Abstract
We used cDNA microarrays in a systematic study of the gene expression responses of HeLa cells and primary human lung fibroblasts to heat shock, endoplasmic reticulum stress, oxidative stress, and crowding. Hierarchical clustering of the data revealed groups of genes with coherent biological themes, including genes that responded to specific stresses and others that responded to multiple types of stress. Fewer genes increased in expression after multiple stresses than in free-living yeasts, which have a large general stress response program. Most of the genes induced by multiple diverse stresses are involved in cell-cell communication and other processes specific to higher organisms. We found substantial differences between the stress responses of HeLa cells and primary fibroblasts. For example, many genes were induced by oxidative stress and dithiothreitol in fibroblasts but not HeLa cells; conversely, a group of transcription factors, including c-fos and c-jun, were induced by heat shock in HeLa cells but not in fibroblasts. The dataset is freely available for search and download at http://microarray-pubs.stanford.edu/human_stress/Home.shtml.
INTRODUCTION
All living organisms need to sense and respond to conditions that stress their homeostatic mechanisms. Many environmental variations impose stresses that can damage or kill the organism in the absence of an appropriate response. For example, heat shock can disrupt protein folding, and changes in redox state can cause temporary or permanent damage to a variety of cellular structures, including proteins, lipids, and DNA. To minimize the effects of stress-related damage, organisms have evolved systems to detect and respond to stress.
For unicellular organisms, the range of expected variations faced by an individual cell can be extreme because such organisms have only limited ability to control environmental variables such as the temperature, extracellular redox state, and nutrient levels. The yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe activate a large transcriptional response, known as the “environmental stress response” (ESR), in response to a variety of stresses (Gasch et al., 2000; Causton et al., 2001; Chen et al., 2003). In S. cerevisiae, this response includes >300 genes with increased expression and 600 genes with reduced expression, encompassing 14% of the genome. This likely reflects the importance for yeast to respond to environmental perturbation to survive. A large general response to environmental stress may not be a ubiquitous characteristic of unicellular organisms; the general stress response of the fungus Candida albicans is considerably smaller than that of Saccharomyces, at least under standard laboratory conditions (Enjalbert et al., 2003).
We might expect that the gene expression programs induced by stresses in human cells will be different from those seen in yeast. Individual human cells in vivo are not subject to the wide variety of environmental conditions experienced by free-living unicellular organisms because mammals have evolved complex buffering and control mechanisms to minimize environmental variation within the organism. In animals, physiological responses typically involve coordinated actions of many cells because the labor involved in maintaining homeostasis is divided among different cell types. Thus, cell communication is likely to be as important as the cell-autonomous response to physiological stimuli in humans, and the diversity of distinct differentiated cell types may be accompanied by a corresponding diversity in their responses to stress. Mammalian cells also have many physiological behaviors not present in yeast that could contribute to their stress responses, such as programmed cell death and complex intercellular interactions.
Biological processes associated with stress responses play important roles in normal development and homeostasis. For example, defects in the heat shock response compromise early development in mice (Xiao et al., 1999), and overexpressing the heat shock transcription factor in Caenorhabditis elegans results in increased longevity (Garigan et al., 2002; Lund et al., 2002). Defects in systems associated with the stress responses contribute to human diseases such as cancer, diabetes (Oyadomari et al., 2002), Alzheimer disease (Terro et al., 2002), Parkinson disease (Imai et al., 2000), Osteogenesis Imperfecta (Lamande and Bateman, 1999), cardiovascular disease (Pockley, 2002), and others. A comprehensive understanding of how cells lines respond to common stresses would also be a useful tool for the interpretation of other large-scale gene expression data.
The response of cell lines to stress has been explored on a limited basis with microarrays, including studies of oxidative stress in breast cancer cells (Chuang et al., 2002), endoplasmic reticulum (ER) stress in HeLa cells (Okada et al., 2002), and UV exposure in keratinocytes (Li et al., 2001; Sesto et al., 2002). However, many of these studies examined less than one-quarter of the genes in the genome and none of them systematically examined the relationships among the gene expression responses to multiple types of stress.
As a first attempt to explore how mRNA levels change in response to various stresses on a genomic scale, we measured the gene expression patterns of ∼25,800 genes in cultured human cells after exposure to heat shock, ER stress, oxidative stress, and confluent growth with cDNA microarrays. We examined the gene expression response to each of these stresses in HeLa cells (which are derived from a cervical carcinoma) and in primary lung fibroblasts. We found that in human cells, unlike in yeast, the set of genes induced by most stresses is smaller than the sets of genes induced by specific stresses. We identified genes expressed in patterns characteristic of known stress responses such as the heat shock response and the unfolded protein response (UPR), as well new expression patterns not previously described in the response of human cells to stress. There were substantial differences in the gene expression responses between the cell lines tested, illustrating that differentiation and cell type influence the biological response of human cells to stress.
MATERIALS AND METHODS
Cell Culture Conditions
Culture conditions were as follows: HeLa S3 cervical carcinoma cells (Puck et al., 1956) were grown in DMEM supplemented with 10% fetal bovine serum, 100 U of penicillin-streptomycin, and 1× nonessential amino acids at 37°C with 5% CO2; normal human diploid lung fibroblasts (American Type Culture Collection, Manassas, VA) were grown under the same conditions as HeLa cells but with 20% serum; and K-562 cells (Lozzio and Lozzio, 1975) were grown in spinner flasks at 37°C in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and 100 U/ml penicillin-streptomycin. All experiments on adherent cells began at least 24 h and at most 48 h after plating and changing the media. Cell density at the first time points was near 2 × 106/150-mm tissue culture dish (Falcon Plastics, Oxnard, CA). Time points for each condition are indicated in Table 1.
Table 1.
Conditions and cell lines used for microarray experiments
| Stress | Cell line | Time points (h) |
|---|---|---|
| Heat shock | ||
| 42° Heat shock | HeLa | 0, 0, 0.5, 1, 2, 3, 4, 6, 8, 24 |
| 42° Heat shock | Fibroblast | 0, 0, 0.5, 1, 2, 4, 10, 16, 24 |
| 42° Heat shock | K-562 | 0, 0, 0, 1, 2, 3, 4, 6, 8, 24 |
| ER stress | ||
| Tunicamycin | HeLa | 0, 0, 0, 0, 0.5, 1, 2, 4, 6, 8, 12, 24, 36 |
| DTT | HeLa | 0, 0.5, 1, 2, 4, 6, 8, 16, 24, 30 |
| DTT | Fibroblast | 0, 0, 0, 0, 0.5, 1, 2, 3, 4, 6, 8, 12, 16, 24, 36 |
| Oxidative stress | ||
| Hydrogen peroxide | HeLa | 0, 0.5, 1, 2, 6, 8, 16, 24, 30 |
| Menadione | HeLa | 0, 0, 0, 0, 0.5, 1, 2, 4, 8, 12, 24, 32 |
| Menadione | Fibroblast | 0, 0, 0, 0, 0.5, 1, 3, 4, 6, 8, 12, 24, 36 |
| Crowding | ||
| Crowding | HeLa | 0, 24, 48, 72, 120, 144, 168 |
| Crowding | Fibroblast | 0, 24, 48, 72 |
Stress Treatments
We induced heat shock by transferring culture flasks from a 37°C incubator to a 42°C incubator. ER stress was induced by treating HeLa cells with 500 nM tunicamycin (Sigma-Aldrich, St. Louis, MO), or by treating HeLa cells or fibroblasts with 2.5 mM dithiothreitol (DTT). Oxidative stress was induced by treating HeLa cells with 600 μM hydrogen peroxide or by treating HeLa cells or fibroblasts with 10 μM menadione bisulfate (Sigma-Aldrich). For the crowding experiments, we seeded HeLa cells at 106 cells/plate and harvested cells at 24-h intervals; we replaced the medium every 24 h to reduce the effect of serum/nutrient starvation. For fibroblasts, cells were seeded at ∼2.5 × 106/plate and collected samples at 24-h intervals, without changing the media.
