Abstract
From the abyss of the ocean to the human gut, bacterial viruses (or bacteriophages) have colonized all ecosystems of the planet earth and evolved in sync with their bacterial hosts. Over 95% of bacteriophages have a tail that varies greatly in length and complexity. The tail complex interrupts the icosahedral capsid symmetry and provides both an entry for viral genome-packaging during replication and an exit for genome-ejection during infection. Here, we review recent progress in deciphering the structure, assembly and conformational dynamics of viral genome-delivery tail machines. We focus on the bacteriophages P22 and T7, two well-studied members of the Podoviridae family that use short, non-contractile tails to infect Gram-negative bacteria. The structure of specialized tail fibers and their putative role in host anchoring, cell-surface penetration and genome-ejection is discussed.
Introduction
The delivery of viral DNA into bacteria requires, in most cases, physical penetration of the bacterial cell envelope, which is impermeable to genome diffusion [1]. To this end, tailed bacteriophages have evolved complex tail machines that extend from a unique capsid vertex, providing both an attachment point to the host surface, and a channel for genome-ejection through the cell envelope [2,3]. In Gram-negative bacteria like Escherichia coli (E. coli) or Salmonella enterica, the cell envelope is a ~150 Å multilayer compartment formed by an inner and an outer membrane (referred to as IM and OM) separated by a peptidoglycan-containing periplasmic space [1]. Both membranes are enriched in integral and peripheral membrane proteins, but differ in lipid composition: the IM contains two symmetric phospholipid leaflets, whereas the OM is asymmetric, with an inner face of phospholipids and an outer face of lipopolysaccharide (LPS) [4]. LPS is a complex glucosamine-based glycolipid unique to Gram-negative bacteria that inserts into the OM via a hydrophobic moiety known as lipid A. Between the OM and IM is a thin layer of peptidoglycans [5], consisting of alternating units of N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) periodically cross-linked by a tetrapeptide. Overall, the cell envelope protects bacteria from environmental insults and is the major obstacle phages must overcome to deliver their genomes inside bacteria.
Architecture of bacteriophage P22 genome-delivery machine
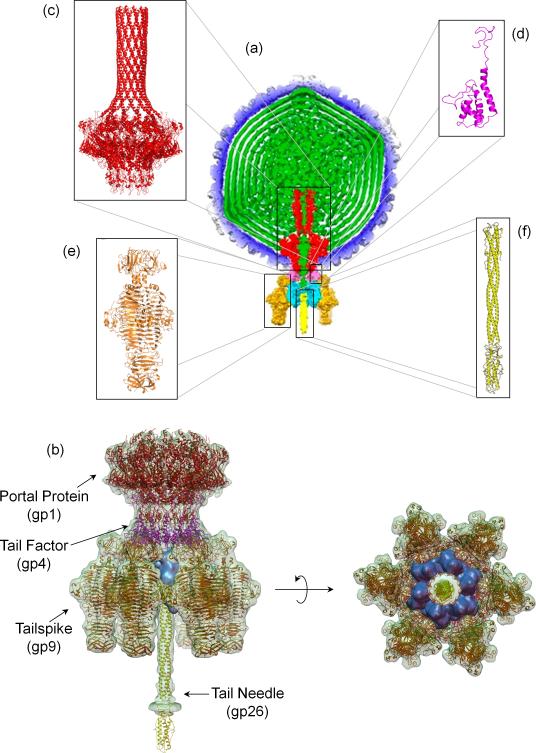
The P22 bacteriophage is one of the best studied Podoviruses [6]. Its short non-contractile tail machine has been visualized in great detail by a series of asymmetric cryo-electron microscopic (cryo-EM) reconstructions of the mature virion [7-9] (Figure 1a). The isolated tail machine extracted from infectious virions was also imaged at 9.4 Å resolution [10,11] using single particle analysis (Figure 1b). This ~2.8 MDa multi-subunit complex is formed by one dodecameric portal protein gp1 (12×83.5 kDa), six trimeric tailspikes gp9 (6×215.4 kDa), twelve copies of the tail accessory factor gp4 (12×18.0 kDa), a hexamer of gp10 (6×52.3 kDa) and a trimer of the tail needle gp26 (3×24.7 kDa). High-resolution crystal structures of four of the five polypeptide chains of P22 were determined [12-15] (Figure 1c-f), which allowed us to generate a pseudo-atomic model of the entire tail machine (Figure 1b) accounting for over 90% of its atoms.
