Abstract
In all eukaryotes, the initiation of DNA replication is regulated by the ordered assembly of DNA/protein complexes on origins of DNA replication. In this report, we examine the role of Cdc6, a component of the prereplication complex, in the initiation of premeiotic DNA replication in budding yeast. We show that in the meiotic cycle, Cdc6 is required for DNA synthesis and sporulation. Moreover, similarly to the regulation in the mitotic cell cycle, Cdc6 is specifically degraded upon entry into the meiotic S phase. By contrast, chromatin-immunoprecipitation analysis reveals that the origin-bound Cdc6 is stable throughout the meiotic cycle. Preliminary evidence suggests that this protection reflects a change in chromatin structure that occurs in meiosis. Using the cdc28-degron allele, we show that depletion of Cdc28 leads to stabilization of Cdc6 in the mitotic cycle, but not in the meiotic cycle. We show physical association between Cdc6 and the meiosis-specific hCDK2 homolog Ime2. These results suggest that under meiotic conditions, Ime2, rather than Cdc28, regulates the stability of Cdc6. Chromatin-immunoprecipitation analysis reveals that similarly to the mitotic cell cycle, Mcm2 binds origins in G1 and meiotic S phases, and at the end of the second meiotic division, it is gradually removed from chromatin.
INTRODUCTION
In all eukaryotes, the firing of origins is restricted to a single round in each cell cycle, and the initiation of this process is dependent on the completion of nuclear division. Any deviation, i.e., incomplete or overreplication, may lead to genome instability or cell death. The regulation of the initiation of DNA replication is accomplished by a sequential assembly of proteins on origins. In Saccharomyces cerevisiae, throughout the cell cycle, origins are bound by a complex of six ORC proteins (Orc1–6) (Aparicio et al., 1997). This complex is essential for the stepwise recruitment of additional proteins to origins. At telophase, the transcription of CDC6 is induced (Zwerschke et al., 1994; Piatti et al., 1995), the stability of the protein is increased due to lack of cyclin-dependent kinase (CDK) activity (Calzada et al., 2000), and Cdc6 is found bound to origins (Cocker et al., 1996; Tanaka et al., 1997). Cdc6 is required for the initiation of DNA replication (Piatti et al., 1995), specifically, to load the six Mcm proteins (Mcm2–7) to origins (Tanaka et al., 1997; Weinreich et al., 1999). In addition, when CDK is active, the Mcm and Cdt1 proteins are excluded from the nucleus (Labib et al., 1999; Nguyen et al., 2000; Tanaka and Diffley, 2002) and may reenter it only upon CDK inhibition (at the end of mitosis), and only when Mcm/Cdt1 are in a complex (Tanaka and Diffley, 2002). The DNA/protein complex formed on origins is referred to as the prereplication complex (preRC).
The second step in preparing cells for DNA replication is the formation of the preinitiation complex (preIC) on origins, a step dependent on the activity of the kinase complexes CDK and Dbf4-dependent kinase (DDK, which is composed of Cdc7 and Dbf4). Phosphorylation of Cdc6 by the Cdc28/Clb CDK complex leads to its ubiquitination and degradation (Elsasser et al., 1996; Elsasser et al., 1999; Sanchez et al., 1999; Calzada et al., 2000). At this point, Cdt1 is absent from the origins and is sequestered from the nuclei (Tanaka and Diffley, 2002). Depending on CDK activation, new proteins, i.e., DDK, Cdc45, and RPA, which are essential for DNA replication, are tethered to origins (Aparicio et al., 1997; Owens et al., 1997; Zou and Stillman, 1998, 2000). The DDK activity is required for the complete loading of Cdc45 and RPA to origins (Zou and Stillman, 2000). Loading of the DNA polymerases and primase to origins depends on Cdc45 and RPA, respectively (Tanaka and Nasmyth, 1998; Zou and Stillman, 2000). During DNA replication, origins are only bound with the six Orc proteins and Mcm10 complexes (Lei and Tye, 2001). The Mcm2–7 proteins that initially move with the replication fork are gradually lost, and after phosphorylation by Cdc28 they are localized in the cytoplasm (Aparicio et al., 1997; Labib et al., 1999). This sequential and regulated assembly and disassembly of proteins on origins ensure that each origin will fire only once during S phase. Phosphorylation of any of Cdc6, Mcm2, or Orc2 by Cdc28/Clb complex suffices to restrict DNA replication to a single round, indicating the presence of at least three redundant mechanisms for this control (Nguyen et al., 2001).
Premeiotic DNA replication is initiated from the same origins as those used in the mitotic cell cycle (Collins and Newlon, 1994). Furthermore, in Schizosaccharomyces pombe, Mcm2 and the Cdc6 homolog Cdc18 are required for this process (Murakami and Nurse, 2001; Lindner et al., 2002), and the Mcm4 protein binds the origin during the premeiotic S phase (Lindner et al., 2002). By contrast, in S. cerevisiae, Cdc6 is apparently not required for meiosis and sporulation (Simchen, 1974). The role of Orc1–6, Mcm2–7, and Cdt1 in initiation of premeiotic DNA replication in S. cerevisiae was not reported. Neither are there currently any reports on the role of the preRC and preIC complexes in regulating premeiotic DNA replication in S. cerevisiae.
In this report, we show that Cdc6 is required for premeiotic DNA replication and that similarly to the mitotic cell cycle, entry into the premeiotic S phase is accompanied by specific degradation of Cdc6. However, the origin-bound Cdc6 is protected from degradation and occupies origins throughout the meiotic cycle. In contrast, Mcm2 binding to origins is similar in mitosis and meiosis; it is gradually removed from origins. Degradation of Cdc6 in the mitotic cycle is dependent on Cdc28, whereas in the meiotic cycle it is Cdc28 independent. We suggest that in the meiotic cycle, the meiosis-specific kinase, the hCDK2 homolog Ime2, is directly involved in Cdc6 degradation, because the two proteins physically associate.
MATERIALS AND METHODS
Strains and Plasmids
The relevant genotype of strains is given in Table 1. All strains used (except Y208) are isogenic to Y422 and were derived from its haploid parents. Y1073: A one-step deletion protocol was used to replace the IME2 allele in the parental haploids of Y422 with an ime2::hisG-URA3-hisG fragment (from plasmid YIp1930). URA+ transformants were patched onto 5-fluoroorotic acid plates to select for derivatives that had recombined out the URA3 gene. Y1384, Y1385, and Y1443 are derivatives of Y422, Y1073, and Y1314, respectively (Guttmann-Raviv et al., 2001), carrying the CDC6–13 × myc chimera. This gene was integrated at the LEU2 locus, by using YIp2668 digested with PpuMI. Y1466 resulted from mating the haploid parents of Y422 and Y208.
Table 1.
S. cerevisiae strains
| Name | Relevant genotype | Remarks |
|---|---|---|
| Y153 | MATa, URA3::gal1-lacZ, LYS2::gal1-HIS3, his3-200 leu2-3,112, trp1-901, gal4Δ, gal80Δ | Harper et al. (1993) |
| Y208 | MATa/MATα, cdc6-1/cdc6-1 | |
| Y422 | MATa/MATα, leu2-3,112/leu2-3,112, ura3-52/ura3-52 | |
| Y1073 | ime2::hisG/ime2::hisG | Isogenic to Y422 |
| Y1314 | MATa/MATα, pCUP1-UBl-HA-CDC28-Degron-URA3/pCUP1-UBI-HA-CDC28-Degron-URA3, ura3-52/ura3-52, leu2,3-112/leu2,3-112 | Guttmann-Raviv et al. (2001) |
| Y1384 | leu2-3,112::LEU2-13xmyc-CDC6/leu2-3,112 | Isogenic to Y422 |
| Y1385 | leu2-3,112::LEU2-13xmyc-CDC6/leu2-3,112 | Isogenic to Y1073 |
| Y1443 | leu2,3-112/leu2,3-112-pCDC6-CDC6-13xmyc | Isogenic to Y1314 |
| Y1466 | MATa/MATα, cdc6-1/CDC6 | Isogenic to parents of Y208 and Y422 |
Plasmids used were as follows: pGAD2F carries pADH1-gal4(768-881)-HA on a LEU2 2μ vector (Fields, personal communication); YIp1930 carries ime2(–268 to –108)-hisG-URA3-hisG-ime2(+1318 to +2231) on pUC vector (Guttmann-Raviv et al., 2001); YEp2053 carries pIME1-CLB1 on a URA3 2μ vector (Guttmann-Raviv et al., 2001); YEp2225 carries pADH1-gal4(dbd)-ime2(1-645)(K97A) on a TRP1 2μ vector; YEp2229 carries pIME1-gal4(dbd)-HA-ime2K97A(1-645)-ADHt on a TRP1 2μ vector; and YIp2668 carries pCDC6-CDC6–13 × myc on pRS405 (Sikorski and Hieter, 1989). These plasmids were constructed in several steps. Details are available upon request.
