Abstract
Cyclase-associated proteins (CAPs) are highly conserved actin monomer binding proteins present in all eukaryotes. However, the mechanism by which CAPs contribute to actin dynamics has been elusive. In mammals, the situation is further complicated by the presence of two CAP isoforms whose differences have not been characterized. Here, we show that CAP1 is widely expressed in mouse nonmuscle cells, whereas CAP2 is the predominant isoform in developing striated muscles. In cultured NIH3T3 and B16F1 cells, CAP1 is a highly abundant protein that colocalizes with cofilin-1 to dynamic regions of the cortical actin cytoskeleton. Analysis of CAP1 knockdown cells demonstrated that this protein promotes rapid actin filament depolymerization and is important for cell morphology, migration, and endocytosis. Interestingly, depletion of CAP1 leads to an accumulation of cofilin-1 into abnormal cytoplasmic aggregates and to similar cytoskeletal defects to those seen in cofilin-1 knockdown cells, demonstrating that CAP1 is required for proper subcellular localization and function of ADF/cofilin. Together, these data provide the first direct in vivo evidence that CAP promotes rapid actin dynamics in conjunction with ADF/cofilin and is required for several central cellular processes in mammals.
INTRODUCTION
The actin cytoskeleton consists of highly dynamic array of filaments, whose assembly and rapid turnover are essential for a large number of motile and morphogenetic processes in eukaryotic cells. The precise form, dynamics, localization, and mechanical properties of the actin cytoskeleton are regulated by an array of actin binding proteins whose activities are controlled by various signaling pathways. One family of proteins central to regulating actin dynamics are the cyclase-associated proteins (CAPs), which are conserved actin monomer binding proteins (mol. wt. = 50–60 kDa) found in all eukaryotic organisms studied (Hubberstey and Mottillo, 2002). CAP (also called Srv2 in yeast) was first identified from budding yeast as a suppressor of the activated ras allele (Fedor-Chaiken et al., 1990) and as factor associated with adenylyl cyclase (Field et al., 1990). Subsequent studies demonstrated that CAPs interact with adenylyl cyclase complex through their N-terminal domain, whereas the C-terminal half of CAPs binds directly to monomeric actin (Gerst et al., 1991; Kawamukai et al., 1992; Freeman et al., 1995; Yu et al., 1999).
Consistent with a role in regulating actin dynamics, the loss of CAP results in cytoskeletal defects in budding yeast, Dictyostelium and Drosophila (reviewed in Hubberstey and Mottillo, 2002). In yeast cells, CAP/Srv2 localizes to the cortical actin patches through an interaction with the actin filament binding protein Abp1 (Lila and Drubin, 1997; Balcer et al., 2003). Deletion of CAP/Srv2 in yeast results in an abnormally large cell size, random budding pattern, and abnormal actin distribution. These defects can be partially suppressed by overexpression of actin monomer binding protein profilin (Gerst et al., 1991; Vojtek et al., 1991). Dictyostelium cells defective in CAP showed enlarged cell size, defects in F-actin organization, fluid phase endocytosis, and directed cell migration (Noegel et al., 1999; Noegel et al., 2003). In Drosophila, the loss of CAP results in various developmental defects and problems in maintaining oocyte polarity. These phenotypes seem to arise from an accumulation of F-actin in CAP-deficient cells (Baum et al., 2000; Benlali et al., 2000; Baum and Perrimon, 2001). The role of CAP in mammalian cells has not been demonstrated and is complicated by the presence of two distinct isoforms, CAP1 and CAP2, whose differences have not been determined (Yu et al., 1994).
Although genetic studies demonstrate that CAPs are involved in regulation of the actin cytoskeleton, an understanding of the mechanism by which CAPs contribute to actin dynamics has evolved slowly. Biochemical studies with purified porcine and Dictyostelium CAPs showed that these proteins bind actin monomers with a 1:1 stoichiometry and suppress the spontaneous polymerization of actin (Gieselmann and Mann, 1992; Gottwald et al., 1996). The role of CAP as actin monomer sequestering protein is further supported by genetic studies on Drosophila suggesting that CAP is required to prevent actin filament polymerization in the eye disk and oocytes (Baum et al., 2000; Benlali et al., 2000). However, recent biochemical studies on yeast and human CAP/Srv2 proteins revealed that their activities are more complex than this. CAP/Srv2 was shown to displace ADF/ cofilins from ADP-actin monomers, recycling ADF/cofilin for new rounds of filament depolymerization and recycling actin monomers to replenish the assembly-competent pool of actin (Moriyama and Yahara, 2002; Balcer et al., 2003).
To elucidate the role of CAP in actin dynamics in cells and to define its importance in actin-dependent cellular processes, we carried out an analysis of mouse CAPs. We found that CAP1 is expressed in most nonmuscle cell types of developing and adult mice, whereas CAP2 expression is mainly restricted to striated muscles and certain brain regions. CAP1 is a highly abundant protein that localizes to dynamic actin structures in mouse nonmuscle cells. Analysis of mouse CAP1 knockdown cells demonstrated that this protein contributes to rapid actin dynamics by promoting cofilin-induced actin filament depolymerization and plays an important role in cell morphogenesis, motility, and receptor-mediated endocytosis.
MATERIALS AND METHODS
Plasmid Construction
The DNA fragments corresponding to residues 1–663 of mouse CAP1 open reading frame, and residues 1–667 and 805-1428 of CAP2 open reading frame were amplified from mouse 17-d embryo cDNA (BD Biosciences Clontech, Palo Alto, CA) or adult mouse brain cDNA. The oligonucleotides created 5′BamHI and 3′HindIII, and 5′BamHI and 3′Xho-sites to CAP1 and CAP2 polymerase chain reaction products, respectively. These fragments were digested and ligated into pBluescript II KS vector to create plasmids pPL157, pPL158, and pPL159. For expressing proteins in Escherichia coli, CAP1 (nucleotides 1–663) and CAP2 (nucleotides 1–667) fragments were subcloned to pGAT2 and pHAT2 expression vectors, respectively (Peränen et al., 1996) to create plasmids pPL155 and pPL218. The accession numbers for the sequences of mouse CAP1 and CAP2 are PIR:I49572 and SPTREMBLNEW:AAH50752, respectively.
Northern Blot Analysis
The [32P]CTP-labeled mouse CAP1 and CAP2 cDNA probes were prepared from plasmids pPL157, pPL158 and pPL159 by using random primed DNA labeling kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. All probes were hybridized to commercial mouse multiple tissue and mouse embryo multiple tissue Northern blot filters (BD Biosciences Clontech) according to manufacturer's instructions. The Northern blot filters were exposed on a PhosphorImager screen for 15 h.
Radioactive In Situ Hybridization
The 35S-labeled antisense probes of ∼700 base pairs were obtained by linearizing the plasmids pPL157 and pPL158 with BamHI and transcribing with T3 RNA polymerase. Plasmid pPL157 was also linearized with XhoI and transcribed with T7 RNA polymerase to create a 35S-labeled control sense probe. In situ hybridizations from paraffin-embedded mouse embryos or adult mouse tissue sections were performed as described previously (Rice et al., 2000).
Proteins
Mouse CAP1 (amino-acids 1–221) was expressed in E. coli BL21 cells as a glutathione S-transferase (GST) fusion protein by using the plasmid pPL155. Cells were grown in Luria broth medium to an optical density of 0.6 at 600 nm, and expression was induced by 0.4 mM isopropyl-β-d-thiogalactosidase. Cells were harvested, washed once with buffer A (20 mM Tris, pH 7.5, 1% Triton X-100, 3 μM phenylmethylsulfonyl fluoride), and lysed by sonication. Cell lysate was centrifuged (all centrifugations are 30 min, 10,000 rpm with a SS-34 rotor if not stated otherwise). Supernatant was further clarified by ultracentrifugation of 30 min, 30,000 rpm, with a T647.5 rotor. The soluble fraction was loaded onto a chelating-Sepharose Ni2+ column (Pharmacia, Peapack, NJ), and the column was washed with 300 ml of buffer B (10 mM Tris, pH 7.5, 50 mM NaCl, 20 mM imidazole). Proteins were eluted with a linear 100-ml gradient from buffer B to C (10 mM Tris, pH 7.5, 50 mM NaCl, 250 mM imidazole), and the fractions containing the CAP1 fragment were pooled and concentrated to 100 μl with concentrator devices (Vivascience, Stonehouse, United Kingdom). The sample was then dissolved in 50 ml of buffer D (50 mM Tris, pH 7.5), concentrated again to 500- to 1000-μl volume, and stored at –70°C. Mouse CAP2 (amino acids 1–227) was expressed as a his-tagged fusion protein from plasmid pPL218 as described above for GST-CAP1. However, because this fragment was insoluble in E. coli, the protein was refolded from inclusion bodies. After cell lysis, the pellet fraction containing the inclusion bodies was resuspended to buffer E (20 mM Tris, pH 7.5) containing 3% Triton X-100 and incubated on ice for 2 h. After centrifugation, the remaining pellet was suspended in buffer E containing 1% Triton and incubated on ice for 30 min. The sample was centrifuged again, and the pellet was suspended to buffer F (40 mM Tris, pH 8.8) containing 8 M urea. Proteins were refolded in buffer F by stepwise decreasing the urea concentration (for 2 h in 6, 4, and 2 M urea, and overnight in 0 M urea). All the steps were carried out at 4°C. The samples were clarified by centrifugation, the refolded CAP2 fragments were concentrated to a volume of 500-1000 μl and stored at –70°C. Mouse cofilin-1 and bovine β/γ actin were purified as described previously (Vartiainen et al., 2002, 2003).
