Abstract
The Escherichia coli NleH1 and NleH2 virulence proteins differentially regulate host transcription of innate immunity genes. The mouse pathogen Citrobacter rodentium encodes one NleH protein, which functions equivalently to E. coli NleH1. We examined the impact of host genetics and intestinal inflammation on the contribution of NleH to C. rodentium colonization of mice differing in LPS responsiveness. NleH expression was detrimental to C. rodentium in C57BL/10ScNJ mice, which do not mount LPS-induced inflammatory responses. This phenotype was reversed if inflammation was induced by chemical means. C. rodentium that expressed both E. coli NleH1 and NleH2 was hypervirulent in C3H/HeJ mice.
Keywords: Escherichia coli, Innate immunity, NF-kappa B (NF-κB), NleH1
1. Introduction
Enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC) are attaching/effacing (A/E) bacterial pathogens that cause infectious diarrhea. Citrobacter rodentium, an A/E pathogen that naturally colonizes the mouse intestine and shares common virulence determinants with EPEC and EHEC, is used as a model with which to study A/E pathogen infections [1]. The type III secretion system (T3SS) is a virulence determinant that translocates bacterial effectors from the pathogen into the host cell. Some of these effectors alter host innate immune responses [2]. Bacterial infection activates the nuclear factor-κB (NF-κB) pathway through Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) proteins, resulting in activation of the κB kinase (IKK) complex. NF-κB binds κB sites within target gene promoters and regulates transcription by recruiting co-activator proteins [3]. The p65 subunit of NF-κB also binds to the non-Rel subunit ribosomal protein S3 (RPS3), which increases the affinity of the NF-κB complex for a subset of gene promoters [3].
E. coli O157:H7 strain EDL933 encodes two copies of the nleH gene, designated nleH1 and nleH2, which both encode T3SS effectors [4]. NleH1 and NleH2 are 84% identical in amino acid sequence, but are encoded on different pathogenicity islands and have different functions. NleH1 and NleH2 share sequence similarity with the Shigella flexneri OspG effector, which inhibits NF-κB by preventing IκBα degradation [5]. NleH1, but not NleH2, inhibits the phosphorylation of RPS3 S209 by IKKb, a post-translational modification that normally promotes RPS3 nuclear translocation [6]. NleH1 and NleH2 thus differentially regulate RPS3/NF-κB-dependent transcriptional activation of innate response genes [7].
C. rodentium DBS100 encodes one NleH protein, which functions equivalently to EHEC NleH1 [7]. Deleting nleH significantly reduces C. rodentium colonization of C57BL/6J mice, a strain of mice considered to be somewhat resistant to C. rodentium infection [7,8]. Complementing C. rodentium ΔnleH with EHEC nleH1, but not with nleH2, restores C. rodentium colonization to wild-type (WT) levels [7]. Infection experiments utilizing C3H/HeJ mice, a strain that is considered susceptible to C. rodentium, revealed that deleting nleH delays and reduces mouse mortality and can be complemented by expressing EHEC nleH1 but not nleH2 [7].
Here we examined the impact of host genetics and intestinal inflammation on the contribution of NleH to C. rodentium colonization of mouse strains differing in susceptibility to C. rodentium. We found that having nleH1 is detrimental to C. rodentium in mice that do not mount an LPS-induced inflammatory response to infection, but this phenotype is reversed if inflammation is induced by chemical means. We also characterized the phenotype of a C. rodentium strain that simultaneously expresses both the EHEC NleH1 and NleH2 effectors. This strain was hypervirulent in C3H/HeJ mice, but was controlled by treating the mice with the anti-NF-κB peptide inhibitor SN-50. Overall, our data suggest that expressing this pair of NleH effectors that differentially regulate the host innate response is beneficial to the pathogen.
