Abstract
The RNA motifs that bind guanidinylated kanamycin A (G Kan A) and guanidinylated neomycin B (G Neo B) were identified via two-dimensional combinatorial screening (2DCS). The results of these studies enabled the “bottom-up” design of a small molecule inhibitor of oncogenic microRNA-10b.
RNA drug targets in the human transcriptome are numerous, however most drugs elicit their effects by modulating protein, not RNA, function.1–3 This is perhaps due to a lack of fundamental understanding about RNA-ligand interactions, particularly the RNA secondary structural elements (motifs) that are targetable and small molecules that are biased for binding RNA motifs. Two-dimensional combinatorial screening (2DCS) has been used to identify the privileged RNA motifs that bind small molecules via selection.4 Two of the most significant concerns for small molecules that target RNA are cell permeability and specificity. Fortuitously, many molecular transporters bind RNA, notably guanidinylated aminoglycosides.5 Herein, we report the identification of the preferred RNA internal loops that bind two guanidinylated aminoglycosides and the development of a bioactive compound targeting a precursor microRNA (miRNA) by using those preferences.
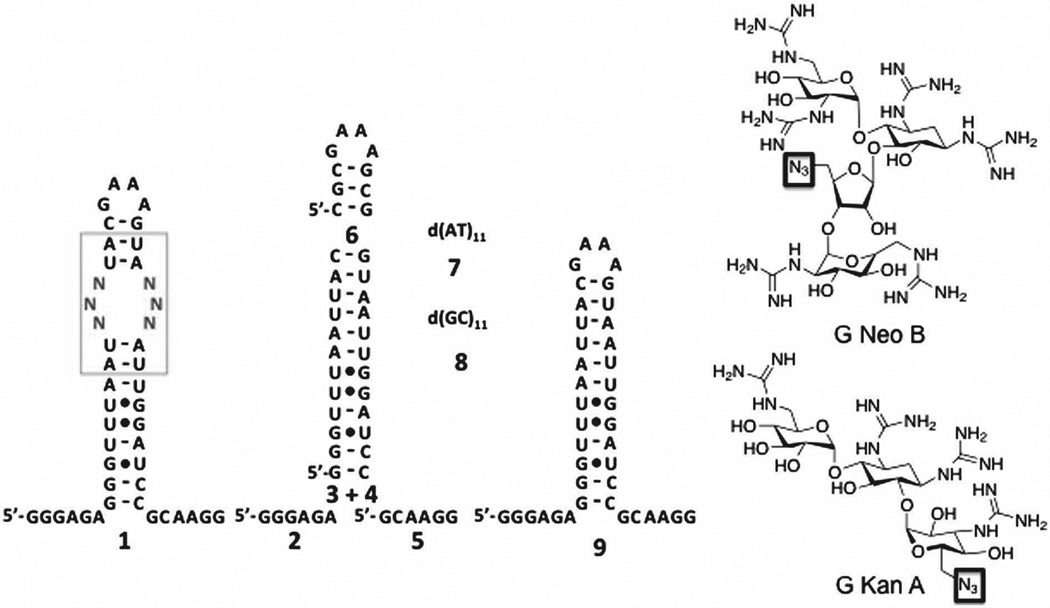
In 2DCS, a small molecule microarray is hybridized with an RNA library of a discrete secondary structural element such as an internal loop (1, for example; Fig. 1). The RNAs bound to each small molecule are excised from the array, amplified, and sequenced. Thus, this approach identifies the privileged RNA motifs for binding a small molecule from thousands of combinations. To enable 2DCS studies of guanidinylated aminoglycosides, G Neo B and G Kan A derivatives (Fig. 1) were synthesized that contain an azide handle for site-specific immobilization onto alkyne-functionalized agarose microarrays (Figs. S-1 – S-9).6 Serial dilutions of the compounds were delivered to the slide surface to afford a dose response after hybridization with 32P-labelled RNA library 1 (Fig. S-10). Hybridization is completed in the presence of unlabeled competitor oligonucleotides 2–8 (Fig. 1) to constrain selected interactions to the randomized regions in 1.4 RNAs bound at the lowest loading above background were harvested, amplified, and sequenced (Tables S-1 and S-2), as interactions captured at lower ligand loading are the highest affinity.4
Fig. 1.