Antibodies
Polyclonal antibodies against CAP1 and CAP2 were generated in guinea pigs and rabbits, respectively. The animals were immunized with purified GST-CAP11–221 and his-tagged CAP21–227 according to standard procedures. For affinity purification of the antisera, recombinant GST-CAP11–221 and his-tagged CAP21–227 proteins were immobilized to CNBr-activated Sepharose 4B beads (Pharmacia), and the antibodies were purified from sera according to manufacturer's instructions. However, to increase the solubility of CAP2 the pH of the coupling buffer was 8.8 in this case. To remove the cross-reactive fraction from the antibodies, the CAP1 antibody was further passed through CAP2 column, and similarly the CAP2 antibody was passed through CAP1 column. The specificity of the antibodies and the amounts of CAP1 and CAP2 proteins in cell extracts were examined by Western blotting as described previously for mouse twinfilins (Vartiainen et al., 2003). Anti-cofilin-1 and anti-actin antibodies are described in Vartiainen et al. (2002) and Mies et al. (1998).
Cell Culture, Small-interfering RNA (siRNA) Transfections, and Immunofluorescence Microscopy
NIH3T3, B16F1, and Neuro2A cells were maintained in DMEM supplemented with 10% fetal bovine serum (Hyclone; B16F1 FCS from PAA Laboratories, Pasching, Austria), 2 mM l-glutamine, penicillin, and streptomycin (Sigma-Aldrich, St. Louis, MO). B16F1 cells stably expressing green fluorescent protein (GFP)-actin (Ballestrem et al., 1998) were maintained in medium supplemented with 1.5 mg/ml Geneticin (Invitrogen, Carlsbad, CA). siRNA duplexes were designed using the algorithm described in Elbashir et al. (2001) and prepared by siRNA construction kit (Silencer; Ambion, Austin, TX). Cofilin-1 and the fluorescein-labeled CAP1 siRNA oligonucleotide (same sequence as 1.CAP1a) duplexes were from Qiagen-Xeragon, Germany. The following oligonucleotides were used in this study: 1) CAP1a (F) 5′-GAACCGAGGCAGCAAGUUUUU-3′, (R) 5′-AAACUUGCUGCCUCGGUUCUU-3′; 2) CAP1b (F) 5′-AAAGCAUGGCAGCCAUCUGUU-3′, (R) 5′-AAAACAGAUGGCUGCCAUGCU-3′; 3) Cofilin-1 (F) 5′-GGAGGACCUGGUGUUCAUCd(TT)-3′, (R) 5′-GAUGAACACCAGGUCCUCCd(TT)-3′; and 4) Control (F) 5′-AGCUUCAUAAGGCGCAUGCUU-3′, (R) 5′-GCAUGCGCCUUAUGAAGCUUU-3′.
The cells were transfected with the siRNA duplexes by using siRNA transfection reagent (GeneSilencer; Gene Therapy Systems, CA). Forty-eight hours after transfections, the cells were detached and replated on coverslips in an appropriate density. For immunofluorescence, B16F1 cells were plated on coverslips coated with 25 μg/ml laminin or 50 μg/ml fibronectin. Cells were fixed with 4% paraformaldehyde (except in AC-15 stainings: 3% paraformaldehyde, 0.2% glutaraldehyde) and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline. For staining with the AC-15 antibody, the CAP1 RNAi cells were also treated with 6 N guanidine hydrochloride (Gdn-HCl) in 50 mM Tris-HCl, pH 7.5, to increase the access of the epitopes to this antibody as described in Peränen et al. (1993). Immunofluorescence was performed as described previously (Vartiainen et al., 2000), and the primary antibodies were used in following dilutions: guinea pig anti-CAP1, 1:200; rabbit anti-Cofilin-1, 1:100; and mouse AC-15, 1:2000. Filamentous actin was visualized by Alexa488-phalloidin 1:100 or rhodamine-phalloidin 1:100 (Molecular Probes, Eugene, OR).
Actin Filament Turnover Assays
B16F1 cells (wild type and CAP1 RNAi transfected) were treated with 5 μM latrunculin-A after 24 h of replating the cells. The cells were fixed at 0, 2, 10, and 30 min after latrunculin-A addition, and CAP1 and filamentous actin were subsequently visualized from the cells by immunofluorescence and rhodamine-phalloidin staining as described above. Fluorescence recovery after photobleaching (FRAP) was applied for measuring the actin filament turnover rates in live B16F1 GFP-actin cells. Confocal imaging was carried out with an LSM 510 confocal microscope equipped with an argon-ion laser (Carl Zeiss, Jena, Germany). For excitation of GFP imaging, the 488-nm line and a 100× oil immersion objective lens (numerical aperture 1.3) were used. The beam-path for GFP contained a 488-nm main dichromatic mirror and a 500- to 530IR-nm bandpass filter for detection of the emitted fluorescence. A temperature-controlled chamber (tempcontrol-37-1) was used to maintain 37°C during all image acquisition. For all data, acquisition was by using LSM 3.0 software. Wild-type and CAP1 knockdown cells were grown for 6–12 h after replating on 50 μg/ml fibronectin-coated glass-bottom dishes (MatTek, Ashland, MA). Region with suitable stress-fibers was bleached with 100 scan iterations of 30 mW argon-ion laser. After bleaching, the cell was scanned automatically 20 times in every 14 s. Recovery of the GFP-actin intensity in the bleached region was measured by TINA software. Changes of the intensity were always compared with the nonbleached control region to diminish the influence of the normal bleaching of the GFP during the assay.
Endocytosis Assay
The efficiency of receptor-mediated endocytosis in NIH3T3 cells was examined by following the uptake of rhodamine-transferrin (Molecular Probes). The cells were replated 48 h after CAP1 siRNA transfection, grown for 24 h, and starved for 30 min in serum-free DMEM. Cells were then incubated for 7 min at 37°C with 20 μg/ml rhodamine-transferrin and fixed for immunofluorescence. To compare the efficiency of endocytosis in wild-type and CAP1 knockdown cells, the rhodamine-transferrin fluorescence intensities of 20 CAP1 knockdown and 20 adjacent wild-type cells were quantified by TINA software.
Cell Motility Assay
The motilities of B16F1 cells were measured by time-lapse videomicroscopy. During the experiment, the cells were kept in a temperature-controlled (37°C) chamber with suitable CO2 pressure in growth medium supplemented with 40 mM HEPES to maintain optimal pH during all image acquisition. Replated B16F1 cells (wild type and CAP1 siRNA transfected) were grown on the 25 μg/ml laminin-coated coverslips for 5 h. To enhance cell migration, final concentrations of 50 μM AlCl3 and 30 mM NaF were added to medium. Immediately after the induction, the migration of the cells were recorded for 100 min with an IX70 inverted microscope (Olympus, Tokyo, Japan) equipped with a Polychrome IV monochromator (TILL Photonics, Martinsried, Germany) by using 20× air objective. After the experiment, the cells were fixed, and CAP1 was visualized by immunofluorescence to distinguish CAP1 knockdown cells from wild-type cells. To confirm the CAP1 siRNA transfection, the intensity of the fluorescein-labeled oligos also was detected. For statistical analysis, migration of the 44 wild-type and 28 CAP1 knockdown B16F1 cells was examined by tracking the position of the nucleus every 20 min. To visualize the CAP1 knockdown defect in migration, time-lapse videos of B16F1 cells expressing GFP-actin were taken. Imaging was processed as described above but without AlF4– treatment and with a 40× water objective.
RESULTS
Tissue-specific Expression Patterns of Mouse CAP1 and CAP2
All unicellular organisms and invertebrates examined have only one CAP protein, whereas mammals seem to have two CAP isoforms. To date, two CAP isoforms have been identified in human and rat, but their functional differences have not been reported (Yu et al., 1994; Swiston et al., 1995). Here, we identified mouse CAP2 from sequence databases and compared its expression pattern to the previously identified mouse CAP1 (Vojtek and Cooper, 1993) by Northern blot and in situ hybridization analyses as well as by using isoform-specific antibodies.
