Abstract
By means of a variety of intracellular scaffolding proteins, a vast number of heterotrimeric G protein–coupled receptors (GPCRs) may achieve specificity in signaling through a much smaller number of heterotrimeric G proteins. Members of the tetraspanin family organize extensive complexes of cell surface proteins and thus have the potential to act as GPCR scaffolds; however, tetraspanin-GPCR complexes had not previously been described. We now show that a GPCR, GPR56/TM7XN1, and heterotrimeric G protein subunits, Gαq, Gα11, and Gβ, associate specifically with tetraspanins and CD81, but not with other tetraspanins. CD9 Complexes of GPR56 with CD9 and CD81 remained intact when fully solubilized and were resistant to cholesterol depletion. Hence they do not depend on detergent-insoluble, raft-like membrane microdomains for stability. A central role for CD81 in promoting or stabilizing a GPR56-CD81-Gαq/11 complex was revealed by CD81 immunodepletion and reexpression experiments. Finally, antibody engagement of cell surface CD81 or cell activation with phorbol ester revealed two distinct mechanisms by which GPR56-CD81-Gαq/11 complexes can be dynamically regulated. These data reveal a potential role for tetraspanins CD9 and CD81 as GPCR scaffolding proteins.
INTRODUCTION
Heterotrimeric G-protein–coupled receptors (GPCRs) are the largest family of cell surface receptors, accounting for >1% of the human genome. GPCRs transduce extracellular signals from odorants, tastants, photons, small molecule and peptide hormones, growth factors, morphogens, and neurotransmitters (Bockaert and Pin, 1999; Marinissen and Gutkind, 2001; Pierce et al., 2002). The critical physiological roles of GPCRs have been repeatedly confirmed in mouse knockout models (Rohrer and Kobilka, 1998) and studies of human heritable diseases (Stadel et al., 1997). GPCRs are also among the most frequent targets of therapeutic drugs (George et al., 2002). In the classical GPCR signal transduction paradigm, ligand binding induces a conformational change in the GPCR that is transmitted to an associated, cytoplasmic heterotrimeric G protein. GDP bound to the G protein Gα subunit dissociates and is replaced with GTP, triggering the dissociation of the Gα subunit from the Gβγ subunits. The dissociated subunits bind and activate downstream effectors until the GTP bound to the Gα subunit is hydrolyzed to GDP, promoting the reassociation of Gα and Gβγ subunits (Pierce et al., 2002).
A major challenge in the study of GPCRs has been to explain how the ∼1000 GPCRs in the human genome are able to achieve specificity in signaling through ∼20 heterotrimeric G proteins. Recent advances highlight the importance of intracellular scaffolding proteins, including PDZ domain, SH2 domain, and polyproline-binding proteins, in organizing GPCRs into GPCR-specific signaling complexes (Hall et al., 1999; Pierce et al., 2002). Scaffolding could add to specificity by linking subsets of GPCRs to specific cytoplasmic signaling proteins. Alternatively, molecular scaffolds might segregate GPCRs in proximity with specific downstream targets.
Members of the tetraspanin family of cell surface proteins may act as molecular scaffolds by forming complexes with other cell surface proteins, including integrins, IgSF proteins, proteoglycans, growth factor receptors, membrane-bound growth factors, and other tetraspanins (Berditchevski, 2001; Boucheix and Rubinstein, 2001; Hemler, 2003). The large number of tetraspanin-associated proteins is envisioned as being organized into a “tetraspanin web” or a network of tetraspanin-enriched microdomains (TEMs). However, despite the wide variety of proteins reported to be in TEMs, GPCR-tetraspanin associations have not yet been reported.
We applied a sensitive mass spectrometry protein sequencing approach to search further for proteins uniquely present in complexes organized by tetraspanins, CD9 and CD81. We previously showed that CD9/CD81 complexes isolated from Brij 96 detergent lysates have a size range significantly <4 million daltons, are fully soluble, and are resistant to cholesterol depletion with methyl-β-cyclodextrin (Stipp et al., 2001b). Our characterization of these CD9/CD81 complexes (Stipp et al., 2001a, 2001b), together with two other independent studies (Charrin et al., 2001; Clark et al., 2001), identified two major proteins within the complexes as EWI-2/PGRL and EWI-F/CD9-P1/FPRP. These novel IgSF protein subfamily members associated specifically with CD9 and CD81 under conditions where associations with other tetraspanins were not detected (Charrin et al., 2001; Stipp et al., 2001a, 2001b). Thus complexes organized by CD9 and CD81 are distinct from other tetraspanin complexes.
We now report that an orphan heterotrimeric G protein–coupled receptor, GPR56, and heterotrimeric G protein subunits, Gαq/11 and Gβ, also associate specifically with tetraspanins CD9 and CD81. Furthermore, we demonstrate that CD81 plays a central role in GPR56-CD81-Gαq/11-Gβ complexes. Finally, the GPR56-CD81-Gαq/11-Gβ complex is dynamically regulated on intact cells, as anti-CD81 antibody triggered dissociation of Gαq/11 and Gβ from GPR56-CD81, whereas phorbol ester induced dissociation and sequestration of GPR56 from CD81-Gαq/11-Gβ. Our results are consistent with a role for CD9 and CD81 as scaffolding proteins in GPCR signal transduction.
MATERIALS AND METHODS
Antibodies
Anti-tetraspanin monoclonal antibodies used were anti-CD81, M38 (Fukudome et al., 1992) and JS64 (Pesando et al., 1986); anti-CD9, ALB6 (Chemicon, Temecula, CA); anti-CD63, 6H1 (Berditchevski et al., 1995); and anti-CD151, 5C11 (Yauch et al., 1998). M2 anti-FLAG epitope antibody, in free form, or conjugated to biotin or agarose, was purchased from Sigma (St. Louis, MO). Anti-integrin monoclonal antibodies used were anti-α6, A6-ELE (Lee et al., 1995), anti-α2, A2-IIE10 (Bergelson et al., 1994), and anti-β1, TS2/16 (Hemler et al., 1984). Other monoclonal antibodies were anti-CD147, 8G6 (Berditchevski et al., 1997a), and anti-CD97, Vim3b (Research Diagnostics, Inc., Flanders, NJ) Polyclonal antibodies used were rabbit anti-Gαq/11 (sc-392), anti-GαI (sc-262), and anti-Gβ (sc-261), all from Santa Cruz Biotechnology (Santa Cruz, CA), horseradish peroxidase-(HRP)-conjugated goat anti-mouse and goat anti-rabbit antibodies (Transduction Labs, Lexington, KY or Sigma), and fluorescein isothiocyanate–conjugated goat anti-mouse antibodies (Biosource International, Camarillo, CA).