2. Materials and methods
2.1. Animal experiments
Animal experiments were performed in accordance with Institutional Animal Care and Use Committee-approved protocols at Kansas State University. Three to six week-old C57BL/6J, C3H/HeJ, and C57BL/10ScNJ mice (Jackson Laboratory) were kept in sterilized cages with filter tops, handled in tissue culture hoods, and fed autoclaved food and water under specific-pathogen-free and controlled temperature and photoperiod conditions. In some cases, 2.5% (w/v) dextran sodium sulfate (DSS, Sigma–Aldrich) was added to drinking water for 7 d ad libitum prior to infection to induce colitis. C. rodentium strains were cultured in Luria–Bertani (LB) broth at 37 °C for 18 h without shaking, with carbenicillin (200 μg/ml) and/or kanamycin (100 μg/ml) when needed. C. rodentium ΔnleH was complemented with E. coli EDL933 NleH1-FLAG (in pFLAG-CTC) and with E. coli EDL933 NleH2-His (in pRSFDuet-1). On the day of infection, bacteria were harvested and washed once in phosphate buffered saline (PBS). Mice were infected by oral gavage with ~6 × 108 CFUs of C. rodentium in 100 μl PBS. In some cases, mice were injected intraperitoneally with 10 μg/kg SN-50 (Enzo Life Sciences) immediately prior to gavage. Mice were monitored twice daily for up to 17 d post-infection and were euthanized if they either lost more than 20% of their initial body weight or showed other clinical signs of infection. At necropsy, 4 cm of each colon, beginning at the anal verge, was collected. Feces were removed before weighing tissue. Colons were homogenized, serially diluted, and plated onto MacConkey agar. Colonization and mouse survival data were analyzed using Kruskal–Wallis and log rank tests, respectively. p values <0.05 were considered significant.
2.2. Real-time PCR
RNA was extracted from mouse colons using the RNeasy PLUS Mini Kit (Qiagen). cDNA was prepared from 1.1 μg RNA by using RT2 First Strand Kit (Qiagen) with oligo(dT) primer. RT-PCR was performed in triplicate using a Rotor-Gene SYBR Green PCR Master Mix (Qiagen) in a Rotor-Gene 6000 (Corbett Research). Relative transcription levels were calculated using the ΔΔCT method using β-actin as a reference gene and further normalized to expression in mock-infected mice. Data were analyzed using Student's t-tests with p values <0.05 considered significant.
3. Results
3.1. The relative contribution of NleH to C. rodentium colonization depends upon the mouse strain
We previously used a gnotobiotic piglet infection model and observed that an E. coli O157:H7 EDL933 strain in which nleH1 was deleted induced more rapid and severe clinical signs of disease, as compared with the wild-type (WT) strain [4]. We also observed in studies of C57BL/6J mice that C. rodentium colonization was reduced ~50-fold upon deleting nleH [7]. Complementing ΔnleH with EHEC nleH1 restored C. rodentium colonization to WT levels, whereas complementing with E. coli NleH2 had no measurable impact [7]. These data prompted us to examine in greater detail the contribution of NleH effectors to C. rodentium colonization.