Media and Genetic Techniques
Minimal acetate medium (PSP2) and sporulation medium (SPM) have been described previously (Kassir and Simchen, 1991). Synthetic dextrose (SD) has been described previously (Sherman, 1991). Meiosis was induced as follows: cells were grown in PSP2 supplemented with the required amino acids to early exponential stage (0.8–1.2 × 107 cells/ml), washed once with water, and resuspended in SPM. The transfer of cells to SPM leads first to accumulation of cells in G1 and then to the entry into meiotic S phase. Under nitrogen depletion, small budded cells do not grow in mass and therefore are delayed in entry into the meiotic cycle. This difference in cell mass between mother and daughter cells is reflected in the fluorescence-activated cell sorting (FACS) analysis. In several cases, the cells accumulated in G1 show a “shoulder” or even a split peak. Nevertheless, the use of this protocol allows us to examine how cells at different cell cycle stages respond to nitrogen depletion and enter meiosis.
β-Galactosidase activity was measured as described previously (Rose and Botstein, 1983). Results are given in Miller units (Miller, 1972) and are an average of at least three independent transformants.
Antibodies
Mouse monoclonal antibodies directed against the myc epitope (9E11, for chromatin-immunoprecipitation [ChIP] assay) were purchased from either BioSource International (Camarillo, CA) or NeoMarkers (Fremont, CA). Goat polyclonal antibodies directed against Mcm2 (yN-19), mouse monoclonal antibodies directed against Gal4(dbd) (RK5C1), mouse monoclonal antibodies directed against the myc epitope (9E10, for Western), and rabbit polyclonal antibodies directed against the PSTAIRE epitope were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Preparation of Yeast Protein Extracts and Western Analysis
Protein extracts were prepared from trichloroacetic acid-treated cells as described previously (Foiani et al., 1995). Western procedure was essentially as described previously (Foiani et al., 1994).
FACS Analysis
Cells were analyzed for DNA content by FACS analysis as described previously (Foiani et al., 1994) by using a FACScan analyzer (BD Biosciences, San Jose, CA). The percentage of cells with more than 2C DNA content was calculated, by using the WinMDI program.
Primers for ChIP Polymerase Chain Reaction (PCR)
Origin specific and nonspecific primers were designed according to Kamimura et al. (2001). ARS305-specific primers were ARS305-S-39.5 kb: 5′ TTTCAGAGCCTTCTTTGGAG 3′ and ARS305-AS-39.5 kb: 5′ CAAACTCCGTTTTTAGCCCC 3′. ARS501-specific primers were ARS501-S 5′ AACTTTTACGATCCAACGCC 3′ and ARS501-SA 5′ GCCTCTACGGGTATTAGCTG 3′. Primers for adjacent sequences were ARS305-S-30.5 kb: 5′ TGCAAACAGTATTCCGGCAC 3′ and ARS305-AS-30.5 kb: 5′ ACACGATCCACGCTGTCCCA 3′.
Chromatin Immunoprecipitation
Formaldehyde was added to 30 ml of 1.2 × 107 cells/ml, to a final concentration of 1%. Cells were incubated for 12 min at room temperature. Cross-linking was stopped by the addition of glycine to a final concentration of 140 mM. ChIP was then carried out essentially as described previously (Tanaka et al., 1997) apart from using Dynabeads protein G (Dynal Biotech, Lake Success, NY) for immunoprecipitation. In cases in which the amount of PCR product was too high (nonlogarithmic reaction), all the samples in that experiment were diluted. This protocol promoted reliable comparisons between samples in a single experiment.
Coimmunoprecipitation
Briefly, cells broken by glass beads were lysed with 1% Triton X-100 in IP buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.1% Nadeoxycholate, 1 mM dithiothreitol, 2.5 mM ortho-vanadate, 10 mM NaF, 10 mM β-glycerol phosphate, and 1 tablet of protease inhibitor mixture [Roche Diagnostics, Indianapolis, IN]). Immunoprecipitation was performed with purified monoclonal anti-myc or anti-Gal4(dbd) antibodies and protein G-conjugated Dynabeads. The washed beads were suspended in Laemmli buffer, and the released protein was subjected to SDS-PAGE.
RESULTS
Cdc6 Is Required for Premeiotic DNA Replication and Spore Formation
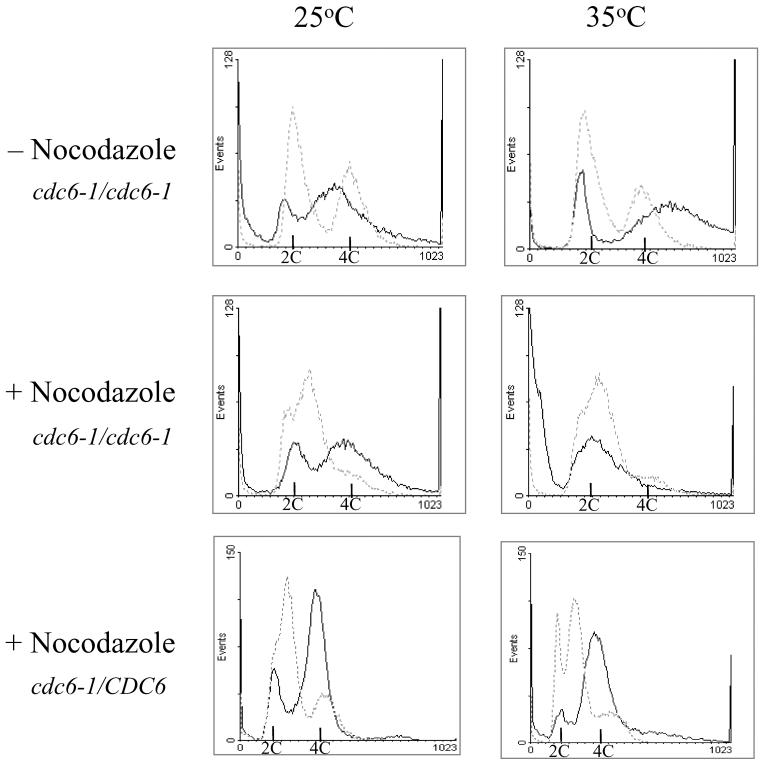
The essential role of Cdc6 in the initiation of DNA replication in the cell cycle prompted us to reexamine its role in meiosis and specifically its requirement for premeiotic DNA replication. The role of Cdc6 in meiosis was previously determined by using the temperature-sensitive allele cdc6-1 and a nonpermissive temperature of 33.5°C (Simchen, 1974). Because this temperature might not be restrictive in the meiotic cycle, in the experiments reported below, we used 35°C as the nonpermissive temperature. We examined the ability of MATa/MATα cdc6-1/cdc6-1 diploid cells transferred to SPM and incubated at either 25 or 35°C to replicate their DNA and sporulate. Figure 1 (top) demonstrates that after 48-h incubation, the fraction of cells with 4C DNA content was similar at both temperatures (81.8 and 76.3% at 25 and 35°C, respectively). However, incubation at 25°C gave rise to 85% asci, whereas at 35°C, no asci were observed. In contrast, the isogenic cdc6–1/CDC6 heterozygous strain gave 73.1% asci at 25°C and 50.4% at 35°C. These results imply that incubation at 35°C had only a minor effect on the sporulation of the isogenic wild-type strain used. We conclude, therefore, that Cdc6 is required for sporulation.
Figure 1.
Cdc6 is required for premeiotic DNA replication. MATa/MATα cdc6-1/cdc6-1 (strain Y208, top two panels) and its isogenic cdc6–1/CDC6 (strain Y1466, bottom panel) cells were grown at 25°C in PSP2 to a titer of 1 × 107 cells/ml. Cells were washed in water, resuspended in warm SPM, and incubated at either 25 or 35°C (time 0) (top). Nocodazole was added to the PSP2 culture to a final concentration of 10 μg/ml. After 2-h incubation, cells were washed and resuspended in SPM as described above (time 0). Samples were taken at 2 h (dashed gray line) and 48 h (solid black line) to process for FACS analysis.