Cell Culture
Human embryonic kidney cells (HEK293), HT1080 fibrosarcoma cells, NT2 embryonic carcinoma cells, and PT67 retroviral packaging cells (CLONTECH, Palo Alto, CA) were cultured in DMEM (Life Technologies, Rockville, MD) supplemented with 10% fetal bovine serum (Life Technologies), 2 mM glutamine, and 100 U/ml penicillin and 100 μg/ml streptomycin. Retinoic acid–differentiated NT2 cells were obtained by treating NT2 cells with 10 μM retinoic acid for 5 weeks, splitting 1:6 into fresh flasks, and treating for 14 days with mitotic inhibitors (Pleasure et al., 1992). U937.7C2, a subclone of U937 monocytic lymphoma cells that do not express CD81, were generously supplied by Shoshona Levy, Stanford University Medical Center. These cells were cultured in RPMI media (Life Technologies) supplemented with 10% fetal bovine serum, glutamine, and antibiotics, as described above.
Purification of CD81 Complexes and Tandem Mass Spectrometry and Sequencing
CD81-associated proteins were prepared as previously described (Stipp et al., 2001b). Briefly, CD81 complexes were affinity-purified from a Brij 96 lysate of NT2RA cells using anti-CD81 mAb JS64 and protein G-Sepharose. CD81-associated proteins were eluted in 1% Triton, resolved by SDS-PAGE, and silver-stained. Individual gel regions were excised, rinsed with 50% HPLC grade acetonitrile, and stored at –20°C until analysis. Proteins were subjected to in-gel reduction, carboxyamidomethylation, and tryptic digestion (Promega, Madison, WI). Peptide sequences were determined by microcapillary reverse-phase chromatography directly coupled to a Finnigan LCQ (Thermo Electron Corp., Mountain View, CA) quadrupole ion trap mass spectrometer equipped with a custom nanoelectrospray source. Interpretation of the resulting MS/MS spectra of the peptides was facilitated by programs developed in the Harvard Microchemistry Facility and by database correlation with the algorithm Sequest (Eng et al., 1994; Chittum et al., 1998).
Construction of GPR56, CD97, and CD81 Expression Vectors
To construct epitope-tagged human GPR56 (also known as TM7XN1) expression vectors, we first obtained a GPR56 cDNA (GenBank accession number AI610263.1, IMAGE clone ID 2187766) in the vector pCMV-SPORT 6.0 from Research Genetics Inc. (Birmingham, AL) Using PCR, this cDNA was repaired in 5′ end of the coding region so that it exactly matched a published GPR56 sequence (Zendman et al., 1999), as verified by complete DNA sequencing. Using the repaired GPR56 cDNA as a template, additional rounds of recombinant PCR with Pfu polymerase were performed to 1) insert a FLAG epitope tag between Glu30 and Asp31, downstream of the putative signal peptide cleavage site at Gly22; 2) in a separate construct, add a FLAG epitope tag to the carboxy terminus of GPR56, after the final residue, Ile687; 3) add a SalI restriction site at the 5′ end of the coding region of each epitope-tagged construct; and 4) add a NotI restriction site at the 3′ end of the coding region of each construct. The resulting amino-terminally tagged (NFLGPR56) and carboxy-terminally tagged (CFLGPR56) cDNAs were cloned into the SalI and NotI sites of the pLXIZ retroviral vector. Final constructs were verified by sequencing.
A full-length human CD97 cDNA (isoform 2) in pCMV-SPORT 6.0 (GenBank accession number BC026690.1; IMAGE ID number 3847171; MGC clone number 9068; Research Genetics Inc.) was obtained and sequenced in its entirety. The CD97 cDNA was then excised from pCMV-SPORT 6.0 with SalI and NotI restriction enzymes and cloned into the pLXIZ retroviral vector, as for the GPR56 constructs above. Human CD81 cDNA (GenBank accession number BE782318; IMAGE ID number 3872952) was obtained from Research Genetics, Inc. After verifying the cDNA by sequencing, XhoI and EcoRI restriction sites were added to the coding 5′ and 3′ ends, respectively, using Pfu polymerase in PCR reactions as above. The resulting PCR product was cloned into the XhoI and EcoR1 restriction sites of the pLXIN retroviral vector and reverified by sequencing.
Retroviral Transduction
Retroviral expression constructs described above were transfected into PT67 packaging cells using the Superfect reagent (QIAGEN, Chatsworth, CA). Forty-eight hours after transfection, PT67 cell supernatants were passed through a 0.45-μm filter, supplemented with 4 μg/ml polybrene (Sigma), and used to infect naïve PT67 packaging cells that had been pretreated with 200 ng/ml tunicamycin (Sigma) for 18 h. Stable virus-producing cells were obtained by selection for 2 weeks in 0.5 mg/ml zeocin (Invitrogen, Carlsbad, CA) or G418 (Gibco BRL, Rockville, MD) as appropriate. The supernatant from these PT67 cells was harvested, filtered, and supplemented with polybrene as above and then was used to infect target 293 or U937 cells. Transduced cells were selected for 2 weeks in 0.3 mg/ml zeocin or 0.5 mg/ml G418 and then maintained in 0.1 mg/ml selective reagent. All cells were maintained in bulk, uncloned populations. For U937 cell lines, immunoselection with magnetic anti-CD81 or anti-FLAG beads was used to separate CD81 and NFLGPR56-expressing cells (∼50% of the uncloned population) from a population of nonexpressing cells.
Immunoprecipitation and Immunoblotting
Cells were lysed in 1% detergent—Brij 96, (Fluka, Ronkonkoma, NY), or Brij 99, CHAPS, or Triton X-100 (all supplied by Sigma)—in 20 mM HEPES, pH 7.3, 150 mM NaCl, and 5 mM MgCl2 (HBSM) with 2 mM phenylmethylsulfonylfluoride (Sigma), 20 μg/ml aprotinin, and 10 μg/ml leupeptin (Roche Molecular Biochemicals, Indianapolis, IN). In some experiments, cells were surface labeled with 0.2 mg/ml sulfo-NHS-LC biotin (Pierce, Rockford, IL) in HBSM for 1 h at room temperature and rinsed thoroughly in phosphate-buffered saline (PBS) before lysis. Cells were extracted on a rocker at 4°C for 1–3 h, and insoluble material was removed by centrifugation at 15,000 × g for 15 min at 4°C. Supernatants were precleared for ≥1 h with protein G-Sepharose (Amersham Pharmacia Biotech, Piscataway, NJ), or, for M2 anti-FLAG agarose immunoprecipitations, with mouse IgG (Sigma) conjugated to agarose beads (Affigel-10; Bio-Rad, Hercules, CA), then centrifuged as above. Specific monoclonal antibodies were added along with protein G-Sepharose, and immune complexes were collected overnight at 4°C with rocking. After four rinses with respective lysis buffer, samples were boiled in Laemmli buffer, resolved by nonreduced SDS-PAGE, and transferred to a nitrocellulose filter (Schleicher & Schuell, Keene, NH).