RNA Preparation
For each time point, we prepared mRNA by using the FastTrack 2.0 mRNA isolation kit (Invitrogen, Carlsbad, CA) from either one plate of HeLa cells, two plates for fibroblasts, or 12 ml of K-562 cells (starting density of ∼8 × 105 cells ml–1), or ∼107 cells/time point. Adherent cells were washed once in 10 ml of phosphate-buffered saline and lysed on the plates; we transferred 12 ml of K-562 cells to a centrifuge tube, washed once in 10 ml of phosphatebuffered saline, and lysed in the tube. We incubated the lysates for 1 h at 45°C and froze on dry ice before continuing the isolations. For the DTT and menadione experiments in fibroblasts, total RNA was prepared with RNeasy kits (QIAGEN, Valencia, CA), and then amplified for use in the microarray experiments with a modified Eberwine protocol (Wang et al., 2000).
Microarray Hybridizations
We labeled RNA isolated from each experiment with Cy5-dUTP (Amersham Biosciences, Piscataway, NJ) and RNA isolated from untreated cells or pooled reference RNA with Cy3-dUTP by reverse transcription and the purified, labeled probe was applied to human cDNA microarrays (Stanford Functional Genomics Facility, http://www.microarray.org/) by using standard protocols (Eisen and Brown, 1999). The cDNA microarrays contained >40,000 spots representing ∼25,802 genes (as estimated using Unigene clusters [build 160]); the microarrays used for the HeLa crowding experiment contained a smaller subset of 21,552 of the clones present on the larger arrays, representing ∼18,325 genes.
RNA from growing HeLa cells was used as the reference (Cy3-labeled sample) for the HeLa heat shock and crowding experiments. For other experiments, pooled RNA from all time points was used. We scanned microarrays by using a GenePix 4000A scanner (Axon Instruments, Foster City, CA), extracted data by using GenePix 3.0 software (Axon Instruments), and stored the data in the Stanford Microarray Database (Sherlock et al., 2001). The entire dataset is available at http://microarray-pubs.stanford.edu/human_stress/Home.shtml.
Data Analysis
To facilitate easy manipulation of the heterogeneous microarray result dataset, we wrote a PERL software package, PCL_analysis, which facilitates the building of heterogeneous datasets from individual experiments and allows the user to individually center or zero-transform each independent time course within a composite dataset, something that is difficult to do with commonly used existing software. This software is available for download at http://pcl-analysis.sourceforge.net/.
We excluded from the analysis all data from spots with inconsistent data (define by a regression R2 < 0.4 for a linear fit between the Cy3 and Cy5 channel pixel intensities) or that had been flagged as defective by visual inspection during data extraction (for example, due to scratches or dust particles). For the subsequent analysis, we used the log2 of the background-subtracted, normalized ratio of the mean Cy5 and mean Cy3 expression values. Normalization was done using the default method of the Stanford Microarray Database, which scales the Cy5 channel signal for each spot by a constant such that the total signal in all well-measured spots is the same as for the Cy3 channel (Sherlock et al., 2001).
We transformed the data for each time course to show expression changes for each gene relative to the zero time point by subtracting the median log2 expression ratios in the time-zero replicates from the corresponding data measured in each time point. The exception was the HeLa crowding time course, where the data were transformed relative to an average of the expression measurements in the zero and 1-d time points. During visualization, we display the transformed zero time points along with the transformed time points because they provide a visual indicator of the consistency of the data for a given gene. We combined the gene expression data from all of the stress experiments with data from a previously published study of HeLa cells synchronized in the cell cycle with double-thymidine and thymidine-nocodazole blocks (Whitfield et al., 2002). For the cell cycle time courses, the data for each gene were mean-centered by subtracting each gene's mean expression value across the entire time course from the expression value for each time point within that time course. Only genes for which at least 70% of expression measurements were considered technically adequate were included in the subsequent analysis. We clustered the full set of genes by using hierarchical clustering with average linkage (Eisen et al., 1998) and chose clusters for further study by visual inspection.
This analysis generated 23 partially overlapping clusters with expression patterns that we believed were biologically relevant based on subjective criteria. Hierarchical clustering organizes the gene expression data such that each gene is assigned to a single cluster of similarly expressed genes. However, genes often have expression profiles related to more than one cluster. We used a secondary cluster growth step to include in our analysis genes with expression patterns similar to one or more of our selected clusters but that clustered elsewhere in the full-genome cluster. To do this, we compared each gene's expression pattern to the mean expression pattern of each of the 23 manually defined hierarchical clusters, and then added the gene to the cluster if it was expressed similarly to that cluster's mean expression profile (based on a Pearson r >0.70). In total, this resulted in 253 additional genes being added to the 23 base clusters. For visualization, we then hierarchically clustered the data for each of these lists of genes separately. We tested the significance of these assignments by repeating the clustering assignments with randomized data; we randomly permuted the expression measurements within rows (genes) to generate 4.6 × 106 randomized expression vectors, performed the “time-zero” transformations and centering as described above, and tested each randomized gene for correlation to the centroid of each cluster. We used the correlations of randomized genes to the cluster centroids to estimate our false positive rate for cluster assignment. An average of 18 randomized genes was assignable to clusters by using the original criteria. The final clustered data subset contains 1673 genes, with an estimated false positive rate of 18/1673 (1.1%).
We compared our dataset with other datasets by first linking the data by using the clone ID and then clustering by using either all genes or subsets corresponding to specific responses, as described in RESULTS. In cases where the comparison data were not generated with the same clone set that is present on our microarrays, we first averaged rows corresponding to the same Unigene identifier and then linked the datasets through these identifiers before clustering.
Comparison of Expression in Yeast-Human Homologues
To directly compare the yeast and human stress response datasets, we used a BLAST-based method to find candidate orthologues between the ENSEMBL-predicted human gene set (Hubbard et al., 2002) and the SGD yeast-predicted protein set (Cherry et al., 1998). We calculated pairwise alignment scores between each yeast-human gene pair by using the WU-BLAST software package (http://blast.wustl.edu) with the options “-postsw –wordmask SEG–W4 –T20 –Z = 1000000000 –lcmask –topcomboN 1.” The human sequences were first masked such that common paralogous domains could not initiate word hits (Sidow, personal communication). We selected pairs with an E-value <10–10 and alignment >80% of the length of both sequences as potential orthologues. Although this method will miss some pairs due to the high divergence between yeast and human, and will also include paralogs predating the yeast-human divergence, it should be enriched for orthologues. Manual browsing of the predicted orthologous pairs confirmed this prediction. We tested for the existence of an ESR orthologous to the yeast response by hierarchically clustering and visualizing all human genes measured that were possible orthologues of genes whose expression increases or decreases in the yeast ESR. The human clones on the arrays were assigned to ENSEMBL genes by linking through RefSeq accession numbers by using tools available at SOURCE (http://source.stanford.edu/) and ENSEMBL (http://www.ensembl.org/).
We compared trends in orthologous gene sets by examining the median expression for all genes in all conditions examined in both species. For yeast, these included heat, hydrogen peroxide, menadione, DTT, and stationary phase; for human cells, they included heat, hydrogen peroxide, menadione, DTT, and crowding.