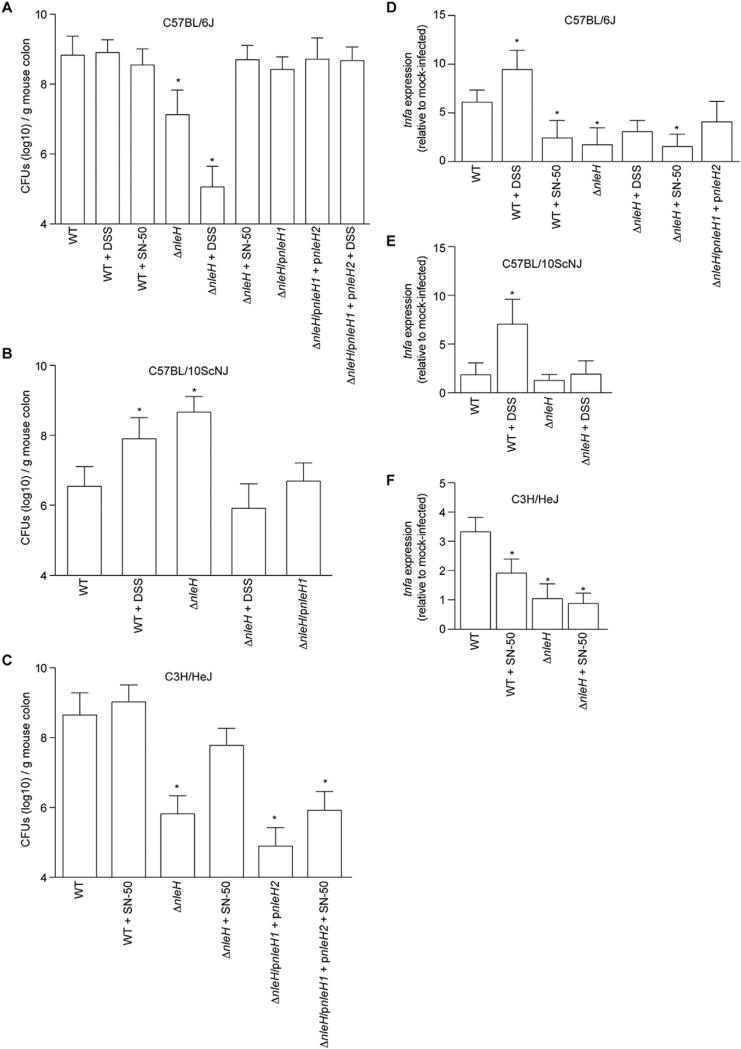
We infected C57BL/6J mice and observed the expected difference [7] in C. rodentium colonization between WT and ΔnleH, 8.8 ± 0.1 vs. 7.1 ± 0.2 (log10) CFUs per g of mouse intestine, respectively (Fig. 1A). Complementing ΔnleH with EHEC nleH1 restored colonization to near WT levels (8.4 ± 0.1 (log10) CFUs/g). We subsequently infected C57BL/10ScNJ mice, which are related to C57BL/6J, but have a deletion in the Tlr4 gene and thus have defective responses to LPS [9]. We observed that expressing NleH was detrimental to C. rodentium colonization in C57BL/10ScNJ mice (Fig. 1B). WT C. rodentium colonized to lower levels than ΔnleH (6.5 ± 0.2 vs. 8.7 ± 0.1 (log10) CFUs/g), respectively. Complementing ΔnleH with EHEC nleH1 reduced colonization of C57BL/10ScNJ mice to 6.7 ± 0.2 (log10) CFUs/g. These data suggested to us that some interplay might exist between NleH and the host genetic background.
Fig. 1.
The contribution of NleH to C. rodentium colonization depends upon the mouse strain. A. C. rodentium colonization of C57BL/6J mice. Colonization (CFUs/g mouse colon) of indicated C. rodentium strains in C57BL/6J mice on days 7–11 post-infection. Where indicated, mice were treated with 2.5% (w/v) dextran sodium sulfate (DSS) for 7 d prior to infection or were injected with 10 μg/kg SN-50 immediately prior to gavage. B. C. rodentium colonization of C57BL/10ScNJ mice on days 7–11 post-infection. C. C. rodentium colonization of C3H/HeJ mice. In all panels, asterisks indicate significantly different colonization magnitude (Kruskal–Wallis test; n = 7–15/group; p < 0.05) as compared with WT C. rodentium on days 1–7 post-infection. D–F. TNF expression. Relative TNF mRNA expression levels from colons obtained from mice (7 days post-gavage) infected with the indicated C. rodentium strains. Asterisks indicate significantly different TNF expression as compared with WT infection (t-test; n = 6; p < 0.05).
3.2. Impact of pro- and anti-inflammatory compounds on C. rodentium colonization
To determine if host inflammation contributed to the differences in C. rodentium colonization of C57BL/6J vs. C57BL/10ScNJ mice, we treated a separate group of C57BL/10ScNJ mice with DSS in their drinking water for 7 d prior to infection. This chemical induces acute colitis in mice, resulting in NF-κB pathway activation [10]. We confirmed the effectiveness of DSS in inducing inflammation by quantifying the expression of the tumor necrosis factor alpha gene (tnfa) using RT-PCR. DSS treatment enhanced tnfa expression in both C57BL/6J and C57BL/10ScNJ mice (Fig. 1D–E).
