SUMMARY
The extent of lung regeneration following catastrophic damage and the potential role of adult stem cells in such a process remains obscure. Sublethal infection of mice with an H1N1 influenza virus related to that of the 1918 pandemic triggers massive airway damage followed by apparent regeneration. We show here that p63-expressing stem cells in the bronchiolar epithelium undergo rapid proliferation after infection and radiate to interbronchiolar regions of alveolar ablation. Once there, these cells assemble into discrete, Krt5+ pods and initiate expression of markers typical of alveoli. Gene expression profiles of these pods suggest that they are intermediates in the reconstitution of the alveolar-capillary network eradicated by viral infection. The dynamics of this p63-expressing stem cell in lung regeneration mirrors our parallel finding that defined pedigrees of human distal airway stem cells assemble alveoli-like structures in vitro and suggests new therapeutic avenues to acute and chronic airway disease.
INTRODUCTION
The 1918 “Spanish” influenza pandemic killed more than 600,000 people in the United States and an estimated 40 million individuals worldwide. Infections by this H1N1 influenza A strain is thought to induce acute respiratory distress syndrome (ARDS) marked by a rapid onset of pneumonia, diffuse alveolar damage and associated hypoxemia, and a massive elevation in inflammatory cytokines (Berthiaume et al., 1999; Matuschak and Lechner, 2010; Ramsey and Kumar, 2011). In recent analyses of influenza pandemics, death was often associated with bacterial co-infections, multiple organ failure, and widespread viral antigen expression in and damage to alveolar as well as to tracheal, bronchial, and bronchiolar epithelia (Lowy, 2003; Gill et al., 2010; Nakajima et al., 2011; Wu et al., 2011). While the terminal pathology of H1N1 influenza and other causes of ARDS is becoming clear, we know less about what role regenerative processes play in recovery from ARDS. Clearly ARDS patients show improved lung function six to twelve months out, but for some both pulmonary and extrapulmonary deficits remain in the longer term (Herridge et al., 2003). How much of the observed improvement in these patients is actually regeneration versus adaptive remodeling remains an area of intense study. Presumably regenerative processes in the airways involve local stem cell populations. Bronchioalveolar stem cells, or BASCs, which express both Clara cell markers (CC10) as well as alveolar type II (AT2) cell markers (SPC), have been described at terminal bronchioles and are proposed to be stem cells for both the bronchiolar as well as the alveolar epithelia (Giangreco et al., 2002; Kim et al., 2005). However, lineage tracing of Scgb1a1+ (CC10) Clara cells demonstrate their role as progenitors in the repair of terminal bronchiolar epithelium but not of the alveolar epithelium (Rawlins et al., 2009). In addition, BASCs lack precise cellular and molecular profiles and may consist of multiple stem cell types with different lineage commitment. For the upper airways, basal cells expressing the stratified epithelial stem cell transcription factor p63 (Yang et al., 1998; Yang et al., 1999; Senoo et al., 2007) have been implicated in regeneration of the tracheobronchial epithelium (Hong et al., 2004; Stripp and Reynolds, 2008; Rock et al., 2009; Giangreco et al., 2009; Rock et al., 2010; Cole et al., 2010).
Whether stem cells for alveolar epithelia also exist in mice and participate in lung regeneration following damage is unknown. Models of lung damage in mice have yet to provide clear evidence for the existence of alveolar regeneration mechanisms. The most common lung injury model involves exposure to bleomycin, which results in widespread bronchiolar and alveolar damage. However, the invariable consequence of bleomycin treatment is parenchymal fibrosis rather than de novo alveolar assembly (Moore and Hogaboam, 2008; Hoshino et al., 2009). The successful adaptation of highly pathogenic human influenza A viruses to mice offers potential insights into both infectious disease and more nuanced models for recovery from ARDS (Mori et al., 1995; Gubareva et al., 1998; Gao et al., 1999; Lu et al., 1999; Besler et al., 2009). For instance, sublethal doses of a murine-adapted H1N1 (PR8) influenza A induces widespread damage to both upper and lower airways marked by epithelial destruction and immune cell infiltrates between four and 14 days post infection (dpi). Remarkably, these mice show viral clearing by eight dpi and a histologically complete recovery of lung tissue over the next several months (Narasaraju et al., 2010). Understanding the extent and molecular sequence of alveolar regeneration and the role of progenitors and stem cells in this process will direct future efforts towards therapeutically enhancing lung regeneration.
In this work we examine the induction and recovery from an ARDS-like syndrome in mice infected with sublethal doses of a murine-adapted H1N1 influenza virus. We show that despite extensive damage to airway epithelial tissues, a p63-expressing population of cells in bronchioles undergoes a massive expansion and dispersion to sites of affected lung parenchyma. These migratory p63-expressing cells form discrete foci or “pods” that expand to a size and shape approximating those of alveoli and express genes linked to alveolar function. In parallel studies we clone three regiospecific stem cells from human airways demonstrate that one of these, the distal airway stem cell (DASC), has the unique potential of differentiating to alveolar lineages. We discuss the possibility that these p63-expressing cells participate in alveolar assembly processes modeled by human DASCs in vitro, and therefore represent key features in lung regeneration in response to ARDS.
RESULTS
H1N1 influenza infection of mouse airways
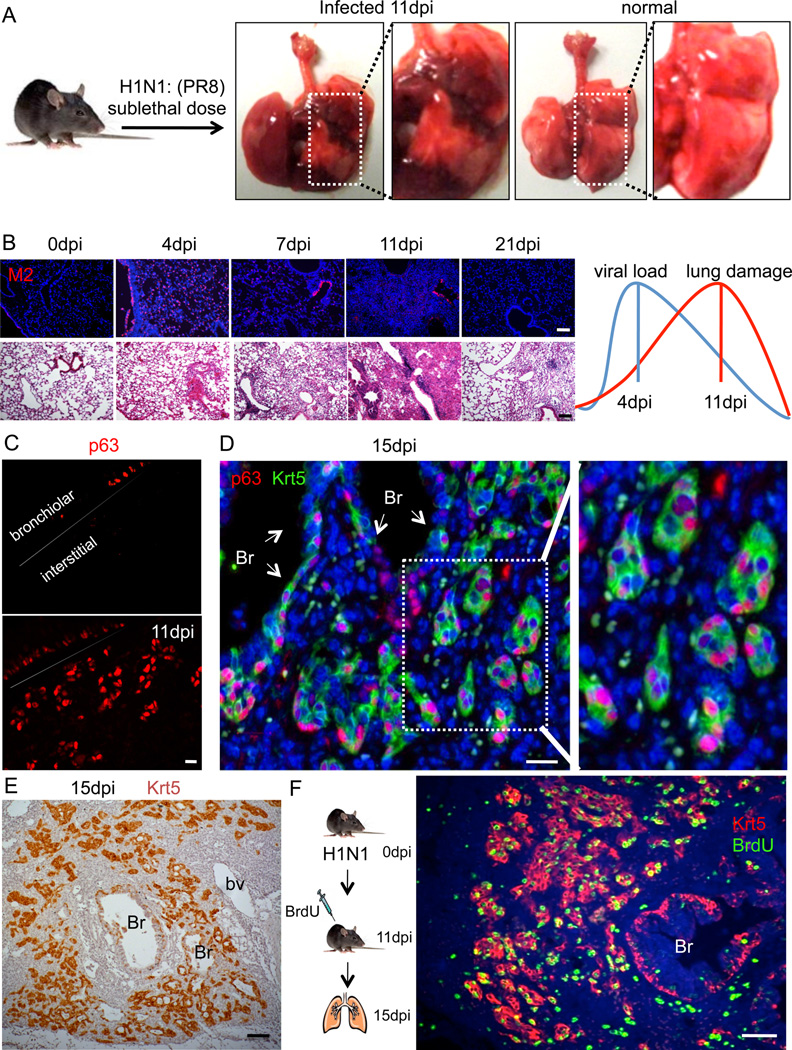
Mice were infected with a murine-adapted H1N1 (PR8) influenza A by intratracheal aspiration at viral titers of 125 to 1×105 PFU to determine an LD50 of approximately 500PFU. We chose 250PFU as a dose to induce significant damage without lethality in our experiments (Fig. 1A; Figure S1A,B). Lungs were harvested at multiple dpi and analyzed by histology and viral protein expression (Fig. 1B). Viral load was estimated by monitoring the expression of the M2 viral ion channel protein by immunofluorescence, which revealed maximal staining at 4dpi and a loss of M2 signal by 11dpi (Fig. 1B). However, tissue damage, as measured by the degree of immune cell infiltration, appeared to peak at 11dpi, was reduced at 21dpi, and was largely cleared across the lung by 60dpi (Fig. 1B). These results are mirrored by weight loss in these animals that reaches an extreme at 10–12dpi and recovers by day 20 (Figure S1C). At the cellular level, we observed widespread destruction of all airway epithelial cells at 7dpi resulting in a significant loss of markers for Clara cells (CC10) and ciliated cells (acetylated α-tubulin, TAp73) in the bronchiolar epithelia and AT2 cells (SPC+) of the alveolar epithelium (Figure S1D). Damage to airway epithelium is consistent with our observations of viral M2 expression in these cells at four days post-infection (Figure S1E). The peak of dense infiltrates of immune cells (CD45+) corresponds to the dense histological appearance of the lung at 11–14dpi (Figure S1F).
Fig. 1. Appearance of p63/Krt5-expressing cells in infected lung.
A. Infected and control lung at 11dpi. Pulmonary edema and hemorrhage are evident in the infected lung. B. Viral M2 protein expression in lung tissue at progressive dpi (top panel). Lung histology at corresponding dpi (bottom panel). Graphics depicts overall trend of viral load versus lung tissue damage. Scale bar, 200µm. C. Detection of p63-expressing cells in sections of lung at 0dpi (top) and at 11dpi. Scale bar, 20µm. D. Co-localization of p63 and Krt5 expression in “pods” in lung parenchyma at 15dpi. Br, bronchiole. Scale bar, 20µm. E. Localization of Krt5 (brown) at 15dpi. Br, bronchiole. Scale bar, 100µm. F. Detection of BrdU (green) and Krt5 (red) at 15dpi after 4 days of BrdU labelling. Br, bronchiole. Scale bar, 100µm.