RESULTS
Overview
We used cDNA microarrays to measure gene expression patterns in cultured human cells after exposure to four types of environmental stress: heat shock, ER stress, oxidative stress, and crowding. The experiments are outlined in Table 1. We examined the responses of two or three cell types to each stress to address the possibility that the responses might differ between cell types. For each stress condition, we measured the temporal responses at several time points over an interval of 24–36 h, allowing us to see the effects of stress over an interval longer than the normal cell cycle duration. For all stresses, we first conducted dose–response studies and chose doses slightly below the threshold where we observed significant lethality.
For the heat shock experiment, we shifted three cell lines, HeLa, primary lung fibroblasts, and K-562 erythroleukemia cells, from 37 to 42°C. We used two agents to induce ER stress: the glycosylation inhibitor tunicamycin and the thiol reducing agent DTT. We also used two agents to induce oxidative stress: menadione, which generates superoxide radicals through a cycling reaction (Monks et al., 1992), and hydrogen peroxide, a source of peroxide radicals. Finally, we performed “crowding” experiments by allowing HeLa cells or fibroblasts to grow past confluence. Although we cannot separate the potential influence of factors such as cell contacts and density, or depletion or degradation of media components on the resulting gene expression changes, these crowding experiments can help us recognize potential confounding effects of differences in cell density and culture time in experiments designed to examine responses to specific stresses.
We analyzed the data by using an extended version of hierarchical clustering (Eisen et al., 1998) to group genes with similar expression changes in response to stress. By analyzing the data from our studies of stress along with a HeLa cell cycle gene expression dataset (Whitfield et al., 2002), we were able to distinguish genes that respond directly to the stress agents from those that change because the fraction of cells in each phase of the cell cycle changes, for example, after cell cycle arrest. After an initial analysis of the entire dataset, representing 25,802 unique Unigene identifiers (putative genes), we selected 23 clusters, containing 1245 distinct Unigene identifiers, for further analysis; these were selected because they had coherent expression patterns for which we could provide a biological explanation. Each individual list of genes (referred to hereafter as a cluster) was then expanded to include other similarly expressed genes (see MATERIALS AND METHODS for details). Our estimated false positive rate for cluster assignment is 1.1%. The complete dataset and the data for each cluster are freely available for download.
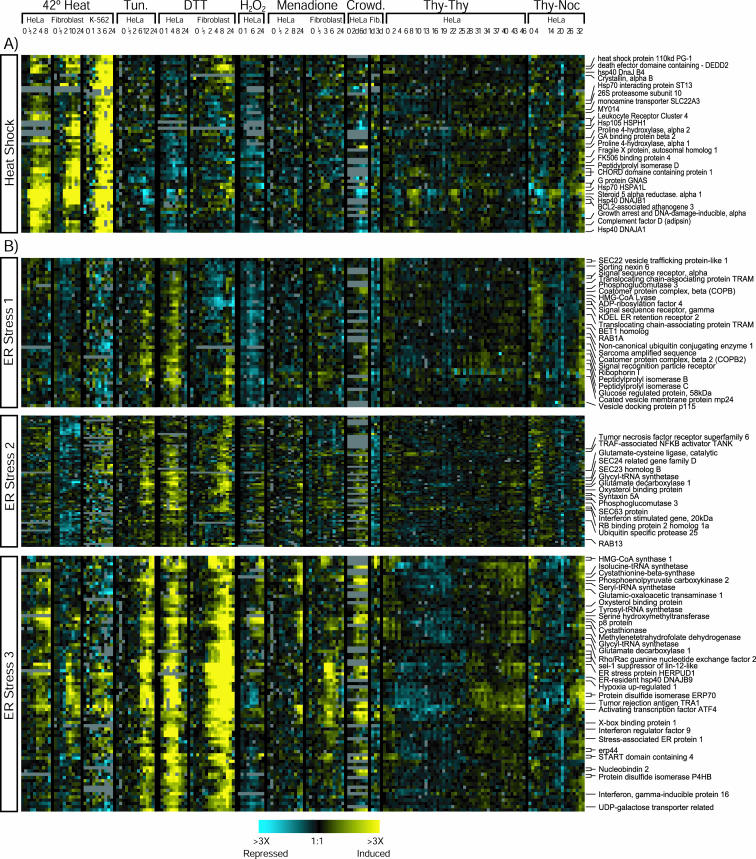
An overview of the major responses is shown in Figure 1, which displays the pooled data for all the genes in the selected individual clusters, organized into one large hierarchical cluster. The experiments yielded two striking results. First, the responses to each type of stress were mostly distinct; there was no dominant general stress response comparable to that of S. cerevisiae. Second, the cell types we tested differ substantially in their gene expression responses to stress.
Figure 1.
Overview of the clusters selected for further analysis. We grouped the 1673 genes present in the 23 clusters that we selected for further analysis (see MATERIALS AND METHODS) and hierarchically clustered them together to demonstrate the relationship between the different expression patterns. Each row represents one gene or clone and each column represents one experiment. Yellow bars represent higher expression than in the zero time points, blue bars reduced lower expression, and gray bars represent missing data. We have indicated on the right the approximate extent of clusters shown in more detail in Figures 3, 4, 5, including genes whose expression was induced by multiple stresses (Figure 3), in specific cell types (Figure 4), or by individual types of stress (Figure 5) (note: because genes can be in more than one cluster, these boundaries are only approximate).
General Responses
Limited Overlap in the Responses of Cultured Human Cells to Different Stresses Gasch et al. (2000) showed that treating S. cerevisiae with any of a wide variety of agents causes a transient increase or decrease in the expression of ∼900 genes, 14% of the genome (the ESR). There was no similarly pervasive general stress response in human cells (Figure 2). Although the expression of some genes increased after multiple stresses, many more transcripts responded to only one or two types of stress. Even the genes whose expression was induced after multiple stresses were usually not completely general. We only found three genes (p8, stearoyl CoA desaturase, and cyclin G2) whose transcripts were induced more than 1.5-fold in response to every stress in each cell type, whereas hundreds of genes passed this threshold in yeast. Similarly, the transcript levels of only 25 genes changed fourfold or more in response to at least three of the four classes of stress in human cells, whereas the expression of 790 genes changed fourfold in at least three of four analogous stress conditions in yeast (heat shock, unfolded protein response, oxidative stress, and stationary phase).
Figure 2.
Comparison of the genome-wide responses to stress between human cell lines and yeast. The full, hierarchically clustered, human stress dataset and yeast stress dataset (Gasch et al., 2000) (all genes) are shown for comparison. Genes for both human and yeast are ordered by hierarchical clustering. The color scheme is the same as for Figure 1; the human data are shown at a higher contrast because of the generally lower magnitude of inductions in that dataset.
The number of genes that responded strongly to each individual stress was smaller in human cell lines than in yeast. For example, expression of 123 human genes consistently (in at least two time points) increased or decreased by at least fourfold after heat shock in human cells, whereas 1042 yeast genes met this criterion. We obtained similar results for the other types of stress. Thus, the transcriptional responses of cultured human cells to individual stresses involve smaller numbers of genes or occur at a lower magnitude than in yeast.