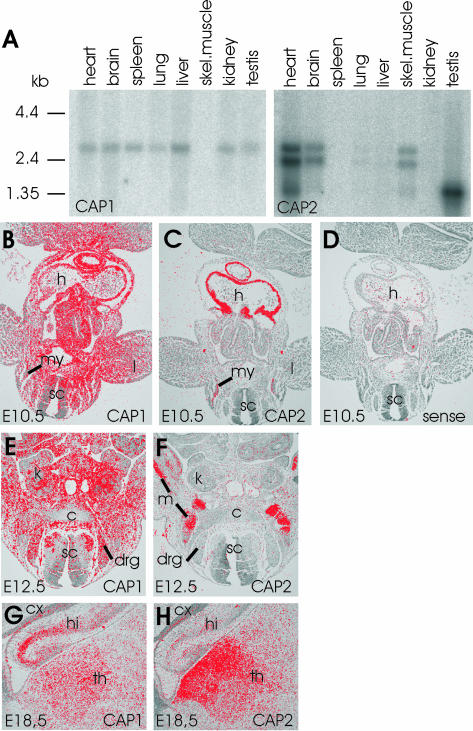
Northern blot analysis showed that CAP1 is strongly expressed already during embryonic development (our unpublished data). CAP1 is also expressed in most adult mouse tissues, but it is excluded from skeletal muscle (Figure 1A). In contrast, only a weak signal was observed with the CAP2 probe in mouse embryos (our unpublished data). In adult mice, strong CAP2 expression was detected in heart, skeletal muscle, and brain, and very weak expression could be detected in lung and liver. In addition, a smaller 1.5-kb band recognized with CAP2 probe was detected in testis (Figure 1A). It is important to note that our CAP2 cDNA probes recognized three different RNAs. The 2.5- and 3-kb RNAs are most likely a consequence of selective use of two polyadenylation signals, because the two RNAs were equally well recognized with probes generated either against 5′ or 3′ half of the CAP2 coding region. The 1.5-kb fragment is probably a splice variant of CAP2, because it was more strongly recognized with a probe generated against 3′ half of CAP2 coding region (our unpublished data).
Figure 1.
Expression patterns of mouse CAP1 and CAP2. (A) Northern blot analysis of CAP1 and CAP2 expression in adult mouse tissues. CAP1 (left) is expressed in all tissues except in skeletal muscle. CAP2 (right) is expressed at high levels in heart, brain, skeletal muscle, and testis and is also found in very low levels in lung and liver. The three different RNA populations recognized by the CAP2 probe are most likely a consequence of alternative mRNA processing (see text for details). (B) In situ hybridization on a transversal section of E10.5 mouse embryo shows that CAP1 expression is widespread. (C) Adjacent section shows that CAP2 expression is restricted to heart and developing muscles. (D) Hybridization of an adjacent section with a control CAP1 sense probe does not give significant background. (E) CAP1 is widely expressed at E12.5. (F) CAP2 is restricted to developing striated muscles at E12.5. (G) Elevated CAP1 expression is detected in hippocampus at E18.5. (H) Highest levels of CAP2 in brain is detected in thalamus at E18.5. These expression patterns indicate that CAP1 plays role as a general isoform, whereas CAP2 is a striated muscle specific isoform during early development. c, cartilage; cx, cortex; drg, dorsal root ganglion; h, heart; hi, hippocampus; k; kidney; l, limb; m; muscle; my, myotome; sc, spinal cord; th, thalamus.
The expression patterns of CAP1 and CAP2 in mouse embryos and adult mouse tissues were examined in more detail by RNA in situ hybridization analysis (Figure 1, B–H). As expected from the Northern blot analysis, CAP1 showed a widespread expression in nearly all cell types throughout the embryonic development (E10.5–18.5; Figure 1, B, E, and G; our unpublished data). At E12.5, especially strong expression was detected in some newborn neuronal populations of the spinal cord and especially weak in the cartilage (Figure 1E). At E14.5 and E18.5, strongest CAP1 expression could be observed in the differentiated neurons of both the central and peripheral nervous system and in the osteoblasts of the developing bone (our unpublished data). In the brain at E18.5, CAP1 was expressed in nearly all areas with especially strong expression in hippocampus (Figure 1G). In adult brain, strongest expression was observed in the cortex, striatum, and hippocampus (our unpublished data). The levels of CAP1 mRNA remained elevated in all other tissues except in skeletal muscle where the expression decreased after birth, and we could not detect any expression in adult muscles (our unpublished data).
The expression of CAP2 was highly restricted to the developing heart and muscle tissues from early on (Figure 1, C and F). At E10.5, very strong expression could be detected in the developing heart and a slightly weaker in the myotomal muscle cells (Figure 1C). Through stages E12.5–14.5, CAP2 expression was very strong and restricted to the developing striated muscles (Figure 1, C and F; our unpublished data). At E18.5, in addition to the strong expression in muscle tissues and in the thalamic area of the brain, a weaker CAP2 expression could be seen in some peripheral ganglia. In adult tissues, strongest CAP2 expression was detected in heart, skeletal muscles, and in some brain areas such as the cortex, striatum, and hippocampus (Figure 1H; our unpublished data).
To examine the expression of CAP1 and CAP2 at protein level, we generated antibodies that differentially recognize these two proteins. A GST-CAP1 (residues 1–221) and his-tagged CAP2 (residues 1–227) were used in the immunizations. Because affinity-purified CAP1 antisera also weakly reacted with CAP2 protein, these antisera were further purified by using a CAP2 affinity column. After adsorbing the cross-reactive fraction by a CAP2 affinity column, we obtained an antibody that was >20-fold more specific for CAP1 than for CAP2 (Figure 2A; our unpublished data). Also, purification of the CAP2 antisera with a similar procedure resulted in an antibody that was >20-fold more specific for CAP2 than for CAP1 (Figure 2A; our unpublished data). Western blot analysis of NIH3T3, B16F1, Neuro 2A, and N18 cell extracts showed that only CAP1 is expressed in these cells at detectable levels (Figure 2B; our unpublished data). The only cells, in which we could detect expression of CAP2 protein, were differentiated myotubes derived from C2C12 myoblasts (our unpublished data). Together with in situ and Northern blot analyses, these data demonstrate that CAP2 is a highly muscle-specific protein, whereas CAP1 is the predominant isoform in most nonmuscle cell types.
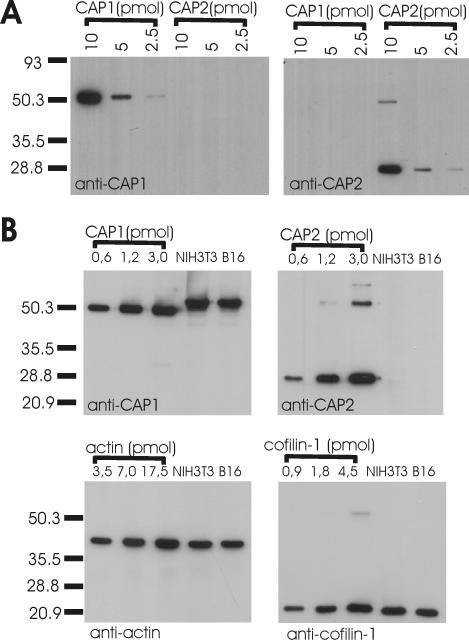
Figure 2.
CAP1 is an abundant protein in NIH3T3 and B16F1 cells. (A) Three different amounts (10, 5, and 2.5 pmol) of purified GST-CAP11–221 (calculated mol. wt. = 51 kDa) and his-tagged CAP21–227 (calculated mol. wt. = 26 kDa) were run on polyacrylamide gels, and the proteins were visualized by Western blotting by using anti-CAP1 (left) or anti-CAP2 (right) antibodies. Purified polyclonal guinea pig anti-CAP1 antibody is specific to CAP1, whereas purified polyclonal rabbit anti-CAP2 antibody only recognizes CAP2. Note that his-tagged CAP21–227 has a small tendency to dimerize on polyacrylamide gels. (B) Mouse NIH3T3 and B16F1 cell extracts and known concentrations of GST-CAP11–221, CAP21–227, β/γ actin, and cofilin-1 were run on polyacrylamide gels, and the proteins were visualized by specific antibodies. CAP2 protein was undetectable in these cells. The CAP1:cofilin-1:β/γ actin ratio in NIH 3T3 and B16F1 cell extracts is ∼1:1:4.