DSS-induced intestinal inflammation fully reversed the colonization phenotype in C57BL/10ScNJ mice, with DSS-treated C57BL/10ScNJ mice displaying the expected benefit to expressing NleH (Fig. 1B). In C57BL/6J mice, which respond to bacterial LPS, DSS treatment did not significantly impact colonization of WT C. rodentium, but further reduced the colonization magnitude of ΔnleH C. rodentium (Fig. 1A). Thus, the inflammatory state of the host appears to dictate the importance of expressing NleH in mouse strains with differing LPS-responses.
To extend the significance of these data, we performed an additional experiment in which we treated C57BL/6J mice, immediately before bacterial gavage, with SN-50, a cell-permeable peptide that blocks NF-κB activity [11]. While NleH was beneficial to C. rodentium colonization of untreated C57BL/6J mice, the pathogen no longer benefited from expressing NleH when infected mice had been pre-treated with SN-50 (Fig. 1A). We observed similar data in studies of C3H/HeJ mice, where SN-50 treatment enhanced the colonization of ΔnleH C. rodentium to near WT levels (Fig. 1C). Despite a reported in vitro half-life of ~12 h [12], we observed that a single IP dose of SN-50 prior to bacterial gavage suppressed host inflammatory responses, as measured by quantifying tnfa expression (Fig. 1D and F). Thus, expressing NleH appears to benefit C. rodentium colonization in environments in which intestinal inflammation and NF-κB activity is relatively high.
3.3. Co-expression of both EHEC NleH1 & NleH2 enhances C. rodentium virulence
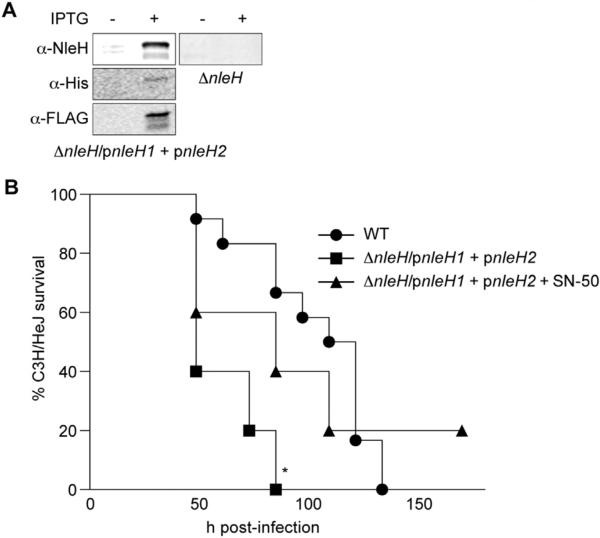
We also performed infection experiments to quantify the benefit of encoding multiple copies of NleH (as is observed in most EHEC serotypes [4]) by determining if expressing both NleH1 and NleH2 enhances C. rodentium virulence. ΔnleH C. rodentium was transformed to express both E. coli EDL933 NleH1 and NleH2 (C. rodentium ΔnleH/pnleH1 + pnleH2; Fig. 2A). This strain was hypervirulent in C3H/HeJ mice (Fig. 2B), suggesting a direct benefit to the pathogen in expressing both effectors. Treating C3H/HeJ mice with SN-50 slowed the rate of mortality in mice infected with C. rodentium ΔnleH/pnleH1 + pnleH2 (Fig. 2B). Thus, expression of both E. coli NleH1 and NleH2 enhances C. rodentium virulence, but is controlled by suppressing NF-κB activity using SN-50. The number of C. rodentium ΔnleH/pnleH1 + pnleH2 recovered from C3H/HeJ mice was less than that of WT C. rodentium (Fig. 1C), despite the increased virulence of this strain (Fig. 2B), somewhat consistent with the inverse relationship we previously observed between E. coli O157:H7 colonization and virulence in gnotobiotic piglets [4]. By contrast, co-expression of NleH1 and NleH2 in C. rodentium did not increase colonization of C57BL/6J (Fig. 1A) or C57BL/10ScNJ mice (data not shown).