Secondary structures of the nucleic acids and small molecules used in this study. Left, 1 is the secondary structure of the 4,096-member RNA 3×3 nucleotide internal loop library; 2–5 are the competitor RNAs used to constrain 2DCS selections to the randomized region in 1. Oligonucleotides 7 and 8 are DNA competitors, and 9 is the cassette into which the randomized region was inserted. Right, structures of azide-functionalized guanidinylated derivatives of kanamycin A and neomycin B
The members of 1 selected for both small molecules were analysed to define features that impart binding affinity using the RNA Privileged Space Predictor program, RNA-PSP, (v 2.0).7 RNA-PSP compares features in 1 (such as a GC step) to the features in selected motifs. A Z-score (which can be converted to the corresponding two-tailed p-value) is computed, which is a measure of statistical confidence that a feature in a selected motif is truly “privileged” for binding to the small molecule (Fig. S-11). A selected RNA motif contains multiple statistically significant features. Previous studies have shown that RNAs with more statistically significant features that contribute positively to molecular recognition are higher affinity.7, 8 In fact, affinity scales with the sum of the Z-scores for each statistically significant feature, or Sum Z-score. The range of Sum Z-scores for G Neo B and G Kan A are shown in Fig. S-11. As expected, RNA motifs selected to bind have the highest Sum Z-scores. Motifs selected to bind G Neo B have low Sum Z-scores scores for G Kan A and vice versa, indicating they are specific for the aminoglycoside they were selected to bind (Figs. S-11 and S-12).
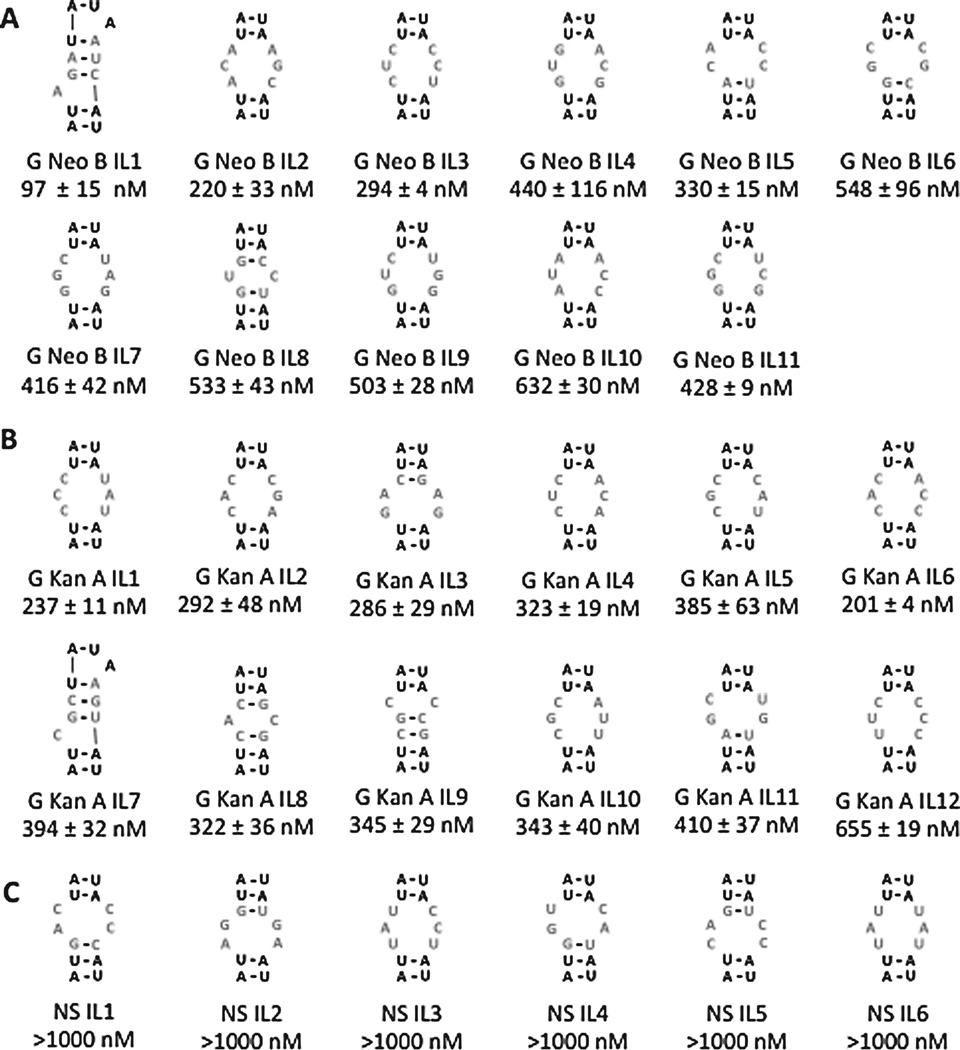
The secondary structures of selected sequences with the highest Sum Z-scores were analyzed,9 and their affinities determined (Fig. 2). The affinities of RNAs selected for G Neo B range from ≈100 to 550 nM while affinities for RNAs selected for G Kan A range from ≈200 to 650 nM (Figs. 2A&B, S-13, and S-14). Importantly, both compounds bind weakly to the entire library 1 and the cassette 9 into which the randomized region was embedded, 9 (Figs. 1 and S-15). Six members of 1 were randomly chosen that have low Sum Z-scores for both compounds (bottom 10%) and their affinities measured. As expected, none of these RNAs motifs (NS IL 1 – NS IL 6) binds G Neo B or G Kan A (Figs. 2C and S-16). Thus, high and low Sum Z-scores can accurately predict affinity of RNA motif-ligand interactions. The effect of the cassette into which an RNA motif is embedded on affinity was also investigated. As expected from previous studies, little difference in affinity is observed when the stem is altered (Figs. 1 and S-17).4, 10
Fig. 2.