CAP1 Is an Abundant Protein That Localizes to Dynamic Actin Structures in Nonmuscle Cells
To understand the role of CAPs in actin dynamics, it is important to know the abundance of these proteins in cells. We compared the molar ratios of CAP1, CAP2, cofilin-1, and actin in mouse NIH3T3 and B16F1 cell extracts. Different concentrations of purified GST-CAP11–221, his-tagged CAP21–227, cofilin-1, and β/γ actin were used as standards in this assay, and specific antibodies against CAP1, CAP2, cofilin-1, and β/γ actin were applied for detecting these proteins from Western blots. The CAP1:cofilin-1:actin ratio in NIH3T3 and B16F1 cells was ∼1:1:4, demonstrating that CAP1 is a highly abundant protein in these cells (Figure 2B). We could not detect any CAP2 protein in NIH3T3 and B16F1 cell extracts, suggesting that CAP1 is the only isoform expressed in these cell lines (Figure 2B).
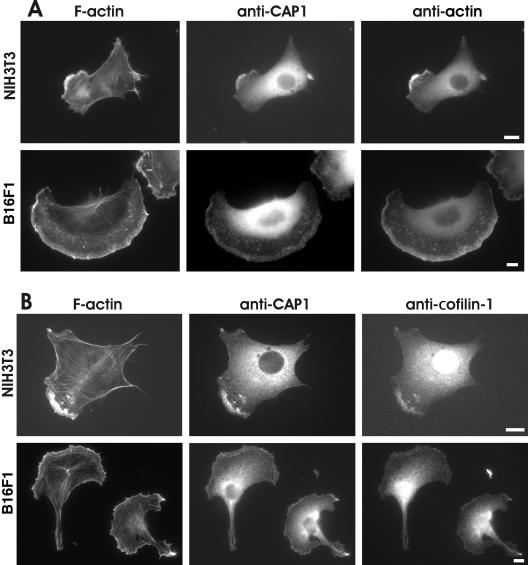
In budding yeast and Dictyostelium Srv2/CAP proteins localize to the cortical actin cytoskeleton (Freeman et al., 1996; Noegel et al., 1999). However, subcellular localization of mammalian CAPs has remained elusive. Studies with polyclonal antibodies and tagged versions of CAP1 showed that it localizes to the dynamic regions of the cortical actin cytoskeleton in C3H-2K fibroblasts (Moriyama and Yahara, 2002). On the other hand, studies with monoclonal human CAP1 antibodies suggested that in addition to cortical actin structures, this protein also localizes to actin stress-fibers in Swiss 3T3 fibroblasts (Freeman and Field, 2000). To clarify these contradictory findings, we examined the localization of CAP1 in cultured NIH3T3 and B16F1 cells by using our isoform-specific CAP1 antibody. CAP1 showed diffuse cytoplasmic localization, but it was also concentrated to actin-rich membrane ruffles. We did not observe detectable CAP1 staining in stress-fibers in B16F1 cells, but we could detect very weak CAP1 staining in stress-fibers of a small fraction of NIH3T3 cells (Figure 3). Interestingly, in polarized cells CAP1 was not concentrated to the F-actin–rich distal edge of the leading lamellipodia, but it showed instead uniform staining across the lamellipodia (Figure 3). The CAP1-rich regions in NIH3T3 and B16F1 cells also colocalized with cofilin-1 staining at the cortical actin structures. However, CAP1 did not show similar nuclear localization than cofilin-1 (Figure 3B). Furthermore, CAP1 also localized to actin monomer-rich regions of these cells (Figure 3A). The actin monomers were visualized by AC-15 antibody that has been reported to recognize actin monomers and certain accessible actin filament structures in cells (Mies et al., 1998).
Figure 3.
Localization of CAP1 in NIH3T3 and B16F1 cells. (A) Cells were stained with Alexa488-phalloidin to visualize F-actin (left), with anti-CAP1 antibody (middle), and with AC15 anti-actin antibody to visualize monomeric actin (right). CAP1 localizes to actin-rich regions of cell cortex and behind the leading edge of the lamellae. (B) F-actin (left) and CAP1 (middle) were visualized in NIH 3T3 and B16F1 cells as described above, and cofilin-1 was visualized by an isoform-specific polyclonal cofilin-1 antibody (right). CAP1 and cofilin-1 show clear colocalization at the cortical actin cytoskeletons of these cells. Bar, 10 μm.
CAP1 Knockdown Cells Show an Accumulation of Abnormal F-Actin Structures
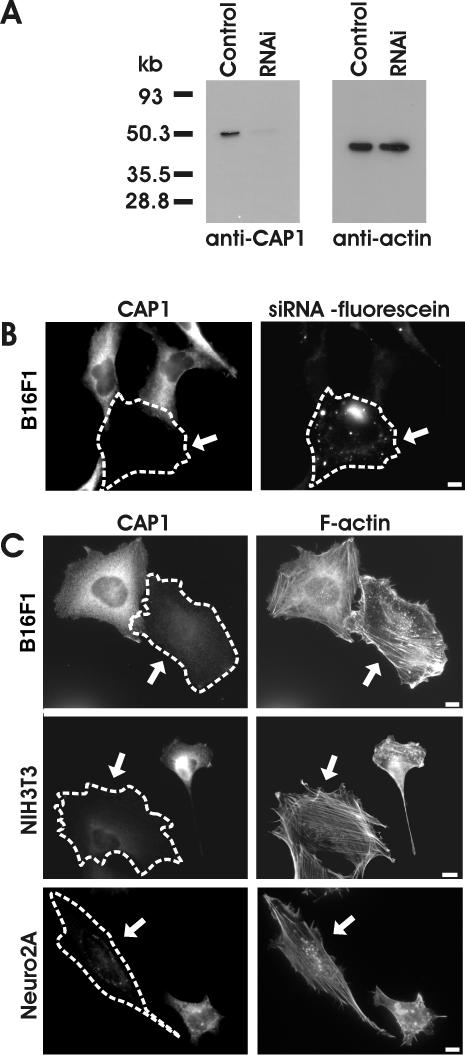
We next examined the cellular roles of CAP1 in mouse NIH3T3, B16F1, and Neuro 2A cell lines by depleting CAP1 from these cells by RNA interference (RNAi). It is important to note that CAP1 is the only isoform that is detectably expressed in these cells (Figure 2B; our unpublished data), and thus simultaneous depletion of CAP2 was not necessary. Transfection of cells with CAP1-specific RNA oligonucleotide duplex resulted in dramatic decrease in the cellular CAP1 levels as detected by Western blotting. However, this siRNA transfection did not affect cellular actin levels or induce CAP2 expression in these cells (Figure 4A; our unpublished data). Immunofluorescence microscopy with CAP1 antibody revealed that the proportion of CAP1 negative cells varied from 30 to 90% between individual experiments. The remaining cells in these experiments seemed to contain similar levels of CAP1 protein as the nontransfected control cells and were thus most likely not transfected with RNAi oligonucleotides. To confirm that this was indeed the case, we transfected NIH3T3 cells with fluorescein-labeled CAP1-specific RNAi oligonucleotides, and compared the CAP1 immunofluorescence with appearance of fluorescein RNAi oligonucleotide dots. The cells with significantly decreased CAP1 staining almost invariably displayed punctate fluorescein labeling, whereas cells with normal CAP1 levels did not contain detectable amounts of the fluorescent oligonucleotide (Figure 4B). Therefore, the cells with dramatically decreased CAP1 levels (and transfected with RNAi oligonucleotides) will be referred as CAP1 knockdown cells, whereas nontransfected cells with normal CAP1 levels will be referred as wild-type cells.
Figure 4.
Depletion of CAP1 by siRNA results in an accumulation of stress-fibers and abnormal morphology in NIH3T3, B16F1, and Neuro2A cells. (A) Western blot analysis demonstrating the CAP1 protein levels in B16F1 control cells and in B16F1 cells transfected with CAP1-specific RNAi oligonucleotide duplex. Equal amounts of cell lysates were run on polyacrylamide gels, and CAP1 (left) and β/γ actin were visualized by Western blotting. (B) Cells transfected with fluorescein-labeled CAP1-siRNA oligonucleotides were stained with anti-CAP1 antibodies. Note, that the cell containing fluorescein-labeled oligonucleotides (arrow) has severely reduced CAP1 levels, whereas the nontransfected cells show normal level of CAP1-staining. (C) siRNA transfected B16F1 (top), NIH3T3 (middle), and Neuro 2A (bottom) cells were stained with anti-CAP1 antibodies (left) and Alexa488-phalloidin (right) to visualize the effect of CAP1 depletion for the actin cytoskeleton. The CAP1 knockdown cells (arrows) showed abnormal accumulation of thick stress-fibers compared with nontransfected wild-type cells. In NIH 3T3 and Neuro 2A cells, the CAP1 depletion also resulted in the loss of membrane ruffles, whereas in B16F1 cells CAP1 depletion resulted in a decreased polarity of existing membrane ruffles. Bar, 10 μm.
