Abstract
Some bacteria under some circumstances swarm; they move rapidly and collectively over a surface. In an effort to understand the molecular signals controlling swarming, we isolated two strains from the same red seaweed – Vibrio alginolyticus B522, a vigorous swarmer, and Shewanella algae B516, which inhibits V. alginolyticus swarming in its vicinity. Plate assays combined with NMR, MS, and X-ray diffraction analyses identified a small molecule, which was named avaroferrin, as a potent swarming inhibitor. Avaroferrin, a previously unreported cyclic dihydroxamate siderophore, is a chimera of two well-known siderophores: putrebactin and bisucaberin. The sequenced genome of S. algae revealed avaroferrin’s biosynthetic gene cluster to be a mashup of putrebactin and bisucaberin biosynthetic genes. Avaroferrin blocks swarming through its ability to bind iron in a form that cannot be pirated by V. alginolyticus, thereby securing this essential resource for its producer.
Keywords: Natural products, siderophore, iron, evolution, motility
Bacteria, which were traditionally studied as single cells in a planktonic environment, are increasingly viewed as multicellular communities on surfaces, and this shift has generated observations, questions, and theories about how and why bacteria move on surfaces.[1] One frequently observed behavior, bacterial swarming motility, is an impressive form of community motility in which founding colonies begin to concentrically expand at rates of ~mm/h. Swarmer cells differ significantly from their non-swarming relatives in morphology and metabolism.[2] Swarmer cells can display increased tolerance for antibiotics[3] and increased virulence in infection models over non-swarming cells.[4–7] In order to gain insights into why bacteria swarm and possibly to discover therapeutic approaches to enhancing antibiotic sensitivity or suppressing virulence, we aimed to identify small molecule regulators of bacterial swarming. Here we report the discovery of a chimeric siderophore produced by Shewanella algae B516 that inhibits swarming of V. alginolyticus B522, annotate its biosynthetic pathway, and identify its likely mechanism of action.
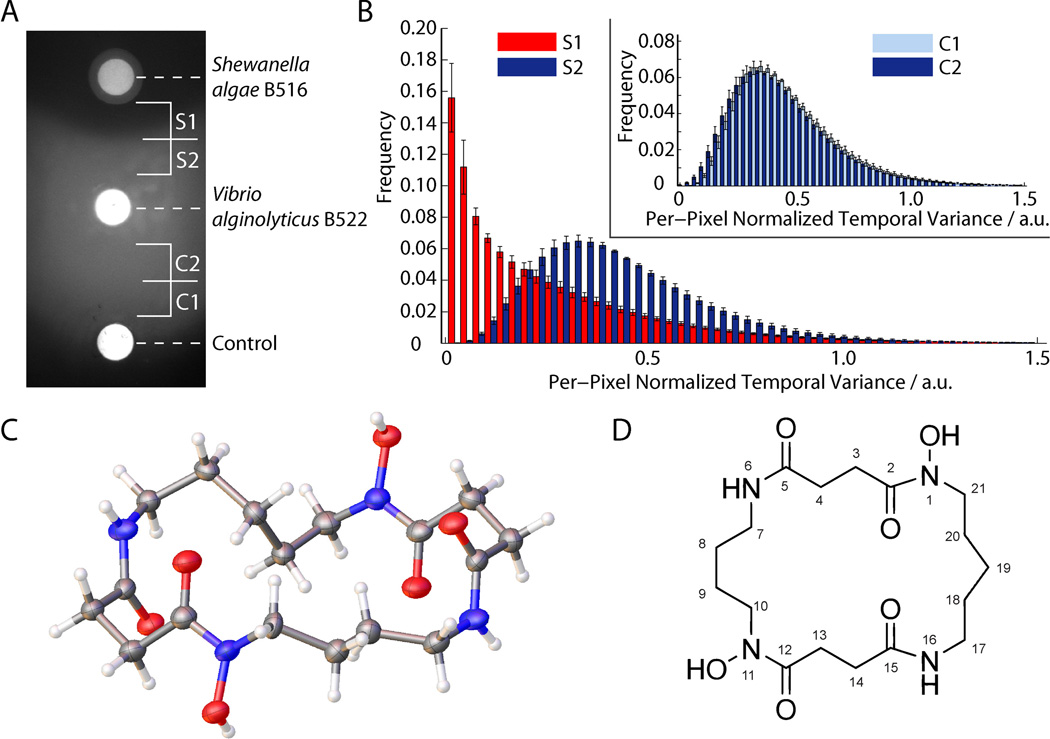
We recently developed an ecologically relevant swarming model based on Vibrio alginolyticus B522, which was isolated from an environmental sample of the carrageenan-producing red seaweed Chondrus crispus, and growth plates prepared with the sulfated polysaccharide κ-carrageenan substituting for agar. The result was a vigorously swarming strain advancing at ~5 mm/h. While screening additional isolates from the same red seaweed we discovered isolate B516, which was surrounded by a clear zone from the swarming V. alginolyticus B522 on carrageenan plates (Figure 1A). This isolate was identified as a Shewanella algae strain using phylogenetic analysis of 16S RNA and gyrB gene sequences (Figure S1).