The above-mentioned results suggest that Cdc6 might not be required for premeiotic DNA replication. However, because Cdc6 is tethered to origins at anaphase, it is possible that the origin-bound Cdc6 is not affected by the increase in temperature, and that consequently Cdc6 is functional in the fraction of M and G1 mitotic cells that were shifted to meiotic conditions. To overcome this problem, cells were first treated with nocodazole, which arrests the cells at G2, a stage with no Cdc6 protein, and then transferred to SPM and incubated at either 25 or 35°C. This treatment allowed better synchronization, at 2 h in SPM most cells were in G1 in comparison to cells that were not treated with nocodazole (Figure 1, compare middle and top). Furthermore, this treatment did not affect the ability of cells to replicate their DNA and form spores when incubated at 25°C (Figure 1, middle). At 2 h in SPM, most cdc6-1/cdc6-1 cells (89.1%) were in G1 with a 2C DNA content, whereas at 48 h in SPM, 74.2% of the cells had 4C DNA content and 79% asci were observed. Incubation at 35°C did not interfere with the response of cells to nitrogen depletion. At 2 h in SPM, 85.6% of the cells had 2C DNA content. However, these arrested cells did not enter premeiotic S, because only 11.6% of the cells had a 4C DNA content at 48 h in SPM (Figure 1, middle). Moreover, a substantial percentage of cells (36%) had less then 2C DNA content. In contrast, in the isogenic cdc6–1/CDC6 heterozygote strain premeiotic DNA replication took place at both the permissive and restrictive temperatures (Figure 1, bottom). We conclude that in the meiotic cycle, Cdc6 is required for initiation of premeiotic DNA replication, and consequently for spore formation.
Cdc6 Protein Is Degraded When Cells Enter Premeiotic DNA Replication
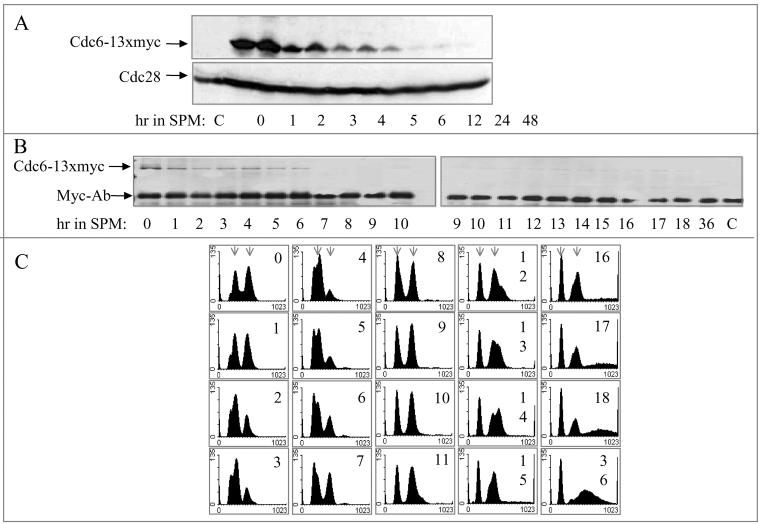
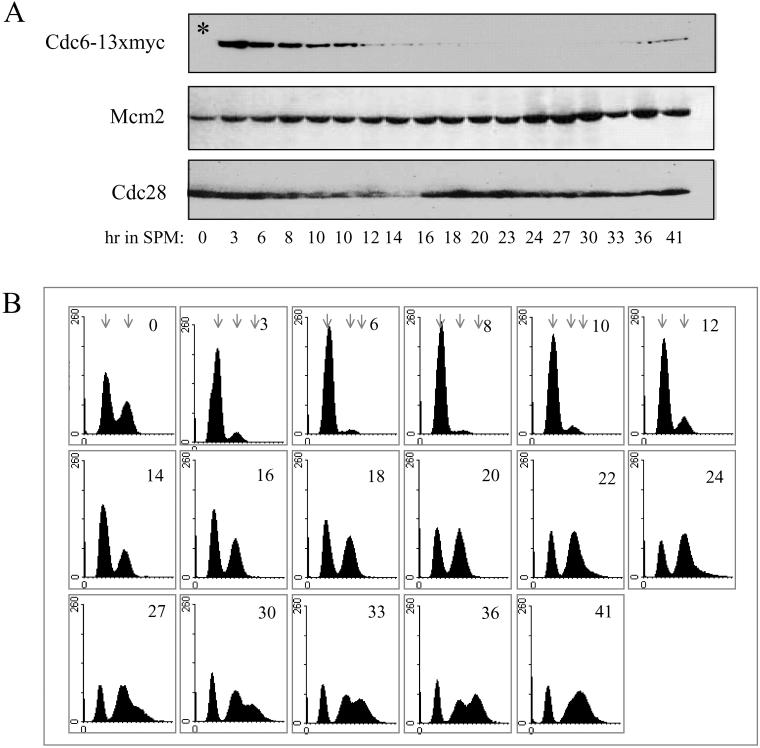
Cdc6 is an unstable protein whose degradation depends on phosphorylation by the Cdc28/Clb kinase (Elsasser et al., 1996; Calzada et al., 2000; Drury et al., 2000). Therefore, in the cell cycle, upon entry into S phase, when this complex is present and functional, Cdc6 is degraded. We wished to determine whether Cdc6 is similarly regulated in the meiotic cycle. In the experiments reported here, Cdc6 was tagged with 13 copies of the myc epitope, and this chimeric gene, expressed from the CDC6 promoter, was integrated at the LEU2 locus. MATa/MATα CDC6/CDC6 leu2-3112::LEU2-CDC6-13 × myc/leu2-3112 (strain Y1384) diploid cells grown in PSP2 to 1×107 cells/ml were transferred to SPM and incubated at 25°C. At various times, proteins were extracted and analyzed by Western blotting, by using antibodies directed against either the myc or the PSTAIRE epitopes, to detect Cdc6-myc and Cdc28, respectively. Figure 2A shows that in the meiotic cycle, Cdc6 is an unstable protein, whose level is decreased after 4- to 6-h incubation in SPM and that at later meiotic times, Cdc6 protein is not detected. On the other hand, the level of Cdc28 remained constant throughout the meiotic cycle. To correlate the decline in Cdc6 with DNA replication, we monitored the progression of premeiotic DNA replication, the level of Cdc6, and the level of origin-bound Cdc6 in the same culture. The level of Cdc6 was measured by immunoprecipitation (Figure 2B), the level of origin-bound Cdc6 was determined by ChIP assay (Figure 5; see below), and DNA replication was observed by FACS analysis (Figure 2C). The IP-Western demonstrates that transfer of cells to meiotic conditions leads to a decline in the steady-state level of Cdc6 and that in cells incubated in SPM for 7 h and onward, it is almost undetectable (Figure 2B). The kinetics of premeiotic DNA replication is revealed by the raw FACS analysis data (Figure 2C). For clarity, the percentage of cells with more than 4C DNA content was calculated and presented in Figure 4A. These results reveal that the transfer of cells to SPM resulted first in an accumulation of G1 cells with 2C DNA content (0–4 h in SPM). Premeiotic DNA replication was initiated at ∼5 h in SPM and completed at ∼11 h (Figures 2C and 4A). The relative level of Cdc6 was calculated from Figure 2B data and was compared with the time of DNA replication. Figure 4A shows the kinetics of DNA replication as the percentage of cells with more than 2C DNA content, in comparison with the level of Cdc6 protein. It shows that in the meiotic cycle, there is a correlation between time of DNA replication and Cdc6 degradation.
Figure 2.
In wild-type cells, Cdc6 is degraded upon entry into premeiotic DNA replication. Wild-type diploid cells (strain Y1384) grown at 25°C in PSP2 to a titer of 1 × 107 were washed in water, resuspended in SPM, and incubated at 25°C. Samples were taken at the indicated times for Western analysis (A), IP-Western (B), and FACS analysis (C). Early and late meiotic times are from two experiments. For comparison, the 9- and 10-h sample were taken from both experiments. The results in B and C are from a single experiment. Cdc6–13 × myc was detected using antibodies directed against the myc epitope. Cdc28 was detected using antibodies directed against the PSTAIRE epitope. C, control, without tagged protein. Sample taken from strain Y422 grown in PSP2 to 1 × 107 cells/ml. Arrows in C indicate the position of cells with 2C and 4C DNA content.