Figure 1. Architecture of bacteriophage P22 genome-delivery tail machine.
(a) Cross-section through the center of the 7.8 Ǻ asymmetric cryo-EM reconstruction of P22 mature virion [7]. The viral genome (in green) is surrounded by a shell of coat protein (blue). The tail complex is formed by portal protein (red), gp4 (pink), gp10 (cyan), gp26 (yellow) and gp9 (orange). (b) Pseudo atomic model of P22 tail machine generated by fitting atomic structures of gp1, gp4, gp26 and gp29 inside the cryo-EM reconstruction of the isolated tail machine [11]. Gp10, whose atomic structure is unknown, is colored in cyan. Ribbon diagrams of individual tail components determined crystallographically are shown in (c) portal protein (pdb 3LJ5) [15], (d) monomeric gp4 (pdb 1VT0) [15], (e) tailspike core (pdb 1TSP) [13] and its N-terminal domain (pdb 1LKT) [12], and (f) tail needle gp26 (pdb 2POH) [14].
The portal protein (gp1) is a hub around which the P22 tail assembles and a ‘connector’ between the tail and coat protein. The portal protein inserts at a five-fold vertex of the icosahedral lattice, where it makes contacts with the coat and scaffolding protein in the procapsid [16] and the coat and viral genome in the mature virion [7,9]. The P22 portal protein consists of a ~0.96 MDa dodecamer [17], which was visualized crystallographically at 3.25 Å resolution as portal protein core (residues 1-602) bound to twelve copies of gp4 and, at lower resolution, as naïve ring (residues 1-725) [15] (Figure 1c). The portal protein has a maximum diameter of ~170 Å and an overall height of ~300 Å, nearly half of which is occupied by a continuous α-helix spanning residues 603-725 that forms the ‘barrel domain’. The X-ray structures of the portal protein core:gp4 complex fits well inside the high resolution asymmetric cryo-EM reconstruction of the mature P22 virion [7], but significant variations exist between the crystal structure and cryo-EM reconstruction at the base of the barrel. In the naïve structure the barrel helices are straight and relaxed [15], whereas this region is twisted in the mature virion [7], possibly as a result of the pressurized genome stored inside the capsid [2]. The barrel is highly susceptible to proteolysis in vitro [18] and is invisible in the reconstructions of P22 procapsid [16] and isolated tail complex [10,11] (Figure 1b), suggesting this helical domain folds specifically upon encapsidation of viral DNA. Accordingly, P22 virions lacking the barrel domain package DNA efficiently but are defective in delivering it into the host, pointing to a role of the barrel in genome-ejecting [7]. In the mature P22 virion, the portal protein is bound to two concentric oligomers formed by tail factors gp4 and gp10. Monomeric gp4 (Figure 1d) assembles upon binding to the portal protein to form a hollow dodecameric ring ~75 Å in height [19,20], while hexameric gp10 binds to a pre-formed portal:gp4 assembly [21] to extend the tail hub. This factor is exceptionally well conserved in P22-like phages [22], but its structure is unknown. Mutants lacking gp10 fail to assemble tailspikes [23,24], suggesting gp10 physically bridges the phage tail to the tailspike protein, as visualized by recent cryo-EM reconstructions [7,11].
The tailspike protein (gp9) is the largest component of P22 tail that accounts for ~46% of its total mass. Each tailspike protomer is built of 13 complete turns of a parallel β-helix domain, which forms a three-stranded interdigitated core [13] and an N-terminal head-binding domain that mediates attachment to the tail machine [12] (Figures 1e, 2a). In the phage tail, six trimeric tailspikes are symmetrically positioned at the vertices of a hexagon ~215 Å in diameter (Figure 1b). The tailspike functions as an adhesion protein that binds to the O-antigenic repeating units of LPS (between 19 - 34 repeating units in Salmonella [25]) and cleaves the α(1,3)-O-glycosidic bond between the rhamnose and galactose components of the LPS using its endorhamnosidase activity [26]. Protruding ~140 Å outside the plane generated by the six tailspikes [7,9] is the tail needle gp26, which forms a 240 Å long trimeric coiled-coil fiber [14,27-29] (Figures 1f, 2b). Deletion studies [30] have revealed that the N-terminal 60 aminoacids of gp26 are essential for binding to hexameric gp10 [21]. Together, the tail factors gp4, gp10, and gp26 are required for the stabilization of newly packaged DNA into the P22 capsids. P22 phages bearing mutations at any of these three genes form procapsids and package DNA but remain unstable, losing their DNA within minutes [31].