For biotinylated samples, blots were blocked with 3% nonfat dry milk (wt/vol) in PBS with 0.01% Tween 20 (PBST; Fisher, Pittsburg, PA) for 1 h at room temperature and rinsed thoroughly with PBST. Blots were developed with a 1-h exposure to HRP-conjugated Extravidin (Sigma) followed by four rinses with PBST and chemiluminescence detection (Western Lightning reagent; Perkin-Elmer Cetus, Boston, MA). For immunoblotting filters were blocked in 5% nonfat milk in PBST for 1 h. The filters were then rinsed twice and treated for 1–2 h with primary antibody diluted in blocking buffer. After four rinses with PBST, blots were developed with a 1-h exposure to HRP-conjugated secondary antibody followed by chemiluminescence as above. FLAG immunoblots were developed with biotinylated M2 anti-FLAG antibody according to the manufacturer's directions. Semiquantitative densitometry was performed using GeneTools software (Syngene, Frederick, MD) on digitized images captured from trans-illuminated films with a CCD camera driven by GeneSnap software (Syngene).
Equilibrium Density Gradient Centrifugation
Cell surface biotinylated NFLGPR56-expressing 293 cells were lysed in 1% Brij 96 as above. Lysate (1 ml) containing ∼9 × 106 cell equivalents was mixed with 1 ml of 90% sucrose, 1% Brij 96 in HBSM, loaded over a 0.5-ml cushion of 50% sucrose, and overlaid with layers of 40% sucrose (1 ml), 20% sucrose (1 ml), and 5% sucrose (0.5 ml) prepared in HBSM without detergent. After centrifugation for 21 h at 45,000 rpm in a Beckman SWTi55 rotor (Fullerton, CA) at 4°C, fractions were collected from the top of the gradient. One milliliter of 1.36% Brij 96 in HBSM was added to each fraction, and NFLGPR56 complexes were immunoprecipitated with anti-FLAG agarose and analyzed by blotting with Extravidin-HRP, as described above.
Immunodepletion
Approximately 1.8 × 107 NFLGPR56-expressing 293 cells were surface-labeled with biotin, rinsed, and lysed in 1% Brij 96, as described above. The lysate was divided into three parts and depleted three times for 1 h with a) protein G-Sepharose alone (mock), b) TS2/16 anti-β1 integrin agarose, and c) M38 anti-CD81 agarose. Depleted samples were further divided into four parts: three parts of 10% each for immunoprecipitation with anti-CD81, FLAG, or β1 integrin antibody followed by detection with ExtrAvidin-HRP, and one part of 70% for immunoprecipitation with FLAG antibody followed by immunoblotting for Gαq/11.
Anti-CD81 Triggering Experiments
Intact cells or membrane fraction (see below) were exposed to anti-CD81 antibodies, M38 or JS64, or to anti-CD151 antibody, 5C11 for 5–15 min at 37 or 0°C. Cells were then rinsed and lysed in 1% Brij 96 as described above; membranes, in 100 μl, were extracted with the addition of 900 μl of 1.1% Brij 96. After a 1-h extraction, insoluble material was removed by centrifugation, and lysates were precleared with CL-6B-Sepharose or with agarose-conjugated mouse IgG; no protein G-Sepharose was used during the preclearing steps of these experiments because specific antibodies had been introduced before lysis. In one experiment, M38 anti-CD81 antibody was added to the lysate during the preclearing step as a control for unintended immunodepletion. After preclearing, samples were immunoprecipitated and analyzed by immunoblotting for Gαq/11, Gβ, CD81, or FLAG epitope as described above.
Membrane Preparation
NFLGPR56-expressing 293 cells, 3.6 × 107, were scraped into 5.5 ml of cold homogenization buffer (20 mM HEPES, pH 7.3, 150 mM NaCl, and 5 mM MgCl2 with 2 mM PMSF, 20 μg/ml aprotinin, and 10 μg/ml leupeptin). Cells were disrupted by 10 strokes in a dounce homogenizer. The homogenate was centrifuged at 500 × g for 5 min at 4°C, and the resulting supernatant was then further centrifuged at 100,000 × g for 40 min in an SwTi55 rotor at 4°C to pellet the membrane fraction. The membranes were resuspended in 500 μl of homogenization buffer; 100 μl of this membrane suspension was either treated with M38 anti-CD81 or left untreated in the antibody triggering experiments described above.
PMA Stimulation
U937 cells were treated with the phorbol ester, 12-tetradecanoyl phorbol 13-acetate (PMA), at 200 nM for 20 min at 37°C. Cells were then rinsed and resuspended in normal growth medium at 37°C for periods up to 2 h. At set intervals, cells were removed, rinsed in ice-cold PBS, and held on ice in 5% goat serum in PBS with 0.01% azide. After the final time point was collected, the cells were stained with M2 anti-FLAG or M38 anti-CD81 monoclonal antibodies, followed by FITC-conjugated goat anti-mouse secondary antibody, and analyzed by flow cytometry. In parallel experiments, PMA-treated cells were extracted with Brij 99 detergent before immunoprecipitation and immunoblotting, as described above.
RESULTS
GPR56 Associates with CD9 and CD81
To search for additional CD81-associated proteins, we used a previously described mass spectrometry protein sequencing approach (Stipp et al., 2001b). Fully solubilized CD81 complexes were affinity purified from a 1% Brij 96 detergent lysate of retinoic-acid–differentiated NT2 teratocarcinoma cells (see MATERIALS AND METHODS). CD81-associated proteins were separated by SDS-PAGE, silver-stained, excised, digested in situ with trypsin, and subjected to tandem mass spectrometry peptide sequencing. As shown in Table 1, we obtained three peptides (from the 70–90-kDa region of the gel) that corresponded to an orphan GPCR named GPR56 (also called TM7XN1 and, in mouse, Cyt28; Liu et al., 1999; Zendman et al., 1999; Terskikh et al., 2001).
Table 1.
Peptides recovered from CD81-associated proteins
| Protein | Peptides | Est cluster; GenBank accession no. | Gel region |
|---|---|---|---|
| TM7XN1/GPR56 | 1. (R) RPSAAPASQQLQSLESK | ||
| 2. (R) DLQLLSQFLK | AJ011001 | 70-90 kDa | |
| 3. (R) LQPTAGLQDLHIHSR | |||
| Gα11 | 4. (K) ILYSHLVDYFPEFDGPQR | AF011497 | |
| 5. (R) IATLGYLPTQQDVLR | 40-60 kDa | ||
| Gαq | 6. (R) VADPAYLPTQQDVLR | U43083 | |
| Gβ | 7. (R) LLVSASQDGK | Multiple (see text) | 20-40 kDa |
Aside from those peptides listed, no other peptides were obtained for GPCRs, heterotrimeric G proteins, or any of their previously described associated proteins.