Figure 1.
Shewanella algae B516 inhibits swarming motility of Vibrio alginolyticus by producing avaroferrin. A) Plate based swarming assay shows a zone of inhibition around the colony of S. algae. B) Cell motion activity given by per-pixel normalized temporal variance of time-lapse image series whereby high temporal variance means high motion activity and the frequency displays the fraction of pixels at a given motion activity. Bacteria in the thin cell layer in the inhibition zone S1 displayed highly reduced motility compared to zone S2 near the colony of V. alginolyticus. Inset: motion activities of the corresponding zones (C1 and C2) in the non-inhibited control (blank) show no distance dependence. Error bars give the standard error of the mean. C) Crystal structure of avaroferrin at 0.84 Å resolution and D) structural drawing of avaroferrin.
High-resolution differential interference contrast (DIC) microscopy showed that the clear zone was not due to toxicity as numerous viable but immobile V. alginolyticus cells could be observed in a thin layer within the zone. Using time-lapse DIC images, we demonstrated that cellular motility was strongly inhibited within the inhibition zone (S1) compared to outside of it (S2) or when compared to negative controls (C1 and C2) at the same distances (Figure 1B, Figure S2). With longer incubation times (several days) the zone eventually disappeared, which led us to conclude that V. alginolyticus 522 was simply being slowed down by S. algae. Our computational analysis of microscopic time-lapse image series confirmed the visual observations, and cells in the inhibition zone spent significantly less time moving (Figure S3). It seemed likely that a diffusible nontoxic swarming inhibitor caused these unusual observations.
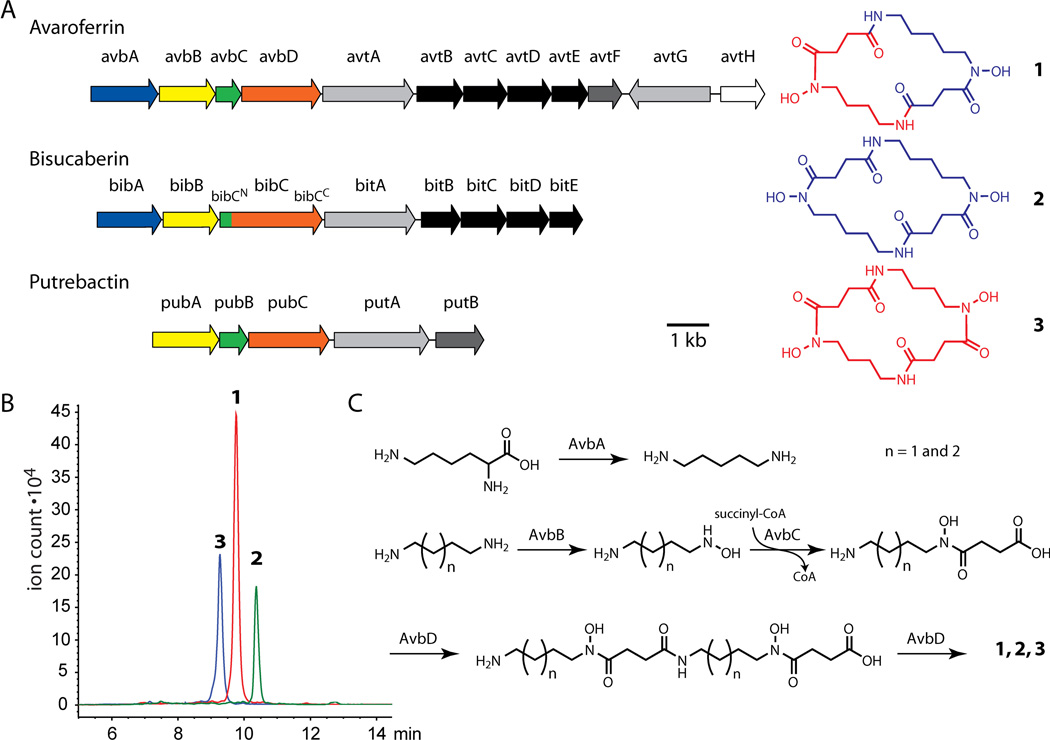
We cultured S. algae B516 on carrageenan plates and extracted the cultures with isopropanol. Fractionation over a C18 Sep-Pak cartridge resulted in an active fraction eluting with 30% methanol, which was purified to homogeneity by HPLC (Figure S4). High-resolution mass spectrometry indicated C17H30N4O4 as a molecular formula. NMR spectroscopic data led to structure 1, which was confirmed by X-ray crystallographic analysis (Figures 1C–1D, Figures S5–12, Table S1). The active compound is a previously unreported macrocyclic dihydroxamate siderophore that we named avaroferrin (1) from the Latin word (avarus) for greedy. Application of 50 nmol of pure avaroferrin and co-cultures with the S. algae strain on a carrageenan plate resulted in the same halo-forming swarming inhibitory activity against V. alginolyticus on the macroscopic scale and the same motility inhibition on the microscopic scale, thereby confirming avaroferrin’s role (Figure S13). Avaroferrin (AF) is the heterodimeric chimera of two homodimeric siderophores: putrebactin (PB)[8] and bisucaberin (BC).