To address the possibility that an ESR like that of yeast might exist, but with a lower amplitude or more discriminately than in yeast, we examined the expression patterns of human orthologues of the genes induced or repressed in the yeast ESR. Visually, the distribution of expression patterns of putative human orthologues of ESR-induced yeast genes was indistinguishable from that of all human genes with yeast orthologues (Supplemental Figure 1). Putative human orthologues of yeast ESR-induced genes in the human stress experiments had a 0.07-fold median decrease, similar to the 0.067-fold median decrease in the expression of the set of all human genes with yeast homologues. Similarly, the median expression of yeast orthologues of broadly stress-induced human genes was similar to the set of all yeast genes with human orthologues (0.11-fold vs. 0.14-fold decreased expression). In contrast, the yeast ESR-induced genes had a median 0.46-fold increased expression in the yeast experiments analogous to our human experiments and the four clusters of human genes with broadly increased expression levels after stress had a median increase of 0.25-fold.
Although genes whose expression increased after stress in our experiments tended to respond selectively to one or a few specific stimuli, expression of many genes was reduced after each type of stress. However, the human homologues of genes reduced in the yeast ESR did not show a significant reduction in expression after stress; they had a 0.079-fold median reduced expression compared with 0.067-fold for all genes with yeast orthologues (p = 0.16). In fact, many of them actually increased in expression after some stresses (Supplemental Figure 1). Yeast homologues of human genes with reduced expression after stress were no more likely than random genes to be part of the repressed component of the yeast ESR; they were equally likely to be part of the induced component of the ESR. Together, these results suggest a high degree of divergence between the human and yeast common stress responses.
To address the possibility that independent general stress responses might exist in each of the different cell types, we searched for genes that were induced or repressed at least twofold in at least one time point for each class of stress in either HeLa cells or fibroblasts. No genes are induced at this threshold when all experiments in either cell type are considered together. We found small sets of genes (15–32) in each class (Supplemental Figure 2). Although these sets of genes have some potentially interesting biological themes, their size and breadth of function are not comparable with the yeast general stress response.
Virtually all of the 25 genes generally induced in HeLa cells also were induced strongly in at least one fibroblast experiment, and thus are not specific to HeLa. However, they are candidates for genes generally induced by stress in HeLa cells. Many of these genes are involved in regulating cell proliferation and cell death [MEN1, p57(Kip2), p21(Cip1/Waf1), c-Fos, c-jun, btg-1, ERBB3, P8, and Caspase 9].
The set of 21 generally fibroblast-inducible genes has many more expressed sequence tags and uncharacterized genes, as well as several genes involved in intercellular signaling and adhesion (angiopoietin-like 1 and 2, BBS2) and the cytoskeleton (katanin A1, myosin regulatory light chain interacting protein). Although these genes are less broadly induced in HeLa cells, most were induced in at least one HeLa time course.
A similar pattern is evident in each cell type's generally repressed genes. The 14 genes with twofold reduced expression after all types of stress in HeLa cells also were repressed in a subset of the fibroblast responses. In contrast, the 32 genes with twofold reduced expression after all types of stress in fibroblasts were mostly unchanged in stressed HeLa cells; they include genes involved in extracellular matrix metabolism and intercellular signaling. These correspond to a cluster, described later, containing genes that most likely cannot be repressed further in HeLa cells because they are basally expressed at very low levels in these cells.
Two Groups of Genes with Increased Expression after Multiple Stresses Although there seems to be no equivalent of the yeast ESR in cultured human cells, expression of some human genes increased after multiple stresses. Seven of the 23 subclusters chosen in our analysis consist of genes induced after multiple stresses. These genes can be divided into two main groups, one with early and transient induction and another with later and more sustained induction (Figure 3). Although many of these genes increased in expression after multiple classes of stresses and in both cell types, very few genes were induced after every condition tested. In addition, many of these broadly induced genes are involved in processes specific to higher organisms such as cell-cell signaling, apoptosis, and control of proliferation. We were able to identify yeast homologues for only 10 of the 216 human genes in these clusters, a much lower fraction than for the other clusters (19.3%) (p = 2.7 × 10–10).
Figure 3.
Genes induced by multiple stresses. Three clusters of early response genes (enriched for immediate early genes) (A) and four clusters with broader general patterns of induction (B) are shown with the same color scheme as in Figure 1.
Transcripts Encoding Many Signal Transduction Proteins Increased Early after Multiple Stresses Three clusters whose transcripts were rapidly and transiently induced by multiple stresses were enriched for genes involved in cell signaling and transcription factors and included many genes known to be induced in the immediate early response to serum (Figure 3A). Of the 83 annotated genes in these clusters, 43% (36) encode transcription factors and 69% (57) encode molecules involved in any step of signal transduction. The presence of many transcription factors and signaling molecules in these clusters is consistent with a role in reprogramming cellular regulatory systems.
The set of the early-response genes differed depending on the type of stress. Expression of all of these genes increased very early after treatment of HeLa cells or fibroblasts with DTT as well as after the addition of fresh medium at the start of each cell cycle time course. Only a subset of the genes in two of the clusters increased expression after oxidative stress. One subset responded to hydrogen peroxide in HeLa cells and heterogeneously to menadione in fibroblasts. These included the AP-1 transcription factors Fos and JunB, and the apoptosis-inducing proteins Caspase 9, p21, tumor necrosis factor-α–induced protein 3, BTG1, and genes involved in intercellular communication. The responses of these genes in crowded cells and after heat shock were more complicated and differed between cell types.
The rapid and transient induction of these genes by could occur through a previously characterized pathway involving IRE1-mediated activation of a tumor necrosis factor-α receptor (TRAF), which then activates c-jun NH2-terminal kinase/stress-activated protein kinase and activator protein 1 (AP-1)–mediated gene expression (Urano et al., 2000). Two of the clusters showed particularly interesting expression patterns after DTT treatment. After reaching a maximum ∼2 h, transcript levels of genes in these clusters dropped dramatically, to a minimum well below the initial expression level. Our data suggest one possible mechanism for this effect; the TRAF inhibitor TANK (Rothe et al., 1996), which was induced by ER stress (see below, Figure 5), could provide negative feedback to TRAF and result in a decreased AP-1 activity.
Figure 5.
Stress-specific gene expression patterns. The heat shock cluster (A) and three clusters containing genes with expression induced by ER stress agents (B) are shown with the same color scheme as in Figure 1.
Transcripts with Sustained Induction by Multiple Stresses Four clusters with sustained increases in expression in most stress time courses contained genes involved in many diverse biological processes (Figure 3B). Some have functions related to secretion and lipid metabolism and could represent a more broadly expressed set of genes responsible for ER stress protection, and others have potential relevance to oxidative stress, including several putative stress-response transcription factors, negative regulators of cell proliferation (including p21cip1/waf1 and GADD153), DNA damage repair proteins, and antioxidants. Many of the genes in these clusters likely play a role in cell signaling, including the transcription factors ATF2, ATF3, and ATF4 and ligands FGF4 and NOTCH2.
Nearly all of the genes induced specifically by both ER stress and oxidative stress agents in fibroblasts were not induced in HeLa cells; we did not identify a cell type-independent response specific to oxidative stress. Many of the generally induced transcripts did not increase in response to one or both oxidative stresses in HeLa cells. They were typically more strongly induced after DTT exposure than in response to classical oxidative stress agents, as was true for the fibroblast-specific redox genes. This could mean that these genes are independently induced by multiple types of stresses or, alternatively, that ER stress, heat, and crowding can induce oxidative stress as a secondary effect.
Genes with Decreased Expression in Response to Diverse Stresses The expression of a large set of genes (>1000) was diminished after most stresses. In general, the maximum magnitudes of these general decreases in transcript levels were less than twofold in human cells. The genes with broadly reduced expression are involved in many aspects of cell biology and metabolism. Some genes encoding cytoskeleton components and motors, nutrient transporters, metabolic enzymes, components of signal transduction pathways, components of transcriptional, RNA metabolism, translational and protein degradation machinery, secretory pathway components, and cell cycle control all had modestly reduced expression after several stresses, although the extent to which they declined varied for each gene and stress. We looked specifically at the expression of genes encoding ribosomal proteins, splicing factors, proteasomal components, and mitochondrial ribosomal proteins (Supplemental Figure 3, B–E). Expression of these genes decreased modestly after most stresses. These could represent direct responses to stress or secondary effects such as stress-induced changes in growth rates.