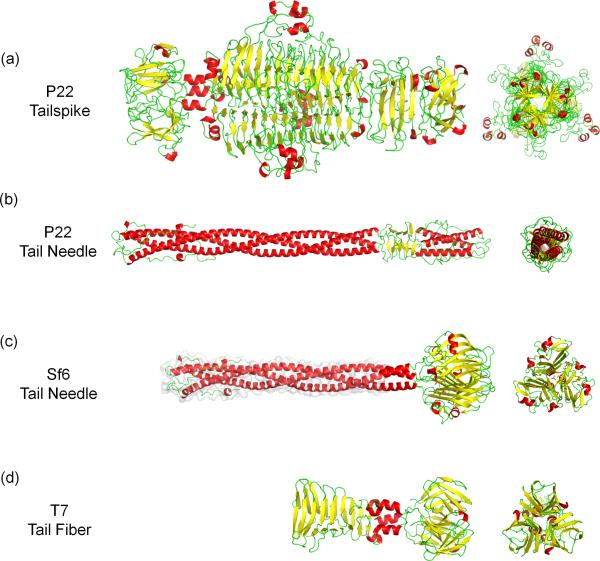
Figure 2. Conservation of tail fibers in Podoviridae.
Ribbon diagrams of (a) P22 tailspike (pdb 1TSP, 1LKT) [13] [12]; (b) P22 tail needle (pdb 2POH) [14]; (c) Sf6 tail needle that includes the crystal structure of its knob domain (pdb 3RWN) [34] and a homology model of the helical core (highlighted with a light gray surface) [32]; (d) T7 tail fiber (pdb A0T) [44]. In all the panels, α-helices, β-strands and loops are colored in red, yellow and green, respectively.
The tail needle super-family and its putative role in genome-delivery
The gene encoding the tail needle has been identified bioinformatically in the genome of over 150 bacteriophages and prophages [32]. While the N-terminal 65 residues are extraordinarily conserved, tail needles can be divided in two subgroups [33] based on the C-terminal domain architecture. In P22-like bacteriophages (Figure 2b), the C-terminus folds into a helical trimer, whose mainchain is inverted with respected to the helical core [14,29,30]. In Sf6-like phages (Figure 2c), the C-terminus of the tail needle ends in a trimeric knob structure [33] similar to the fiber knob of bacteriophage PRD1 and Adenovirus [34]. Ultra-high resolution crystal structures of the phage Sf6 [34] and HS1 [35] tail needle knobs identified three L-glutamate molecules bound to a trimeric knob, however the functional significance of this ligand remains unknown. All tail needles analyzed experimentally display remarkable structural stability and denature in solution with an apparent melting temperature between 85-90°C [33].