[9, 10] All three molecules are dimers of polyamines, the four-carbon cadaverine and the five-carbon putrescine, joined to succinic acid through a hydroxamate linkage (Figure 2). To investigate the biosynthesis of avaroferrin, we sequenced the genome of S. algae B516 and found only a single gene cluster with homology to the bisucaberin and putrebactin clusters, which have been described previously for other species (Figure 2A, Figure S14).[11, 12] This cluster, (avbA-D), had highest homologies with genes in the bisucaberin cluster bibA-C of Vibrio salmonicida[12] or genes in the putrebactin cluster pubA-C of Shewanella putrefaciens (Table S2).[11]
Figure 2.
Biosynthesis of avaroferrin. A) Annotated gene cluster for avaroferrin biosynthesis and transport in comparison to bisucaberin and putrebactin clusters of V. salmonicida and S. putrefaciens, respectively. Functional and sequence homologies of gene products are represented by the same color. Structures of all three compounds, which are aligned with their biosynthetic clusters, illustrate the chimeric nature of avaroferrin. B) HPLC traces with the respective [M+H]+ ions extracted show that all three siderophores are produced by S. algae B516 in 2:1:1 ratio. C) Proposed route for the promiscuous biosynthesis of avaroferrin (1), bisucaberin (2), and putrebactin (3) by the V. alginolyticus biosynthetic cluster.
The biosynthesis of putrebactin and bisucaberin requires putrescine or cadaverine, which are produced by decarboxylases operating on ornithine or lysine, respectively (Figure 2). One of the amines on the polyamine is converted to the hydroxylamine, which is then coupled to succinyl-CoA to form the corresponding hydroxamic acid. This unit is dimerized by coupling the free amine of one monomer to the free carboxyl of another monomer to form the linear precursor. The same enzyme forms the amide bond that macrocyclizes the linear precursor into the cyclic homodimer.[11, 12]
In avaroferrin, the biosynthetic cluster comprises avbA, a close homolog of the decarboxylase bibA for cadaverine biosynthesis in the bisucaberin pathway and avbB-D that most closely resemble pubA-C of in putrebactin biosynthesis. The enzymes encoded by avbC and avbD in the avaroferrin cluster correspond to the single fused two-domain protein BibC in the bisucaberin cluster. Eight trailing genes were identified with various transport related functions (avtA-H), and closer analysis also revealed their mixed parentage. While a series of ABC transporter (avtB-E) for siderophore export exhibited highest homologies to the bisucaberin exporters (bitB-E), the TonB-dependent siderophore receptors (avtA and avtG) and a reductase (avtF) shared homology with that of putrebactin cluster (putA and putB) in Shewanella. A fragment of a similar gene cluster has been reported from a metagenomic library of an unknown species from deep sea sediments, and that cluster cloned into E. coli produced bisucaberin.[13] This observation suggested that avaroferrin might not be the only siderophore produced by S. algae B516. Re-examination of the inactive fractions of the S. algae B516 extracts using LC-MS analysis showed that both homodimers, putrebactin and bisucaberin, were present (Figure 2A).