The H1N1 influenza-infected mice show widespread cytopathic effects and extreme weight loss, and yet both these effects are mitigated between 21–60dpi. Remarkably, these mice recover without the acquisition of lung fibrosis that accompanies the induction of lung damage by bleomycin (Figure S1G), suggesting the possibility that epithelial regeneration underlies recovery from influenza.
Emergence of p63-expressing cells during influenza infection
Given the identification of p63-expressing basal cells as stem cells for nasal and tracheal epithelia of the upper airways (Rock et al., 2009), we asked whether p63-expressing cells might also participate in lung regeneration following influenza infection. We found little evidence of p63-expressing basal cells in the bronchioles of normal mice. However, by 7dpi, cells expressing p63 were evident in bronchioles (Figure S2A,B). By 11dpi, both p63 cells and Clara cells were found intermingled in the bronchiolar epithelium, and by 21 days most of the bronchiolar epithelium appeared restored at the level of Clara cells while those with p63-expression in the bronchioles were less evident. This rise and fall of p63 expressing cells is reflected in overall p63 protein expression in distal airways (Figure S2C). Unexpectedly, p63-expressing cells were also found in large numbers in the highly damaged lung parenchyma at 11dpi (Fig. 1C, Figure S2B). On closer inspection, these p63-expressing cells in the damage lung appeared to be clustered in small groups (Fig. 1D). Using other markers of basal cells, such as antibodies to keratin 5 (Krt5), it was evident that the p63-expressing cells formed discrete clusters or pods (hereafter “Krt5 pods”) in interstitial lung (Fig. 1D). On a gross level, Krt5 pods were distributed in a concentric pattern about bronchioles (Fig. 1E). BrdU labeling of proliferating cells at 11dpi revealed robust cell division of the Krt5+ cells in the bronchioles as well as in the Krt5 pods (Fig. 1F). Direct quantification of BrdU+/Krt5+ cells revealed a progressive decrease in intrabronchiolar regions from 11dpi and an increase in interbronchiolar regions from 11dpi (Figure S2D). The appearance of Krt5+ pods in the peribronchiolar regions of lung parenchyma coincides with the pinnacle of influenza-induced lung damage. Significantly, Krt5+ pods were not observed in the bleomycin-dependent lung fibrosis model (Figure S2E).
Molecular analysis of human clonogenic airway cells
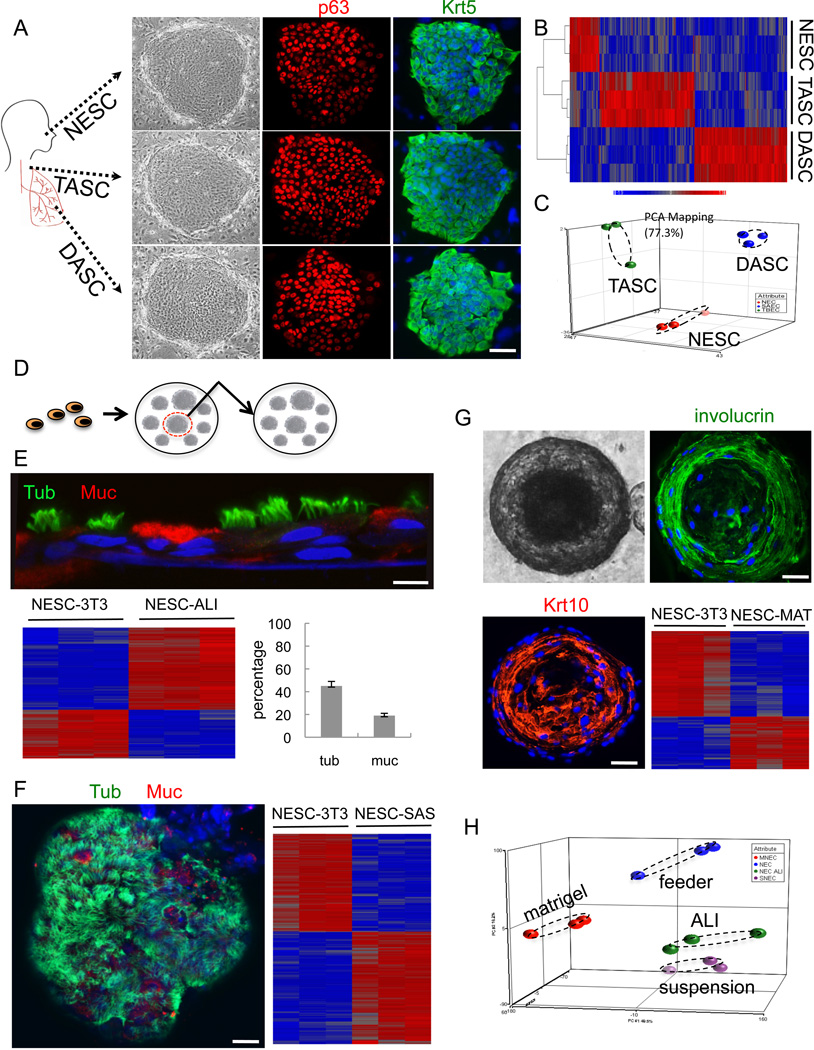
To identify distal airway stem cells and assess their relationship to stem cells of the upper airways, we employed single cell cloning methods on populations of human epithelial cells derived from nasal turbinate, tracheobronchial epithelia, and distal airway tissue including bronchioles and alveoli (Rheinwald and Green, 1975; Barrandon and Green, 1987; Senoo et al., 2007). We obtained immature colonies from approximately 1:500 to 1:2000 cells, and all of these stained uniformly for p63 and for keratin 5 (Krt5) (Fig. 2A). These immature clones were provisionally designated nasal epithelial stem cells (NESCs), tracheal airway stem cells (TASCs), and distal airway stem cells (DASCs). About 80 percent of these clones could be propagated further for at least an additional estimated 50 doublings while maintaining an immature phenotype (not shown). Despite the fact that the original starting cell populations were obtained from disparate regions of the airways, the immature stem cell clones appeared indistinguishable by morphology and staining with basal cell markers. Significantly, gene expression datasets from these clones are binned in regiospecific group by unsupervised clustering and by PCA of the whole genome expression patterns despite sharing gene expression of approximately 99 percent of the 17,500 hybridizing genes (Fig. 2B,C).
Fig. 2. Cloning and pedigree tracking of regiospecific stem cells from human lung.
A. Schematic of human airways as source of cells for stem cell cloning. NESCs, nasal epithelial cells; TASCs, tracheobronchial epithelial cells; DASCs, small airway epithelial cells. Left panel, Epithelial cell clones on irradiated Swiss 3T3 cells. Middle panel, p63 immunofluorescence. Right panel, Keratin 5 (Krt5) immunofluorescence. Scale bar, 50µm. B. Comparative heatmap of NESC, TASC and DASC expression profiles. C. Principle Component Analysis (PCA) of expression microarrays. D. Schematic showing pedigree tracking and expansion. E. Air-liquid interface (ALI) differentiation of NESCs indicated by antibodies to tubulin and mucin 5A to mark ciliated cells and goblet cells, respectively. Histogram of counts of ciliated cells (tub) and goblet cells (muc). Expression heatmap comparing NESCs grown on 3T3 feeder cells (NESC-3T3) with those differentiated in ALI culture. Scale bar, 20µm. F. Self-assembly sphere (SAS) assay of NESCs. Staining SASs with antibodies to tubulin (green) and Muc5A (red) revealing ciliated cells and goblet cells, respectively. Heatmap depicting differential gene expression of NESCs and those differentiated in SAS culture. Scale bar, 50µm. G. 21 day 3-D Matrigel cultures of NESCc (phase contrast, upper left panel) and confocal immunofluorescence using antibodies to involucrin and Krt10 (upper right panel and lower left panel). Scale bar, 50µm. Gene expression heatmap of NESC and those in Matrigel (lower right panel). H. PCA of whole genome expression microarrays of NESCs and those differentiated by indicated methods.