GPR56 is a member of a GPCR subfamily termed LN-TM7 (Zendman et al., 1999) or LNB-TM7 (Stacey et al., 2000). This subfamily is characterized by transmembrane domain homology to the B clan of GPCRs and by unusually large N-termini (LN denotes “large N-terminus”) that can contain modules from other protein families, including EGF, immunoglobulin, cadherin, lectin, laminin, and thrombospondin-like domains (Stacey et al., 2000; Krasnoperov et al., 2002; Fredriksson et al., 2003). The GPR56 N-terminus contains none of these modules but does contain a GPCR proteolytic site (GPS), a putative cleavage site that is characteristic of the LNB-TM7 subfamily and is defined by a conserved pattern of cysteine residues (Krasnoperov et al., 2002). The GPR56 mRNA is present in certain poorly metastatic tumor cell lines (Zendman et al., 1999), the thyroid gland, adult and developing rodent brain (Liu et al., 1999), and hematopoietic stem cells and neuronal precursors (Terskikh et al., 2001), but the GPR56 protein had not been studied.
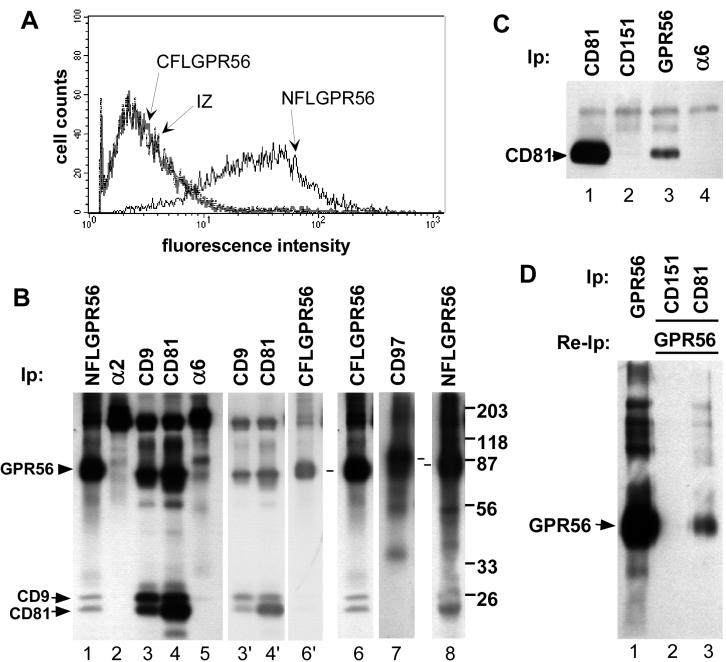
To verify that GPR56 is a CD81-associated protein, we transduced two different GPR56 cDNAs into the human embryonic kidney 293 cell line to obtain 293-NFLGPR56 cells with an N-terminal FLAG epitope tag, and 293-CFLGPR56, with a C-terminal FLAG tag. Using anti-FLAG antibody, 293-NFLGPR56 cells were positive by flow cytometry (Figure 1A), whereas CFLGPR56 cells were indistinguishable from 293 IZ control cells, which had been infected with an empty retroviral vector. These results are consistent with an extracellular N-terminus and an intracellular C-terminus, as predicted for a GPCR.
Figure 1.
Specific association of CD9 and CD81 with GPR56. (A) Nonpermeabilized 293-NFLGPR56 cells (solid black line), 293-CFLGPR56 cells (solid gray line), or 293 IZ empty vector control cells (broken black line) were stained with M2 anti-FLAG antibody followed by FITC-goat anti-mouse 2o antibody and analyzed by flow cytometry. (B) The indicated proteins were immunoprecipitated from a Brij 96 lysate of biotinylated 293-NFLGPR56 cells (lanes 1–5 and 8), 293-CFLGPR56 cells (lane 6), or 293-CD97 cells (lane 7). Dashes between lanes 7 and 8 designate CD97 and GPR56, respectively. For lanes 3, 4, and 6, lighter exposures are shown (lanes 3′, 4′ and 6′) to allow better resolution of the 70–80-kDa region. (C) The indicated proteins were immunoprecipitated from a 1% Brij 96 detergent extract of 293-NFLGPR56 cells, followed by immunoblotting with the M38 anti-CD81 mAb. Cell equivalents, 4.5 × 105, were used for the immunoprecipitation in lane 1, and 4.3 × 106 cell equivalents were used in lanes 2–4. Analysis of the blot by densitometry indicated that ∼2% of total CD81 may be GPR56-associated. (D) Because of the limited sensitivity of the FLAG antibody in immunoblotting, coprecipitation of GPR56 with CD81 was analyzed by reimmunoprecipitation. About 1.8 × 107 biotinylated 293-NFLGPR56 cells were lysed in 1% Brij 96 and GPR56 was immunoprecipitated directly from 5% of the lysate (lane 1). The remainder of the lysate was divided equally between CD151 and CD81 immunoprecipitations (lanes 2 and 3). Next, CD151- and CD81-associated proteins were eluted with Triton X-100 from protein G–bound CD151 and CD81 immune complexes and reimmunoprecipitated for FLAG-GPR56. Analysis by densitometry indicated that ∼2% of total GPR56 may be CD81-associated. In A through D, antibodies used were M2 anti-FLAG agarose (for GPR56), A2-IIE10 anti-α2 integrin, Alb-6 anti-CD9, M38 anti-CD81, A6-ELE anti-α6 integrin, Vim3B anti-CD97, and 5C11 anti-CD151. Material in GPR56 lanes that is >150 kDa varied between experiments and may be SDS-resistant oligomers of GPR56.