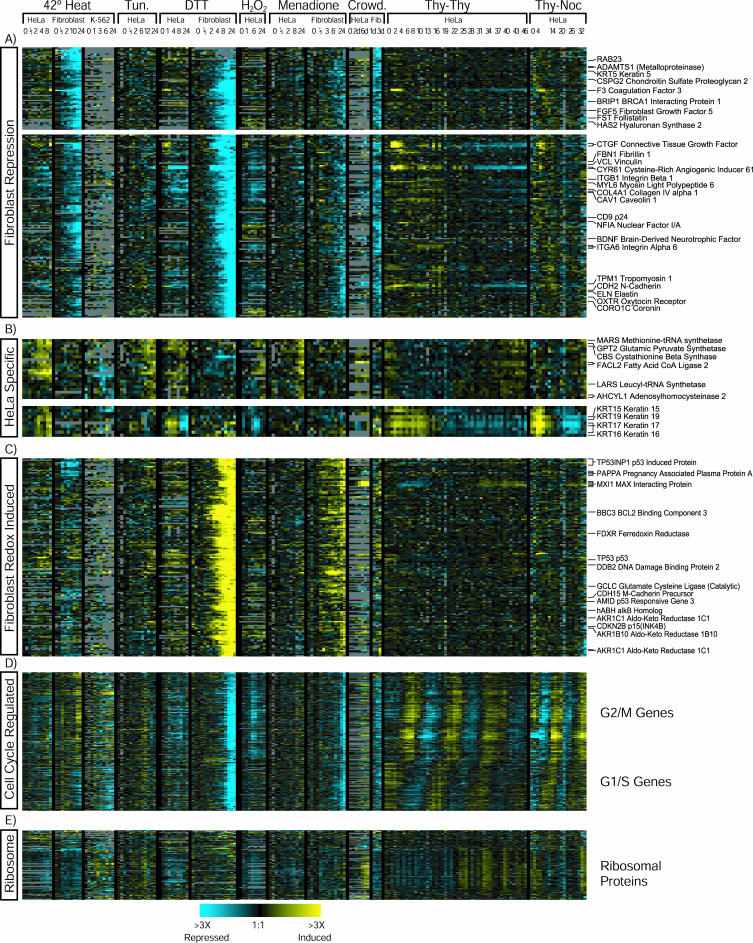
Transcripts That Respond to Multiple Stresses Differently in HeLa Cells and Fibroblasts One of the most striking results of this study was the extent to which fibroblasts and HeLa cells differed in their responses to stress (Figure 4). These differences are seen, for example, in two related clusters, containing genes that were not cell cycle regulated and whose transcript levels decreased after all stresses in fibroblasts but not HeLa cells (Figure 4A). Many of these genes are involved in cell adhesion (vinculin, keratin 5, CD9, N-cadherin, coronin 1C, caveolin 1, and integrin β-1) and synthesis of extracellular matrix (collagens COL4A1, COL5A2, elastin, hyaluronan synthetase, fibrillin 1, and chondroitan sulfate proteoglycan 2). All of these genes are expressed at a much higher in fibroblasts than in HeLa cells in a dataset comparing gene expression patterns in multiple cell types and tissues (Whitfield et al., 2003). The expression of relatively few transcripts changed in response to multiple stresses specifically in HeLa cells. A few are represented in two small clusters of genes with HeLa-specific induction (Figure 4B).
Figure 4.
Cell line-dependent gene expression patterns. Data for cell line specific clusters are shown with the same color scheme as in Figure 1. Clusters of genes whose expression was repressed by stress in fibroblasts (A), induced by stress in HeLa cells (B), or induced by redox stress in fibroblasts (C) were produced using the two-step clustering method described in MATERIALS AND METHODS. (D) We extracted and clustered the data for 1134 clones found to be cell cycle regulated by (Whitfield et al., 2002). The genes shown are the core of the resulting cluster and contain the most strongly cell cycle-regulated genes. (E) Data for all of the genes encoding ribosomal proteins present on the arrays were extracted and the genes were ordered by hierarchical clustering.
Condition-specific Responses to Stress
The responses of cultured human cells to each type of stress were dominated by condition-specific patterns of gene expression that overlapped only slightly. We describe the responses to each stress condition in more detail here. For each type of stress, there were many genes whose expression increased or decreased in one cell type but not another; in some cases, these cell type-specific responses dominated the cell type-independent responses.
Heat Shock
The Specific Heat Shock Response Involves Many Unexpected Genes We found a cluster of 50 unique genes whose expression increased after heat shock in all three of the cell lines we examined, but not in response to the other types of stress (Figure 5A). The genes with the strongest increase in expression, those that changed at least 10-fold in response to heat, include many familiar heat shock genes; genes encoding chaperones that assist in protein folding and their cofactors, including hsp70, hsp40, Hsp105, and Hsp110, hsp47 (SERPINH2), and a modulator of hsp70 activity (BAG3).
Several other genes were induced principally by heat shock but less strongly (4- to 10-fold) than those described above. These genes include additional regulators of chaperone function (Hsp70-interacting protein ST13, Hsp90 cochaperone TEBP, and FK506 binding protein 4) as well as several genes important in posttranslational modification of proteins (transglutaminase and proline 4-hydroxylase α 1 and 2). Also, transcripts for genes involved in protein degradative pathways (proteasomal subunit PSMD10 and Siah-interacting protein), signal transduction (G protein GNAS, transcription factor GABPB2 and GADD45 α), membrane transport and signaling (phospholamban, monoamine transporter SLC22A3, and leukocyte receptor cluster 4), and metabolic processes (arylamine N-methyltransferase and a cytochrome b5 reductase) increased specifically after heat shock. The expression of several genes of unknown function increased after heat shock. The newly identified induction of these genes in response to heat shock suggests that the physiology of the mammalian heat shock response is broader than just regulation of protein folding and may include aspects related to cell-cell signaling and growth control.
Different Cell Types Differ Significantly in Their Transcriptional Responses to Heat Primary fibroblasts but neither HeLa nor K-562 cells increased expression of genes characteristically expressed in the G2/M phases of the cell cycle genes after 10 h of heat shock, suggesting a G2/M cell cycle arrest (Figure 4D). It was previously reported that heat shock causes a transient G2/M arrest in primary cell lines, but not in many cancer cell lines (Nakahata et al., 2002).
We saw no such evidence for cell cycle arrest in the gene expression programs of heat-shocked HeLa and K-562 cells. Instead transcripts for a large set of signal transduction molecules and transcription factors (such as c-fos and c-jun), many of which are induced in the “immediate-early” response to serum (Iyer et al., 1999), accumulated after 8–24 h of heat shock in these cancer cells but not in fibroblasts (Figure 3A). This response differed between K-562 and HeLa lines: three distinct clusters of immediate early genes were induced in heat shocked HeLa cells, whereas genes in only one of these clusters responded similarly in K-562 cells. None of these transcripts were induced by heat shock in fibroblasts; indeed, they actually decreased moderately in fibroblasts after heat shock.