Pedigree tracking of airway stem cells
The long-term self-renewal potential of the putative human stem cell clones allowed us to isolate independent pedigrees for the analysis of the progeny of a single cell (Fig. 2D). Expansion of these lines yielded abundant immature cells of known pedigree for a range of differentiation assays to assess lineage potential. For instance, we analyzed pedigree lines of NESCs and posted these through multiple differentiation assays. The air-liquid interface model (ALI; Schmidt et al., 1996) has been a powerful tool in airway epithelial differentiation and supports the differentiation of goblet cell and ciliated cells from immature populations of nasal epithelial cells (Usui et al., 2000). Significantly, all of our pedigree-defined lines of NESCs showed similar distributions of goblet cells and ciliated cells in ALI cultures (Fig. 2E), supporting the concept the pedigree lines we have developed from single cells in fact have lineage potential ascribed to NESCs. Whole genome expression analysis of immature, pedigree-defined lines and ALI-differentiated cells support this notion and showed an increased expression of genes involved in ciliogenesis (e.g. DYNLRB2, 22.7×;TUBA4b, 9.8×; and DNAH6, 7.2×; all p<0.05) and in goblet cell function (e.g. MUC1, 3.8×; MUC13, 5.6×; and MUC20, 4.51×; all p<0.05) (Fig. 2E). We also performed self-assembling sphere (SAS; Jorissen et al., 1989) cultures of NESCs. SAS cultures, like the ALI cultures, induce both goblet cell and ciliated cell differentiation from NESCs, and involve the self-assembly of epithelial cells in conditions where cells cannot adhere to a solid support. NESC pedigrees readily assemble into spheres under such conditions within 24 hours, and efficiently undergo both goblet cell and ciliated cell differentiation over the following 15 days upon transfer to differentiation media (Fig. 2F). Unlike the ALI cultures, which stratify into basal cells and suprabasal differentiated cells, the SAS cultures show a monolayer of ciliated cells and goblet cells polarized to the exterior and lack basal cells altogether (not shown). Again, whole genome expression analysis supports the concept that SAS cultures promote ciliogenesis (e.g. increased expression of DYNLRB2, 68.4×; DNAH7, 28.6×; TEKT1, 25.1×; all p<0.05) and goblet cell formation (increased expression of MUC15, 6.7×; MUC20, 7.6×; SCGB2A1, 6.1×; all p<0.05) from our NESCs (Fig. 2F). In contrast to their similar differentiation programs in ALI and SAS cultures, NESCs behaved very differently in 3-D Matrigel cultures. NESCs showed robust formation of solid spheres in Matrigel between days 5 and 10, which subsequently hollow with the addition of differentiation media, and by day 21 show immature cells at the periphery and cells with squamous differentiation towards the lumen (Fig. 2G). This squamous metaplasia is supported by the comparison between gene expression of undifferentiated NESCs and those differentiated for 21 days in Matrigel, which show strong expression of squamous epithelial genes (e.g. LCE2B, 173.1×; KRT10, 6.0×; and SPRR2A, 4.5×; all p<0.05) (Fig. 2G). Similar development of a squamous metaplasia has been seen with the differentiation of nasal turbinate epithelial cell populations (Endres et al., 2005), suggesting that our pedigree-defined cells retain the capacity for this pathway of differentiation. Overall gene expression PCA indicates that each of these differentiation assays yields different outcomes marked by a stratified airway epithelium in ALI cultures, a non-stratified airway epithelium in SAS cultures, and a squamous metaplasia in 3-D Matrigel cultures (Fig. 2H).
DASC pedigrees assemble into alveolar-like structures in vitro
Defined pedigrees of TASCs and DASCs were grown in ALI cultures to compare their differentiation potential with that of NESC lines. Like the NESC lines, the TASC pedigrees showed robust differentiation into ciliated cells and mucin-producing goblet cells during the 21-day period of ALI culture (Fig. 3A). In contrast, the DASC pedigrees showed only minor indications of mucin expression, rare ciliated cell formation, and occasional CC10 expression indicative of Clara cells (Figure S3A). The TASC lines further differed from the DASC lines in their degree of stratification in ALI cultures. The TASCs presented a multilayered epithelium with p63-positive basal cells (not shown) underneath differentiated goblet and ciliated cells, while the DASC pedigrees retain a monolayer appearance over the differentiation period (Fig. 3B). Gene expression analysis of the ALI cultures of TASC and DASC lineages confirmed these observations, with the differentiated TASC cultures showing high expression of genes involved in ciliogenesis, mucin production, and epithelial stratification compared to DASCs grown in ALI culture (Fig. 3A).
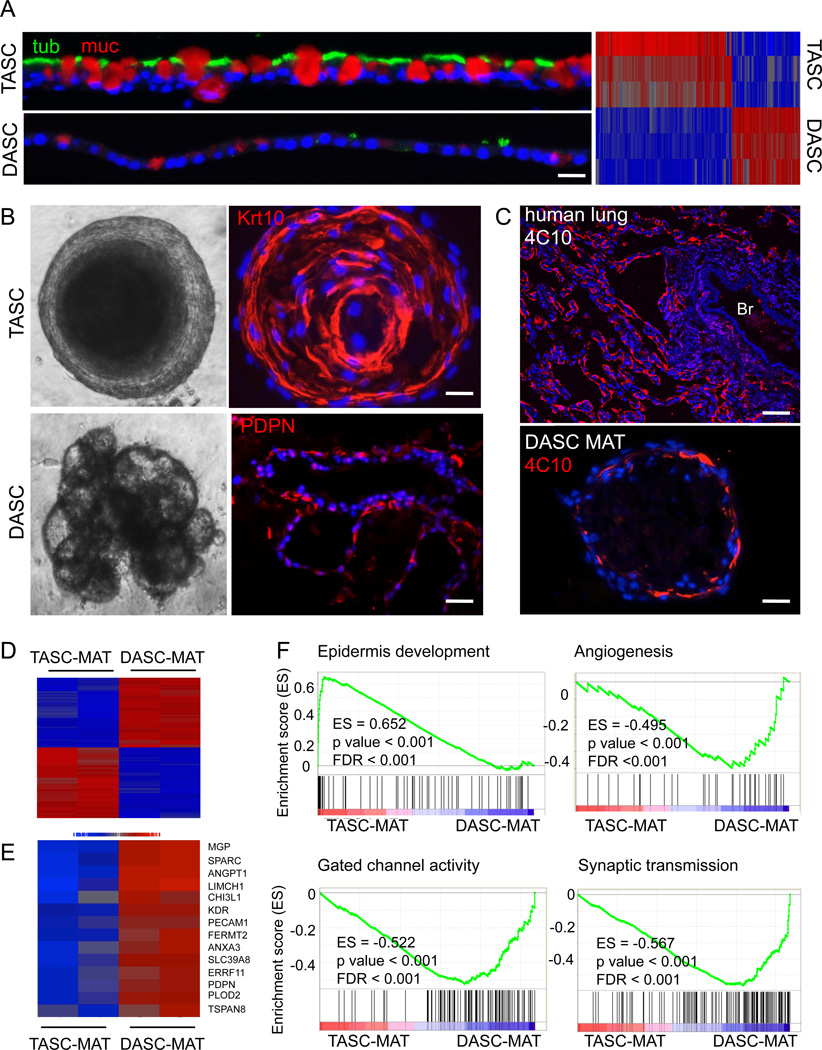
Fig. 3. Alveolar-like structures from DASCs in 3-D Matrigel cultures.
A. TASCs and DASCs following 21 days of ALI culture using antibodies to tubulin (green) and Muc5A (red) to reveal ciliated cells and goblet cells, respectively. Scale bar, 20µm. Heatmap of gene expression differences in DASC and TASC lines upon differentiation in ALI cultures. B. Comparison of structures formed by TASCs and DASCs following 21 days in 3-D Matrigel. Top panel, Phase contrast and confocal images of TASC spheres. Confocal immunoflourescence with antibodies to the squamous marker Krt10 (red). Scale bar, 20µm. Bottom panel, Multivesicular, unilaminar structures formed by DASCs following 21 days in Matrigel and confocal immunofluorescence image showing PDPN immunofluorescence. Scale bar, 50µm. C. Top panel, Immunofluorescence micrographs of staining patterns on human deep lung with anti-alveoli monoclonal antibody 4C10. Scale bar, 100µm. Bottom panel, Immunofluorescence micrographs of monoclonal antibody 4C10 on the structures developed from DASC lines in 21-day Matrigel cultures. Scale bar, 50µm. D. Heatmap of differentially expressed genes from microarrays of DASC and TASC lines differentiated in 3-D Matrigel. E. Heatmap of DASC genes that are both differentially expressed in Matrigel and expressed in interstitial regions of mouse lung. F. Gene Set Enrichment Analysis of datasets derived from TASC and DASC differentiation in 3-D Matrigel culture.
In 3-D Matrigel cultures, the TBEC lines, like the NESC pedigrees, underwent squamous metaplasia (Fig. 3B). Curiously, the DASC pedigree lines also assemble into spheres but ultimately these hollow and collapse into multispherical structures by day 21 (Fig. 3B). The hollowing effect appears to be the direct consequence of apoptosis as these cells within the spheres show robust activated caspase-3 staining (Figure S3B). The multispherical entities formed from DASC lines do not stratify but appear to be comprised of unilaminar cellular assemblies that express the alveolar type 1 marker PDPN (Fig. 3B). These same structures are labeled with the 4C10 monoclonal antibody that recognizes a ca. 300kDa protein specific to human alveoli (Fig. 3C).
Consistent with the divergent differentiation of the TASC and DASC pedigrees in Matrigel cultures, datasets of whole genome microarrays of the structures assembled in 3-D cultures by these lines showed large differences (Fig. 3D) including a host of genes implicated in alveolar structure or formation (Fig. 3E and Table S1). These findings were supported by wider Gene Set Enrichment Analysis (GSEA), which showed the squamous metaplasia of the TASC Matrigel structures expressed genes associated with epidermal development (Fig. 3F). In contrast, the DASC-derived structures showed high expression of genes associated with angiogenesis regulation, monocation transport (Matthay et al., 2002; Eaton et al., 2009), an important physiological function of AT1 cells, and synaptic transmission, a possible reflection of the importance of neurogenic control of alveolar function (Sakuma et al., 2006).
