [PubMed] [Google Scholar]
- Harding, T.M., Morano, K.A., Scott, S.V., and Klionsky, D.J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema, E.H., Lewis, M.J., Black, M.W., and Pelham, H.R.B. (2003). Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 22, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L. (1999). Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9, 107–112. [DOI] [PubMed] [Google Scholar]
- Hicke, L., Zanolari, B., Pypaert, M., Rohrer, J., and Riezman, H. (1997). Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol. Biol. Cell 8, 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis, J.C., Nichols, B.J., Dhruvakumar, S., and Pelham, H.R.B. (1998). Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.-P., and Klionsky, D.J. (2002). Autophagy in yeast: a review of the molecular machinery. Cell Struct. Funct. 27, 409–420. [DOI] [PubMed] [Google Scholar]
- Huang, W.-P., Scott, S.V., Kim, J., and Klionsky, D.J. (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845–5851. [DOI] [PubMed] [Google Scholar]
- Ishihara, N., Hamasaki, M., Yokota, S., Suzuki, K., Kamada, Y., Kihara, A., Yoshimori, T., Noda, T., and Ohsumi, Y. (2001). Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell 12, 3690–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L., and Casanova, J.E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10, 60–67. [DOI] [PubMed] [Google Scholar]
- Jungmann, J., and Munro, S. (1998). Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 17, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C.A., and Schekman, R. (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst, M., and Emr, S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat Rev Mol. Cell. Biol. 3, 893–905. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., Stromhaug, P.E., and Klionsky, D.J. (2002). Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P.E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S.V., Ohsumi, Y., Dunn, W.A., Jr., and Klionsky, D.J. (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Scott, S.V., Oda, M.N., and Klionsky, D.J. (1997). Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 137, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J. (2004). Autophagy, Georgetown, TX: Landes Bioscience.
- Klionsky, D.J., Cueva, R., and Yaver, D.S. (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J., and Emr, S.D. (2000). Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M.J., Nichols, B.J., Prescianotto-Baschong, C., Riezman, H., and Pelham, H.R.B. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M.J., and Pelham, H.R.B. (1996). SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell 85, 205–215. [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., and Pelham, H.R.B. (2002). A new yeast endosomal SNARE related to mammalian Syntaxin 8. Traffic 3, 922–929. [DOI] [PubMed] [Google Scholar]
- Lybarger, S.R., and Maddock, J.R. (2000). Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 97, 8057–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo, D.B., and Colombo, M.I. (2002). Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 3, 472–482. [DOI] [PubMed] [Google Scholar]
- Nara, A., Mizushima, N., Yamamoto, A., Kabeya, Y., Ohsumi, Y., and Yoshimori, T. (2002). SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 27, 29–37. [DOI] [PubMed] [Google Scholar]
- Nice, D.C., Sato, T.K., Stromhaug, P.E., Emr, S.D., and Klionsky, D.J. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to PtdIns(3)P at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, B.J., Ungermann, C., Pelham, H.R.B., Wickner, W.T., and Haas, A. (1997). Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature 387, 199–202. [DOI] [PubMed] [Google Scholar]
- Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D.J. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132. [DOI] [PubMed] [Google Scholar]
- Noda, T., Suzuki, K., and Ohsumi, Y. (2002). Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12, 231–235. [DOI] [PubMed] [Google Scholar]
- Novick, P., Field, C., and Schekman, R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S.D. (1998). Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95, 847–858. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis, E., and Codogno, P. (2003). Autophagy: a barrier or an adaptive response to cancer. Biochim. Biophys. Acta 1603, 113–128. [DOI] [PubMed] [Google Scholar]