Morphologically, the tail needle is the most distal moiety of the phage exposed to the outside and is likely responsible for first contacting the host outer surface. This idea is supported by the observation that during P22 infection, the tail needle is released from the virion into the host, possibly to open a channel for DNA release [36]. The slender structure of gp26, reminiscent of a “drill bit” (Figure 2b), is consistent with this putative function. Chimeric phages carrying swapped tail needle knobs efficiently infect Salmonella, suggesting that the knob does not confer host specificity, at least under laboratory conditions [35]. However, genetic changes to the needle tip that replace entirely the knob with the short foldon domain of bacteriophage T4 fibritin [37] reduce infectivity and significantly slow down potassium release from the host during infection. This suggests that the needle plays a role in DNA delivery by controlling the kinetics of DNA ejection into the host. Intriguingly, aggregates of purified Salmonella LPS are sufficient to trigger P22 tail needle ejection in vitro, followed by slow release of viral DNA over a period of about 5 hours [38]. Although in vitro ejection is significantly slower than in vivo genome-ejection, which occurs within minutes, the cross-talk between LPS and gp26 suggests that the signal for tail needle ejection and genome release may be transmitted by the tailspike to the tail needle through a series of conformational changes within the tail hub (Figure 1b). In support of this idea, we found that the tail needle can adopt two conformations in response to pH changes (Bhardwaj, A., personal communication). At neutral pH, the N-terminus of gp26 (residues 1-60) adopts a random coil conformation that binds gp10 and closes the DNA-ejection channel. Acidic pH drives folding of gp26 N-termini into a trimeric helical bundle [14,29], possibly representative of a post-ejection conformation. Gp26 ejection is followed by ejection of three essential proteins gp7, gp16 and gp20 (or ejection proteins) [31], that are thought to form a conduit through the cell envelope [39].
Conformational changes accompany genome-ejection of bacteriophage T7
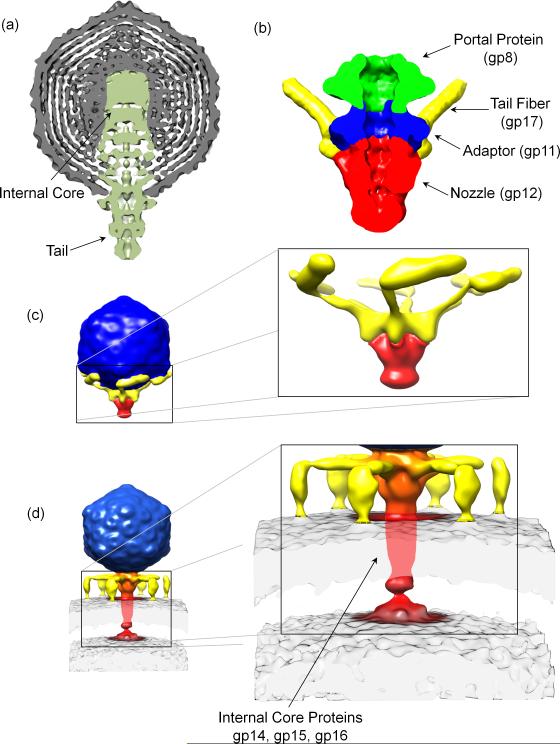
T7 is another well-studied member of the Podoviridae family. A medium resolution asymmetric reconstruction of T7 mature virion [40] revealed a T=7 icosahedral capsid ~600 Å in diameter (Figure 3a). The tail complex occupies a unique five-fold vertex and projects 185 Å outside the capsid layer and ~265 Å inside the virion where it forms a ~175 Å wide ‘internal core’. Docking the 8 Å resolution connector structure (gp8) [41] into the internal core unambiguously assigned the position of the dodecameric portal vertex. Above the connector, internal core proteins gp14, gp15, and gp16 form 12-, 8- and 4-fold symmetric rings [42]. Minimal density is observed in this reconstruction for the tail fibers (gp17) that are disordered due to their flexibility. These fibers were partially resolved in a 16 Å reconstruction of T7 tail complex extracted from mature virion [43] (Figure 3b). This ~2.7 MDa assembly is built by a dodecameric portal (gp8), six trimeric fibers (gp17) and two rings of a dodecameric adaptor (gp11) and a hexameric nozzle protein (gp12). The crystal structure of a C-terminal fragment of gp17 (residues 317-553) was recently determined [44] (Figure 2d). Surprisingly, this trimeric fiber consists of a tapered pyramid of interlocked β-sheets, similar to P22 tailspike (Figure 2a), that is connected by two short α-helices to a C-terminal knob similar to the Sf6 phage tail needle knob (Figure 2c) [34]. The T7 tail fiber binds reversibly to E. coli LPS followed by a secondary irreversible attachment of the tail to an unknown primary receptor [26].
Figure 3. Structural changes in T7 tail machine upon genome ejection.