Assembly of alveoli-like structures conserved in rodent-derived DASC pedigrees
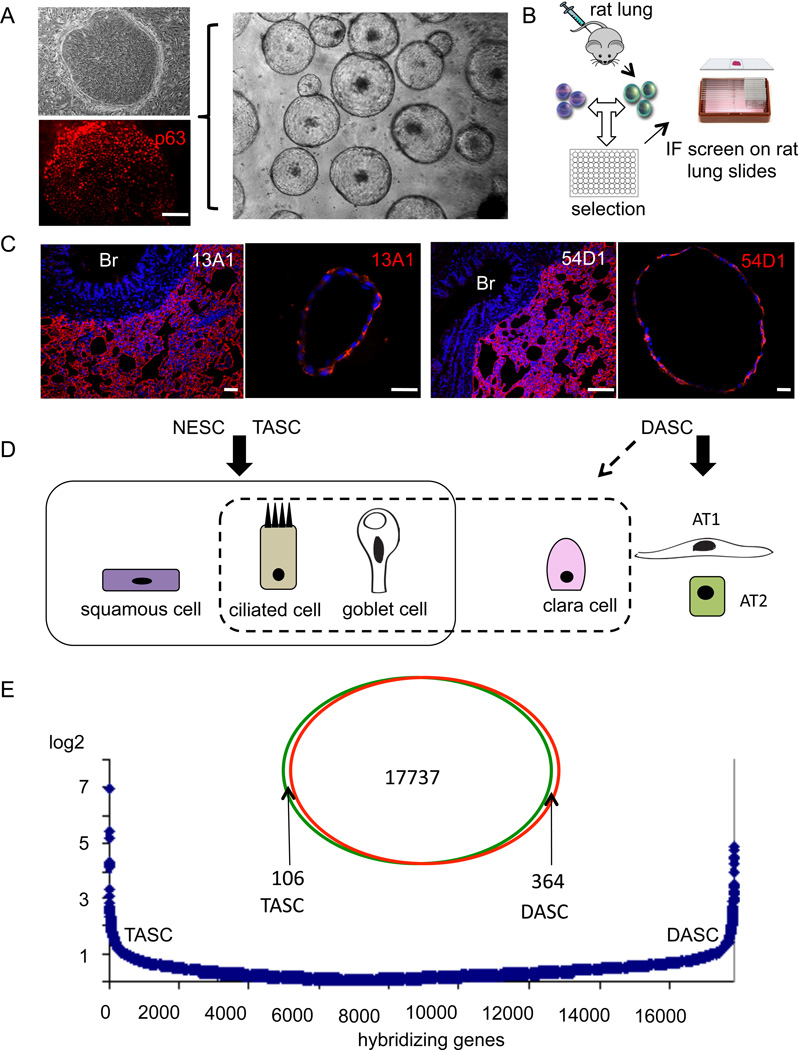
To test whether similar p63-expressing stem cells could be derived from the deep lung of non-human species, we obtained multiple immature clones from distal airways of six-week-old rats (Fig. 4A). Defined pedigrees were developed from several of these that were subsequently grown in 3-D Matrigel cultures. All of the rat pedigree-specific DASC lines formed uniform solid spheres after 10 days of growth that subsequently hollowed over the next 11 days (Fig. 4A). To determine whether these structures contain proteins linked to alveoli in vivo, we generated a panel of monoclonal antibodies from mice immunized to rat distal airway tissue (Fig. 4B,C). Monoclonal antibodies 13A1 and 54D1 are specific to rat alveoli and recognize proteins with molecular masses of 45 and 25kDa, respectively. These antibodies, which do not stain rat immature rat DASC clones, efficiently stain the unilaminar structures produced by the DASC pedigree lines (Fig. 4D). These data suggest that these alveolar-like structures express genes found in alveoli. Further, the DASCs we describe here are fully committed to alveolar lineages with additional potential to form Clara, ciliated, and mucin producing cells and therefore distinct from the TASCs and NESCs that are committed to ciliated cells, goblet cells, and as well can undergo squamous metaplasia (Fig. 4D). Despite these differences in lineage commitment, DASCs show only minor differences in gene expression compared with TASCs and NESCs in the range of 100–200 genes (Fig. 4E, Figure S4).
Fig. 4. Conservation of alveoli-like differentiation in rat DASC pedigrees.
A. Rat DASCs on 3T3 cells derived from single cell suspension of deep lung tissue. Top left, Phase contrast; Bottom left, Immunofluorescence with anti-p63 antibodies (red). Middle panel, Image of unilaminar structures produced by rat DASC pedigree-specific lines after 21 days in 3-D Matrigel culture. Scale bar, 50µm. B. Schematic showing immunization of mice with rat lung and screening for interstitial lung-specific monoclonal antibodies. C. Immunofluorescence on rat deep lung and unilaminar structures developed from rat DASC lines in Matrigel cultures with anti-alveoli monoclonal antibodies 13A1 and 54D1. Scale bar, 100µm. D. Schematic of the differentiation potential of regiospecific airway stem cells derived from multiple in vitro models. E. Venn diagram depicting differentially expressed genes between TASC and DASC stem cell pedigrees, while the graphics below indicate the absolute fold-change in gene expression between TASC and DASC among more than 17,000 informative genes.
Influenza-infected lung displays a massive increase in clonogenic basal cells
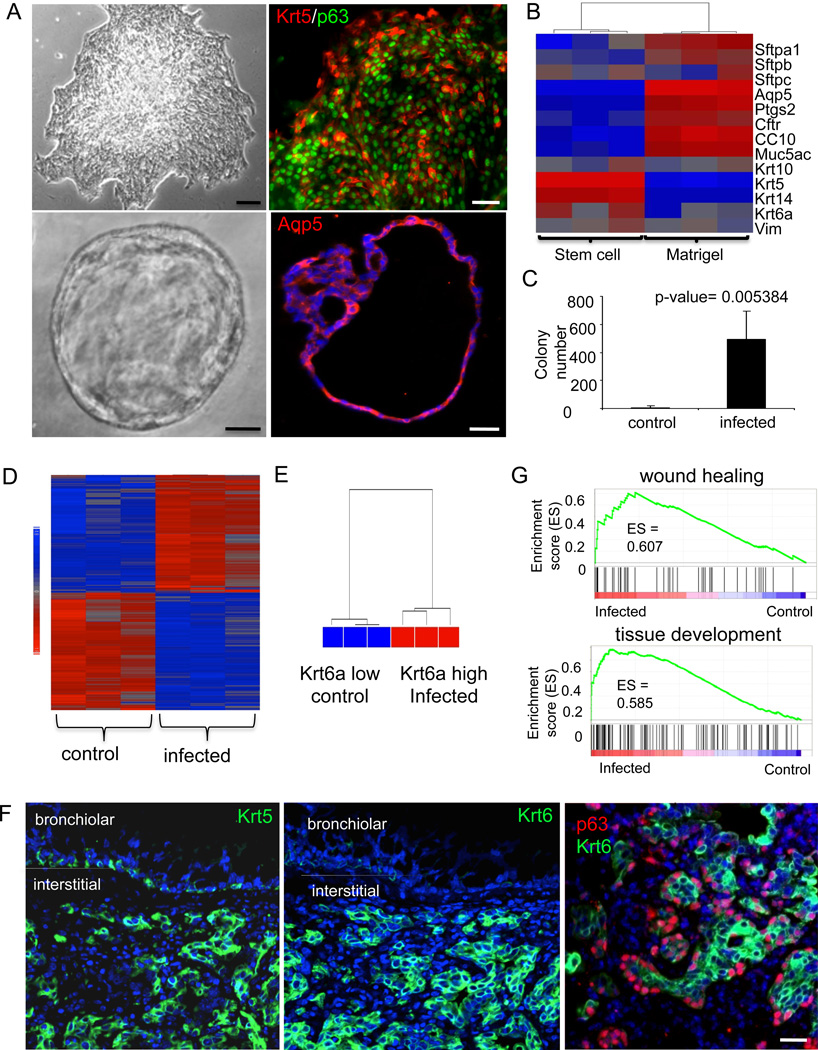
To probe the basal-like cells observed in infected lung, we plated dissociated distal airway tissues in clonogenic assays (Fig. 5A, top panel). The colonies that arose were uniform in appearance from the infected and control lungs and were composed of small, p63-expressing immature cells and expressed Krt5 (Fig. 5A, top panel). When grown Matrigel cultures, cells from these colonies from solid spheres that hollow through cell death to yield unilaminar structures similar to those generated by human DASCs (cf. Fig. 3B). These unilaminar structures stain for antibodies to aquaporin 5 (Aqp5), a marker of alveoli (Fig. 5A, lower panel). A comparison of expression performed on RNA amplified from individual clones and the Matrigel structures formed from them showed reproducible differences in gene expression (Fig. 5B). The immaturity markers Krt5 and Krt14 are lost in during differentiation in Matrigel, while markers of alveoli, such as Aqp5 and surfactant proteins Sftpa1, Sftpb, and Sftpc, are all upregulated in these structures. Consistent with the appearance of large numbers of p63-positive cells in lung parenchyma following H1N1 influenza infection (cf. Fig. 1), we observed a several hundred-fold increase in p63-expressing clonogenic cells in infected lungs (Fig. 5C).
Fig. 5. Expansion of basal-like cell population in lung following infection.
A. Upper left, Image of typical colony of basal-like cells derived from three-week-old mouse lung. Scale bar, 200µm. Upper right, Immunofluorescence image of typical colony stained with anti-Krt5 (green) and anti-p63 (red). Scale bar, 50µm. Lower left, Image of spheres generated by growth of cloned cells in 3-D Matrigel culture for 10 days showing hollow lumen. Scale bar, 50µm. Lower right, Section of 3-D sphere stained with anti-Aqp5 antibodies. Scale bar, 50µm. B. Heatmap showing relative gene expression pattern of selected genes in cloned stem cells and 3-D Matrigel spheres. C. Quantification of colony numbers obtained from control and infected mice at 12 dpi. Number of mice n=7. Error bars, SD of mean. D. Heatmap of differentially expressed genes (> two-fold) between three different colonies obtained from control and infected lungs. E. Unsupervised clustering analysis using whole transcriptome for control and infected colonies. F. Immunofluorescence micrographs showing distribution of Krt5 (left panel; green), Krt6 (middle panel; green), and p63 (red) and Krt6 (green) in sections of 12dpi lung (right panel). Bronchiolar and interstitial lung regions are marked. Scale bar, 20µm. G.GSEA survey data showing enrichment of wound healing gene expression in colonies derived from infected lung.
Whole genome microarray revealed reproducible differences in 358 genes among three clones from control and infected lungs (Fig. 5D). One of the genes proved to be keratin 6A (Krt6a), a known marker of migrating keratinocytes during wound healing in the epidermis (Wojcik et al., 2000). Expression of this gene alone differentiates between the clones derived from control lung and 12dpi lung (Fig. 5E). In vivo, Krt6 antibodies differentiate between basal-like cells in Krt5 pods of interstitial regions from basal cells of bronchioles whereas Krt5 antibodies recognize both these ectopic basal cells and those in the bronchiolar epithelium (Fig. 5F). GSEA of the expression data sets derived from the distal airway colonies revealed a bias for genes involved in wound healing, tissue development, and regulation of growth in those from infected mice (Fig. 5G and Figure S5).