Anti-FLAG epitope immunoprecipitations from 1% Brij 96 lysates revealed comparable levels of N- and C-terminally FLAG-tagged GPR56 proteins on the surface of 293 cells, and comparable association with CD9 and CD81-like proteins (Figure 1B, lanes 1 and 6). CD9 and CD81 also strongly associate with each other, as previously observed (Stipp et al., 2001a, 2001b). The identities of the individual CD9 and CD81 bands have been repeatedly established by immunoprecipitation in stringent buffers that disrupt CD9-CD81 interactions (cf. Berditchevski et al., 1996). In contrast, no CD9- or CD81-like bands coprecipitated with the highly expressed integrin α2or α6 subunits in 293 cells (lanes 2 and 5), as expected from previous results (Stipp et al., 2001a, 2001b). Within the CD9 and CD81 immunoprecipitates (lanes 3 and 4), surface-labeled 78-kDa GPR56 was not obvious. Instead, there appears to be abundant EWI-2/PGRL, a 70-kDa IgSF protein and major CD9 and CD81 partner (Clark et al., 2001; Stipp et al., 2001a). Lighter exposures (lanes 3′, 4′, and 6′) more clearly show that labeled proteins associated with CD9 and CD81 are indeed slightly smaller than GPR56. For comparison, we also expressed CD97, another LNB-TM7 GPCR subfamily member, in 293 cells (293-CD97 cells). In the same experiment, comparable levels of cell surface biotin-labeled CD97 and GPR56 were recovered by immunoprecipitation (Figure 1B, lanes 7 and 8). However, only GPR56 coprecipitated CD9- and CD81-like bands (lane 9). Hence, not all LNB-TM7–subtype GPCR proteins associate with CD9 and CD81.
Confirming the identity of CD81 in GPR56 immunoprecipitations, CD81 was detected by immunoblotting both in the CD81-positive control immunoprecipitate (Figure 1C, lane 1) and the GPR56 immunoprecipitate (lane 3), but not in CD151 or α6 integrin immunoprecipitates (lanes 2 and 4). To complement results in Figure 1, B and C, a reciprocal experiment was carried out (Figure 1D). GPR56, CD151, or CD81 were immunoprecipitated from a Brij 96 extract of biotinylated 293-NFLGPR56 cells. We then eluted tetraspanin-associated proteins from bead-bound CD81 and CD151 using Triton X-100, reimmunoprecipitated with anti-FLAG agarose, and recovered GPR56 from the CD81 (lane 3), but not from the CD151 (lane 2) immunoprecipitation.
Biochemical Characterization of GPR56-CD9/CD81 Complexes
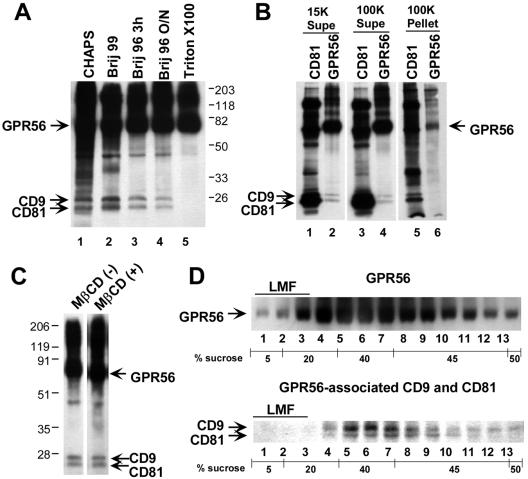
The amount of CD9 and CD81 that coprecipitated with GPR56 from biotinylated 293-NFLGPR56 cells was no greater in a mild CHAPS detergent extract (Figure 2A, lane 1) than in Brij 96 extracts (lanes 3 and 4), although a large number of other cell surface–biotinylated species were observed in CHAPS. This is consistent with a previous report in which tetraspanin complexes solubilized in CHAPS were quite large (in excess of 20 million daltons; Skubitz et al., 2000). In a Brij 99 extract, somewhat more CD9 and CD81 coprecipitated with GPR56 than in either CHAPS or Brij 96 extracts (Figure 2A, lane 2), but far fewer additional species coprecipitated than in CHAPS. This is consistent with the intermediate stringency of Brij 99 for preserving tetraspanin interactions and the intermediate size of Brij 99–solubilized complexes (Claas et al., 2001). A significant fraction (∼50–60%) of the GPR56-associated CD9 and CD81 observed in Brij 99 was preserved in Brij 96 lysates (lanes 3 and 4), regardless of whether the complex was captured in a 3- or 18-h overnight immunoprecipitation. The stability of GPR56-CD9/CD81 complexes in Brij 96, which solubilizes tetraspanin complexes of <4 million daltons (Claas et al., 2001; Stipp et al., 2001b), suggests that a significant fraction of the complexes is stable outside the context of large insoluble raft-like domains. In Triton X-100, a detergent frequently used to prepare classical detergent-insoluble membrane microdomains, the association of GPR56 with CD9 and CD81 was completely disrupted (lane 5).
Figure 2.
Fully soluble GPR56 complexes with CD9 and CD81. (A) Cell surface–biotinylated 293-NFLGPR56 cells were extracted with the indicated detergents, and GPR56 complexes were immunoprecipitated with anti-FLAG agarose (∼2.6 × 106 cells per IP). (B) A 1% Brij 96 extract of biotinylated 293-NFLGPR56 cells was centrifuged at 15,000 × g for 15 min. The resulting supernatant was used for M38 anti-CD81 or M2 anti-FLAG (GPR56) immunoprecipitation (lanes 1 and 2) or was further centrifuged for 40 min at 100,000 × g. The 100K supernatant (lanes 3 and 4) and the resuspended pellet (lanes 5 and 6) were then immunoprecipitated for CD81 or GPR56. Supernatant from ∼5.75 × 105 cell equivalents was analyzed in lanes 1–4, and pelleted material from ∼2.9 × 106 cell equivalents was analyzed in lanes 5 and 6. (C) A 1% Brij 96 extract of biotinylated 293-NFLGPR56 cells was divided into two aliquots and left untreated or treated with methyl-β-cyclodextrin (13 mg/ml final concentration). After a 10-min incubation at room temperature, GPR56 complexes were immunoprecipitated overnight at 4°C(∼1 × 106 cell equivalents per IP). (D) A 1% Brij 96 lysate of ∼ 9 × 106 biotinylated 293-NFLGPR56 cells was loaded in 45% sucrose over a 50% sucrose cushion and overlaid with a detergent-free step gradient of 40, 20, and 5% sucrose. After overnight centrifugation, Brij 96 was added to a final concentration of 1% to each gradient fraction, and GPR56 complexes were immunoprecipitated. Top panel: the distribution of the 70–80-kDa GPR56 band in the gradient; bottom panel: the distribution of coprecipitating CD9 and CD81 bands, shown separately to allow for a darker exposure. LMF, “light membrane fraction,” the region of the gradient where cholesterol-dependent detergent-insoluble membrane microdomains are localized.
To test further whether CD9 and CD81 complexes with GPR56 were fully solubilized in Brij 96, biotinylated 293-NFLGPR56 cell lysate was ultracentrifuged at 100,000 × g for 45 min, to pellet detergent-insoluble membrane vesicles before CD81 or GPR56 immunoprecipitation. As shown in Figure 2B, the vast majority of the CD81 complexes in general and the GPR56-CD9/CD81 complexes in particular remained in the supernatant after the 100,000 × g spin (compare lanes 3 and 4 with lanes 1 and 2). Relatively little CD9, CD81, and GPR56 appeared in the 100,000 × g pellet (lanes 5 and 6).