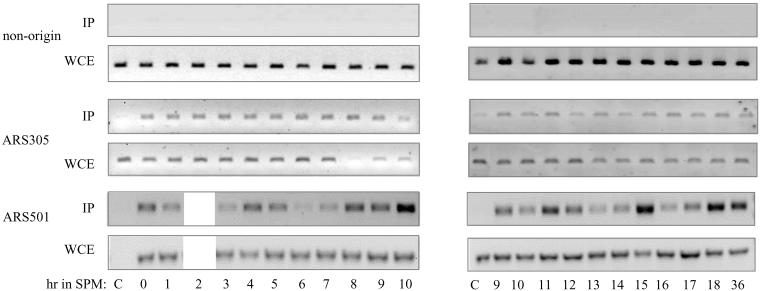
Figure 5.
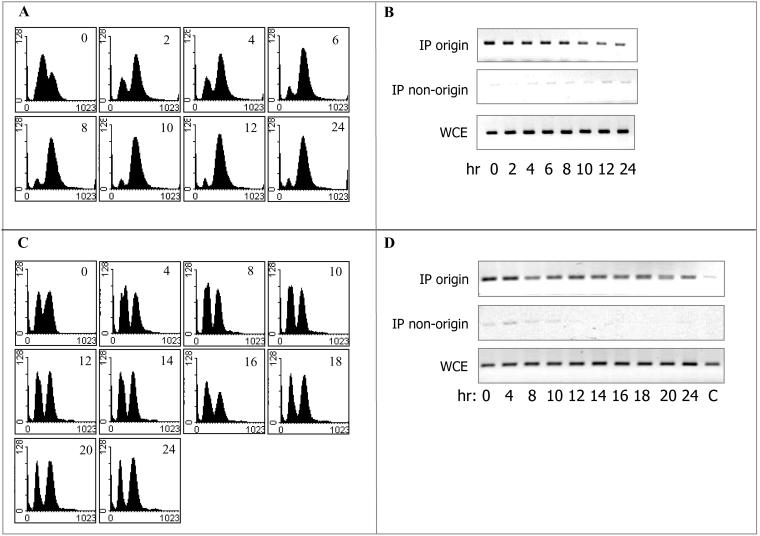
Origin-bound Cdc6 is stable throughout the meiotic cycle. Wild-type diploid cells (strain Y1384) grown at 25°C in PSP2 to a titer of 1 × 107 were washed in water, resuspended in SPM, and incubated at 25°C. Samples were taken at the indicated times for immunoprecipitation (Figure 2B), FACS (Figure 2C), and ChIP analysis (this figure). For comparison, the 9- and 10-h sample were taken from both experiments. c, the strain without the myc tagged Cdc6 was Y422. WCE, whole cell extract before IP. The primers used to amplify the indicated DNA are described in MATERIALS AND METHODS.
Figure 4.
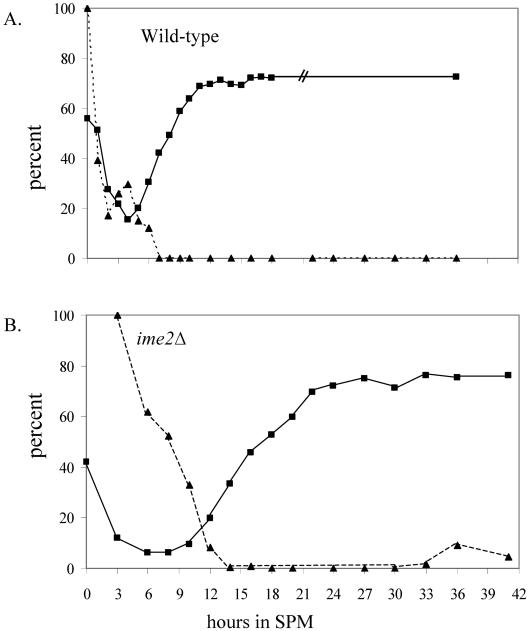
Cdc6 is degraded upon entry into premeiotic S phase. The percentage of cells with 4C DNA content was calculated from Figures 2 C and 3B (squares, filled line). The relative level of Cdc6 was calculated from Figures 2B and 3A (triangles, dashed line). (A) Wild-type strain. (B) ime2Δ strain.
To determine whether the correlation between DNA replication and Cdc6 degradation reflects interdependent events, we examined the level of Cdc6 in diploid cells deleted for IME2. IME2 encodes a meiosis-specific CDK-like kinase required for setting the time for the initiation of premeiotic DNA replication, for restricting premeiotic DNA replication to a single round, and for spore formation (Foiani et al., 1996). Figure 3B shows that in ime2Δ/ime2Δ diploid cells initiation of DNA replication is delayed by 8 h in comparison with its isogenic wild-type strain (DNA replication is initiated at 12 instead of 4 h in SPM, in the wild-type strain). Furthermore, at 27 h in SPM, these ime2Δ/ime2Δ cells enter a second round of DNA replication, and cells with more than 4C DNA content are observed (Figure 3B). Samples were also taken at the same times for Western analysis to measure the level of Cdc6. Figure 3A shows that from 12 h in SPM and onward, the level of Cdc6 is decreased, between 20 and 30 h in SPM Cdc6 is not detected, and at later meiotic times, namely, 33–41 h, it reoccurs. In contrast, the levels of Mcm2 and Cdc28 are constitutive throughout the meiotic cycle (Figure 3A). The decrease in the level of Cdc6 is correlated with the initiation of DNA replication (Figures 3 and 4B). The increase in the level of Cdc6 at late meiotic times might reflect the need for Cdc6 protein for the second round of DNA replication in these cells. The correlation between the second round of DNA replication and Cdc6 degradation is probably masked by the slight asynchrony of the culture. We conclude that similarly to the cell cycle, Cdc6 degradation and initiation of premeiotic DNA replication are two interdependent events.
Figure 3.
In IME2-deleted cells, Cdc6 is degraded upon entry into premeiotic DNA replication. ime2Δ/ime2Δ diploid cells (strain Y1385) grown at 25°C in PSP2 to a titer of 1 × 107 were washed in water, resuspended in SPM, and incubated at 25°C. Samples were taken at the indicated times for Western (A) and FACS analysis (B). Early and late meiotic times are from two experiments. For comparison, the 10-h sample was taken from both experiments. Cdc6–13 × myc, Cdc28, and Mcm2 were detected using antibodies directed against the myc epitope, the PSTAIRE epitope, and Mcm2 N-terminus peptide, respectively. *, control, without tagged protein. Sample taken from strain Y1073 grown in PSP2 to 1 × 107 cells/ml. Arrows in B indicate the position of cells with 2C, 4C, and >4C DNA content.
The Origin-bound Cdc6 Is Stable throughout the Meiotic Cycle
In the cell cycle, the stability of the Cdc6 protein is independent of its binding state, namely, whether it is bound to chromatin or free (Tanaka et al., 1997; Seki and Diffley, 2000). We used ChIP to determine the level of origin-bound Cdc6 throughout the meiotic cycle, in wild-type cells. MATa/MATα diploid cells carrying the CDC6-13 × myc chimera (Y1384) grown in PSP2 to 1×107 cells/ml were transferred to SPM, and at various times samples were taken to determine DNA content (Figure 2C) and after cross-linking with formaldehyde, to process for ChIP analysis by using ARS-specific and -nonspecific probes. Figure 2B (discussed above) shows the IP-Western, whereas Figure 5 shows the ChIP results. Figure 5 shows that IP of Cdc6–13 × myc promoted the specific PCR enrichment of DNA origins, in comparison with nonorigin DNA, or to cells lacking the myc-tagged Cdc6 (c). By contrast, without IP, in the whole cell extract (WCE), the level of PCR products was essentially the same in all samples. To our surprise, the origin-bound Cdc6 was present in all samples. At ARS305 the level of the origin-bound Cdc6 was constitutive, whereas at ARS501, the level of the PCR products fluctuated, but Cdc6 was certainly present throughout the meiotic cycle (Figure 5). By contrast, in the same samples, the IP-Western analysis reveals that the free Cdc6 was specifically degraded upon entry into premeiotic S phase (Figure 2B). Similar results, reflecting the stability of the origin bound Cdc6 was also observed in the ime2Δ diploid strain (our unpublished data; Figure 6D). The different behavior of ARS305 and ARS501 might reflect differences in chromatin structure, which can affect the accessibility of the chromatin-bound Cdc6 to different enzymes (see below). Thus, in comparison with the cell cycle, in the meiotic cycle, the origin-bound Cdc6 is protected from phosphorylation and consequent degradation, ubiquitination, or degradation.