(a) Cross-section through the center of the 21 Ǻ 5-fold symmetrized cryo-EM reconstruction of T7 mature virion [40]. (B) Cryo-EM reconstruction of T7 tail machinery isolated from mature virions [43]. (C-D) Cryo-ET reconstructions of the pre- and post-adsorption conformation of bacteriophage T7. Tail fibers undergo dramatic remodeling and a new density, likely corresponding to internal core proteins gp14, gp15 and gp16 appears in the host periplasm.
Cryo-electron tomography (cryo-ET) was recently used to capture T7 in the act of ejecting its genome inside E. coli minicells [45]. Tomographic reconstructions of T7 mature virions resolved tail fibers folded against the capsid and symmetrically organized around the tail hub (Figure 3c). Engagement of T7 tail tip with a specific receptor present in E. coli results in two dramatic quaternary structure conformational changes (Figure 3d). First, tail fibers detach from the capsid and undergo a large rotation to grab the host surface, adopting an ‘adsorption structure’. Second, the tail tip inserts a ~450 Å long ‘DNA ejection conduit’ inside the host cell envelope [45] that likely contains internal core proteins gp14, gp15 and gp16 [46]. This reconstruction supports a model whereby the tail fibers are dynamic structures that by adopting a folded-back conformation allow a higher rate of virion diffusion. The idea of dynamic tail fiber accompanying genome ejection was also emphasized by a recent cryo-EM analysis of the marine phage P-SSP7, a far-relative of T7 and ocean most abundant phage [47]. Overall, transient extension of flexible tail fibers upon contact with a surface may allow a phage to explore a large volume in search for a cell, or a specific receptor.
Conclusions
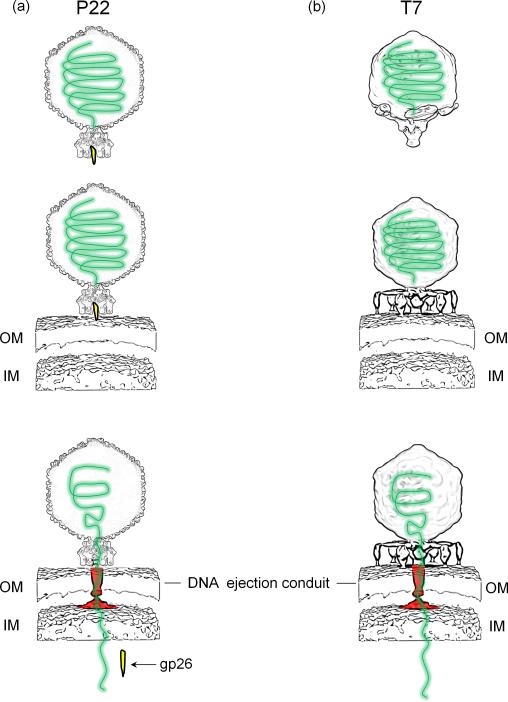
In recent years, the synergy of ‘top-down’ cryo-EM asymmetric reconstructions of infectious virions and ‘bottom-up’ high resolution crystal structures of individual components has allowed to visualize the structure of large viral genome-delivery machines with unprecedented accuracy. In parallel, the advent of cryo-electron tomography is allowing scientists to put these machines in their biological context and begin deciphering the mechanisms of genome-ejection at the single virion level. The tail machines of P22 and T7 described in this review present significant differences and some analogies. Both tails are too short to span a bacterial cell envelope and use tailspikes to attach to a ‘primary receptor’ on the host surface [26]. P22 virions (Figure 4a) adsorb onto the Salmonella surface using tailspikes, which recognize and cleave the O-antigen polysaccharide portion of the surface LPS [26]. In all cryo-EM reconstructions obtained thus far, the P22 tailspikes are rigid and there is no experimental evidence of tailspikes squatting onto the host surface or undergoing major conformational changes (Parent, K., personal communication). Tailspikes form a static crown that attaches to the host OM and, using its endorhamnosidase activity, brings the phage in close proximity to the host surface and mediates tail needle ejection through the cell envelope. It is also possible that the tail needle tip contacts an unknown ‘secondary receptor’ [48] to facilitate, or perhaps simply accelerate DNA-ejection. In either case, tail needle ejection is followed by delivery of ejection proteins gp7, gp16 and gp20 that have been hypothesized to assemble in the host cell envelope to form a conduit for DNA translocation. In contrast, T7 (and likely P-SSP7) (Figure 4b) have dynamic tail fibers that undergo dramatic conformational changes upon contact to the host cell surface allowing the virus to ‘walk’ onto the host surface searching for a receptor. T7 absorption is also accompanied by ‘puncturing’ of the host OM, followed by insertion of three internal core proteins gp14, gp15, and gp16 into the infected cell envelope to generate a conduit for genome-delivery. In summary, different yet fundamentally analogous molecular machines mediate ejection of bacteriophage P22 and T7 genome into their Gram-negative hosts. Although entropically driven, this process is energetically costly [2] and the exact energetics of genome-ejection remains a topic of controversy.