Classical lipid rafts can be disrupted with the cholesterol-depleting reagent, methyl-β-cyclodextrin (MβCD; Yancey et al., 1996). However, CD9 and CD81 association with GPR56 was unaffected by MβCD at 13 mg/ml in cell lysates (Figure 2C). Also, classical lipid rafts float at lower densities in sucrose density gradients. In Brij 96 lysates, these “light membrane fractions” occur at the top of the gradient, in fractions 1–3 (Claas et al., 2001; Stipp et al., 2001b). However, most of the GPR56 and all of the detectable GPR56-CD9/CD81 complexes appeared in higher density fractions (nos. 4–9), as expected for detergent-solubilized “nonraft” proteins (Figure 2D). In sum, the majority of GPR56-CD9/CD81 complexes do not depend upon large, detergent-insoluble membrane microdomains for stability.
Specific Association of Gαq/11 and Gβ Subunits with CD9, CD81, and GPR56
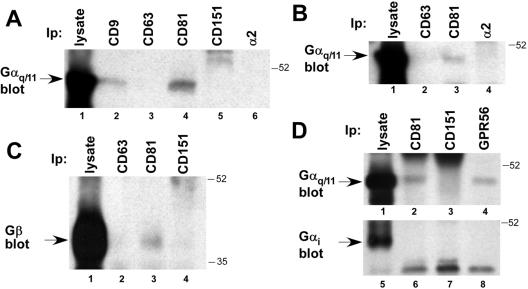
Mass spectroscopic protein sequencing of CD81-associated proteins revealed not only GPR56, but also closely related heterotrimeric G protein subunits, Gα11 and Gαq (from the 40–60-kDa gel region), and Gβ (from the 20–40-kDa gel region; see Table 1). Confirming these results, Gαq/11 was detected by blotting in CD9 (Figure 3A, lane 2) and CD81 (lane 4) immunoprecipitates, but not in immunoprecipitates of tetraspanins CD63 (lane 3) and CD151 (lane 4), or α2 integrin (lane 6) from a Brij 96 lysate of 293 cells. Unfortunately, the quality of commercially available antisera specific for Gαq or Gα11 did not allow us to assess their individual associations with CD9 and CD81. Using HT1080 cells, which express little or no CD9, we again detected Gαq/11 specifically associated with CD81 (Figure 3B, lane 3), but not with CD63 or the α2 integrin subunit (lanes 2 and 4). Hence CD9 is unlikely to be required for CD81-Gαq/11 complex formation. Similarly, we detected Gβ subunit associated with CD81 (Figure 3C, lane 3), but not with CD63 or CD151 (lanes 2 and 4). The Gβ peptide identified by mass spectrometry is a sequence common to β subunits 1–4, and our anti-Gβ antiserum recognizes all four subunits. Thus, the precise identity of the CD81-associated Gβ subunit(s) remains to be determined.
Figure 3.
Specific association of Gαq/11 and Gβ with CD9, CD81, and GPR56. (A) A Brij 96 detergent lysate of 293 cells was immunoprecipitated for the indicated molecules, followed by blotting for Gαq/11. Lysate from ∼4 × 106 cells was used for each immunoprecipitation in lanes 2–6 and was compared with ∼2 × 105 cell equivalents analyzed directly in lane 1. (B) A Brij 96 detergent extract of HT1080 cells was analyzed by immunoprecipitation and Gαq/11 immunoblotting, as in A. About 1.7 × 105 cell equivalents of lysate were used in lane 1, and 3.3 × 106 in each of lanes 2–4. (C) A Brij 96 lysate of A431 cells was immunoprecipitated for the indicated molecules, followed by blotting for the Gβ subunit. About 4 × 106 cell equivalents were used for the immunoprecipitations in lanes 2–4, compared with ∼ 2 × 105 analyzed directly in lane 1. (D) A Brij 96 lysate from ∼1.8 × 107 293-NFLGPR56 cells was immunoprecipitated for the indicated proteins, followed by blotting for Gαq/11 (lanes 1–4) or Gαi (lanes 5–8). In each experiment 2% of the lysate was analyzed directly (lanes 1 and 5), and the remainder was divided evenly among lanes 2–4 and 6–8. Antibodies used were as in Figure directly (lanes 1 and 5), and the remainder was divided evenly among lanes was analyzed 1, along with polyclonal anti-Gαq/11, Gαi, and Gβ antisera.
Because CD81 and CD9 associated with both GPR56 and Gαq/11, we next tested whether a GPR56-Gαq/11 complex could be detected. As shown in Figure 3D, we did indeed observe Gαq/11 associated with both CD81 and GPR56 (lanes 2 and 4), but not with CD151 (lane 3), in a Brij 96 lysate of 293-NFLGPR56 cells. Subsequently, we also confirmed the specific association of the Gβ subunit with CD9, and GPR56 (see Figure 6, below). Consistent with the fact that no other CD81-associated Gα subunits were identified by mass spectrometry (see Table 1), we did not detect the heterotrimeric G protein subunit, Gαi, associated with either CD81 or GPR56 (Figure 3D, lanes 5–8).
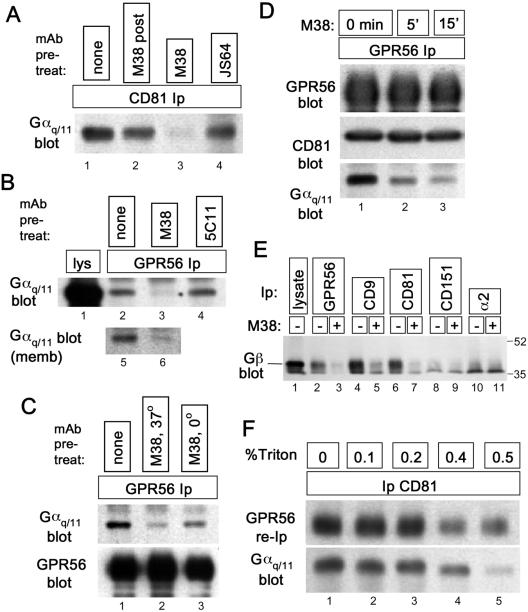
Figure 6.