Secondary structures of the internal loops that were studied for binding ligands in this study. The nomenclature for each loop is an abbreviated ligand name (G Kan A or G Neo B) followed by IL (refers to internal loop) and a unique number. The dissociation constant is reported in nM below the loop identifier. A, the internal loops selected to bind G Neo B. B, the internal loops selected to bind G Kan A. C, loops that were not selected to bind either ligand. As predicted, these loops bind weakly to both G Kan A and G Neo B with K s >1000 nM.
Sum Z-scores can also be used to predict selectivity.11 The affinities of two RNAs selected to bind G Neo B and two RNAs selected to bind G Kan A were measured for binding both small molecules (labelled in Fig. S-11). The RNAs were chosen based on their Sum Z-scores; loops with large Sum Z-scores for their cognate ligand and small Sum Z-scores for the non-cognate ligand were selected. As expected, this analysis allowed for prediction of compound selectivity. For example, no saturation was observed when up to 10 µM RNA G Neo B IL 8 was added to 50 nM G Kan A (Figs. 2 and S-18). Likewise, the Kd for RNA motif G Neo B IL 5 and G Kan A is 1400 nM (approximately 5-fold selective for G Neo B) (Figs. 2 and S-18). The observation that G Neo B IL5 exhibits lower selectivity is expected as is Sum Z-score for Kan A is higher than that for G Neo B IL 8 (Fig. S-11). The selectivity of RNA motifs selected to bind G Kan A were measured for binding G Neo B. No saturation is observed when up to 10 µM G Kan A IL 1 or G Kan A IL 2 is added to 50 nM G Neo B (Figs. 2 and S-18). Collectively, these studies illustrate that higher and lower Sum Z-scores indicate tight and weak binding, respectively, and can be used to predict ligand selectivity and highlight that interactions identified by 2DCS are selective.
Traditional drug or chemical probe discovery is accomplished by screening small molecules for modulating a specific target. By using the information obtained via 2DCS herein, a different, “bottom-up” route was implemented. In this approach, RNA motif-small molecule interactions identified by 2DCS were mined against the RNA folds in the human transcriptome, namely miRNA precursors. MiRNAs function by binding to the 3’ untranslated regions (UTRs) of mRNAs, negatively regulating translation. MiRNAs are transcribed as primary (pri-) miRNAs that are processed in the nucleus by Drosha, and then exported to the cytoplasm as pre-miRNAs that are processed by Dicer to form the mature miRNA (Fig. 3A).12 Many miRNAs cause or contribute to many diseases.13 Binding of a small molecule to Drosha or Dicer processing sites could inhibit biogenesis of aberrantly expressed miRNAs, thereby alleviating disease.
Fig. 3.