CAP1 knockdown cells were typically slightly larger and less polarized than wild-type cells. Phalloidin staining demonstrated that CAP1 knockdown cells contained more and thicker stress-fibers than nontransfected wild-type cells (Figure 4C). Instead of typical crisscross pattern, the stress-fibers in CAP1 knockdown cells were often aligned in parallel with each other. It is important to note that in NIH3T3 and Neuro 2A cells, the CAP1 depletion typically resulted in the disappearance of actin-rich membrane ruffles and accumulation of the majority of F-actin to stress-fibers (Figure 4C). In contrast, many CAP1-depleted B16F1 cells still contained membrane ruffles, but they were significantly less polarized than the ones in wild-type B16F1 cells.
To ensure that the phenotypes were specific to CAP1 depletion, we transfected these cells with a second CAP1-specific duplex oligonucleotide and with a scrambled control oligonucleotide (for details, see MATERIALS AND METHODS). Also, the second CAP1 oligonucleotide resulted in a depletion of CAP1 and in similar accumulation of actin stress-fibers as described above. In contrast, transfection of the cells with the control RNA oligonucleotide duplex did not result in either depletion of CAP1, accumulation of parallel stress-fibers, or decrease in the cell polarity (our unpublished data).
CAP1 Is Involved in Cell Motility and Endocytosis in Mammalian Cells
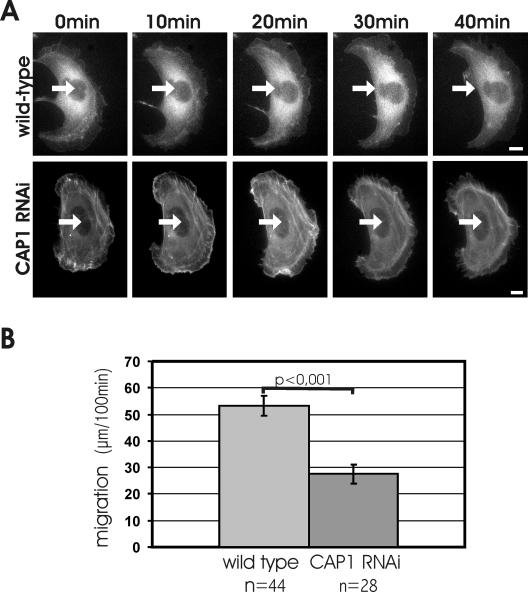
Because CAP1 depletion resulted in an accumulation of stress-fibers and decrease in cell polarity, we investigated the role of CAP1 in the migration of B16F1 cells. We first quantified the migration rates of wild-type and CAP1 knockdown B16F1 cells. Because these cells displayed only relatively short motility tracks during the 100-min measurement period, the migration was further induced by addition of AlF –4 to the medium (Hahne et al., 2001). Under these conditions, wild-type B16F1 cells often displayed directional migration tracks. Although also some CAP1 knockdown cells were motile, they were typically unable to perform directed migration, but instead projected multiple lamellipodia and constantly changed their direction of motility. Quantification of the motility tracks from 44 wild-type and 28 CAP1 knockdown cells demonstrated that CAP1 depletion decreased the motility of these cells to one-half of that of the wild-type cells (Figure 5B). Although there was some variation in the migration distances within each group, the analysis of the data showed that the difference between the migration distances of wild-type and CAP1 knockdown cells was statistically highly significant (p < 0.001). It is important to note that in these experiments as well as the ones described below, the cells were fixed after the assay and stained with CAP1 antibody to distinguish CAP1 knockdown cells from wild-type cells (our unpublished data).
Figure 5.
CAP1 is important for cell migration. (A and Supplementary Videos 1 and 2) Representative examples of motilities of wild-type (top) and CAP1 knockdown (bottom) B16F1 cells expressing GFP-actin. White arrows indicate the locations of the nuclei in the first frame. Wild-type cells typically displayed directional migration. CAP1 knockdown cells were capable to extend and retract their lamellipodia, but they were generally unable to migrate to certain direction. Note that also the reorganization of actin filament structures is much slower in CAP1 knockdown cells than in wild-type cells. Bar, 10 μm. (B) Migration of 44 wild-type and 28 CAP1 knockdown B16F1 cells were monitored for 100 min. The average motility distances of wild-type cells and CAP1 knockdown cells were 53.4 and 27.6 μm, respectively. SEM and statistical significance of the data are indicated in the graph.
To visualize the cytoskeletal defects in CAP1 knockdown cells during migration, we followed by videomicroscopy the motility of B16F1 cells expressing GFP-actin. Wild-type B16F1 cells typically displayed polarized migration, and the actin cytoskeletons of these cells showed rapid and constant reorganization (Figure 5A; Supplementary Video 1). In contrast, the actin filament structures in CAP1 knockdown cells were significantly less dynamic, and these cells did not display polarized migration. However, although the stress-fibers of CAP1 knockdown cells displayed only very slow reorganization, these cells were still capable of retracting and extending their lamellipodia (Figure 5A; Supplementary Video 2).
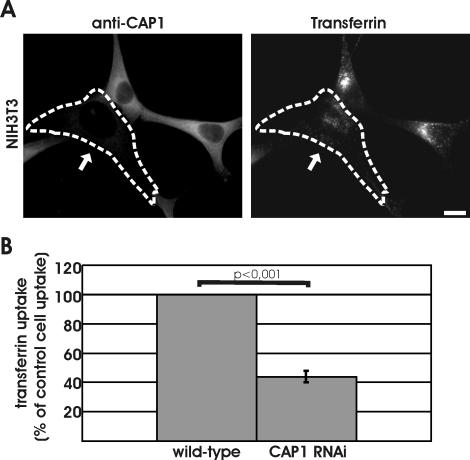
The actin cytoskeleton also plays an important role in endocytosis (Engquist-Goldstein and Drubin, 2003). We thus examined the role of CAP1 in receptor-mediated endocytosis by measuring the capability of CAP1 knockdown cells to endocytose rhodamine-transferrin. After 7 min of uptake, a strong perinuclear accumulation of transferrin was observed in wild-type NIH3T3 cells (Figure 6A). In contrast, CAP1 knockdown cells showed significantly weaker transferrin uptake. Instead of strong perinuclear accumulation of transferrin, these cells showed a more uniform cytoplasmic dot-like localization pattern of transferrin (Figure 6A). Quantification of the intensities of rhodamine-transferrin from 20 wild-type and 20 CAP1 knockdown cells demonstrated that CAP1 depletion decreased the efficiency of transferrin uptake to ∼45% of that of wild-type cells (Figure 6B).
Figure 6.
Depletion of CAP1 results in defects in receptor-mediated endocytosis. (A) Uptake of transferrin was compared between wild-type and CAP1 knockdown cells. Cells were incubated for 7 min in 20 ng/ml rhodamine-transferrin and stained with anti-CAP1 antibody (left) to visualize CAP1 knockdown cells (white arrow). Depletion of CAP1 from cells resulted in a decrease in transferrin uptake. Instead of strong perinuclear accumulation, transferrin (right) showed uniform punctate cytoplasmic staining in CAP1 knockdown cells. Bar, 10 μm. (B) Intensity of rhodamine-transferrin fluorescence was quantified by TINA software from 20 knockdown cells and compared with the intensity of rhodamine fluorescence of an adjacent wild-type cell from the same frame. In CAP1 knockdown cells, transferrin uptake was reduced to 44% of the one in wild-type cells. SEM = 0.04 and statistical significance of the data (p < 0.001) are indicated in the graph.
Depletion of CAP1 Results in Mislocalization of Cofilin and Decreased Actin Filament Depolymerization Rates
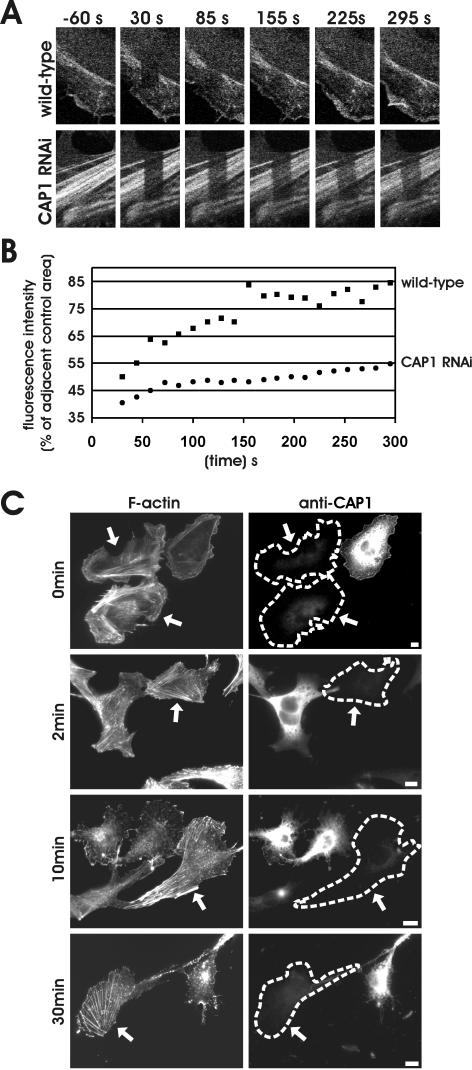
To elucidate the mechanism by which CAPs contribute to cytoskeletal dynamics, we examined the effects of CAP1 depletion on actin filament turnover and depolymerization in cultured mammalian cells. For actin filament turnover assays, we used FRAP and bleached by intense laser irradiation a region of B16F1 cells expressing GFP-actin. After bleaching, the exchange of GFP-actin between bleached and unbleached region was then monitored for 295 s by taking frames every 14 s. This assay was performed for several wild-type and CAP1 knockdown cells, representative examples of which are shown (Figure 7A). After the experiment, the cells were fixed and stained with CAP1 antibody to distinguish wild-type cells from CAP1 knockdown cells.