An anti-CD81 antibody triggers dissociation of Gαq/11 from GPR56-CD81. (A) About 5 × 106 HT1080 cells were treated for 20 min at 37°C with 5 μg/ml anti-CD81 mAbs M38 (lane 3) or JS64 (lane 4) or were left untreated (lanes 1 and 2). Cells were rinsed with ice-cold PBS and lysed on ice in 1% Brij 96. After centrifugation, lysates were precleared with mouse IgG directly conjugated to agarose. For the sample in lane 2, 5 μg/ml M38 was added during this preclearing step. After preclearing, JS64 and M38 concentrations in each lysate were adjusted to 5 μg/ml, protein G was added, and CD81 immune complexes were collected. The presence of Gαq/11 in each immunoprecipitate was measured by immunoblotting. (B) Top panel: ∼7 × 106 293-NFLGPR56 cells were treated for 15 min at 37°C with 5 μg/ml anti-CD81 mAb, M38 (lane 3), anti-CD151 mAb, 5C11 (lane 4), or no antibody (lane 2), before rinsing in cold PBS and lysing in 1% Brij 96. GPR56 was immunoprecipitated with anti-FLAG agarose, followed by blotting for Gαq/11. In lane 1, lysate from ∼ 1.5 × 105 cell equivalents was loaded. Lanes 5 and 6: cell membrane fraction (see MATERIALS AND METHODS) was treated with M38 (lane 2) or left untreated (lane 1) and then extracted with Brij 96. GPR56 was immunoprecipitated from the extracts, and Gαq/11 was blotted, as in lanes 1–4. (C) 293-NFLGPR56 cells were left untreated (lane 1), treated with M38 at 37°C for 15 min (lane 2), or prechilled on ice and treated with M38 in ice-cold buffer for 15 min (lane 3). Cells were then lysed in Brij 96, and GPR56-Gαq/11 association was analyzed as in B. By densitometry, Gαq/11 association was reduced by 84% (lane 2) and 63% (lane 3) after correction for small variations in recovered GPR56. (D) About 7 × 106 293-NFL-GPR56 cells were treated with the M38 antibody for the indicated times (0 min was not treated) before lysis in Brij 96 and GPR56 immunoprecipitation as in B. The levels of GPR56, CD81, and Gαq/11 in each immunoprecipitate were assayed by immunoblotting. (E) The indicated proteins were immunoprecipitated from 1% Brij 99 lysates of 293-NFLGPR56 cells that had been treated with M38 anti-CD81 (15 min. at 37°C) or left untreated. Coprecipitating Gβ subunit was detected by immunoblotting. About 2 × 106 cell equivalents were analyzed in lanes 2–11, compared with ∼ 2 × 105 in lane 1. (F) A clarified Brij 99 extract of ∼5.4 × 107 biotinylated 293-NFLGPR56 cells was divided into five equal pools, and Triton X-100 was added to the indicated concentrations. After CD81 immunoprecipitation and rinsing with the appropriate detergent mixture, 90% of the bead-bound CD81 immune complexes were resuspended in 1% Triton X-100 (no Brij 99) to elute CD81-associated proteins, and then GPR56 was reimmunoprecipitated and detected by blotting with HRP-ExtrAvidin (top panel). The remaining 10% of the CD81 immune complexes were blotted for Gαq/11 (bottom panel).
Central Role of CD81 in Promoting/Stabilizing GPR56-Gαq/11 Association
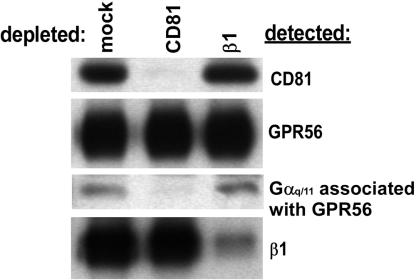
The existence of GPR56-CD81, CD81-Gαq/11, and GPR56-Gαq/11 complexes raised the possibility of a GPR56-CD81-Gαq/11 complex. To test this, biotinylated 293-NFLGPR56 cells were lysed in Brij 96 and immunodepleted with agarose beads alone (mock depletion) or with anti-CD81 or anti-β1 integrin agarose. Depleted pools were then immunoprecipitated with anti-CD81, anti-GPR56 (FLAG), or anti-β1 integrin antibodies. As shown in Figure 4, the CD81 immunodepletion removed all of the detectable CD81, but left behind a substantial pool of GPR56, as expected because of the relatively low fraction of GPR56 associated with CD81. Remarkably, however, this pool of GPR56 contained essentially no associated Gαq/11. Thus, all of the Gαq11 associated with GPR56 could be removed by CD81 immunodepletion, strongly supporting the existence of GPR56-CD81-Gαq/11 complexes and suggesting a possible role for CD81 in promoting or stabilizing GPR56-Gαq/11 association. In the β1 integrin control immunodepletion, neither CD81, nor GPR56, nor the GPR56-associated pool of Gαq/11 was removed, despite the fact that nearly all of the β1 integrin subunit was depleted. In a converse experiment, depletion of exogenously expressed GPR56 left behind a substantial pool of CD81-associated Gαq/11 (unpublished data). This was expected because we had previously observed CD81-Gαq/11 complexes in 293 cells that did not express exogenous GPR56 (Figure 3A).
Figure 4.
Evidence for GPR56-CD81-Gαq/11 complexes. 293-NFL-GPR56 cells, 1.8 × 107, were biotinylated and extracted with Brij 96. Equal portions of the lysate were depleted with protein G-Sepharose (mock), M38 anti-CD81, or TS2/16 anti-β1 integrin agarose. After each depletion, CD81 (top panel), GPR56 (second panel), and β1 integrin (bottom panel) were detected by immunoprecipitation followed by ExtrAvidin-HRP chemiluminescence. GPR56-associated Gαq/11 (third panel) was detected by FLAG immunoprecipitation followed by Gαq/11 blotting. Lysate from 6 × 105 cell equivalents was used for each immunoprecipitation in panels 1, 2, and 4, and lysate from 4.2 × 106 cell equivalents was used for each immunoprecipitation in panel 3.