The three siderophores were found in the ratio AF:PB:BC of 2:1:1, which would be the expected outcome if the various steps in the pathway were indifferent to the number of methylene groups in the polyamine substrate (Figure 2B). This apparent lack of sensitivity might be expected for the virtually identical chemical transformations involved (Fig. 2C),[11, 12] especially in light of an earlier study showing that the AvbD homolog PubC catalyses both dimerization and cyclization.[11]
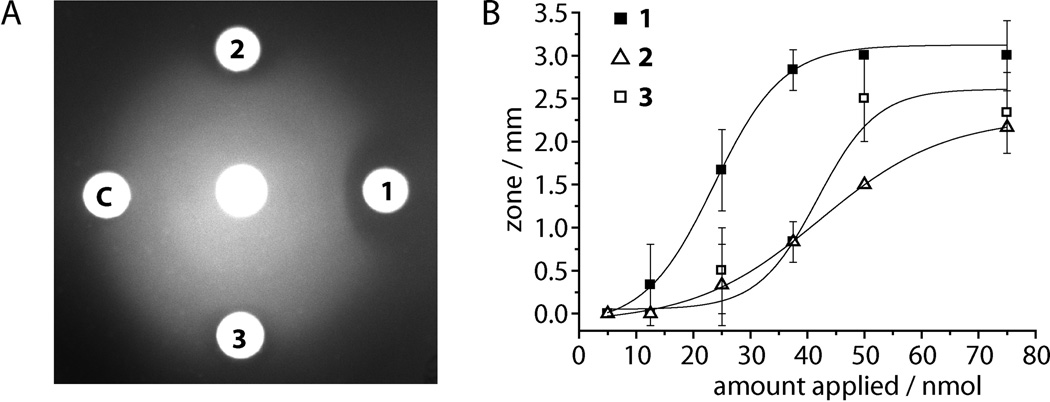
We also isolated and characterized putrebactin and bisucaberin (Figures S15–16) and compared their swarming inhibitory activity for Vibrio alginolyticus with that of avaroferrin. In addition to being produced in two-fold greater amount, avaroferrin is also roughly twice as active as bisucaberin and putrebactin in producing zones of inhibited motility, and the zones it produces persist longer (Figure 3A and B). Their relative lack of activity both explains why they were missed in the original bioactivity-guided fractionation and raises the issue of how such similar molecules could have such different anti-swarming activity.
Figure 3.
Avaroferrin (1) is more potent than bisucaberin (2) and putrebactin (3). A) After prolonged (30 h) incubation swarming of V. alginolyticus B522 from the central disc is still significantly inhibited by 1 while the zones of 2 and 3 have been largely overrun by swarming cells (75 nmol of each compound). C: DMSO control. B) Dose response curve for all three compounds with inhibition zone size as readout with half effective doses (ED50) of 24 nmol for avaroferrin and 41 nmol and 43 nmol for bisucaberin and putrebactin, respectively.
Neither putrebactin nor bisucaberin have been implicated in swarming before. Both however are well-known siderophores, the small diffusible molecules that many bacteria synthesize to scavenge iron, as its poorly soluble ferric form, from the environment. To distinguish whether avaroferrin inhibited swarming through its ability to act as a siderophore or through its ability to act as some sort of signal, we tested whether addition of ferric iron could antagonize its swarming inhibitory activity. Avaroferrin co-applied with excess of ferric iron no longer inhibited swarming. Ferric iron alone had no effect on swarming (Figure S17). This antagonism between added iron and avaroferrin suggested that either V. alginolyticus needed iron to swarm and that avaroferrin limited its ability to acquire it, or alternatively that avaroferrin in its iron bound form could not function as the putative signal.