Figure 6.
Ectopic, elevated activation of Cdc28 results in premature entry into premeiotic S phase without affecting the stability of the origin-bound Cdc6. Wild-type (Y1384; A and B) and isogenic ime2Δ/ime2Δ (Y1385; C and D) diploid cells carrying pIME1-CLB1 ona2μ vector (YEp2053) grown at 25°C in PSP2 to a titer of 1 × 107 were washed in water, resuspended in SPM, and incubated at 25°C. Samples were taken at the indicated times for FACS (A and C) and ChIP analysis (B and D). c, the strain without the myc tagged Cdc6 was Y422 (B) or Y1073 (D). WCE, whole cell extract before IP. Primers used to amplify origin (ARS305) and the nonorigin DNA are described in MATERIALS AND METHODS.
Ectopic, Elevated Activation of Cdc28 Results in Premature Entry into Premeiotic S Phase without Affecting the Stability of the Origin-bound Cdc6
The stability of the origin-bound Cdc6 protein implies that the chromatin-bound Cdc6 is inaccessible to the enzymes promoting its degradation. This might result from either the meiosis-specific change in chromatin structure or the use of a meiosis-specific enzyme that cannot reach the chromatin-bound Cdc6. We hypothesized that in the meiotic cycle, phosphorylation of Cdc6 by Ime2, rather than Cdc28, marks it for degradation and that Ime2 is inaccessible to the chromatin-bound Cdc6. This hypothesis is based on the following observations: 1) diploid cells carrying either the cdc28-4 or ime2Δ mutations arrest after premeiotic DNA replication, whereas the double mutant cdc28-4 ime2Δ arrests in G1 (Guttmann-Raviv et al., 2001); and 2) ectopic overexpression of Clb1 at early meiotic times leads to premature entry into premeiotic S phase (Guttmann-Raviv et al., 2001). This hypothesis predicts that premature, elevated activation of Cdc28 at early meiotic times will lead to destabilization of the origin-bound Cdc6. Early meiotic activation of Cdc28 was accomplished by fusing CLB1 to the IME1 promoter. As reported, when wild-type and ime2Δ diploid cells carrying pIME1-CLB1 on a multicopy plasmid were transferred to SPM, cells did not arrest in G1, and directly entered the premeiotic S phase (Figure 6, A, and C, respectively). Nevertheless, in these cells, the origin-bound Cdc6 remained stable (Figure 6, B and D). The observation that overexpression of Clb1 at early meiotic times advanced premeiotic DNA replication suggests, that at least in this experimental system, Cdc28 was responsible for controlling entry into premeiotic S phase. Nevertheless, it had no effect on the stability of the origin-bound Cdc6. We conclude, therefore, that the stabilization of the chromatin-bound Cdc6 does not result from replacing Cdc28 by a different kinase. We suggest that the change in chromatin structure, which takes place during premeiotic DNA replication, is most probably responsible for the protection of the chromatin-bound Cdc6.
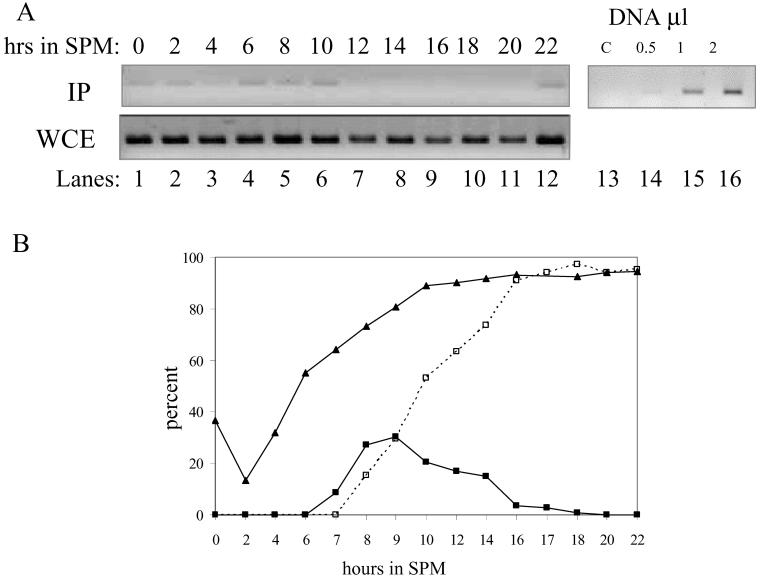
In the Meiotic Cycle, the Mcm2 Protein Is Transiently Released from Origins
The stability of the origin-bound Cdc6 protein calls for the presence of an additional mechanism that prevents refiring of origins. In the mitotic cycle, phosphorylation of any of Cdc6, Mcm2, or Orc2 suffices to restrict DNA replication to a single round in each cell cycle (Nguyen et al., 2001). As described, in the mitotic cycle, phosphorylation of Mcm2 results in its gradual release from origins and its exclusion from nuclei (Aparicio et al., 1997; Tanaka et al., 1997; Labib et al., 1999; Nguyen et al., 2000). Western analysis reveals that the level of Mcm2 is constitutive throughout the meiotic cycle (our unpublished data; Figure 3A). The level of the origin-bound Mcm2 protein throughout the meiotic cycle was determined by ChIP analysis. Figure 7A shows that Mcm2 is present on origins at early meiotic times (0–10 h in SPM), between 14 and 20 h in SPM it is absent, and reoccurs after 22 h in SPM (similar results were obtained when samples for ChIP were taken up to 36 h; our unpublished data). To correlate the exclusion of Mcm2 from origins to DNA replication and nuclear divisions, samples were also taken for FACS analysis and 4,6-diamidino-2-phenylindole staining. Figure 7B reveals that DNA replication took place between 3 and 10 h in SPM. At 7 h in SPM cells have entered meiosis I, giving rise to the accumulation of binucleate cells, whereas 1 h later the second meiotic division took place, and tetranucleate cells were observed (Figure 7B). Thus, Mcm2 is excluded from origins after the completion of premeiotic S phase and the first nuclear division, at a time corresponding to the middle/end of meiosis II. Furthermore, at later meiotic times, i.e., at 22 h and onward, Mcm2 reoccupies the origins (Figure 7A), probably reflecting a loss of CDK activity in cells exiting meiosis II.
Figure 7.
Mcm2 is excluded from DNA origins during meiotic nuclear divisions. Diploid cells (strains Y1384) grown at 30°C in PSP2 to a titer of 1 × 107 were washed in water, resuspended in SPM, and incubated at 30°C. Samples were taken at the indicated times for ChIP (A) and FACS analysis and DAPI staining (B). (A) Antibodies directed against Mcm2 were used. C, control without formaldehyde cross-linking. WCE, whole cell extract before IP. The PCR amplification was done using 1 μl of DNA from each sample. Top right panel shows that this quantity gives rise to a linear range PCR. (B) Percentage of cells with 4C DNA content calculated from the FACS analysis is illustrated (triangles). Percentage of binucleated (closed squares) and tetranucleated (open squares, dashed line) cells was determined after DAPI staining.