Krt5 pods linked to lung regeneration?
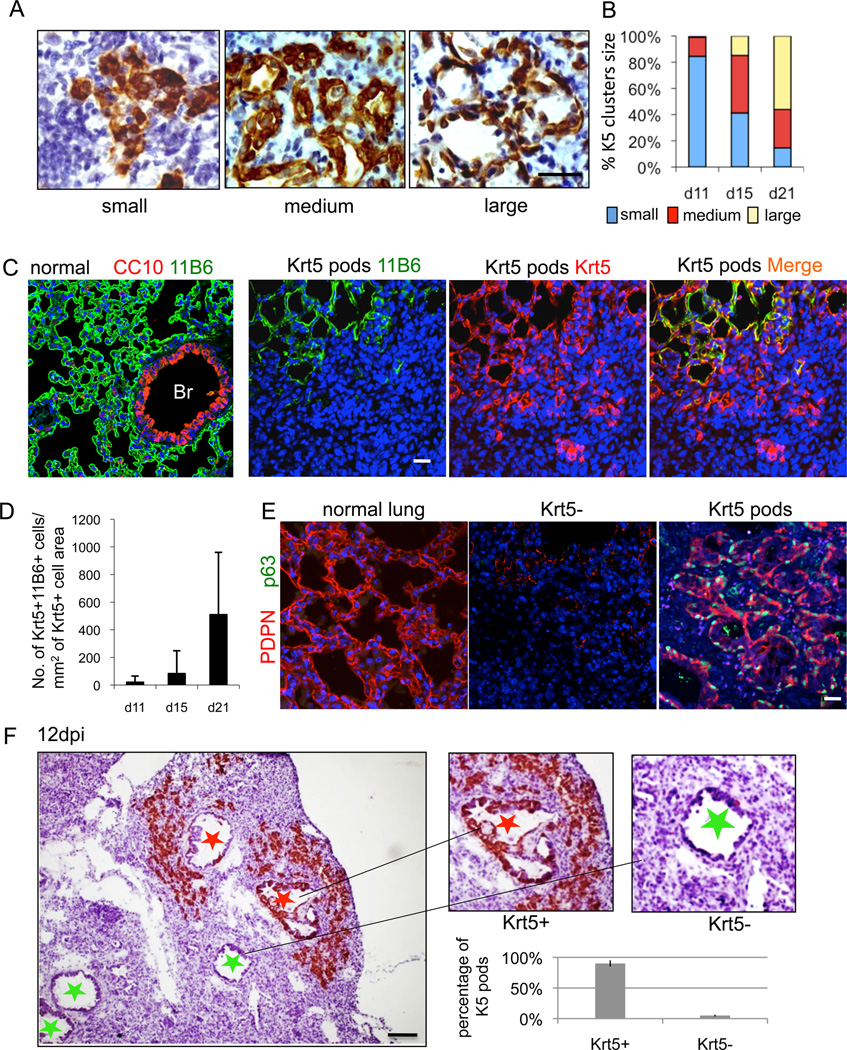
The ability of human distal airway stem cells to form alveoli-like structures in vitro made it tempting to predict that the Krt5 pods of basal-like cells seen in the influenza-damaged lung were components of a regenerative process. Consistent with this, the general appearance of individual Krt5 pods at 11dpi was one of tight clusters of cells, while at 15 and 21dpi many of these pods had lumens reminiscent of alveoli (Fig. 6A). Direct quantification of these Krt5 pod “types” over time confirmed this notion, suggesting a progression over 10 days from a tight group of Krt5+ cells to structures resembling alveoli (Fig. 6B). We asked whether the Krt5 pods would stain with a new monoclonal antibody we generated from mice immunized with human fetal lung designated here as “11B6”. 11B6 reacts specifically to alveolar regions of mouse lung but not to bronchioles (Fig. 6C, Figure S6A). Staining of regions of damaged lung containing Krt5 pods with the 11B6 monoclonal antibody revealed co-localization of the two antigens especially in those pods with larger diameters, suggesting a specificity for those Krt5 pods with larger lumens (Fig. 6C). Quantification of this link between 11B6 and Krt5+ pods also correlated with dpi (Fig. 6D). We also probed the Krt5 pods with antibodies to PDPN, which shows an alveoli-specific pattern in normal lung (Fig. 6E). PDPN antibodies also co-localize with Krt5 pods (Fig. 6E; Figure S6B).
Fig. 6. Dynamics of Krt5 pods in infected lung parenchyma.
A. Sections of infected lung showing variable appearance of Krt5 pods revealed by Krt5 immunohistochemistry. Scale bar, 20µm. B. Quantification of the Krt5 pod size categories at 11, 15, and 21dpi. Number of clusters (n) counted at 11, 15 and 21 dpi are 200, 424 and 572, respectively. C. Left panel, Staining of normal lung with the 11B6 monoclonal antibody showing alveolar-specific signal. Middle panels, Staining of Krt5 pods in damaged lung with the 11B6 monoclonal antibody (green) and Krt5 antibodies (red), respectively. Right panel, A merge of 11B6 and Krt5 antibody signals of the middle panels. D. Quantification of Krt5+11B6+ cells per Krt5+ cell area over indicated dpi. Number of mice n=4. Error bars indicate standard deviation of the mean. Scale bar, 50µm. E. Staining of normal lung with antibodies to PDPN (red) (left panel). Middle panel, Staining of region of damaged lung lacking Krt5 pods with antibodies to PDPN (red) and p63 (green) showing an absence of signal. Right panel, Co-localization of PDPN (red) and p63 (green) in region of damaged lung that has clusters of Krt5 pods. Scale bar, 50µm. F. Section of 12dpi lung showing clusters of Krt5 pods about bronchioles that possess Krt5-expressing cells (red star) and the absence of Krt5 pods surrounding bronchioles lacking Krt5-expressing cells (green star). Scale bar, 100µm. Histogram reports direct counts of peribronchiolar clusters of Krt5 pods adjacent to bronchioles with (93.8%) and without Krt5-expressing cells. Number of sections n=32. Error bars, SD of mean.
In assessing the influenza-infected lung from a panoramic view, we observed three distinct areas: normal appearing lung parenchyma with histologically normal alveoli, damaged regions marked by semi-dense infiltrate and Krt5 pods, and damaged regions marked by highly dense infiltrates but without Krt5 pods (Figure S6C). Krt5+ pods were never observed in regions of histologically normal lung tissue. The presence of Krt5 pods in less-densely infiltrated regions is that these regions are undergoing regenerative repair and clearing dense immune cell infiltrates in the process. Indeed the regions marked by Krt5+ pods show intermingled, CD45-positive immune cells in the immediate proximity (Figure S6D). We also noticed that within regions of dense infiltrate were bronchioles that lacked Krt5-positive basal cells, whereas bronchioles associated with satellite Krt5-positive pods also had Krt5-positive basal cells (Fig. 6F). While speculative, this observation might reflect the possibility that if the infection eradicates the bronchiolar epithelium, it may be incapable of spawning a peribronchiolar population of Krt5 pods.
Krt5 pods at sites of lung regeneration
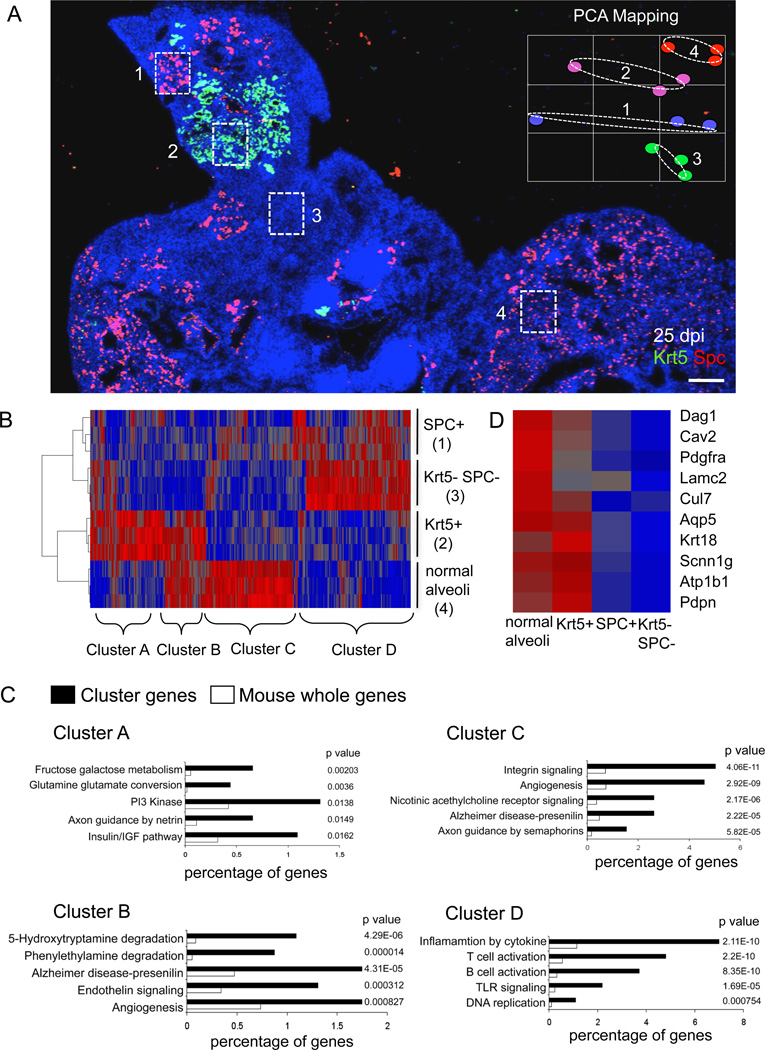
We used laser capture microdissection (LCM) to isolate RNA for expression microarray analysis from frozen sections of 25 dpi lung regions rich in Krt5 pods, normal or repaired lung, or regions of high-density infiltrates without obvious repair. The regions of high-density infiltrates without obvious alveoli were further divided into regions with and without SPC, a marker of alveolar type II cells (Fig. 7A). PCA of these datasets revealed that the regions rich in Krt5 pods were most closely related to the regions of normal or repaired lung (Fig. 7A, inset). This impression is reinforced by a heatmap of differentially expressed genes in these datasets which shows the similarity between the densely infiltrated regions devoid of alveolar markers and the densely infiltrated, SPC+ regions marked as Cluster D genes (Fig. 7B). Cluster D genes include strong representations by immune cell functions and innate immune signaling consistent with the persistence of a dense infiltration of immune cells (Fig. 7C; Table S2). In contrast, the regions with Krt5 pods show significant overlap with normal lung in a gene set designated Cluster B including genes involved with angiogenesis and endothelin signaling (Khimji and Rockey, 2010), as well as aromatic amine degradation known to be functionally linked to lung endothelial cells (Gillis and Pitt, 1982) (Fig. 7C, Table S2). Additionally, Cluster B shows an overrepresentation by genes involved in Wnt, Hedgehog, and nicotinic acetylcholine receptor signaling. Together, these data suggest that the Krt5 pods are in association with elements of endothelial cells involved in capillary formation. Moreover, Cluster B genes include an array of alveolar genes not expressed in regions of damaged lung lacking Krt5 pods (marked by the SPC+/Krt5− or SPC−/Krt5−) (Fig. 7D; Table S2), including PDPN, caveolin 2, Aqp5, and PDGFRα. Cluster C genes, those overrepresented only in regions non-damaged lung, are characterized by gene sets associated with integrin signaling, angiogenesis, nicotinic acetylcholine receptor signaling, and axon guidance by semaphorins (Fig. 7C; Table S2). Consistent with the link between Krt5 pods and lung regeneration, we see robust co-localization of Krt5 and the alveolar-specific monoclonal antibody 11B6, but no 11B6 staining of damaged regions regardless of whether they have SPC staining (Figure S7). On the broadest level, these data support the notion that regions with Krt5 pods express genes similar to apparently normal or repaired lung, and very different from regions marked by severe damage. They also suggest a dynamic process involving a host of pathways whose significance for the recovery from ARDS will require extensive empirical validation.