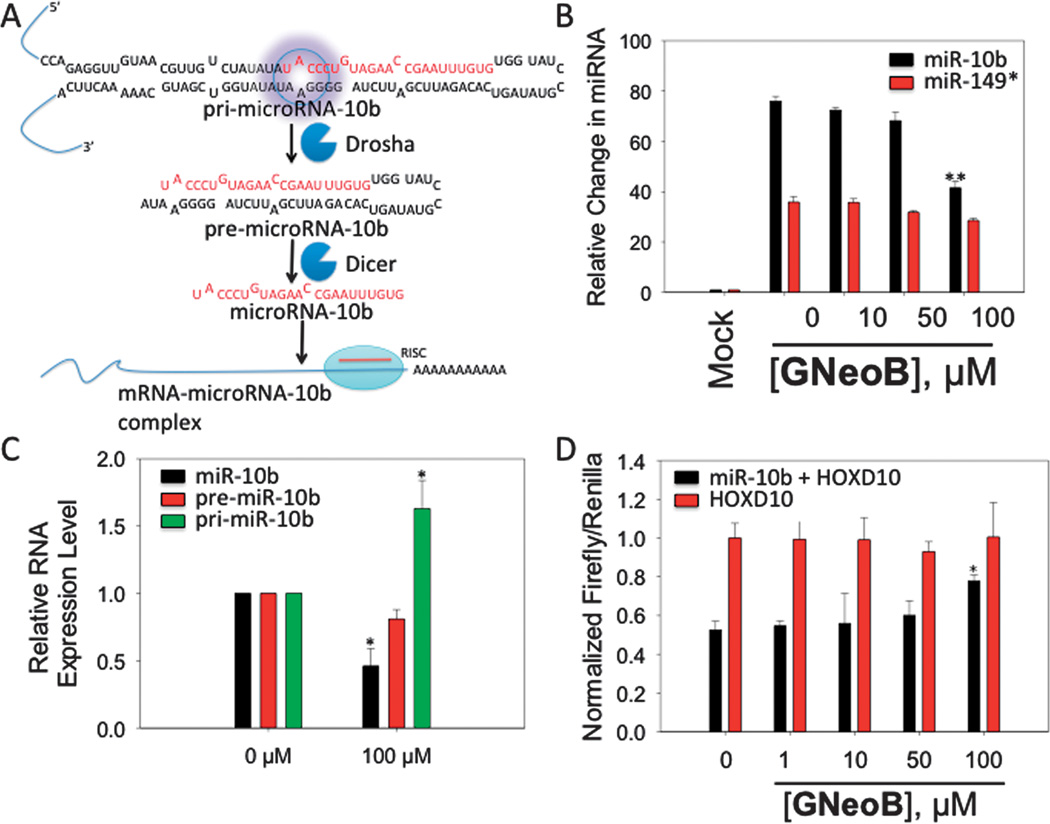
G Neo B inhibits miR-10b biogenesis and boosts production of a downstream target. A, miR-10b is processed to produce the mature miRNA in two steps, first by Drosha in the nucleus and then by Dicer in the cytoplasm. The mature miRNA modulates protein expression. The target site for G Neo B is highlighted with a purple circle. B, G Neo B inhibits production of mature miR-10b as determined by qRT-PCR. C, G Neo B increases the amount of pri-miR-10b and diminishes pre- and mature miR-10b levels, as expected if G Neo B binds to the Drosha site and inhibits processing. D, G Neo B affects production of luciferase when a luciferase mRNA is under control of miR-10b, validating that G Neo B affect miRNA biogenesis and downstream protein targets (*, p < 0.05; **, p < 0.01 as determined by a two-tailed student t-test; n≥3).
By mining the RNA motif-small molecule pairs identified herein against the secondary structures of miRNA precursors in miRBase,14 we identified that G Neo B IL 12 is the Drosha processing site in the miR-10b precursor. Overexpression of miR-10b has been implicated in numerous cancers with invasive and metastatic characteristics,15 and there are no known small molecule effectors of miR-10b. Inhibition of miR-10b biogenesis could increase production of downstream proteins that it regulates. We validated that G Neo B binds the internal loop in miR-10b, including its non-nearest neighbour closing pairs, by embedding 5’AUACC/3’UAAGG in 9 and measuring affinity, affording a Kd of 417 ± 60 nM (Fig. S-19).
Next, we studied the effect of G Neo B on miR-10b biogenesis. HeLa cells were transfected with a construct that allows overproduction of pri-miR-10b and then treated with G Neo B. As shown in Fig. 3B, G Neo B inhibits production of mature miR-10b by 50% at 100 µM dosage. Importantly, G Neo B does not affect biogenesis of a control miRNA that is not predicted to bind the small molecule (miR-149) as determined by a two-tailed student t-test (Figs. 3B and S-20).16 A decrease in mature miRNA levels could occur through various mechanisms. G Neo B was designed to bind miR-10b’s Drosha processing site, thereby inhibiting processing. If G Neo B inhibits Drosha processing, then there should be a boost in the amount of primiR-10b and concomitant decreases in the amount of pre- and mature forms. We therefore determined the expression levels of pri-, pre-, and mature miR-10b by qRT-PCR (Figs. 3C & S-21). G Neo B increases the amount of pri-miRNA by ~60% and decreases the pre- and mature forms by ~30% and ~60%, respectively (Fig. 3C and S-21).