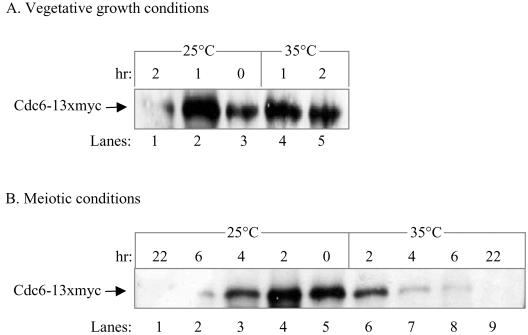
Cdc28 Is Not Required for the Degradation of Cdc6 in Meiosis
In the cell cycle, phosphorylation of Cdc6 by Cdc28/Clb targets it to degradation. We examined, therefore, whether Cdc28 is also required for the degradation of Cdc6 in the meiotic cycle. We used the temperature degradable cdc28-degron allele (Labib et al., 2000; Guttmann-Raviv et al., 2001). Diploid cells homozygous for the pCUP-UB-DHFRts-HA-CDC28 allele grown in PSP2 medium at 25°C to 1 × 107 cells/ml were transferred to 37°C for 2 h. Cells were then washed, resuspended in either prewarmed PSP2 or SPM media, and incubated at either 25 or 35°C (time 0). This treatment causes complete degradation of Cdc28, and, as a consequence, abolishes its kinase activity (Guttmann-Raviv et al., 2001). At various times, samples were taken for IP-Western and FACS analysis, to measure the levels of Cdc6-13 × myc and DNA content, respectively. Under vegetative growth conditions (PSP2 medium), incubation at either 25 or 35°C for 1 h resulted in an increase in the number of cells in G1. At time 0, the level of cells with 2C DNA content was 73.6%, and after an additional 1-h incubation, it reached 84.4 and 90.9% at 25 and 35°C, respectively. In these samples, as was expected from the accumulation of cells in G1, there was a concomitant increase in the level of Cdc6 protein (Figure 8A, lanes 2–4). Further incubation at 25°C resulted in a drastic reduction in the level of Cdc6, whereas at 35°C, Cdc6 remained stable (Figure 8A, compare lanes 1 and 5). These results confirm previous reports demonstrating that under conditions promoting vegetative growth, Cdc28 is responsible for the degradation of Cdc6 (Elsasser et al., 1996; Elsasser et al., 1999; Sanchez et al., 1999; Calzada et al., 2000). A different result was obtained for Cdc28-depleted cells transferred to SPM. The release of cells to SPM and incubation at 25°C for 2 h caused an increase in the level of Cdc6 (Figure 8B, compare lanes 4 and 5). Further incubation at either 25 or 35°C led to the gradual destabilization of Cdc6 (Figure 8B, lanes 1–3, 6–9). The kinetics of Cdc6 degradation at 35°C is even faster than that at 25°C, probably reflecting a shortening of all meiotic events at the higher temperature. Thus, in the meiotic cycle, the stability of Cdc6 is independent of Cdc28.
Figure 8.
Cdc28 is not required for the degradation of Cdc6 in the meiotic cycle. Diploid cells homozygous for cdc28-degron (strain Y1443) grown at 25°C in PSP2 to a titer of 1 × 107 cells/ml were shifted to 37°C for 2 h (time 0). (A) G1-arrested cells were then shifted to either 25 or 35°C in PSP2 medium, for vegetative growth conditions. (B) G1-arrested cells were then washed, resuspended in SPM, and incubated at either 25 or 35°C for meiotic conditions. At the indicated hours, samples were taken to process for IP-Western by using anti-myc antibodies.
Cdc6 and Ime2 Physically Associate
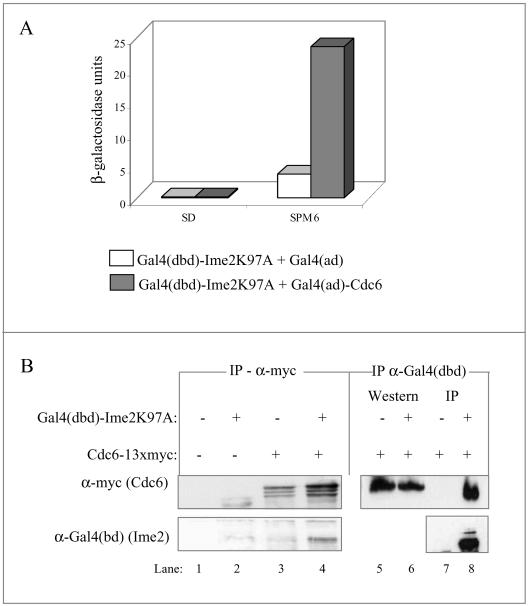
The observation that under meiotic conditions Cdc28 is not required for destabilization of Cdc6 suggests that another kinase may substitute for this job. Because Ime2 is required for the timing of Cdc6 degradation (Figures 3 and 4B), and because in the double mutant cdc28-4 ime2Δ premeiotic DNA replication is absent, we hypothesized that Ime2 might be directly involved with phosphorylation, and consequently degradation, of Cdc6. This hypothesis predicts that these two proteins will physically associate. We used the two-hybrid and coimmunoprecipitation assays to explore this possibility. For these experiments, we used a kinase dead allele of IME2 (Ime2K97A) that gives rise to an increase in the stead-state level of Ime2 (Guttmann-Raviv and Kassir, 2002). Figure 9A shows that in cells expressing both Gal4(dbd)-Ime2K97A and Gal4(ad)-Cdc6 there is a substantial increase in the expression of the gal1-lacZ reporter gene in comparison with cells expressing Gal4(dbd)-Ime2K97A and Gal4(ad). Interestingly, this two-hybrid interaction was observed when cells were incubated in SPM for 6 h and was absent from exponentially cells grown in SD medium. Lack of interaction in SD may result from either a competition between Ime2 and Cdc28 for the binding to Cdc6 or from the requirement for meiosis-specific modifications on either Ime2 or Cdc6.
Figure 9.
Cdc6 and Ime2 physically associate. (A) Two-hybrid interaction. Level of β-galactosidase (given in Miller units) in a haploid strain (Y153) carrying pADH1-gal4(1-147)-HA-ime2K97A (YEp2225) with either pADH1-gal4(768-881)-HA (pGAD2F) or pADH1-gal4(768-881)-HA-CDC6 (YEp1349). Proteins were extracted from 1 × 107 cells/ml grown in SD medium or from cells grown in SD medium to stationary, washed in water, and incubated in SPM medium at a titer of 1 × 107 cells/ml for 6 h. (B) Coimmunoprecipitation. Proteins were extracted from cells incubated for 3 h in SPM. Anti-myc (lanes 1–4) or anti-Gal4(dbd) (lanes 5–8) immune complexes were prepared from the following strains: wild-type Y422 (lanes 1 and 2) or Y1384 (lanes 3 and 4) and ime2Δ (Y1385, lanes 5–8), diploid cells, carrying pIME1-gal4(dbd)-HA-ime2K97A (YEp2229) (lanes 2, 4, 6, and 8). Strains Y1384 and Y1385 (lanes 3–8) also carried CDC6–13 × myc. Proteins were separated on SDS-PAGE, and immunoblotting was done with anti-Gal4(dbd) or anti-myc, as indicated.
We used coimmunoprecipitation assay to verify the physical association between Ime2 and Cdc6. For these experiments, Cdc6 was tagged with 13 × myc and Ime2 with Gal4(dbd). To detect association under meiotic conditions, Ime2 was expressed from the meiosis-specific IME1 promoter and Cdc6 from its own promoter. Figure 9B shows that in diploid cells expressing both proteins, Gal4(dbd)-Ime2 was immunoprecipitated along with Cdc6-myc in an anti-myc immune complex (Figure 9B, lane 4). However, in cells expressing Gal4(dbd)-Ime2 alone or Cdc6-myc alone, Gal4(dbd)-Ime2 was not recovered (Figure 9B, lanes 2 and 3, respectively). In the reciprocal experiment, Cdc6-myc was immunoprecipitated along with Gal4(dbd)-Ime2 in an anti-Gal4(dbd) immune complex (Figure 9B, lane 8). In cells expressing Cdc6-myc alone, Gal4(dbd)-Ime2 was not recovered (Figure 9B, lane 7), whereas it was efficiently detected, in both strains by the Western blot analysis (Figure 9B, lanes 5 and 6). Ime2 was specifically recovered when the same antibody was used for both immunoprecipitation and detection (Figure 9B, lane 8). We conclude, therefore, that Cdc6 and Ime2 physically associate.
DISCUSSION
The machinery of DNA replication is identical in the mitotic and meiotic cycles (Simchen, 1974; Budd et al., 1989; Longhese et al., 1993), and uses the same origins (Collins and Newlon, 1994). Nonetheless, the length of premeiotic DNA synthesis is extended (Williamson et al., 1983; Padmore et al., 1991) and during this phase cells become committed to meiotic recombination (for review, see Kupiec et al., 1997). In addition, the coordination between DNA replication and nuclear division is lost, as seen by the two nuclear divisions that follow a single round of DNA replication. These observations yield the prediction that premeiotic DNA replication might be subject to specific regulation. This report initiated a study on the role of the preRC component Cdc6 in controlling premeiotic DNA synthesis.