ER Stress (DTT and Tunicamycin)
Cell Type-independent Patterns of Gene Expression in Response to Endoplasmic Reticulum Stress The responses of human cell lines to agents that interfere with ER protein homeostasis have been studied extensively on a gene-by-gene basis. The response to these stimuli is known as the UPR (for review, see Kaufman, 2002). Many types of stimuli are known to induce this response, including disulfide bond-reducing agents, glycosylation inhibitors, drugs that interfere with calcium transport, and glucose starvation. We found three clusters of coordinately expressed genes whose expression increased relatively specifically after treatment with both DTT and the glycosylation inhibitor tunicamycin (Figure 5B). Genes in these clusters play important roles in ER physiology; many are involved in processes such as secretion, ER protein folding, and membrane synthesis. It is notable that many of the genes in these clusters have not previously been characterized as part of the ER stress response. In general, genes in the UPR clusters responded more rapidly to DTT treatment (3–4 h in HeLa) than to tunicamycin (12–18 h), perhaps because DTT causes immediate accumulation of misfolded proteins in the ER, by reducing existing disulfide bonds, whereas tunicamycin causes a more gradual accumulation of misfolded proteins as old proteins are replaced by newly synthesized proteins that have not been properly glycosylated.
The first ER stress response cluster consisted primarily of genes involved in targeting proteins to the ER. Most of their transcripts were only subtly induced (∼2-fold), suggesting that modest changes in the levels of their transcripts is sufficient for an adaptive response to ER stress. This cluster included genes important for transport between the ER and the Golgi, including both structural components of vesicle coats as well as regulators of vesicle formation and targeting. It also included ER cotranslational translocation complex subunits, an ER KDEL retention receptor, and ER chaperones (the nonconventional ubiquitin NCUBE1, an ER hsp70 and peptidylprolyl isomerases). Several of the genes in the UPR clusters had not previously been linked to ER stress.
The second cluster of ER stress-responsive genes showed similarly low magnitudes of change, but their transcript levels increased slightly earlier. This cluster included genes involved in vesicle trafficking, lipid transport, signal transduction as well as metabolic genes that had not previously been shown to be responsive to ER stress, including phosphoglucomutase 3, glutamate decarboxylase, glutamate-cysteine ligase, and glycyl tRNA synthetase.
The third ER stress response cluster was also enriched for known unfolded protein response genes. These included a large number of ER chaperones and also one of the transcription factors important for UPR expression, XBP-1. This cluster was less specific to ER stress than the other two; many genes in this cluster also increased in expression after crowding, oxidative stress, or heat shock. However, these genes were not induced as strongly or consistently by non-UPR stresses as were the genes in the “general response” clusters and are categorized here as ER stress response on the basis of this and previous literature.
Differences between HeLa Cells and Fibroblasts in Their Responses to ER Stress Agents Although there was good concordance between the genes induced by ER stress in HeLa cells and fibroblasts, the kinetics and patterns of expression differed. Induction of most of the genes peaked 3–4 h after treatment of HeLa cells with DTT, but 12–16 h after treatment of fibroblasts with DTT. Also, the transcript levels of many genes, primarily those involved in the translocation of proteins into the ER, were initially reduced in fibroblasts for the first 4–6 h after treatment with DTT, after which they increased to a similar total increase as in HeLa cells.
A large number of genes were specifically induced in DTT treatment in fibroblasts but not by DTT or tunicamycin treatment in HeLa cells. In fibroblasts, DTT treatment caused a decrease in the relative abundance of most cell cycle-regulated transcripts, which suggests that the cells are exiting the cell cycle. However, alteration of the cell cycle alone is insufficient to explain most of the observed response of fibroblasts to DTT. We could not detect decreases in cell cycle-regulated transcripts until long after DTT treatment (∼24 h), whereas levels of many of these transcripts specifically increased or decreased soon after DTT treatment of fibroblasts (as early as 2 h). Many of the genes whose expression was altered by DTT showed a reciprocal response after treatment with serum (Iyer et al., 1999). Because DTT reduces disulfide bonds (e.g., in cell surface receptors or their ligands), it may have disrupted the ability of the cell to respond to factors in the serum-containing medium we used. The relative specificity of this effect to fibroblasts might reflect the greater dependence of fibroblasts on serum factors compared with HeLa cells.
Oxidative Stress
A Putative Response to Oxidative Stress and ER Stress in Fibroblasts but Not HeLa Cells We were unable to find any clusters of genes whose transcripts increase specifically after treatment with oxidative stress agents. Many of the genes whose transcripts were induced by multiple stresses had functions relevant to oxidative stress as described above, perhaps because oxidative damage can occur after diverse types of stress.
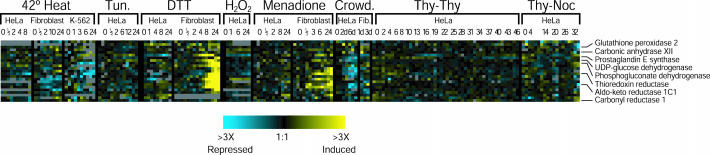
A second candidate oxidative stress response was a large cluster containing genes whose expression was induced in fibroblasts treated with DTT or menadione, but not under the same conditions in HeLa cells (Figure 4C). This cluster contains many genes important in proliferation control and oxidative stress. This cluster may represent a component of the oxidative stress response that is present in fibroblasts but absent or lost in HeLa cells.
Expression of some of the genes in this cluster may change as a result of cell cycle arrest. Some human cell types arrest in the cell cycle after treatment with oxidative stress agents (Pani et al., 2000; Bilodeau et al., 2002). We looked for evidence of cell cycle arrest in our cells by examining the expression of 1134 cell cycle-regulated genes (Whitfield et al., 2002) (Figure 4D). Expression of many cell cycle-regulated genes, and virtually all of those regulated most strongly, was greatly diminished in fibroblasts 12–24 h after treatment with DTT or menadione, which is consistent with a cell cycle arrest. We saw little or no decrease in the expression of these genes in HeLa cells after the same stresses. About one-half of the genes with specific increases in expression after DTT and menadione treatment in fibroblasts are expressed at a higher level in serum-starved (nondividing) fibroblasts than in fibroblasts grown in the presence of serum (Chang et al., 2002), suggesting that the increased expression of these genes might simply reflect cell cycle arrest. Some of these genes, however, could play an active role in arresting the cell cycle; indeed, several of the genes in this cluster are suspected to regulate proliferation (p53, MAX interacting protein 1, p53-inducible nuclear protein 1, cellular repressor of E1A-stimulated genes, and RB-inhibiting gene 1). However, other genes in this cluster are not differentially expressed in resting and dividing fibroblasts and thus may respond directly to oxidative stress.
Ribosomal Protein Transcripts Increase in “Crowded” HeLa Cells The crowding experiments are difficult to interpret because a complex set of changes including confluence itself, depletion of components of the media, and pH changes occur as cells become confluent. In addition, because HeLa cells and fibroblasts differ substantially in how fast they exhaust the media, the HeLa crowding experiment included media changes not included in the fibroblast crowding experiments. However, we did see an intriguing increase in transcripts encoding ribosomal proteins in HeLa cells after 5–6 d of crowding (Figure 4E), ∼2 d after the cells reached confluence; this did not occur in confluent fibroblasts. Additional targeted experiments will be needed to determine the significance of this result.