Figure 4. Modes of genome-ejection in Podoviruses.
Schematic diagram depicting the adsorption and genome-ejection strategy used by bacteriophages P22 (A) and T7 (B). In both cases, internal core proteins are thought form a DNA conduit (colored in red) in the host cell envelope that mediates ejection of viral DNA (in green) inside the host cytoplasm.
Highlights.
Structural overview of bacteriophages P22 and T7 genome-delivery tail machines
Use of hybrid structural methods to study viral genome-delivery machines
Cryo-electron tomography snapshots the conformational dynamics of genome-ejection
Evidence that Podoviruses insert a DNA-ejection conduit in the host cell envelope
Acknowledgements
This work was supported by the National Institute of Health grant 1R01GM100888 to G.C. A.B. is supported by the National Cancer Institute Cancer Center Support Grant P30 CA56036.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2○○.Molineux IJ, Panja D. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol. 2013;11:194–204. doi: 10.1038/nrmicro2988. [An exceptionally well-written review that describes physical principles and biological mechanisms of viral genome-ejection.] [DOI] [PubMed] [Google Scholar]
- 3.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 4.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 6.Teschke CM, Parent KN. ‘Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–130. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7○○.Tang J, Lander GC, Olia A, Li R, Casjens S, Prevelige P, Jr., Cingolani G, Baker TS, Johnson JE. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [A 7.8 Å asymmetric cryo-EM reconstruction of P22 mature virion showing exceptional detail for the tail complex, including a full view of the barrel domain of portal protein.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The Structure of an Infectious p22 Virion Shows the Signal for Headful DNA Packaging. Science. 2006 doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 10.Tang L, Marion WR, Cingolani G, Prevelige PE, Johnson JE. Three-dimensional structure of the bacteriophage P22 tail machine. Embo J. 2005;24:2087–2095. doi: 10.1038/sj.emboj.7600695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lander GC, Khayat R, Li R, Prevelige PE, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 14.Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007;14:1221–1226. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- 15○○.Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [The first high-resolution crystal structure of P22 portal protein, solved in complex with twelve copies of the tail factor gp4 and, to a lower resolution, as naïve dodecameric ring. This structure revealed the existence of a C-terminal helical tube named ‘barrel domain’.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16○○.Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [A high-resolution symmetrized reconstruction of P22 mature virion and P22 procapsid revealing the position of scaffolding protein.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJ. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J Mol Biol. 2008;379:385–396. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cingolani G, Moore SD, Prevelige PE, Jr., Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139:46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- 19.Zheng H, Olia AS, Gonen M, Andrews S, Cingolani G, Gonen T. A conformational switch in bacteriophage p22 portal protein primes genome injection. Mol Cell. 2008;29:376–383. doi: 10.1016/j.molcel.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olia AS, Al-Bassam J, Winn-Stapley DA, Joss L, Casjens SR, Cingolani G. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J Mol Biol. 2006;363:558–576. doi: 10.1016/j.jmb.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Olia AS, Bhardwaj A, Joss L, Casjens S, Cingolani G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry. 2007;46:8776–8784. doi: 10.1021/bi700186e. [DOI] [PubMed] [Google Scholar]
- 22.Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 23.Poteete AR, Botstein D. Purification and properties of proteins essential to DNA encapsulation by phage P22. Virology. 1979;95:565–573. doi: 10.1016/0042-6822(79)90509-9. [DOI] [PubMed] [Google Scholar]
- 24.Poteete AR, Jarvik V, Botstein D. Encapsulation of phage P22 DNA in vitro. Virology. 1979;95:550–564. doi: 10.1016/0042-6822(79)90508-7. [DOI] [PubMed] [Google Scholar]