Fig. 2.
Co-expression of both EHEC NleH1 & NleH2 enhances C. rodentium virulence. A. C. rodentium ΔnleH was modified to express both E. coli EDL933 nleH1 and nleH2 from pFLAG-CTC and pRSFDuet-1, respectively. Protein expression was confirmed using α-NleH (Genscript), α-His (Sigma), and α-FLAG (Sigma) primary antibodies. B. C3H/HeJ survival. Survival (percentage of the initial population) of C3H/HeJ mice is plotted as a function of time after oral gavage with the indicated C. rodentium strains. Asterisk indicates significantly different survival rate (log-rank test; n = 10–12/group; p < 0.05) as compared with WT infection.
4. Discussion
Overall, our data suggest that a benefit to expressing NleH is in the control of host inflammation. This may manifest in either a direct manipulation of the host response or in altering the intestinal microbiota, two alternative hypotheses that will be tested in future experiments. We suggest that NleH function may somehow be activated by TLR4 signaling and is therefore only beneficial to the pathogen when the host is competent to generate a pro-inflammatory response to LPS.
Host genetic variation plays an important role in susceptibility to enteric bacterial infection. The C3H/HeJ mouse strain, an LPS non-responder, suffers high mortality from C. rodentium infection. However, increased susceptibility is not directly due to LPS responsiveness, as genetically related mouse strains with normal LPS responses are also susceptible to infection [13]. C. rodentium spread is delayed in TLR4 deficient mice, though the duration of infection is unaltered, suggesting TLR4-mediated responses are not necessarily protective and may instead contribute to inflammation and tissue damage [14].
TLR4-dependent signaling may reduce the abundance of commensal organisms in the gut, some species of which provide a barrier against non-resident bacteria and can help regulate inflammation [15]. Microbial transplantation between mouse strains can modulate the severity of infection by altering pro-inflammatory responses [16]. Thus, the composition of the intestinal microbiota not only plays a role in altering susceptibility to infection but also influences the extent of tissue damage and mortality, independent of host genetics [15]. C. rodentium colonization stimulates robust intestinal inflammation which depletes overall microbial density and diversity of the microbiota in the colon [17].
While acute intestinal inflammation had been commonly associated with inducing host defenses to limit microbial dissemination, recent studies instead have shown that acute inflammation is exploited by pathogens [18]. This inflammatory response and the ability of Salmonella to access nutrients [19] and host-derived electron acceptors generated during colitis [18], allow Salmonella to outcompete the resident microbiota for colonization sites [20]. The inflammatory response also results in antimicrobial peptide production, to which microbiota are often more susceptible than pathogens [21].
NleH1 targets the NF-κB pathway to suppress inflammation [4,6,7]. A/E pathogens possess other T3SS effectors, including EspT, which induce inflammation [22]. The pathogen may induce inflammation to disrupt the normal microbial flora, thus allowing the pathogen to adhere and colonize host tissues and enable the organism to spread [20,23]. However, a prolonged immune response may not be desirable, as this inhibits pathogen survival [23]. Spatial and temporal control of host innate immune responses exerted by effectors is essential to the success of pathogens. The functional difference between NleH1 and NleH2 is an example of how pathogens have evolved mechanisms to fine-tune the activity of the host NF-κB pathway.
The data presented here are observational and do not explicitly suggest a mechanism as to why the importance of NleH expression to C. rodentium colonization differs among mouse strains. However, a recent report provided a mechanism for the counterintuitive benefit of host inflammation for a bacterial pathogen [24]. IL-22 induces the production of antimicrobial peptides that sequester essential metal ions from pathogenic bacteria, thus inhibiting their growth. Though both WTand IL-22−/− mice have comparable levels of inflammation after Salmonella infection, Salmonella colonization is reduced in IL-22−/− mice. Salmonella is uninhibited by IL-22-induced inflammation because it has alternative pathways to obtain essential metals from the environment, giving it the ability to outcompete resident microbiota. How bacteria have integrated virulence proteins into host signal transduction pathways in specific spatiotemporal contexts to regulate host inflammatory responses remains an intriguing area of investigation.