Figure 7.
Actin filament assembly and disassembly rates are severely diminished in CAP1 knockdown cells. (A) Actin filament treadmilling rates in wild-type and CAP1 knockdown cells were measured by using FRAP. Selected regions of B16F1 cells expressing GFP-actin were bleached by intense laser irradiation, and recovery of the bleached area was monitored by taking images every 14 s, starting 30 s after bleaching. Figure shows representative example of a wild-type cell (top) and CAP1 knockdown cell (bottom). (B) Rate of the fluorescence recovery was analyzed from the image series by TINA software. In each frame, the fluorescence intensity of the bleached region was compared with the fluorescence of the control region (in same picture next to bleached one) to diminish the error caused by normal photobleaching during the monitoring period. Recovery of the bleached regions in the CAP1 RNAi cells were significantly slower than in wild-type cells. (C) Actin filament depolymerization in wild-type and CAP1 knockdown cells were measured by using actin monomer-sequestering drug latrunculin-A. Cells were treated with 2 μm latrunculin-A, fixed at different time points (0, 2, 10, and 30 min), and F-actin and CAP1 were visualized by immunofluorescence. Actin filament structures were rapidly disrupted in wild-type cells, whereas in CAP1 knockdown cells (arrows) the disappearance of stress-fibers was significantly slower.
In wild-type cells, we observed almost complete recovery of the GFP-actin fluorescence at the cortical actin cytoskeleton within 30 s after photobleaching. A nearly complete recovery of stress-fibers occurred within ∼150 s (Figure 7, A and B). In contrast, the fluorescence recovery of stress-fibers in CAP1 knockdown cells was significantly slower and was not completed during the 295-s monitoring period (Figure 7, A and B). These results show that the turnover of stress-fibers is reduced in CAP1 knockdown cells. Because the actin filament turnover rates are very rapid at the cortical actin structures, this method could not be applied for comparing the rates of actin dynamics between the cortical actin cytoskeletons of wild-type and CAP1 knockdown cells.
Having established that the actin filament turnover rates are diminished in CAP1 knockdown cells, we next compared the actin filament depolymerization rates of wild-type and CAP1 knockdown cells. This was assessed by treating the cells with the actin monomer-sequestering drug latrunculin-A and measuring the timing of loss of actin filament structures. Because latrunculin-A does not interact with actin filaments, but functions by sequestering actin monomers, the disappearance rates of actin structures are thought to reflect the rate of actin monomer dissociation from filament ends (Coue et al., 1987; Ayscough et al., 1997). The actin filament depolymerization assay was carried out for B16F1 cells, and F-actin was visualized by phalloidin staining these cells after 0, 2, 10, and 30 min of latrunculin-A addition. Typical examples of these cells are shown in (Figure 7C). After addition of 5 μM latrunculin-A, the majority of the wild-type cells rapidly lost their stress-fibers, most of the stress-fibers disappearing during the first 2 min of latrunculin-A treatment. After 5 min, only ∼50% of the wild-type cells displayed detectable stress-fibers, and after 30 min stress-fibers were present in only ∼15% of wild-type cells. In contrast, the stress-fibers in CAP1 knockdown cells were much more resistant to depolymerization, and after 5 and 30 min of latrunculin-A addition, the proportion of stress-fiber containing cells were 80 and 50%, respectively (Figure 7C; our unpublished data).
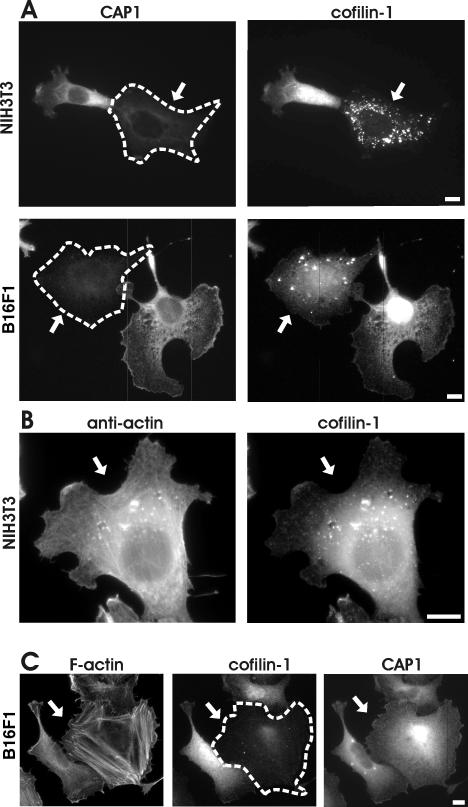
FRAP and latrunculin-A studies show that actin filament turnover rates are diminished in CAP1 knockdown cells. However, CAPs are actin monomer binding proteins that do not depolymerize actin filaments. Because a recent biochemical study reported that CAP1 can promote ADF/cofilin-dependent actin filament turnover at least in vitro (Moriyama and Yahara, 2002), we examined the possible effect of CAP1-depletion on localization of the major ADF/cofilin isoform, cofilin-1, in B16F1 and NIH3T3 cells. In wild-type cells CAP1 and cofilin-1 show diffuse cytoplasmic staining but are also concentrated at the cortical actin cytoskeleton (Figure 3). Interestingly, in CAP1 knockdown cells the cofilin-1 staining at the cortical actin cytoskeleton was diminished significantly, and a large proportion of cofilin-1 localized to punctate cytoplasmic structures. The abnormal cofilin-1 localization was seen in both NIH3T3 and B16F1 CAP1 knockdown cells, although the amount and intensity of these abnormal cofilin-1 aggregates varied between individual cells (Figure 8A). These abnormal cofilin aggregates also contain monomeric actin, because they co-localized with anti-actin (AC-15 antibody) staining (Figure 8B). However, the actin staining in these aggregates became visible only after Gdn-HCl treatment of the cells, suggesting that the actin epitopes are mostly nonaccessible to the antibodies unless these aggregates are denatured.
Figure 8.
CAP1 is important for correct subcellular localization and function of cofilin-1. (A) NIH 3T3 (top) and B16F1 (bottom) cells transfected with CAP1 siRNA oligonucleotides were stained with anti-CAP1 (left) and anti-cofilin-1 (right) antibodies. In CAP1 knockdown cells (arrows), cofilin-1 no longer localized to dynamic actin-rich structures, but instead it accumulated to the dot-like structures in the cytoplasm. (B) NIH3T3 cells transfected with CAP1 siRNA were treated with Gdn-HCl to denature proteins and stained with an anti actin (AC-15) (left) and cofilin-1(right) antibodies. The abnormal cofilin aggregates in CAP1 knockdown cells contain also actin. (C) F-actin (left), cofilin-1 (middle), and CAP1 (right) were visualized in B16F1 cells transfected with cofilin-1–specific RNA oligonucleotide duplexes. Cofilin-1 knockdown cell (arrow) shows similar accumulation of actin stress-fibers as seen in CAP1 knockdown cells. However, cofilin-1 depletion does not significantly affect the subcellular localization of CAP1. Bar, 10 μm.
Because CAP1 has a significant effect on the localization of cofilin-1 in these cells, we next examined the reciprocal effect of cofilin-1 depletion on localization of CAP1. Cofilin-1 depletion resulted in a similar accumulation of actin stress-fibers as described above for CAP1-depletion (Figure 8C). However, no significant differences in CAP1 localization were detected between wild-type and cofilin-1 knockdown cells. Therefore, these data suggest that CAP1 plays an important role in regulating the correct localization and function of cofilin-1 in mammalian cells but that cofilin-1 does not contribute to CAP1 localization.