Next, we determined if G Neo B can remove repression of miR-10b’s downstream targets using a model system. MiR-10b directly suppresses HomeoboxD10 (HOXD10) mRNA, which functions as a tumor suppressor17 by inhibiting genes involved in cell migration and extracellular matrix remodelling such as RhoC, α3-integrin and Mt1-MMP.18 The HOXD10 3’ UTR was fused to luciferase; therefore, luciferase activity is inversely proportional to mature miR-10b levels. The construct was co-transfected with the pri-miR-10b construct into HeLa cells, followed by treatment with G Neo B. In agreement with the decrease in mature miR-10b observed by qRT-PCR (Figs. 3A & B), G Neo B stimulates production of luciferase by 1.5-fold (Fig. 3D). Importantly, G Neo B does not affect luciferase production in the absence of miR-10b, as determined by co-transfection of the luciferase-HOXD10 construct and a control miRNA plasmid that does not regulate HOXD10 (miR-149) (Fig. 3D).
Streptomycin is the only other small molecule known to affect miRNA biogenesis in cells and targets miR-21;19 other compounds have been shown to affect miR-21 and miR-122 production by targeting transcription factors.20, 21 Although G Neo B has modest activity, it can be optimized. For example, modular assembly is a robust approach that improves the bioactivity of small molecules that target repeating transcripts.22–24 G Neo B’s azide handle makes it amendable to such an approach. Although modular assembly increases molecular weight, which is generally considered unfavourable, it is possible that this potential issue could be assuaged because G Neo B is a molecular transporter. Importantly, these studies highlight that small molecules can be designed to target RNA by using the output of 2DCS, rather than using high throughput screening.
Supplementary Material
Acknowledgements
We thank Matthew Belair and Pavel Tsitovich for studies on the synthesis of G Neo B. This work was funded by the National Institutes of Health (R01-GM097455). MDD is a Research Corporation Cottrell Scholar and a recipient of the Camille & Henry Dreyfus Teacher-Scholar Award.
Footnotes
Electronic Supplementary Information (ESI) available contianing, synthetic methods, and additional data. See DOI: 10.1039/b000000x/
Notes and references
References
- 1.Thomas JR, Hergenrother PJ. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Chow CS, Bogdan FM. Chem. Rev. 1997;97:1489–1514. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 3.Guan L, Disney MD. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 4.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. ACS Chem. Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 5.Luedtke NW, Carmichael P, Tor Y. J. Am. Chem. Soc. 2003;125:12374–12375. doi: 10.1021/ja0360135. [DOI] [PubMed] [Google Scholar]
- 6.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 7.Velagapudi SP, Seedhouse SJ, Disney MD. Angew. Chem. Int. Ed. Engl. 2010;49:3816–3818. doi: 10.1002/anie.200907257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Velagapudi SP, Seedhouse SJ, French J, Disney MD. J. Am. Chem. Soc. 2011;133:10111–10118. doi: 10.1021/ja200212b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL. J. Am. Chem. Soc. 2008;130:11185–11194. doi: 10.1021/ja803234t. [DOI] [PubMed] [Google Scholar]
- 11.Velagapudi SP, Pushechnikov A, Labuda LP, French JM, Disney MD. ACS Chem. Biol. 2012;7:1902–1909. doi: 10.1021/cb300213g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Calin GA, Croce CM. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW, Weinberg RA. Nat. Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, Hersey P, Zhang XD, Wu M. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15840–15845. doi: 10.1073/pnas.1019312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrio M, Arderiu G, Myers C, Boudreau NJ. Cancer Res. 2005;65:7177–7185. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Teruya-Feldstein J, Weinberg RA. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 19.Bose D, Jayaraj G, Suryawanshi H, Agarwala P, Pore SK, Banerjee R, Maiti S. Angew. Chem. Int. Ed. Engl. 2012;51:1019–1023. doi: 10.1002/anie.201106455. [DOI] [PubMed] [Google Scholar]
- 20.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Angew. Chem. Int. Ed. Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young DD, Connelly CM, Grohmann C, Deiters A. J. Am. Chem. Soc. 2010;132:7976–7981. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 22.Childs-Disney JL, Hoskins J, Rzuczek S, Thornton C, Disney MD. ACS Chem. Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Childs-Disney JL, Parkesh R, Nakamori M, Thornton CA, Disney MD. ACS Chem. Biol. 2012;7:1984–1993. doi: 10.1021/cb3001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. J. Am. Chem. Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.