Fig. 7. Gene expression profiles of regionally distinct infected lung.
A. Section of 25dpi lung stained with antibodies to Krt5 (green), SPC (red), and counterstained with DAPI (blue). Four regions are demarcated with boxes as laser capture microdissection targets 1) SPC+ cells in densely infiltrated zones, 2) Krt5 pods, 3) SPC−/Krt5− zones with dense infiltrates, and 4) SPC+ cells in normal lung. Inset shows PCA of three independent LCM samples corresponding to regions 1–4. Scale bar, 200µm. B. Heatmap of 2205 differentially expressed genes with p value < 0.05 derived from LCM samples of regions 1–4. Gene clusters A–D are indicated. C. Gene Ontology analysis of gene clusters A–D indicated in heatmap together with associated p values. D. Heatmap indicating relative expression of individual genes linked to alveoli in the datasets corresponding to regions 1–4.
Lineage tracing of alveolar progenitors from bronchioles to alveoli
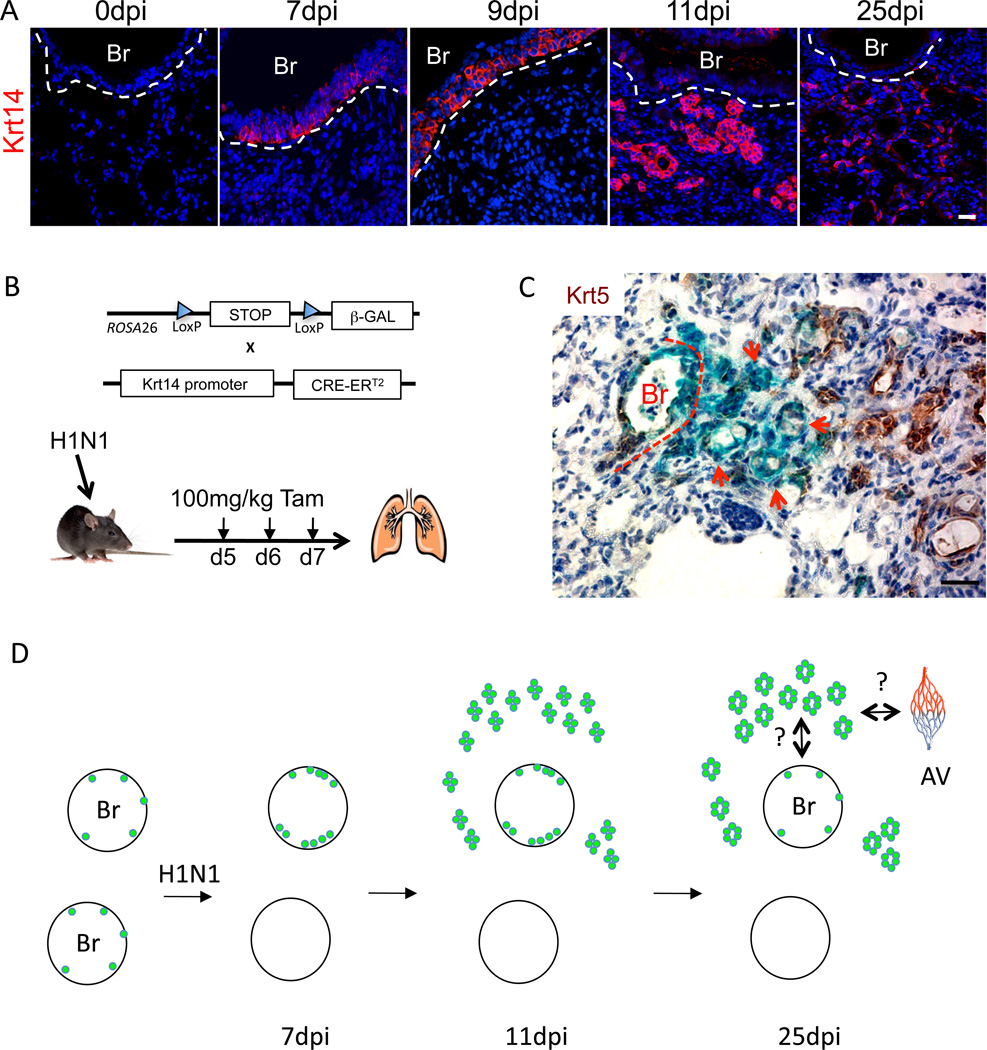
While clusters of Krt5+ pods always appear in a peribronchiolar pattern in influenza-infected lung about bronchioles with Krt5+ cells, their origins remained to be established. To test whether the Krt5 cells that appear in the bronchioles are indeed the origins of the parenchymal Krt5+ pods, we used standard lineage tracing methods based on Tamoxifen-dependent LacZ expression driven by Cre recombinase from the keratin 14 (Krt14) promoter (Mao et al., 1999; Vasioukhin et al., 1999). Krt14-postive cells are not present in bronchioles prior to infection but become evident at 4 days post-infection as the same cells expressing Krt5 and increase in numbers through day 9 while remaining restricted to the bronchioles (Fig. 8A). At or around 11dpi, there appears to be a concerted migration of these Ktr5/Krt14 positive cells to interstitial lung and the appearance of Krt5 pods (Fig. 8A; Figure S8). Treatment of these mice with daily injections of Tamoxifen at 5, 6, and 7dpi resulted in labeling of both the bronchioles and the Krt5+ pods at 25dpi (Fig. 8B,C). These data are consistent with the notion that the Krt5+ pods arise from cells that migrate from the bronchioles to local sites of interbronchiolar lung damage.
Fig. 8. Lineage tracing and schematic of response of Krt5 cells to influenza infection.
A. Localization of Krt14 expression in influenza-infected lung at indicated dpi. Br, bronchiole. Scale bar, 20µm. B. Schematic of alleles for conditional expression of β-galactosidase via Krt14-Cre recombinase activation. C. LacZ staining (blue) and Krt5 immunochemistry (brown) in 25dpi mouse lung. Scale bar, 20µm. D. Schematic depicting two bronchioles (Br) with rare Krt5-expressing cells at the basement membrane that proliferate in response to infection (top) or are killed by the infection (bottom). Early Krt5 pods are evident by 11dpi, and mature with luminal formation by day 25. Major questions for how they link to alveolar capillary (av) beds and to the bronchioles are indicated.