Use of the Stress Dataset to Interpret Other Datasets
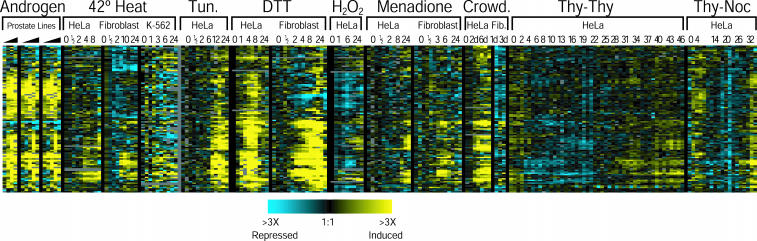
One of the principal uses of a systematic characterization of a physiological gene expression is to provide a framework for interpretation of other gene expression patterns. Two examples illustrate this application. DePrimo et al. (2002) studied androgen-regulated gene expression in prostate cancer cell lines. They noted that androgens induced the expression of genes involved in vesicle trafficking and secretion of proteins in these cell lines paralleling the effect of androgens on the secretion of prostatic fluid. We found that the major clusters of genes that we found to be ER stress responsive were similarly induced by androgen treatment of prostate cancer cells (Figure 6). This raises the possibility that the responses of cells to ER stress and the response of prostate cells to androgen share some biological features. For example, changes resulting from androgen exposure may trigger ER stress, or these responses could proceed through shared regulatory mechanisms. (Garber et al., 2001) identified molecular subtypes of lung adenocarcinoma based on global gene expression patterns. We found that many of the genes that were characteristically expressed at high levels in a molecular subtype with a poor prognosis had increased expression in DTT- and menadione-treated fibroblasts (Figure 7), suggesting that there may be a connection between oxidative stress and the pathogenesis of lung cancer.
Figure 6.
Expression of ER stress response genes in androgen-treated prostate cell lines. We clustered the genes corresponding to the three putative unfolded protein response clusters (Figure 4B) by using data from time courses of androgen-treated prostate cell lines (DePrimo et al., 2002) and from the stress data. The color scheme is the same as in Figure 1.
Figure 7.
Expression of genes associated with poor prognosis adenocarcinoma of the lung in the stress experiments. We clustered genes expressed in a poor-prognosis subtype of adenocarcinoma of the lung (Garber et al., 2001) by using the data from the stress experiments. The color scheme is the same as in Figure 1.
DISCUSSION
Systematic examination of the global transcriptional response to stresses in human cells, although limited in the variety of stresses and number of cell types examined, revealed that the responses are highly specific to both the type of cell and the nature of the stress. In a complex multicellular organism, different cell types are likely to experience different types of stress in vivo. For example, endothelial cells are exposed to shear stress, epidermal cells to light and temperature fluctuations, and transitional epithelial cells to varying pH and osmotic pressures. Stress responses could be more intense in cell types whose natural history is associated with extreme environmental variation, and particularly for cells that carry a disproportionate burden in maintaining homeostasis for the entire organism in response to a particular stress.
One striking feature of the transcriptional responses of human cells to stress is the apparent importance of cell-cell communication. In complex metazoans such as humans, specialized tissues and organs bear the major burden in maintaining homeostasis with respect to distinct stressful conditions. The prominent role of cell-cell signaling in the gene expression responses of individual cell types to specific stimuli highlights the division of labor among specialized cells in these responses. This is a recurring feature of physiological responses in mammalian cells (Iyer et al., 1999; Boldrick et al., 2002). In metazoans, homeostatic responses to changes in temperature, pH, osmolarity, and many other perturbations are largely delegated to specialized organs and cells. It is likely that the in vivo responses of tissues to stress have many features that depend on cell-cell communication and thus cannot be observed in homogeneous cell culture. Indeed, many genes were induced in mouse hearts in vivo after oxidative stress (Edwards et al., 2003) that were not induced by oxidative stress in cultured fibroblasts or HeLa cells. Compartmentalization of function might influence how cells respond to stress. For example, one feature of the yeast ESR is the altered expression of genes involved in carbohydrate storage. Because specific human cell types perform this function, other cell types probably do not need to induce expression of genes important for energy storage.
The fact that two of the cell types we studied were cancer cells may have played an important part in the cell type-specific differences we observed. We already noted that a possible checkpoint defect might have led to the induction of immediate early genes in the cancer cell lines but not fibroblasts after heat shock. Other differences that might be associated with the cancer phenotype include the muted response of HeLa cells to DTT and menadione and the induction of ribosomal proteins in crowded HeLa cells. More data will be needed to determine the relative importance of cancer-related regulatory defects and cell type of origin in cell type-specific stress-induced gene expression differences.
The absence of a strong general stress response in human cells is in contrast with free-living yeasts and bacteria, which do have large common stress responses. Several key differences between these distant species might play a role in this divergence. Single cells in multicellular organisms experience different selective pressures than unicellular organisms; human cells may have a more controlled response because they may decide to undergo apoptosis. These experiments were performed in somewhat artificial cell culture conditions, including atmospheric oxygen and serum-containing medium. Because these conditions are different from the in vivo state normally experienced by these cells and thus might be stresses themselves, a general stress response could be constitutively active in cultured cells and incapable of responding further. Moreover, human cells probably experience a less variable environment than free-living organisms, which also could explain the diminished general stress response
Many genes important for basal metabolism and core cellular components, such as ribosomal proteins splicing factors and cytoskeletal proteins, whose expression may be broadly associated with cell growth, were repressed after multiple stresses. This feature of the transcriptional response to stress might represent a general slowing of the cellular growth rate in stressed cells and a secondary coupling of the growth rate to expression of various metabolic and cell type specific genes, rather than a direct stress-mediated repression.
The majority of genes whose transcripts were induced by oxidative stress also were induced after treatment with the reducing agent DTT. Many of these transcripts were independently shown to be oxidative stress responsive in breast cancer cell lines (Chang et al., 2002). These results are consistent with the idea that ER stress causes intracellular oxidative stress; studies in yeast suggest that ER stress induces oxidative damage to proteins (Sagt et al., 2002) and that the response to oxidative and reductive stress overlap (Trotter and Grant, 2002). An alternative possibility is that DTT and menadione both perturb the cellular redox balance, and the shared response serves to increase the cell's redox buffering capacity, which might protect from reducing or oxidative stress.
Three clusters enriched for known ER stress-responsive genes whose transcripts were induced most strongly by ER stress agents had subtly different kinetics and varied in their specificity to ER stress. This seems to indicate multiple modes of regulation of the UPR. Indeed, human cells are known to have three parallel ER stress-responsive regulatory pathways (reviewed in Ma and Hendershot, 2001). One proceeds through the cleavage-mediated activation of the transcription factor ATF6, another proceeds through IRE1-mediated alternative splicing of the transcription factor XBP-1 (which is present in the third ER stress cluster), and a third proceeds through a translational control step and ultimately results in the activation of the transcription factor ATF4. The separate patterns of regulation detected here may correspond to distinct sets of genes regulated through these parallel pathways.
In conclusion, we have described the gene expression responses of cultured normal and neoplastic human cells to four classes of extrinsic stress: heat shock, ER stress, oxidative stress, and crowding or starvation. These responses are much more distinct and specific to the individual agents than the corresponding responses in the yeast S. cerevisiae. In addition, different cultured cell types vary significantly in their responses to stress. The dataset is intended as a resource for further exploration and discovery and to help provide a framework for the interpretation of global gene expression patterns in human cells and tissues. The full dataset, as well as detailed versions of all figures, are available for download and search at http://microarray-pubs.stanford.edu/human_stress/Home.shtml.