- 25.Palva ET, Makela PH. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107:137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- 26.Casjens SR, Molineux IJ. Short noncontractile tail machines: adsorption and DNA delivery by podoviruses. Adv Exp Med Biol. 2012;726:143–179. doi: 10.1007/978-1-4614-0980-9_7. [DOI] [PubMed] [Google Scholar]
- 27.Berget PB, Poteete AR. Structure and functions of the bacteriophage P22 tail protein. J Virol. 1980;34:234–243. doi: 10.1128/jvi.34.1.234-243.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews D, Butler JS, Al-Bassam J, Joss L, Winn-Stapley DA, Casjens S, Cingolani G. Bacteriophage P22 tail accessory factor GP26 is a long triple-stranded coiled-coil. J Biol Chem. 2005;280:5929–5933. doi: 10.1074/jbc.C400513200. [DOI] [PubMed] [Google Scholar]
- 29.Olia AS, Casjens S, Cingolani G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 2009;18:537–548. doi: 10.1002/pro.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhardwaj A, Olia AS, Walker-Kopp N, Cingolani G. Domain organization and polarity of tail needle GP26 in the portal vertex structure of bacteriophage P22. J Mol Biol. 2007;371:374–387. doi: 10.1016/j.jmb.2007.05.051. [DOI] [PubMed] [Google Scholar]
- 31.Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- 32.Leavitt JC, Gilcrease EB, Wilson K, Casjens SR. Function and horizontal transfer of the small terminase subunit of the tailed bacteriophage Sf6 DNA packaging nanomotor. Virology. 2013;440:117–133. doi: 10.1016/j.virol.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhardwaj A, Walker-Kopp N, Casjens SR, Cingolani G. An evolutionarily conserved family of virion tail needles related to bacteriophage P22 gp26: correlation between structural stability and length of the alpha-helical trimeric coiled coil. J Mol Biol. 2009;391:227–245. doi: 10.1016/j.jmb.2009.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhardwaj A, Molineux IJ, Casjens SR, Cingolani G. Atomic structure of bacteriophage Sf6 tail needle knob. J Biol Chem. 2011;286:30867–30877. doi: 10.1074/jbc.M111.260877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavitt JC, Gogokhia L, Gilcrease EB, Bhardwaj A, Cingolani G, Casjens SR. The tip of the tail needle affects the rate of DNA delivery by bacteriophage p22. PLoS One. 2013;8:e70936. doi: 10.1371/journal.pone.0070936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Israel V. E proteins of bacteriophage P22. I. Identification and ejection from wild-type and defective particles. J Virol. 1977;23:91–97. doi: 10.1128/jvi.23.1.91-97.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 38.Andres D, Hanke C, Baxa U, Seul A, Barbirz S, Seckler R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem. 2010;285:36768–36775. doi: 10.1074/jbc.M110.169003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 40.Agirrezabala X, Martin-Benito J, Caston JR, Miranda R, Valpuesta JM, Carrascosa JL. Maturation of phage T7 involves structural modification of both shell and inner core components. EMBO J. 2005;24:3820–3829. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8A resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W. Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A. 2013;110:6811–6816. doi: 10.1073/pnas.1215563110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuervo A, Pulido-Cid M, Chagoyen M, Arranz R, Gonzalez-Garcia VA, Garcia-Doval C, Caston JR, Valpuesta JM, van Raaij MJ, Martin-Benito J, et al. Structural characterization of the bacteriophage t7 tail machinery. J Biol Chem. 2013;288:26290–26299. doi: 10.1074/jbc.M113.491209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44○.Garcia-Doval C, van Raaij MJ. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc Natl Acad Sci U S A. 2012;109:9390–9395. doi: 10.1073/pnas.1119719109. [This paper reports the first crystal structure of a C-terminal fragment of bacteriophage T7 tail fiber gp17.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45○○.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [An exceptional tomographic analysis of bacteriophage T7 prior and during infection that reveals dramatic quaternary structure conformational changes in the tail complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012;427:177–188. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]