DISCUSSION
CAPs are ubiquitous actin monomer binding proteins, but their roles in actin dynamics and various cellular processes have been largely unclear. Here, we made an important step forward by defining the cellular roles of mammalian CAPs. We show that 1) mammals have two differentially expressed CAP isoforms from which CAP1 is the predominant isoform in nonmuscle cells, and CAP2 is expressed mainly in striated muscle cells; 2) CAP1 contributes to cytoskeletal dynamics by increasing cellular actin filament depolymerization rates; 3) CAP1 is required for the correct subcellular localization of cofilin-1; and 4) CAP1 plays an important role in morphogenesis, polarized migration, and receptor-mediated endocytosis in mammalian nonmuscle cells.
CAP1 Recycles ADF/Cofilin for New Rounds of Depolymerization
Our FRAP analysis and studies with latrunculin-A provided direct evidence that actin filament depolymerization rates are diminished in CAP1 knockdown cells. In these experiments, we concentrated on measuring the actin filament turnover rates of stress-fibers, because the actin filament turnover rates at the cortical actin structures were too fast for the time resolutions of these methods. The abnormal accumulation of cofilin-1 to cytoplasmic dot-like structures suggests that CAP1 plays an important role in regulating ADF/cofilin localization in these cells. Furthermore, because the phenotypes of CAP1 knockdown cells are very similar to the ones in cofilin-1 knockdown cells and in cells from which ADF/cofilins were inactivated by overexpression of LIM kinase (Arber et al., 1998), our data suggest that CAP1 is important for the proper cellular activity of ADF/cofilins. We speculate that, as suggested by recent biochemical studies (Moriyama and Yahara, 2002; Balcer et al., 2003), CAPs play an important role in recycling ADF/cofilins from actin monomers for new rounds of filament depolymerization. In the absence of CAPs, ADF/cofilins accumulate to cytoplasmic aggregates, and this will consequently result in a decrease in the actin filament depolymerization rates as observed in CAP1 knockdown cells. It is also important to note that our studies show that CAPs are highly abundant proteins in cells. The CAP1:cofilin-1:β/γ actin ratio in cultured mouse cells was ∼1:1:4. Cofilin-1 is the most abundant ADF/cofilin isoform in most mammalian nonmuscle cells (Vartiainen et al., 2002). Similarly, it was recently reported that CAP/Srv2:actin ratio in budding yeast is ∼1:10 (Balcer et al., 2003). Because of its high abundance, CAP1 is likely to directly contribute to actin filament turnover in cells rather than being an intermediate component of a signaling cascade.
The increase in the amount and intensity of stress-fibers observed here in CAP1 knockdown cells is consistent with previous studies showing that in Drosophila and Dictyostelium, mutations in CAP gene result in an accumulation of filamentous actin (Noegel et al., 1999; Baum et al., 2000; Benlali et al., 2000). However, our studies demonstrated that in mouse CAP1 knockdown cells, the F-actin accumulation results from a decrease in actin filament depolymerization rates rather than from diminished actin monomer-sequestering activity as suggested previously for Drosophila and Dictyostelium CAP mutants (Noegel et al., 1999; Baum et al., 2000; Benlali et al., 2000; Wills et al., 2002). We thus speculate that, similarly to mouse CAP1 knockdown cells, also the Drosophila and Dictyostelium CAP mutant phenotypes arise from an actin filament depolymerization defect rather than from lack of actin monomer-sequestering activity as suggested previously. It was recently shown that in budding yeast CAP/Srv2 displays genetic interactions with certain cofilin and profilin mutant alleles, suggesting that it is intimately involved in actin dynamics (Balcer et al., 2003). However, further studies are required to clarify whether yeast CAP/Srv2 also plays a similar important role in ADF/cofilin activity and localization as demonstrated here for mouse CAP1. It will be also important to determine how the activity and localization of CAPs are regulated in cells. At least in budding yeast, the actin filament binding protein, Abp1, seems to be essential for correct localization of CAP/Srv2 to cortical actin patches (Lila and Drubin, 1997; Balcer et al., 2003). Thus, it will be important to elucidate whether the mammalian homologues of Abp1 or related proteins contribute to CAP1 activity and localization in mouse nonmuscle cells.
Role of CAP1-induced Actin Dynamics in Cell Morphogenesis, Motility, and Endocytosis
Here, we show for the first time that in mammalian cells CAP plays an important role in cell polarity, motility, and receptor-mediated endocytosis. The motility defects agree with previous studies in Dictyostelium, where strains expressing diminished levels of CAP were defective in chemotaxis migration (Noegel et al., 2003). How does CAP contribute to cell migration? Because our studies demonstrated that CAP1 knockdown cells typically display a loss of polarized lamellipodia, we speculate that CAP1-promoted actin dynamics is required for the maintenance of cell polarity essential for productive cell migration. It is important to note that inactivation of ADF/cofilin by overexpressing LIM kinase results in similar lack of lamellipodia polarization and cell migration as demonstrated here for CAP1 knockdown cells (Dawe et al., 2003). Because our studies showed that CAP1 is necessary for the correct activity and localization of cofilin-1, it is likely that the motility defects in CAP1 cells result from diminished ADF/cofilin activity/recycling. Recent studies demonstrated that also in Drosophila S2 cells the inactivation of CAP causes defects in lamellipodia formation, suggesting that CAP may be important for cell motility also in Drosophila (Rogers et al., 2003).
In Dictyostelium, mutations in CAP result in a reduction in fluid phase endocytosis, whereas in budding yeast CAP/Srv2 deletion does not result in endocytic defects (Wesp et al., 1997; Noegel et al., 1999). Thus, CAP activity seems to contribute differentially to endocytosis in different organisms. However, also yeast CAP/Srv2 is somehow linked to endocytic processes, because it has been shown to be synthetically lethal with endocytic protein SlaII and a randomly generated CAP/Srv2 mutant allele unexpectedly disrupts endocytosis (Lila and Drubin, 1997; Wesp et al., 1997). Our studies showed that in cultured mouse NIH3T3 cells, the lack of CAP1 leads to an ∼50% decrease in receptor-mediated endocytosis. Similar endocytic defects have been reported in some mammalian cells after disruption of the actin cytoskeleton by cytochalasin-D or latrunculin-A (Fujimoto et al., 2000), and in NIH3T3 cells after inactivation of ADF/cofilins by RNAi (Hotulainen, Paunola, Vartiainen, and Lappalainen, unpublished data). Together, these studies suggest that the actin cytoskeleton does not play an obligatory role in endocytosis in mammalian cells but that the actin dynamics are required for efficient endocytosis in many mammalian cell types. Consequently, actin dynamics have been shown to be involved in the inward movement of endosomes from the plasma membrane in mammalian cells and in budding yeast (Merrifield et al., 2002; Kaksonen et al., 2003). We thus speculate that, due to decreased actin dynamics in CAP1 knockdown cells, the rate of endocytic internalization is diminished. This is also in agreement with the observed decreased perinuclear localization of endocytic vesicles in these cells.
Differential Distribution of Mammalian CAP1 and CAP2 Suggests Specific Roles during Development and in Adult Tissues
Previous studies have shown that humans and rats have two different CAP proteins (Yu et al., 1994; Swiston et al., 1995). Here, we demonstrate that also mice have two CAPs, suggesting that most mammals likely have two CAP isoforms. Possible biochemical differences between the two mammalian CAP isoforms have not been reported, and differences in their expression patterns were examined previously only by reverse transcription-polymerase chain reaction. These studies suggested that both isoforms are widely expressed in rat tissues (Swiston et al., 1995). Here, we show by Northern blot, RNA in situ hybridization analysis, and by using isoform-specific antibodies, that CAP1 is widely expressed in various nonmuscle cell types but is absent from differentiated skeletal muscle cells. In contrast, CAP2 is a striated muscle-specific protein during early mouse development. At later stages, CAP2 becomes strongly expressed also in specific areas of the central nervous system, and in adult mice, expression is especially strong in heart, skeletal muscle, and brain. It is also important to note that our in situ hybridization analyses indicate that at least one of these two CAP proteins is expressed in nearly all cells of developing and adult mice. This supports our cell biological findings demonstrating that CAPs are central regulators of actin dynamics and that they play important roles in many fundamental cellular processes such as morphogenesis, polarization, migration, and endocytosis.
Similar differential expression specificities have been observed previously for other mammalian actin binding proteins such as ADF/cofilin, twinfilin, and capping protein (Ono et al., 1994; Schafer et al., 1994; Vartiainen et al., 2003). Typical for all these protein families is the situation where only one “muscle-isoform” is expressed in skeletal muscle cells, whereas other isoform(s) are present in nonmuscle cell types. This may reflect the fact that actin dynamics in muscle cells is significantly slower than in most nonmuscle cell types. It has been shown that nonmuscle ADF/cofilin isoforms promote faster actin filament depolymerization than the muscle-specific isoforms (Ono and Benian, 1998; Vartiainen et al., 2002). In the future, it will be important to elucidate whether also CAP1 and CAP2 have different effects on actin filament turnover in vitro. Finally, because our studies demonstrate that CAPs play an important role in localization and recycling ADF/cofilins in cells, it is also possible that CAP1 has evolved to interact with nonmuscle ADF/cofilins, whereas CAP2 may have evolved to specifically promote interactions with the muscle-isoform of ADF/cofilin.