Supplementary Material
Acknowledgments
We thank members of the Botstein and Brown laboratories for helpful discussions; May C. Chen and James Ford for fibroblasts and assistance in thecollection of time points; Arend Sidow for assistance with the yeast-human BLAST; Mike Fero and the Stanford Functional Genomics Facility for human microarrays; and Audrey Gasch for critical reading of the manuscript. J.I.M. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. M.L.W. was supported by a National Research Service Award Postdoctoral Fellowship from the National Human Genome Research Institute (HG00220-02) and by funds from the Scleroderma Research Foundation. N.D.T. is supported by the Stanford Genome Training Program (National Human Genome Research Institute grant #5 T32 HG00044). This work was supported by grants from the National Cancer Institute to D.B. and P.O.B. (CA-77097, CA-85129). P.O.B. is an Investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–11–0799. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–11–0799.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Bilodeau, J.F., Patenaude, A., Piedboeuf, B., Carrier, C., Petrov, P., Faure, R., and Mirault, M.E. (2002). Glutathione peroxidase-1 expression enhances recovery of human breast carcinoma cells from hyperoxic cell cycle arrest. Free Radic. Biol. Med. 33, 1279–1289. [DOI] [PubMed] [Google Scholar]
- Boldrick, J.C., Alizadeh, A.A., Diehn, M., Dudoit, S., Liu, C.L., Belcher, C.E., Botstein, D., Staudt, L.M., Brown, P.O., and Relman, D.A. (2002). Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99, 972–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton, H.C., Ren, B., Koh, S.S., Harbison, C.T., Kanin, E., Jennings, E.G., Lee, T.I., True, H.L., Lander, E.S., and Young, R.A. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.Y., Chi, J.T., Dudoit, S., Bondre, C., van de Rijn, M., Botstein, D., and Brown, P.O. (2002). Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA 99, 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Toone, W.M., Mata, J., Lyne, R., Burns, G., Kivinen, K., Brazma, A., Jones, N., and Bahler, J. (2003). Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., et al. (1998). SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, Y.Y., et al. (2002). Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 62, 6246–6254. [PubMed] [Google Scholar]
- DePrimo, S.E., Diehn, M., Nelson, J.B., Reiter, R.E., Matese, J., Fero, M., Tibshirani, R., Brown, P.O., and Brooks, J.D. (2002). Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol. 3, RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, M.G., Sarkar, D., Klopp, R., Morrow, J.D., Weindruch, R., and Prolla, T.A. (2003). Age-related impairment of the transcriptional responses to oxidative stress in the mouse heart. Physiol. Genomics 13, 119–127. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., and Brown, P.O. (1999). DNA arrays for analysis of gene expression. Methods Enzymol. 303, 179–205. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert, B., Nantel, A., and Whiteway, M. (2003). Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, M.E., et al. (2001). Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. USA 98, 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan, D., Hsu, A.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Kenyon, C. (2002). Genetic analysis of tissue aging in Caenorhabditis elegans. A role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, T., et al. (2002). The Ensembl genome database project. Nucleic Acids Res. 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, Y., Soda, M., and Takahashi, R. (2000). Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664. [DOI] [PubMed] [Google Scholar]
- Iyer, V.R., et al. (1999). The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. (2002). Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110, 1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamande, S.R., and Bateman, J.F. (1999). Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin. Cell Dev. Biol. 10, 455–464. [DOI] [PubMed] [Google Scholar]
- Li, D., Turi, T.G., Schuck, A., Freedberg, I.M., Khitrov, G., and Blumenberg, M. (2001). Rays and arrays: the transcriptional program in the response of human epidermal keratinocytes to UVB illumination. FASEB J. 15, 2533–2535. [DOI] [PubMed] [Google Scholar]
- Lozzio, C.B., and Lozzio, B.B. (1975). Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 45, 321–334. [PubMed] [Google Scholar]
- Lund, J., Tedesco, P., Duke, K., Wang, J., Kim, S.K., and Johnson, T.E. (2002). Transcriptional profile of aging in C. elegans. Curr. Biol. 12, 1566–1573. [DOI] [PubMed] [Google Scholar]
- Ma, Y., and Hendershot, L.M. (2001). The unfolding tale of the unfolded protein response. Cell 107, 827–830. [DOI] [PubMed] [Google Scholar]
- Monks, T.J., Hanzlik, R.P., Cohen, G.M., Ross, D., and Graham, D.G. (1992). Quinone chemistry and toxicity. Toxicol. Appl. Pharmacol. 112, 2–16. [DOI] [PubMed] [Google Scholar]
- Nakahata, K., Miyakoda, M., Suzuki, K., Kodama, S., and Watanabe, M. (2002). Heat shock induces centrosomal dysfunction, and causes non-apoptotic mitotic catastrophe in human tumour cells. Int. J. Hyperthermia 18, 332–343. [DOI] [PubMed] [Google Scholar]
- Okada, T., Yoshida, H., Akazawa, R., Negishi, M., and Mori, K. (2002). Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyadomari, S., Koizumi, A., Takeda, K., Gotoh, T., Akira, S., Araki, E., and Mori, M. (2002). Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. [see comments]. J. Clin. Investig. 109, 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani, G., Colavitti, R., Bedogni, B., Anzevino, R., Borrello, S., and Galeotti, T. (2000). A redox signaling mechanism for density-dependent inhibition of cell growth. J. Biol. Chem. 275, 38891–38899. [DOI] [PubMed] [Google Scholar]
- Pockley, A.G. (2002). Heat shock proteins, inflammation, and cardiovascular disease. [see comments]. Circulation 105, 1012–1017. [DOI] [PubMed] [Google Scholar]
- Puck, T.T., Marcus, P.I., and Cieciura, S.J. (1956). Clonal growth of mammalian cells in vitro. J. Exp. Med. 103, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe, M., Xiong, J., Shu, H.B., Williamson, K., Goddard, A., and Goeddel, D.V. (1996). I-TRAF is a novel TRAF-interacting protein that regulates TRAF-mediated signal transduction. Proc. Natl. Acad. Sci. USA 93, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagt, C.M., Muller, W.H., van der Heide, L., Boonstra, J., Verkleij, A.J., and Verrips, C.T. (2002). Impaired cutinase secretion in Saccharomyces cerevisiae induces irregular endoplasmic reticulum (ER) membrane proliferation, oxidative stress, and ER-associated degradation. Appl. Environ. Microbiol. 68, 2155–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesto, A., Navarro, M., Burslem, F., and Jorcano, J.L. (2002). Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 99, 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock, G., et al. (2001). The Stanford Microarray Database. Nucleic Acids Res. 29, 152–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terro, F., Czech, C., Esclaire, F., Elyaman, W., Yardin, C., Baclet, M.C., Touchet, N., Tremp, G., Pradier, L., and Hugon, J. (2002). Neurons overexpressing mutant presenilin-1 are more sensitive to apoptosis induced by endoplasmic reticulum-Golgi stress. J. Neurosci. Res. 69, 530–539. [DOI] [PubMed] [Google Scholar]
- Trotter, E.W., and Grant, C.M. (2002). Thioredoxins are required for protection against a reductive stress in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 46, 869–878. [DOI] [PubMed] [Google Scholar]
- Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H.P., and Ron, D. (2000). Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. [DOI] [PubMed] [Google Scholar]
- Wang, E., Miller, L.D., Ohnmacht, G.A., Liu, E.T., and Marincola, F.M. (2000). High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol. 18, 457–459. [DOI] [PubMed] [Google Scholar]
- Whitfield, M.L., Finlay, D.R., Murray, J.I., Troyanskaya, O.G., Chi, J.T., Pergamenschikov, A., McCalmont, T.H., Brown, P.O., Botstein, D., and Connolly, M.K. (2003). Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc. Natl. Acad. Sci. USA 100, 12319–12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., et al. (2002). Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X., Zuo, X., Davis, A.A., McMillan, D.R., Curry, B.B., Richardson, J.A., and Benjamin, I.J. (1999). HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18, 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.