Cdc6 Is Required for Initiation of Premeiotic DNA Replication
In the cell cycle, simple temperature shift-up experiments did not reveal the requirement of Cdc6 for DNA replication (Hartwell, 1976). However, when G2-arrested cells were released to the nonpermissive temperature, cells did not initiate DNA replication in the mitotic cell cycle (Piatti et al., 1996). These “contradictory results” are due to the fact that Cdc6 is tethered to origins during anaphase and early G1 (Cocker et al., 1996; Tanaka et al., 1997). In this report, using the same rationale, we determined that Cdc6 is required for the initiation of premeiotic DNA synthesis. A concomitant shift of cells to SPM and to the restrictive temperature resulted in entry into premeiotic S phase. By contrast, release of cells from G2 arrest to SPM and incubation at the restrictive temperature arrested cells in G1, before premeiotic DNA replication (Figure 1). The requirement of Cdc6 for premeiotic DNA replication suggests that Cdc6 might fulfill the same function in both cycles.
When cdc6-1 cells were directly shifted to SPM and incubated at a nonpermissive temperature of 35°C, asci were not observed, although premeiotic DNA replication took place (Figure 1). These results suggest that either Cdc6 is required for an additional event in meiosis or that the DNA synthesis observed was incomplete or defective. The latter hypothesis is supported by the behavior of this mutant in the cell cycle. cdc6-1 cells arrest in the mitotic cell cycle as large budded cells, before nuclear division, and the cells are sensitive to hydroxyurea (Hartwell, 1976).
Cdc6 Is Degraded upon Entry into Premeiotic DNA Replication
In the cell cycle, when cells enter S phase, Cdc6 is targeted for degradation (Elsasser et al., 1996, 1999; Sanchez et al., 1999; Calzada et al., 2000). Similarly, in this report we show that entry into premeiotic DNA replication is also accompanied by the degradation of Cdc6 (Figures 2 and 3). In the cell cycle, Cdc28 activity is required for the phosphorylation of Cdc6, which targets the latter to degradation. In this report, we show that depletion of Cdc28 leads, indeed, to the stabilization of Cdc6 in the mitotic cycle (Figure 8A). However, in the meiotic cycle, depletion of Cdc28 did not stabilize Cdc6 (Figure 8B).
The role of Cdc28 in the initiation of premeiotic DNA replication remains ambiguous. The use of the cdc28-degron allele led to the conclusion that Cdc28 is not essential for entry into premeiotic DNA replication (Guttmann-Raviv et al., 2001). By contrast, the use of the analog sensitive cdc28-as1 allele led to the conclusion that Cdc28 is required for premeiotic DNA replication (Benjamin et al., 2003). These authors suggested that the difference between the two reported experiments might be due to residual, undetected activity of Cdc28 (Benjamin et al., 2003). However, in this report the same restrictive temperature was used in the mitotic and meiotic cycles. Thus, a temperature that suffices for depletion of Cdc28, and by that prevents degradation of Cdc6 in the cell cycle, has no effect on the parallel process in meiosis (Figure 8), which is a temperature-sensitive process by its own. We conclude that in the meiotic cycle destabilization of Cdc6 is independent of Cdc28. This conclusion is in agreement with the suggestion that in the meiotic cycle Ime2 can substitute for Cdc28 (Guttmann-Raviv et al., 2001). This conclusion is based on the observation that in cdc28-4, cdc28-degron, and ime2Δ diploids premeiotic DNA replication takes place in cells shifted to the nonpermissive temperature, whereas the cdc28-4 ime2Δ double mutant arrest before premeiotic DNA replication (Guttmann-Raviv et al., 2001). Furthermore, elevated expression of Clb1 at early meiotic times leads to premature initiation of DNA replication (Figure 6; Guttmann-Raviv et al., 2001), suggesting that under normal conditions increased activation of Cdc28 is deleterious or that Cdc28 is not required for entry into premeiotic S phase.
The following observations suggest that Ime2 is the kinase responsible for Cdc6 degradation in the meiotic cycle. 1) In diploid cells homozygous for an IME2 null allele, premeiotic DNA replication and Cdc6 degradation are concomitantly delayed (Figures 3 and 4). 2) Using the two-hybrid assay, we show interaction between Ime2 and Cdc6 (Figure 9A). 3) Coimmunoprecipitation assays reveal that under meiotic conditions Ime2 and Cdc6 associate (Figure 9B). These results suggest that Ime2 is directly involved with destabilization of Cdc6. It further suggests that under meiotic conditions Ime2 phosphorylates Cdc6. These results also support the hypothesis that Cdc28 is not required for the initiation of premeiotic DNA synthesis.
Throughout Meiosis, Origin-bound Cdc6 Is Protected from Degradation
We used ChIP analysis to determine whether the origin-bound Cdc6, like the unbound fraction, is degraded when cells enter premeiotic S phase. We show that the origin-bound Cdc6 fraction is protected and that its level remains roughly constant throughout the meiotic cycle (Figure 5). By contrast, in yeast mitotic cell cycle, both the chromatin-bound and the free Cdc6 fractions are similarly degraded (Tanaka et al., 1997). We postulated that the meiosis-specific protection might result from the use of Ime2 rather than Cdc28. This hypothesis was based on the assumption that the chromatin-bound Cdc6 might be inaccessible to Ime2, which is a larger protein (73.6 KDa, in comparison to 34 KDa of Cdc28). This hypothesis predicts that “artificial activation” of premeiotic DNA replication by Cdc28 will result in concomitant degradation of the origin-bound Cdc6. Premature activation of Cdc28 was accomplished in wild-type and ime2Δ diploids carrying the pIME1-CLB1 gene (Figure 6). However, the origin-bound Cdc6 remained protected (Figure 6). We suggest that the change in chromatin structure that takes place under meiotic conditions, protect Cdc6 from either phosphorylation or the ubiquitin/proteosome machinery.
The availability of origin-bound Cdc6 in spores might be required for efficient DNA replication when spores are germinated. This suggestion is based on the observation that in Xenopus, the translation of cdc6 mRNA during oocyte maturations is essential to promote DNA replication competence for early embryo divisions (Lemaitre et al., 2002; Whitmire et al., 2002; Kubota and Takisawa, 2003). In human cells, the chromatin-bound Cdc6 is stable in S and G2 cells, whereas the soluble Cdc6 is destroyed (Coverley et al., 2000). This phenomenon, in which meiosis in yeast is regulated in a manner similar to the mammalian cell cycle rather than similar to the yeast cell cycle, was reported previously for the behavior of the spindle pole body (SPB). In all eukaryotes, SPB duplication occurs when cells enter S phase. However, SPB separation occurs in mammalian and in yeast meiosis after the completion of S phase, whereas in the yeast cell cycle it occurs during S phase (Kupiec et al., 1997).
In the mitotic cell cycle, loading of Cdc45 onto origins is correlated with the time of origin-bound Cdc6 release from chromatin (Aparicio et al., 1997; Zou and Stillman, 2000). This result suggests that either degradation of the origin-bound Cdc6 is required for Cdc45 loading, or that this “time correlation” stems from their similar dependence on CDK activity. The observation that in the meiotic cycle the origin-bound Cdc6 is not degraded suggests that the latter is the correct hypothesis.
Prevention of Origin Refiring during the Meiotic Cycle
The observation that in the meiotic cycle Cdc6 occupies origins constitutively, raises the question on how refiring of origins is precluded. In S. pombe, overexpression of Cdc18/Cdc6 leads to the refiring of origins (Nishitani and Nurse, 1995). In S. cerevisiae, however, stabilization of Cdc6 does not lead to a reinitiation of DNA synthesis, because additional mechanisms, which rely on Orc2 and Mcm2 phosphorylation by Cdc28/Clb, suffice to restrict DNA replication to a single round (Nguyen et al., 2001). We show that in the meiotic cycle, similarly to the mitotic cycle, Mcm2 is removed from the chromatin at MII, which suggests that Mcm2 absence is responsible for preventing the reinitiation of DNA synthesis at this stage. At later meiotic times, Mcm2 reoccupies origins, probably reflecting a lack of CDK activity when cells exit MII (Petronczki et al., 2003). Restriction of DNA replication to a single round in the meiotic cycle might also be attributed to the specific degradation of the catalytic subunit and the B subunit of DNA polymerase α that takes place after the completion of premeiotic DNA replication (Foiani et al., 1996). Moreover, regulation through additional proteins, such as Orc2 (Nguyen et al., 2001) and Cdt1 (Yanow et al., 2001), may also contribute to the restriction of DNA replication to a single round.