Supplementary Material
Acknowledgments
We thank Drs. Bruce Goode and Keith Kozminski for critical reading of the manuscript. This study was supported by grants from Academy of Finland, Sigrid Juselius Foundation, Biocentrum Helsinki, and European Molecular Biology Organization Young Investigator Program.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–01–0048. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–01–0048.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Arber, S., Barbayannis, F.A., Hanser, H., Schneider, C., Stanyon, C.A., Bernard, O., and Caroni, P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805–809. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcer, H.I., Goodman, A.L., Rodal, A.A., Smith, E., Kugler, J., Heuser, J.E., and Goode, B.L. (2003). Coordinated regulation of actin filament turnover by a high molecular weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159–2169. [DOI] [PubMed] [Google Scholar]
- Ballestrem, C., Wehrle-Haller, B., and Imhof, B.A. (1998). Actin dynamics in living mammalian cells. J. Cell Sci. 111, 1649–1658. [DOI] [PubMed] [Google Scholar]
- Baum, B., Li, W., and Perrimon, N. (2000). A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10, 964–973. [DOI] [PubMed] [Google Scholar]
- Baum, B., and Perrimon, N. (2001). Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol. 3, 883–890. [DOI] [PubMed] [Google Scholar]
- Benlali, A., Draskovic, I., Hazelett, D.J., and Treisman, J.E. (2000). act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281. [DOI] [PubMed] [Google Scholar]
- Coue, M., Brenner, S.L., Spector, I., and Korn, E.D. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316–318. [DOI] [PubMed] [Google Scholar]
- Dawe, H.R., Minamide, L.S., Bamburg, J.R., and Cramer, L.P. (2003). ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 13, 252–257. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A.E., and Drubin, D.G. (2003). Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell. Dev. Biol. 19, 287–332. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken, M., Deschenes, R.J., and Broach, J.R. (1990). SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340. [DOI] [PubMed] [Google Scholar]
- Field, J., et al. (1990). Cloning and characterization of C.A.P., the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 61, 319–327. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., Chen, Z., Horenstein, J., Weber, A., and Field, J. (1995). An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 270, 5680–5685. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., and Field, J. (2000). Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil. Cytoskeleton 45, 106–120. [DOI] [PubMed] [Google Scholar]
- Freeman, N.L., Lila, T., Mintzer, K.A., Chen, Z., Pahk, A.J., Ren, R., Drubin, D.G., and Field, J. (1996). A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol. Cell. Biol. 16, 548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, L.M., Roth, R., Heuser, J.E., and Schmid, S.L. (2000). Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic 1, 161–171. [DOI] [PubMed] [Google Scholar]
- Gerst, J.E., Ferguson, K., Vojtek, A., Wigler, M., and Field, J. (1991). CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol. Cell. Biol. 11, 1248–12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann, R., and Mann, K. (1992). ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 298, 149–153. [DOI] [PubMed] [Google Scholar]
- Gottwald, U., Brokamp, R., Karakesisoglou, I., Schleicher, M., and Noegel, A.A. (1996). Identification of a cyclase-associated protein (CAP) homologue in Dictyostelium discoideum and characterization of its interaction with actin. Mol. Biol. Cell 7, 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne, P., Sechi, A., Benesch, S., and Small, J.V. (2001). Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492, 215–220. [DOI] [PubMed] [Google Scholar]
- Hubberstey, A.V., and Mottillo, E.P. (2002). Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16, 487–499. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487. [DOI] [PubMed] [Google Scholar]
- Kawamukai, M., Gerst, J., Field, J., Riggs, M., Rodgers, L., Wigler, M., and Young, D. (1992). Genetic and biochemical analysis of the adenylyl cyclase-associated protein, cap, in Schizosaccharomyces pombe. Mol. Biol. Cell. 3, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila, T., and Drubin, D.G. (1997). Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8, 367–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield, C.J., Feldman, M.E., Wan, L., and Almers, W. (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4, 691–698. [DOI] [PubMed] [Google Scholar]
- Mies, B., Rottner, K., and Small, J.V. (1998). Multiple immunofluorescence microscopy of the cytoskeleton. In: Cell Biology: A Laboratory Handbook, 2nd ed., ed. J.E. Celis, New York: Academic Press, 469–476.
- Moriyama, K., and Yahara, I. (2002). Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115, 1591–1601. [DOI] [PubMed] [Google Scholar]
- Noegel, A.A., Blau-Wasser, R., Sultana, H., Muller, R., Israel, L., Schleicher, M., Patel, H., and Weijer, C.J. (2004). The cyclase associated protein CAP as regulator of cell polarity and cAMP signaling in Dictyostelium. Mol. Biol. Cell 15, 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel, A.A., Rivero, F., Albrecht, R., Janssen, K.P., Kohler, J., Parent, C.A., and Schleicher, M. (1999). Assessing the role of the ASP56/CAP homologue of Dictyostelium discoideum and the requirements for subcellular localization. J. Cell Sci. 112, 3195–3203. [DOI] [PubMed] [Google Scholar]
- Ono, S., Minami, N., Abe, H., and Obinata, T. (1994). Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 269, 15280–15286. [PubMed] [Google Scholar]
- Ono, S., and Benian, G.M. (1998). Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273, 3778–3783. [DOI] [PubMed] [Google Scholar]
- Peränen, J., Rikkonen, M., Hyvönen, M., and Kääriäinen, L. (1996). T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373. [DOI] [PubMed] [Google Scholar]
- Peränen, J., Rikkonen, M., and Kääriäinen, L. (1993). A method for exposing hidden antigenic sites in paraformaldehyde-fixed cultured cells, applied to initially unreactive antibodies. J. Histochem. Cytochem. 41, 447–454. [DOI] [PubMed] [Google Scholar]
- Rice, D.P., Aberg, T., Chan, Y., Tang, Z., Kettunen, P.J., Pakarinen, L., Maxson, R.E., and Thesleff, I. (2000). Integration of FGF and TWIST in calvarial bone and suture development. Development 127, 1845–1855. [DOI] [PubMed] [Google Scholar]
- Rogers, S.L., Wiedemann, U., Stuurman, N., and Vale, R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D.A., Korshunova, Y.O., Schroer, T.A., and Cooper, J.A. (1994). Differential localization and sequence analysis of capping protein beta-subunit isoforms of vertebrates. J. Cell Biol. 127, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiston, J., Hubberstey, A., Yu, G., and Young, D. (1995). Differential expression of CAP and CAP2 in adult rat tissues. Gene 165, 273–277. [DOI] [PubMed] [Google Scholar]
- Vartiainen, M., Ojala, P.J., Auvinen, P., Peränen, J., and Lappalainen, P. (2000). Mouse A6/twinfilin is an actin monomer-binding protein that localizes to the regions of rapid actin dynamics. Mol. Cell. Biol. 20, 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen, M.K., Mustonen, T., Mattila, P.K., Ojala, P.J., Thesleff, I., Partanen, J., Lappalainen, P. (2002). The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen, M.K., Sarkkinen, E.M., Matilainen, T., Salminen, M., and Lappalainen, P. (2003). Mammals have two twinfilin isoforms whose subcellular localizations and tissue distributions are differentially regulated. J. Biol. Chem. 278, 34347–34355. [DOI] [PubMed] [Google Scholar]
- Vojtek, A., Haarer, B., Field, J., Gerst, J., Pollard, T.D., Brown, S., and Wigler, M. (1991). Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell 66, 497–505. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B., and Cooper, J.A. (1993). Identification and characterization of a cDNA encoding mouse CAP: a homolog of the yeast adenylyl cyclase associated protein. J. Cell Sci. 105, 777–785. [DOI] [PubMed] [Google Scholar]
- Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A.L., and Riezman, H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills, Z., Emerson, M., Rusch, J., Bikoff, J., Baum, B., Perrimon, N., and Van Vactor, D. (2002). A Drosophila homolog of cyclase-associated proteins collaborates with the Abl tyrosine kinase to control midline axon pathfinding. Neuron 36, 611–622. [DOI] [PubMed] [Google Scholar]
- Yu, G., Swiston, J., and Young, D. (1994). Comparison of human CAP and CAP2, homologs of the yeast adenylyl cyclase-associated proteins. J. Cell Sci. 107, 1671–1678. [DOI] [PubMed] [Google Scholar]
- Yu, J., Wang, C., Palmieri, S.J., Haarer, B.K., and Field, J. (1999). A cytoskeletal localizing domain in the cyclase-associated protein, CAP/Srv2p, regulates access to a distant SH3-binding site. J. Biol. Chem. 274, 19985–19991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.