DISCUSSION
Recovery from lung damage that has advanced to ARDS is highly variable and poorly understood at present. Critical questions remain to understand the differential fates of ARDS patients, the potential of lung regeneration versus fibrosis, and whether therapies can sway clinical outcomes. In this work we addressed the recovery from ARDS in mice infected by H1N1 influenza. The histological evidence suggests a largely complete lung restoration several months following severe influenza infection (this study; Narasaraju et al., 2009). In fact we show that, unlike bleomycin-induced ARDS, which invariably leads to fibrosis without evidence of regeneration (Moore and Hogaboam, 2008; Hoshino et al., 2009), mice recovering from influenza infections lack detectable lung fibrosis even following viral doses approaching the LD50. These data suggest that considerable regeneration of lung tissue, including complex alveolar-capillary networks, must be acting in this recovery. Indeed for the conducting, upper airways of mice, such as nasal passages, trachea, and bronchi, there are abundant data for regeneration after severe damage involving p63-expressing basal cells (Stripp and Reynolds, 2008; Rock et al., 2010). Despite this progress in understanding stem cells of the conducting airways, definitive evidence for a stem cell that can contribute to lung regeneration has been more elusive. Bronchioalveolar stem cells, or BASCs (Kim et al., 2005), have been a useful model for such stem cells but have not been cloned nor characterized beyond a limited marker set. More recently, a c-Kit-positive stem cell from the human airways expressing many markers of embryonic stem (ES) cells have been described to give rise to both epithelial and endothelial components of alveolar capillary complexes in xenograft experiments (Kajstura et al., 2011). The stem cells we describe are fundamentally different from BASCs or the c-Kit, ES-like stem cells presented earlier and first drew our attention as massive numbers of p63-expressing cells in the damaged lung parenchyma at the height of influenza-induced damage. p63-expressing cells are not found in normal interstitial lung, and rarely even detected in normal bronchioles. However, p63 cells increase dramatically in bronchioles in the first several days of influenza infection, and appear in nearby damaged interstitial lung at 11 days post infection where they continue proliferation and assemble into pods. Remarkably, the number of clonogenic p63-expressing cells in the distal airways increases several hundred-fold within seven days of influenza infection, and these cells assume aspects of gene expression patterns seen in p63-expressing stem cells in the epidermis during wound repair. Pods containing these cells are almost always found in a radial pattern about a bronchiole that also has p63-expressing cells, and not about bronchioles that lack p63 cells. One interpretation of these data is that bronchioles are the source of these cells, a concept supported by our lineage tracing experiments with the Krt14-Cre/Rosa26-stop-LacZ mice.
Our efforts to clone and characterize three regiospecific stem cells of the airways has provided a foundation for understanding the nature of the p63-expressing cells that comprise the Krt5 pods following influenza infection. The role of p63-expressing basal cells as stem cells for distal lung was largely discounted because they proved to be rare in the small bronchioles compared with the upper airways. Thus we were surprised with the ease by which we could generate immature clones of p63-expressing cells from populations of human distal airway epithelial cells. Despite the nearly 99% overlap in gene expression between DASCs and the upper airway TASCs and NESCs, DASCs displayed commitment to a unique program of differentiation that includes alveolar epithelium.
What is the evidence that these Krt5 pods are intermediates in alveolar regeneration of influenza-damaged lung, and not a pathogenic pathway akin to bronchiectasis? Part of the argument is a kinetic one, as 11dpi Krt5 pods are solid spheres of cells that develop lumen and expand in size over the next 10 days to form alveoli-like structures. This hollowing and unilaminar appearance is strikingly similar in timing and appearance to the alveoli-like structures formed by human DASCs in 3-D cultures. These data were further supported by the co-staining of the Krt5 pods with antibodies directed to antigens found exclusively in alveoli such as PDPN and the target of the 11B6 monoclonal antibody. Neither of these antibodies reacts with regions of influenza-damaged lung that lack Krt5 pods.
Perhaps the most intriguing evidence favoring a role for Krt5 pods in alveolar regeneration was the direct comparison of gene expression profiles of discrete regions of lung at 25dpi. Four regions are evident in these lungs: 1) normal lung marked by ordered alveolar networks, 2) highly infiltrated zones without alveolar markers whatsoever, 3) zones of immune cell infiltration and disordered SPC staining, and 4) clusters of Krt5 pods showing intermediate infiltration. The expression microarray data revealed that the high-density regions (SPC+ or SPC−) showed similar patterns of gene expression marked by inflammation, innate immune functions, and B and T cell profiles. From these data, which are consistent with the high level of infiltration of both regions and the very low expression of alveolar markers such as PDPN or the 11B6 monoclonal antibody, it is likely that both regions are damaged and not undergoing active repair. Regions marked by clusters of Krt5 pods, in contrast, had gene expression profiles that showed significant overlap with those of normal regions of interstitial lung. Within the set of genes overlapping between the clusters of Krt5 pods and the normal lung were genes common to AT1 cells. Interestingly, Gene Ontology analysis showed that these overlapping genes contained genes involved in angiogenesis, endothelin signaling, and aromatic amine degradation, all processes or functions attributed to lung endothelial cells and the formation of new blood vessels. Thus regions occupied by Krt5 pods are likely associated with, in addition to AT1 cell markers, the initiation of new blood vessels. It is tempting to speculate that such angiogenesis is related to the need to bring capillary beds into direct contact with regenerating alveolar structures.
The study presented here leaves key questions unresolved. What, for instance, are the signals that trigger the proliferation and radiation of p63-expressing cells from bronchioles to sites of influenza-damaged lung? How do incipient alveolar structures assembled in damaged lung link up to the conducting airway? What are the mechanisms that merge microvasculature assemblies with the nascent alveoli? Why is SPC expression, a marker of AT2 cells, absent from the alveolar-like structures formed by the Krt5 pods? Answers to these questions, as well as those concerning the dynamics of alveolar assembly, will be essential for determining the kinetics and regulation of lung regeneration. The work presented here focuses on alveoli formation in what is likely to be the simplest aspect of lung regeneration (Fig. 8D). How these incipient structures link up to the conducting airways as well as the pleural capillary network represents an immense challenge but one that is likely to generate new therapies for an array of presently incurable airway diseases.
EXPERIMENTAL PROCEDURES
Animal Models
C57/Bl6 adult mice were infected with a sublethal dose of Influenza H1N1 A/PR/8/34 mouse adapted virus by intratracheal inhalation and lungs were harvested at various time points post infection. For BrdU incorporation assays 30 mg/kg BrdU in sterile PBS was administered IP and mice were sacrificed at different time points post injection. All procedures were conducted under IACUC guidelines and approved protocols. For bleomycin experiments, mice were treated with 6U/kg bleomycin by intratracheal instillation and were sacrificed at different time points post treatment to harvest lungs.
Histology and Immunofluorescence
Mice were sacrificed and lungs were inflated and fixed with 4% formaldehyde prior to paraffin embedding. Antibodies included influenza virus A M2 protein (Abcam), p63 (4A4 clone), alveolar markers (13A1, 54D1, 4C10,11B6 clone), Krt5 (Neomarkers, Lifespan Biosciences), Muc5Ac, CC10, SPC, Pdpn, Aqp5, Cd45 (Santa Cruz), BrdU (Accurate Chemical), p73 (3A6 clone), Ivl, Krt14, Krt10, Krt6 (Covance), pan-Keratin, SMA and acetylated alpha tubulin (Sigma). Appropriate Alexa flour 488 or 594 conjugated secondary antibodies (Invitrogen) were used for IF and Vector Labs ABC kit with DAB substrate (Vector laboratories) were used for IHC. Murine monoclonal antibodies were generated to human 22 wk fetal lung tissue under IRB approval using standard methods (Kohler and Milstein, 1975).
Laser Capture Microdissection
Mice were euthanized at various time points post infection and lungs were harvested. To preserve RNA quality and tissue morphology the tissues were quickly embedded in OCT® (TissuTek™) in a cryomold, frozen on dry ice, and stored at −80 until further use. 10 µm sections of the tissue were cut using a cryotome and sections were mounted on poly-L-lysine-coated glass foiled polyethylene naphthalate (PEN) slides for LCM (Leica Microsystems, Bannock Burn, IL). Sections were stained for several markers and consecutive sections were stained using HistoGene® LCM Frozen section staining kit (Arcturus) staining solution. The PALM® Robot Microbeam laser microdissection system (P.A.L.M. GmbH, Bernried, Germany) in combination with a Zeiss microscope was used to dissect out desired cells. Isolated cells were collected in adhesive caps (Carl Zeiss, Inc.) and RNA was extracted using the Pico Pure RNA extraction kit (Arcturus).
Microarray and Bioinformatics
RNAs obtained from LCM and colonies were amplified using the WT Pico RNA Amplification System, WT-Ovation Exon Module and Encore Biotin Module (NuGEN Technologies) and hybridized onto GeneChip® Mouse Exon 1.0 ST Array. GeneChip operating software was used to process all the Cel files and calculate probe intensity values. To validate sample quality probe hybridization ratios were calculated using Affymetrix Expression Console software. The intensity values were log2-transformed and imported into the Partek Genomics Suite 6.5(beta). Exons were summarized to genes and a 1-way ANOVA was performed to identify differentially expressed genes. P values and fold-change were calculated for each analysis. Heatmaps were generated using Pearson’s correlation and Ward’s method and Principal Component Analysis was conducted using all probe sets. Pathway analyses were performed using Gene Set Enrichment Analysis (GSEA) software and PANTHER database (Subramanian et al., 2005; Yuan et al., 2009).
Clonogenic Assays
Control and infected mouse lungs were harvested at 12dpi, and the trachea and lungs were separated and placed on ice. The tissues were washed in cold wash buffer (DMEM:F12, 1:1, 100µg/ml pen strep, 5% fetal bovine serum). The lungs were then cut into small pieces and digested with dissociation buffer (DMEM:F12,1:1, 100µg/ml pen strep, 5%fetal bovine serum, DNase I, 0.1% protease XIV (Sigma)) overnight at 4°C with gentle rocking. Cells were resuspended and passed through a 40µm nylon cell strainer (BD Falcon). The dissociated cells from each lung were plated on irradiated J2-3T3 feeder layers in 10cm plates (BD Falcon) and cultured in cFAD medium at 37°C with 5% CO2.
Lung Epithelial Cell Cloning and In Vitro Differentiation
Mouse lung cells were cultured on irradiated 3T3 feeder layers in cFAD medium (Rheinwald and Green, 1975). The cells were then transferred to growth factor-reduced Matrigel (BD Biosciences) and cultured in growth medium (DMEM/F12, 1:1, 1% Pen-Strep, 10µg/ml insulin, 5µg/ml transferrin, 0.1µg/ml cholera toxin, 25ng/ml EGF, 30µg/ml BPE, 5%FBS, 50ng/ml FGF10 and 30ng/ml HGF for four days after which differentiation was induced until day 12 with differentiation medium (DMEM/F12, 1:1, 1%penstrep, 5µg/ml insulin, 5µg/ml transferrin, 0.025µg/ml cholera toxin, 5ng/ml EGF, 30µg/ml BPE, 1mg/ml BSA, 50ng/ml FGF10 and 30ng/ml HGF). The cells were then frozen in OCT for cryosectioning and staining or dissolved in Trizol for standard RNA isolation, cDNA conversion, and Syber green qPCR.