In this report, focusing on Cdc6, we reached the conclusion that the preRC complex may function in the mitotic and meiotic cell cycles in a similar manner. This conclusion suggests that preIC complex may also be required for entry into the premeiotic S phase. Previous reports, however, showed that a key regulator of the preIC assembly, Cdc7, is not required for premeiotic DNA replication (Schild and Byers, 1978; Horesh et al., 1979; Hollingsworth and Sclafani, 1993). The meiotic characterization of Cdc7 was based on temperature-sensitive alleles. It is possible that the low restrictive temperature used in these experiments masked the fact that it is required, similarly to the effect observed here for Cdc6. Future analysis will reveal whether preIC formation is regulated in a similar or dissimilar manner in the mitotic and meiotic cell cycles. Furthermore, future analysis will uncover the reasons for the fundamental differences between DNA replication in the mitotic and meiotic cycles.
Acknowledgments
We thank K. Benjamin, I. Herskowitz, and M. Foiani for helpful discussions. We thank M. Foiani for technical assistance. We thank J. Diffley and S. Fields for kindly providing plasmids. This work was supported by a grant from the United States-Israel Binational Science Foundation.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–08–0617. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–08–0617.
References
- Aparicio, O.M., Weinstein, D.M., and Bell, S.P. (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Benjamin, K.R., Zhang, C., Shokat, K.M., and Herskowitz, I. (2003). Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 17, 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M.E., Wittrup, K.D., Bailey, J.E., and Campbell, J.L. (1989). DNA polymerase I is required for premeiotic DNA replication and sporulation but not for X-ray repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada, A., Sanchez, M., Sanchez, E., and Bueno, A. (2000). The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J. Biol. Chem. 275, 9734–9741. [DOI] [PubMed] [Google Scholar]
- Cocker, J.H., Piatti, S., Santocanale, C., Nasmyth, K., and Diffley, J.F. (1996). An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Collins, I., and Newlon, C.S. (1994). Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol. Cell. Biol. 14, 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley, D., Pelizon, C., Trewick, S., and Laskey, R.A. (2000). Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113, 1929–1938. [DOI] [PubMed] [Google Scholar]
- Drury, L.S., Perkins, G., and Diffley, J.F. (2000). The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10, 231–240. [DOI] [PubMed] [Google Scholar]
- Elsasser, S., Chi, Y., Yang, P., and Campbell, J.L. (1999). Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell 10, 3263–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser, S., Lou, F., Wang, B., Campbell, J.L., and Jong, A. (1996). Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell 7, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., Liberi, G., Lucchini, G., and Plevani, P. (1995). Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol. 15, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., Marini, F., Gamba, D., Lucchini, G., and Plevani, P. (1994). The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani, M., Nadjar-Boger, E., Capone, R., Sagee, S., Hashimshoni, T., and Kassir, Y. (1996). A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253, 278–288. [DOI] [PubMed] [Google Scholar]
- Guttmann-Raviv, N., Boger-Nadjar, E., Edri, I., and Kassir, Y. (2001). Cdc28 and Ime2 Possess Redundant Functions in Promoting Entry Into Premeiotic DNA Replication in Saccharomyces cerevisiae. Genetics 159, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann-Raviv, N., and Kassir, Y. (2002). Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hartwell, L. (1976). Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol. 104, 803–817. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, R.E.J., and Sclafani, R.A. (1993). Yeast pre-meiotic DNA replication utilizes mitotic origin ARS1 independently of CDC7 function. Chromosoma 102, 415–420. [DOI] [PubMed] [Google Scholar]
- Horesh, O., Simchen, G., and Friedmann, A. (1979). Morphogenesis of the synapton during yeast meiosis. Chromosoma 75, 101–115. [DOI] [PubMed] [Google Scholar]
- Kamimura, Y., Tak, Y.S., Sugino, A., and Araki, H. (2001). Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir, Y., and Simchen, G. (1991). Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 194, 94–110. [DOI] [PubMed] [Google Scholar]
- Kubota, Y., and Takisawa, H. (2003). Block to DNA replication in meiotic maturation: a unified view for a robust arrest of cell cycle in oocytes and somatic cells. Bioessays 25, 313–316. [DOI] [PubMed] [Google Scholar]
- Kupiec, M., Byers, B., Esposito, R.E., and Mitchell, A.P. (1997). Meiosis and Sporulation in Saccharomyces cerevisiae, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Labib, K., Diffley, J.F., and Kearsey, S.E. (1999). G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1, 415–422. [DOI] [PubMed] [Google Scholar]
- Labib, K., Tercero, J.A., and Diffley, J.F. (2000). Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Lei, M., and Tye, B.K. (2001). Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Lemaitre, J.M., Bocquet, S., and Mechali, M. (2002). Competence to replicate in the unfertilized egg is conferred by Cdc6 during meiotic maturation. Nature 419, 718–722. [DOI] [PubMed] [Google Scholar]
- Lindner, K., Gregan, J., Montgomery, S., and Kearsey, S. (2002). Essential role of MCM proteins in premeiotic DNA replication. Mol. Biol. Cell 13, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese, M.P., Jovine, L., Plevani, P., and Lucchini, G. (1993). Conditional mutations in the yeast DNA primase genes affect different aspects of DNA metabolism and interactions in the DNA polymerase alpha-primase complex. Genetics 133, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. (1972). Experiments in Molecular Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Murakami, H., and Nurse, P. (2001). Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat. Genet. 28, 290–293. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.Q., Co, C., Irie, K., and Li, J.J. (2000). Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr. Biol. 10, 195–205. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.Q., Co, C., and Li, J.J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068–1073. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., and Nurse, P. (1995). p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83, 397–405. [DOI] [PubMed] [Google Scholar]
- Owens, J.C., Detweiler, C.S., and Li, J.J. (1997). CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc. Natl. Acad. Sci. USA 94, 12521–12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore, R., Cao, L., and Kleckner, N. (1991). Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66, 1239–1256. [DOI] [PubMed] [Google Scholar]
- Petronczki, M., Siomos, M.F., and Nasmyth, K. (2003). Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423–440. [DOI] [PubMed] [Google Scholar]
- Piatti, S., Bohm, T., Cocker, J.H., Diffley, J.F., and Nasmyth, K. (1996). Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Piatti, S., Lengauer, C., and Nasmyth, K. (1995). Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a `reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M., and Botstein, D. (1983). Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101, 167–180. [DOI] [PubMed] [Google Scholar]
- Sanchez, M., Calzada, A., and Bueno, A. (1999). The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem. 274, 9092–9097. [DOI] [PubMed] [Google Scholar]
- Schild, D., and Byers, B. (1978). Meiotic effects of DNA-defective cell division cycle mutations of Saccharomyces cerevisiae. Chromosoma 70, 109–130. [DOI] [PubMed] [Google Scholar]
- Seki, T., and Diffley, J.F. (2000). Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc. Natl. Acad. Sci. USA 97, 14115–14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen, G. (1974). Are mitotic functions required in meiosis? Genetics 76, 745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, S., and Diffley, J.F. (2002). Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nat. Cell Biol. 4, 198–207. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Knapp, D., and Nasmyth, K. (1997). Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., and Nasmyth, K. (1998). Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 17, 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, M., Liang, C., and Stillman, B. (1999). The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA 96, 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire, E., Khan, B., and Coue, M. (2002). Cdc6 synthesis regulates replication competence in Xenopus oocytes. Nature 419, 722–725. [DOI] [PubMed] [Google Scholar]
- Williamson, D.H., Johnston, L.H., Fennell, D.J., and Simchen, G. (1983). The timing of the S phase and other nuclear events in yeast meiosis. Exp. Cell Res. 145, 209–217. [DOI] [PubMed] [Google Scholar]
- Yanow, S.K., Lygerou, Z., and Nurse, P. (2001). Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., and Stillman, B. (1998). Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou, L., and Stillman, B. (2000). Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol. Cell. Biol. 20, 3086–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke, W., Rottjakob, H.W., and Kuntzel, H. (1994). The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem. 269, 23351–23356. [PubMed] [Google Scholar]