- Panek, H.R., Conibear, E., Bryan, J.D., Colvin, R.T., Goshorn, C.D., and Robinson, L.C. (2000). Identification of Rgp1p, a novel Golgi recycling factor, as a protein required for efficient localization of yeast casein kinase 1 to the plasma membrane. J. Cell Sci. 113, 4545–4555. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B. (2001a). SNAREs and the specificity of membrane fusion. Trends Cell Biol. 11, 99–101. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R.B. (2001b). Traffic through the Golgi apparatus. J. Cell Biol. 155, 1099–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., Bryant, N.J., and Stevens, T.H. (1997). The membrane protein alkaline phosphatase is delivered to the vacuole by a route that is distinct from the VPS-dependent pathway. J. Cell Biol. 138, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescianotto-Baschong, C., and Riezman, H. (1998). Morphology of the yeast endocytic pathway. Mol. Biol. Cell 9, 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg, A., Clermont, Y., and Kepes, F. (1993). Modulation of the Golgi apparatus in Saccharomyces cerevisiae sec7 mutants as seen by three-dimensional electron microscopy. Anat. Rec. 237, 441–452. [DOI] [PubMed] [Google Scholar]
- Rambourg, A., Clermont, Y., Nicaud, J.M., Gaillardin, C., and Kepes, F. (1996). Transformations of membrane-bound organelles in sec14 mutants of the yeasts Saccharomyces cerevisiae and Yarrowia lipolytica. Anat. Rec. 245, 447–458. [DOI] [PubMed] [Google Scholar]
- Raymond, C.K., Howald-Stevenson, I., Vater, C.A., and Stevens, T.H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Black, M.W., and Pelham, H.R.B. (2000). Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell 11, 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Klionsky, D.J. (2002). Autophagy in the eukaryotic cell. Eukaryotic Cell 1, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Pelham, H.R.B. (2001). Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Tucker, K.A., Stromhaug, P.E., and Klionsky, D.J. (2004). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Wang, C.-W., Stromhaug, P.E., Shintani, T., and Klionsky, D.J. (2003). Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 278, 5009–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S.E., Banta, L.M., Kohrer, K., McCaffery, J.M., and Emr, S.D. (1996). Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell 7, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.S., Klionsky, D.J., Banta, L.M., and Emr, S.D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, T., and Mayer, A. (2000). Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 151, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P.V., Takegawa, K., Fry, M.J., Stack, J.H., Waterfield, M.D., and Emr, S.D. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91. [DOI] [PubMed] [Google Scholar]
- Scott, S.V., Baba, M., Ohsumi, Y., and Klionsky, D.J. (1997). Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 138, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S.V., Guan, J., Hutchins, M.U., Kim, J., and Klionsky, D.J. (2001). Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S.V., Hefner-Gravink, A., Morano, K.A., Noda, T., Ohsumi, Y., and Klionsky, D.J. (1996). Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA 93, 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., Marcusson, E.G., Cereghino, J.L., and Emr, S.D. (1997). Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J. Cell Biol. 137, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N., McCaffery, J.M., and Emr, S.D. (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J. Cell Biol. 142, 665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seron, K., et al. (1998). A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell 9, 2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Huang, W.-P., Stromhaug, P.E., and Klionsky, D.J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahama, K., Noda, T., and Ohsumi, Y. (1997). Mutational analysis of Csc1/Vps4p: involvement of endosome in regulation of autophagy in yeast. Cell Struct. Funct. 22, 501–509. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Peak-Chew, S.Y., and Pelham, H.R.B. (2000). Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19, 4885–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., and Pelham, H.R.B. (2001). An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 20, 5991–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., and Pelham, H.R.B. (2002). Vps51p links the VFT complex to the SNARE Tlg1p. J. Biol. Chem. 277, 48318–48324. [DOI] [PubMed] [Google Scholar]
- Sollner, T., Whiteheart, S.W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., and Rothman, J.E. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.-W., Kim, J., Huang, W.-P., Abeliovich, H., Stromhaug, P.E., Dunn, W.A., Jr., and Klionsky, D.J. (2001). Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J. Biol. Chem. 276, 30442–30451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr, A., Avaro, S., Prescianotto-Baschong, C., Haguenauer-Tsapis, R., and Riezman, H. (2000). The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149, 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D.W., Wilcox, C.A., Flynn, G.C., Chen, E., Kuang, W.J., Henzel, W.J., Block, M.R., Ullrich, A., and Rothman, J.E. (1989). A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature 339, 355–359. [DOI] [PubMed] [Google Scholar]