Cloning and Differentiating Human and Rat Airway Stem Cells
Please see Supplemental Methods.
HIGHLIGHTS.
Lung regeneration after H1N1 influenza infection involves p63-expressing DASCs
Pedigrees of cloned DASCs are intrinsically committed to form alveoli in vitro
DASCs radiate to damaged lung to form “Krt5 pods” that mature to incipient alveoli
Expression profile of Krt5+ pods reflects dynamic intermediates in lung regeneration.
Supplementary Material
ACKNOWLEDGEMENTS
This work is dedicated to Howard Green. We thank Thomas Lufkin and Petra Kraus for their advice and help with mouse models, Jianzhu Chen and his laboratory as well at the staff of the ABSL2 laboratory at National University of Singapore for their assistance with the viral model, and Susan and Keith Rogers for help with histology. We thank the staff at the Nikon Imaging Facility at the Harvard Medical School, and Graham Daniel Wright of the Institute of Medical Biology imaging facility. We thank all the members in the Xian-McKeon laboratory for helpful discussions and support, and Edison Liu and Barbara Knowles for their critical review of the manuscript. This work was supported by the NIH (RC1 HL100767, RO1-GM083348, and R21CA124688), the Defense Agency Research Projects Agency (DARPA; Project N66001-09-1-2121), the Singapore-MIT Alliance for Research and Technology, the European Research Council, Agence de Nationale, the Institute of Medical Biology and the Genome Institute of Singapore of the Agency for Science, Technology and Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. U.S.A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Szretter KJ, Katz JM, Tumpey TM. Use of animal models to understand the pandemic potential of highly pathogenic avian influenza viruses. Adv. Virus Res. 2009;73:55–97. doi: 10.1016/S0065-3527(09)73002-7. [DOI] [PubMed] [Google Scholar]
- Berthiaume Y, Lesur O, Dagenais A. Treatment of adult respiratory distress syndrome: plea for rescue therapy of the alveolar epithelium. Thorax. 1999;54:150–160. doi: 10.1136/thx.54.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, Reynolds SD. Tracheal basal cells: a facultative progenitor cell pool. Am. J. Pathol. 2010;177:362–376. doi: 10.2353/ajpath.2010.090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DC, Helms MN, Koval M, Bao HF, Jain L. The contribution of epithelial sodium channels to alveolar function in health and disease. Annu Rev Physiol. 2009;71:403–423. doi: 10.1146/annurev.physiol.010908.163250. [DOI] [PubMed] [Google Scholar]
- Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J. Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Reynolds SD, Stripp BR. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am. J. Pathol. 2002;161:173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, Stripp BR. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc. Natl. Acad. Sci. USA. 2009;106:9286–9291. doi: 10.1073/pnas.0900668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JR, Sheng ZM, Ely SF, Guinee DG, Beasley MB, Suh J, Deshpande C, Mollura DJ, Morens DM, Bray M, Travis WD, Taubenberger JK. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch. Pathol. Lab. Med. 2010;134:235–243. doi: 10.5858/134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis CN, Pitt BR. The fate of circulating amines within the pulmonary circulation. Annu. Rev. Physiol. 1982;44:269–281. doi: 10.1146/annurev.ph.44.030182.001413. [DOI] [PubMed] [Google Scholar]
- Green H. The birth of therapy with cultured cells. Bioessays. 2008;30:897–903. doi: 10.1002/bies.20797. [DOI] [PubMed] [Google Scholar]
- Gubareva LV, McCullers JA, Bethell RC, Webster RG. Characterization of influenza A/HongKong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J. Infect. Dis. 1998;178:1592–1596. doi: 10.1086/314515. [DOI] [PubMed] [Google Scholar]
- Herridge MS, Cheung AM, Tansey CM, Matte-Martyn A, Diaz-Granados N, Al-Saidi F, Cooper AB, Guest CB, Mazer CD, Mehta S, Stewart TE, Barr A, Cook D, Slutsky AS Canadian Critical Care Trials Group. One-year outcomes in survivors of the acute respiratory distress syndrome. N. Engl. J. Med. 2003;348:683–693. doi: 10.1056/NEJMoa022450. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: Evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol. 2004;286:L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- Hoshino T, Okamoto M, Sakazaki Y, Kato S, Young HA, Aizawa H. Role of proinflammatory cytokines IL-18 and IL-1beta in bleomycin-induced lung injury in humans and mice. Am. J. Respir. Cell. Mol. Biol. 2009;41:661–670. doi: 10.1165/rcmb.2008-0182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Rota M, Hall SR, Hosoda T, D'Amario D, Sanada F, Zheng H, Ogórek B, Rondon-Clavo C, Ferreira-Martins J, Matsuda A, Arranto C, Goichberg P, Giordano G, Haley KJ, Bardelli S, Rayatzadeh H, Liu X, Quaini F, Liao R, Leri A, Perrella MA, Loscalzo J, Anversa P. Evidence for human lung stem cells. N Engl J Med. 2011;364:1795–1806. doi: 10.1056/NEJMoa1101324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Khimji AK, Rockey DC. Endothelin- biology and disease. Cell Signal. 2010;22:1615–1625. doi: 10.1016/j.cellsig.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Lowy RJ. Influenza virus induction of apoptosis by intrinsic and extrinsic mechanisms. Int Rev Immunol. 2003;22:425–449. doi: 10.1080/08830180305216. [DOI] [PubMed] [Google Scholar]
- Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay MA, Folkesson HG, Clerici C. Lung epithelial fluid transport and the resolution of pulmonary edema. Physiol Rev. 2002;82:569–600. doi: 10.1152/physrev.00003.2002. [DOI] [PubMed] [Google Scholar]
- Mendelsohn MG, Dilorenzo TP, Abramson AL, Steinberg BM. Retinoic acid regulates, in vitro, the two normal pathways of differentiation of human laryngeal keratinocytes. In Vitro Cell Dev Biol. 1991;27A:137–141. doi: 10.1007/BF02630999. [DOI] [PubMed] [Google Scholar]
- Matuschak GM, Lechner AJ. Acute lung injury and the acute respiratory distress syndrome: pathophysiology and treatment. Mo. Med. 2010;107:252–258. [PMC free article] [PubMed] [Google Scholar]
- Moore BB, Hogaboam CM. Murine models of pulmonary fibrosis. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2008;294:L152–L160. doi: 10.1152/ajplung.00313.2007. [DOI] [PubMed] [Google Scholar]
- Mori I, Komatsu T, Takeuchi K, Nakakuki K, Sudo M, Kimura Y. Viremia induced by influenza virus. Microb. Pathog. 1995;19:237–244. doi: 10.1016/s0882-4010(95)90290-2. [DOI] [PubMed] [Google Scholar]
- Nakajima N, Sato Y, Katano H, Hasegawa H, Kumasaka T, Hata S, Tanaka S, Amano T, Kasai T, Chong JM, Iiduka T, Nakazato I, Hino Y, Hamamatsu A, Horiguchi H, Tanaka T, Hasagawa A, Kanaya Y, Oku R, Oya T, Sata T. Histopathological and immunohistochemical findings of 20 autopsy cases with 2009 H1N1 virus infection. Mod Pathol. 2011;2011 doi: 10.1038/modpathol.2011.125. (ahead of print). [DOI] [PubMed] [Google Scholar]
- Narasaraju T, Ng HH, Phoon MC, Chow VT. MCP-1 antibody treatment enhances damage and impedes repair of the alveolar epithelium in influenza pneumonitis. Am. J. Respir. Cell. Mol. Biol. 2010;42:732–743. doi: 10.1165/rcmb.2008-0423OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey C, Kumar A. H1N1: viral pneumonia as a cause of acute respiratory distress syndrome. Curr. Opin. Crit. Care. 2011;17:64–71. doi: 10.1097/MCC.0b013e3283427259. [DOI] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model Mech. 2010;3:545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Gu X, Wang Z, Maeda S, Sugita M, Sagawa M, Osanai K, Toga H, Ware LB, Folkesson G, Matthey MM. Stimulation of alveolar epithelial fluid clearance in human lungs by exogenous epinephrine. Crit Care Med. 2006;34:676–681. doi: 10.1097/01.CCM.0000201403.70636.0F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Hubsch U, Wurzer H, Heppt W, Aufderheide M. Development of an in vitro human nasal epithelial (HNE) cell model. Toxicol Lett. 1996;88:75–79. doi: 10.1016/0378-4274(96)03720-4. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 is essential for the proliferative potential of stem cells of stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Stripp BR, Reynolds SD. Maintenance and repair of the bronchiolar epithelium. Proc. Am. Thorac. Soc. 2008;5:328–333. doi: 10.1513/pats.200711-167DR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad, Sci. USA. 2005;43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui S, Shimizu T, Kishioka C, Fujita K, Sakakura Y. Secretory cell differentiation and mucus secretion in cultures of human nasal epithelial cells: use of a monoclonal antibody to study human nasal mucin. Ann Otol Rhinol Laryngol. 2000;109:271–277. doi: 10.1177/000348940010900307. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Metcalf JP, Wu W. Innate immune response to influenza virus. Curr Opin Infect Dis. 2011;24:235–240. doi: 10.1097/QCO.0b013e328344c0e3. [DOI] [PubMed] [Google Scholar]
- Wojcik SM, Bundman DS, Roop DR. Delayed wound healing in keratin 6a knockout mice. Mol. Cell. Biol. 2000;20:5248–5255. doi: 10.1128/mcb.20.14.5248-5255.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillette E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Y Loh YH, Yaw LP, Robson P, Lim B, Ng H-H. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.