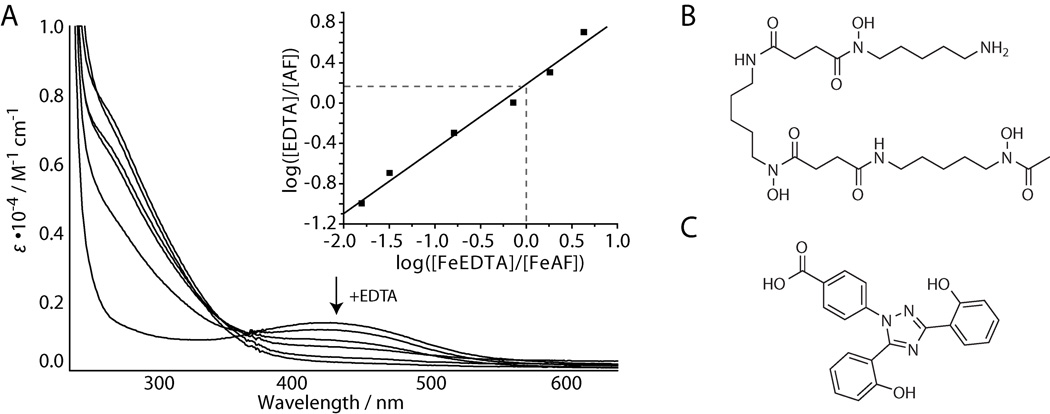
We compared the iron binding abilities of avaroferrin, bisucaberin, and putrebactin by measuring their pM(FeIII) values as described previously.[14] All three formed red iron complexes with λmax = 430 nm and had very similar pM(FeIII) values at pH 7.4 (23.5 for AF, 23.3 for BC and 22.8 for PB), that agree with previously published data (Figure 4A, Figure S18).[15] These small differences are unlikely to be responsible for avaroferrin’s unique activity, and we thus explored the potency of diverse iron chelators and siderophores. Desferoxamine, a bacterially produced structural relative of bisucaberin (Figure 4B) with an even higher pM(FeIII)-value, did not inhibit swarming at the highest dose (500 nmol) tested – a dose 100-fold higher than the minimum inhibitory dose of avaroferrin (Table S3). These findings aren’t completely unexpected as many previous reports on Vibrio have noted their ability to use siderophores that they do not produce, xenosiderophores, from other species including deferoxamine in V. vulnificus[16] and V. furnissii.[17] The siderophore ferrichrome and the natural iron chelators 2,3-dihydroxybenzoic acid and quercetin were either inactive in swarming inhibition or had only very low activity (Table S3, Figure S19). The only chelator with comparable potency in swarming inhibition was the synthetically optimized high efficiency iron chelator deferasirox, which had a MSID value (5 nmol) comparable to avaroferrin’s (Table S3, Figure 4C). The activities of siderophores and chelators did not correlate with their binding affinities for iron (Table S3), which argues that the ability of V. alginolyticus to use siderophore-bound iron, not the siderophore’s ability to bind iron determines swarming inhibitory activity. In addition, liquid culture studies showed that avaroferrin did not affect the viability of V. alginolyticus at concentration of up to 1 mM but reduced growth rates at concentrations higher than 100�µM, which indicates that avaroferrin is competing with the iron uptake mechanisms of V. alginolyticus (Figure S20). Genome analysis of V. alginolyticus B522 revealed at least 20 different proteins homologous to known siderophore receptors along with other iron-uptake and storage proteins, which attest to both its iron dependence and its ability to import iron via xenosiderophores (Table S4).
Figure 4.
Complex formation with ferric iron. A) Spectrophotometric titration of avaroferrin with EDTA at pH 7.4 with [Fe3+] = [AF] = 0.1 mM. AF: avaroferrin. Inset: plot for pM(FeIII) determination. The ΔpM to EDTA is given by the y intercept. B) Structure of deferoxamine and C) deferasirox.
Why bacteria swarm remains largely a mystery, and theories reflect familiar mass migration themes – escaping from a challenging environment and/or exploring a new and more favourable environment – and it is likely that swarming signals, like other chemical signals, are highly context dependent. Iron limitation induces swarming of V. parahaemolyticus[18] and other studies have shown that swarming coincides with the upregulation of iron metabolism, siderophore production, and expression of iron acquiring mechanisms in many bacteria.[6, 19, 20]
While iron is one of the most common elements in the Earth’s crust, it largely occurs in its minimally soluble ferric form. At neutral pH values, ferric ion solubility is ~10−18 M.[21] Bacterially produced siderophores can complex ferric ions and then return them to cells through specialized receptors. Most bacteria have receptors for siderophores that they do not produce which allow them to engage in iron (or siderophore) piracy, and bacteria are under selective pressure to continuously evolve new siderophores that thwart piracy. The ability to compete for ferric iron through varying siderophore repertoires might inform strategies for treating bacterial infections as was recently illustrated by the ability of probiotic strain Escherichia coli Nissle to outcompete pathogenic Salmonella through superior iron acquisition.[22]
This study illustrates how S. algae B516 produces avaroferrin to defeat piracy by V. alginolyticus B522 by developing a modest twist on a familiar structure. The discovery of avaroferrin also provides an important insight into the connection between iron and swarming motility. The biosynthetic cluster that creates avaroferrin also illustrates an important consequence of the modular pathways used to create so many bacterially produced small molecules: they facilitate the evolutionary recombination of modules to create new structures with new functions.[23]
Supplementary Material
Footnotes
We are grateful to Jennifer Waters and the Nikon Imaging Center at the Harvard Medical School for using the high resolution microscope and to Dr. Shao-Liang Zheng of the Center for Crystallographic Studies, Harvard University for solving the crystal structure of avaroferrin. Image analysis and software tool development was performed at the Image and Data Analysis Core (IDAC) at Harvard Medical School with the help of Hunter Elliott. This research was supported by a Leopoldina Research Fellowship (LPDS 2009-45) of the German Academy of Sciences Leopoldina (TB), NIH grant GM086258 (JC), and NERCE-BEID through 5U54 AI057159 (JC).