Acknowledgments
The project described was supported by grant number AI099002 from the National Institute of Allergy and Infectious Diseases (NIAID). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Deng W, Li Y, Vallance BA, Finlay BB. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect Immun. 2001;69:6323–35. doi: 10.1128/IAI.69.10.6323-6335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman MM, McFadden G. Modulation of NF-kappaB signalling by microbial pathogens. Nat Rev Microbiol. 2011;9:291–306. doi: 10.1038/nrmicro2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–39. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, et al. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 2009;5:e1000708. doi: 10.1371/journal.ppat.1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–51. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat Immunol. 2011;12:335–43. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham TH, Gao X, Tsai K, Olsen R, Wan F, Hardwidge PR. Functional differences and interactions between the E. coli type III secretion system effectors NleH1 and NleH2. Infect Immun. 2012;80:2133–40. doi: 10.1128/IAI.06358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Angulo VA, Deng W, Thomas NA, Finlay BB, Puente JL. Regulation of expression and secretion of NleH, a new non-locus of enterocyte effacement-encoded effector in Citrobacter rodentium. J Bacteriol. 2008;190:2388–99. doi: 10.1128/JB.01602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutinho A, Forni L, Melchers F, Watanabe T. Genetic defect in responsiveness to the B cell mitogen lipopolysaccharide. Eur J Immunol. 1977;7:325–8. doi: 10.1002/eji.1830070517. [DOI] [PubMed] [Google Scholar]
- 10.Marrero JA, Matkowskyj KA, Yung K, Hecht G, Benya RV. Dextran sulfate sodium-induced murine colitis activates NF-kappaB and increases galanin-1 receptor expression. Am J Physiol Gastrointest Liver Physiol. 2000;278:G797e804. doi: 10.1152/ajpgi.2000.278.5.G797. [DOI] [PubMed] [Google Scholar]
- 11.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 12.Liu RY, Fan C, Olashaw NE, Wang X, Zuckerman KS. Tumor necrosis factor-alpha-induced proliferation of human Mo7e leukemic cells occurs via activation of nuclear factor kappaB transcription factor. J Biol Chem. 1999;274:13877–85. doi: 10.1074/jbc.274.20.13877. [DOI] [PubMed] [Google Scholar]
- 13.Vallance BA, Deng W, Jacobson K, Finlay BB. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun. 2003;71:3443–53. doi: 10.1128/IAI.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan MA, Ma C, Knodler LA, Valdez Y, Rosenberger CM, Deng W, et al. Toll-like receptor 4 contributes to colitis development but not to host defense during Citrobacter rodentium infection in mice. Infect Immun. 2006;74:2522–36. doi: 10.1128/IAI.74.5.2522-2536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergstrom KS, Sham HP, Zarepour M, Vallance BA. Innate host responses to enteric bacterial pathogens: a balancing act between resistance and tolerance. Cell Microbiol. 2012;14:475–84. doi: 10.1111/j.1462-5822.2012.01750.x. [DOI] [PubMed] [Google Scholar]
- 16.Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. Altering host resistance to infections through microbial transplantation. PLoS ONE. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16:107–14. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond B, Crepin VF, Collins JW, Frankel G. The WxxxE effector EspT triggers expression of immune mediators in an Erk/JNK and NF-kappaB-dependent manner. Cell Microbiol. 2011;13:1881–93. doi: 10.1111/j.1462-5822.2011.01666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haneda T, Ishii Y, Shimizu H, Ohshima K, Iida N, Danbara H, et al. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell Microbiol. 2012;14:485–99. doi: 10.1111/j.1462-5822.2011.01733.x. [DOI] [PubMed] [Google Scholar]
- 24.Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014 Feb 20;40:262–73. doi: 10.1016/j.immuni.2014.01.003. http://dx.doi.org/10.1016/j.immuni.2014.01.003. Epub 2014 Feb 6. PMID: 24508234. [DOI] [PMC free article] [PubMed] [Google Scholar]