Supplementary Material
Refer to Web version on PubMed Central for supplementary material.
References
- 1.Kearns DB. Nat Rev Microbiol. 2010;8:634. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarter L, Silverman M. Mol Microbiol. 1990;4:1057. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 3.Butler MT, Wang Q, Harshey RM. Proc Natl Acad Sci U S A. 2010;107:3776. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison C, Lai HC, Hughes C. Mol Microbiol. 1992;6:1583. doi: 10.1111/j.1365-2958.1992.tb00883.x. [DOI] [PubMed] [Google Scholar]
- 5.Allison C, Emody L, Coleman N, Hughes C. J Infect Dis. 1994;169:1155. doi: 10.1093/infdis/169.5.1155. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Frye JG, McClelland M, Harshey RM. Mol Microbiol. 2004;52:169. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 7.Macfarlane S, Hopkins MJ, Macfarlane GT. Infect Immun. 2001;69:1120. doi: 10.1128/IAI.69.2.1120-1126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledyard KM, Butler A. J Biol Inorg Chem. 1997;2:93. [Google Scholar]
- 9.Takahashi A, Nakamura H, Kameyama T, Kurasawa S, Naganawa H, Okami Y, Takeuchi T, Umezawa H, Iitaka Y. J Antibiot (Tokyo) 1987;40:1671. doi: 10.7164/antibiotics.40.1671. [DOI] [PubMed] [Google Scholar]
- 10.Winkelmann G, Schmid DG, Nicholson G, Jung G, Colquhoun DJ. Biometals. 2002;15:153. doi: 10.1023/a:1015206419613. [DOI] [PubMed] [Google Scholar]
- 11.Kadi N, Arbache S, Song L, Oves-Costales D, Challis GL. J Am Chem Soc. 2008;130:10458. doi: 10.1021/ja8027263. [DOI] [PubMed] [Google Scholar]
- 12.Kadi N, Song L, Challis GL. Chem Commun (Camb) 2008:5119. doi: 10.1039/b813029a. [DOI] [PubMed] [Google Scholar]
- 13.Fujita MJ, Kimura N, Yokose H, Otsuka M. Mol Biosyst. 2012;8:482. doi: 10.1039/c1mb05431g. [DOI] [PubMed] [Google Scholar]
- 14.Abergel RJ, Zawadzka AM, Raymond KN. J Am Chem Soc. 2008;130:2124. doi: 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- 15.Hou Z, Raymond KN, O'Sullivan B, Esker TW, Nishio T. Inorg Chem. 1998;37:6630. doi: 10.1021/ic9810182. [DOI] [PubMed] [Google Scholar]
- 16.Kim CM, Park YJ, Shin SH. J Infect Dis. 2007;196:1537. doi: 10.1086/523108. [DOI] [PubMed] [Google Scholar]
- 17.Tanabe T, Funahashi T, Miyamoto K, Tsujibo H, Yamamoto S. Biol Pharm Bull. 2011;34:570. doi: 10.1248/bpb.34.570. [DOI] [PubMed] [Google Scholar]
- 18.McCarter L, Silverman M. J Bacteriol. 1989;171:731. doi: 10.1128/jb.171.2.731-736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. J Bacteriol. 2007;189:950. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogales J, Dominguez-Ferreras A, Amaya-Gomez CV, van Dillewijn P, Cuellar V, Sanjuan J, Olivares J, Soto MJ. BMC Genomics. 2010;11:157. doi: 10.1186/1471-2164-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neilands JB. J Biol Chem. 1995;270:26723. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 22.Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Cell Host Microbe. 2013;14:26. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischbach MA, Walsh CT, Clardy J. Proc Natl Acad Sci U S A. 2008;105:4601. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.