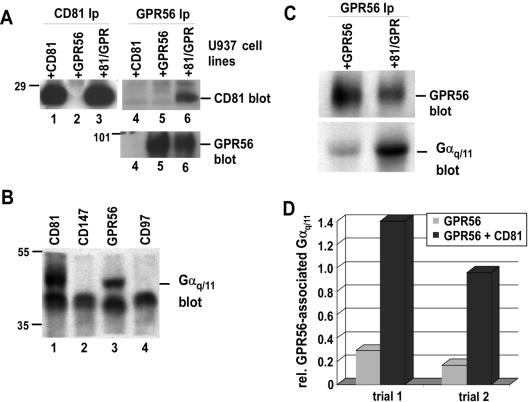
For a complementary test of CD81 function in supporting GPR56-Gαq/11 association, we turned to U937 monocytic leukemia cells, selected to lack detectable CD81 expression (Hamaia et al., 2001). We reconstituted these cells with human CD81, NFLGPR56, or both proteins. Flow cytometry confirmed the expression of the transduced proteins as well as the relative expression levels of two endogenous proteins, CD97 and CD147 (Table 2). In a separate experiment, we observed no CD9 expression in these cells, using flow cytometry (unpublished data). CD81 and GPR56 (FLAG) immunoblotting also confirmed i) the expression of CD81 specifically in the CD81-transduced cells (Figure 5A, lanes 1 and 3); ii) the expression of GPR56 specifically in the GPR56-transduced cells (Figure 5A, bottom panel, lanes 5 and 6); and iii) the association of CD81 with GPR56 in the dually transduced cells (Figure 5A, top panel, lane 6). In the CD81+ GPR56+ cells, we detected Gαq/11 specifically associated with CD81 (Figure 5B, lane 1) and GPR56 (lane 3), but not with another abundant cell surface protein CD147 (lane 2) or with the LNB-TM7 family member, CD97 (lane 4). The lack of CD97-Gαq/11 association is consistent with our previous observation that CD97 fails to associate with CD81 (Figure 1B).
Table 2.
Flow cytometry of U937 cell lines
| Cell line | GPR56 | CD97 | CD81 | CD147 |
|---|---|---|---|---|
| U937 GPR56 | 189 | 102 | 1 | 353 |
| U937 CD81 | 0 | 114 | 502 | 449 |
| U937 GPR56/CD81 | 115 | 107 | 285 | 419 |
Mean fluorescence intensity units (mfi) after subtraction of background staining from an irrelevant antibody. GPR56 staining was with anti-FLAG antibody.
Figure 5.
CD81 enhances GPR56-Gαq/11 association. U937 cells were transduced with CD81 (+CD81), GPR56 (+GPR56), or both proteins (+81/GPR). (A) U937 cells were lysed with 1% Brij 99 and immunoprecipitated for CD81 (lanes 1–3; 1 × 107 cells/lane) or GPR56 (lanes 4–6; 2 × 107 cells/lane), followed by blotting with the M38 anti-CD81 antibody (top panel) or with the M2 anti-FLAG antibody, to detect GPR56 (bottom panel). (B) The indicated proteins were immunoprecipitated from a Brij 99 extract of 4 × 107 U937 + 81/GPR cells, followed by immunoblotting Gαq/11. (C) 2 × 107 of the indicated U937 cell types were lysed in 1% Brij 99, and GPR56 was immunoprecipitated with anti-FLAG agarose. GPR56 and coprecipitating Gαq/11 were blotted with biotinylated M2 anti-FLAG antibody or with Gαq/11 polyclonal antibody respectively. (D) Semiquantitative densitometry of results in C above and of a second independent trial. The y-axis equals the Gαq/11:GPR56 ratio. GeneTools software (Syngene) was used to analyze digitized images captured from trans-illuminated films.
Next we compared immunoprecipitated GPR56 from lysates of equal numbers of CD81– and CD81+ cells. In the CD81+ cells, dramatically more Gαq/11 coprecipitated with GPR56 (Figure 5C, bottom panel), despite the fact that the mean GPR56 expression level was lower in the CD81+ cells (top panel, and see also Figure 5A, and Table 2). In two separate experiments, the relative amount of Gαq/11 associated with GPR56 was enhanced about fivefold in CD81+ cells, as determined by semiquantitative densitometry (Figure 5D).
Dynamic Regulation of the GPR56-CD81-Gαq/11 Complex: CD81 Antibody Effects
Because the GPR56 ligand has not been identified, it was not feasible to test directly CD81's role in signaling through GPR56. Therefore, we tested the effects of engaging the GPR56-CD81-Gαq/11 complex with an anti-CD81 antibody, M38. When intact HT1080 cells were treated with the M38 antibody for 15 min before lysis in Brij 96 and CD81 immunoprecipitation (Figure 6A), there was greatly diminished Gαq/11 association with CD81 (compare treated cells in lane 3 to untreated control in lane 1). Pretreating the cells with a different anti-CD81 mAb, JS64, did not reduce the amount of CD81-associated Gαq/11 (lane 4). Furthermore, M38 antibody added after lysis reduced the CD81-Gαq/11 association to a much lesser extent (lane 2). Similar results were obtained in three independent experiments.
We hypothesized that if CD81 is central to GPR56-Gαq/11 association, then perturbing CD81-Gαq/11 interaction should interaction. also perturb GPR56-Gαq/11 Indeed pretreatment of intact 293-NFLGPR56 cells with M38 caused dissociation of Gαq/11 from GPR56 (Figure 6B, lane 3), just as we had observed for CD81. A control anti-CD151 antibody had no effect on GPR56-Gαq/11 association (lane 4). Although the antibody treatment time was brief, it was possible that M38-induced CD81 internalization was the basis for disrupted Gαq/11 interaction. However, using cell membranes (Figure 6B, lanes 5 and 6) or prechilled cells in ice-cold buffer (Figure 6C, lanes 2 and 3), we again found that M38 antibody triggered dissociation of Gαq/11 from GPR56. Thus M38 retained the ability to disrupt the complexes even under conditions where the endocytic machinery had presumably been disabled by subcellular fractionation or was impaired because of cell incubation at 0°C.
M38 treatment of intact 293-NFLGPR56 cells had no effect on the amount of GPR56 subsequently recovered, nor was the amount of CD81 associated with GPR56 reduced (Figure 6D). However, the amount of Gαq/11 associated with the GPR56-CD81 complex was dramatically reduced, even after 5 min of antibody exposure, and continued to decline at the 15-min time point. Thus, unlike the previous immunodepletion experiments, in which the pool of CD81-associated GPR56 was removed, leaving behind a pool of GPR56 that did not associate with Gαq/11 (Figure 4), treatment of intact cells with the M38 antibody triggered the loss of Gαq/11 without depleting or disrupting the CD81-GPR56 complex. As shown in Figure 6E, M38 treatment also disrupted association of the Gβ subunit with GPR56, CD9, and CD81 (compare lanes 2 and 3, lanes 4 and 5, and lanes 6 and 7, respectively). No Gβ was detected in association with CD151 or α2 integrin, regardless of the presence of M38 (lanes 8–11).
The preservation of the GPR56-CD81 association, even as Gαq/11 dissociated, suggested that Gαq/11 is not required for GPR56-CD81 interaction. To test this further, we immunoprecipitated CD81 complexes from Brij 99 lysates containing increasing amounts of Triton X-100. As shown in Figure 6F, CD81 associations with GPR56 and Gαq/11 decreased in parallel as Triton X-100 concentration increased, until, at 0.5% Triton X-100, the CD81-Gαq/11 association fell off more sharply. This result was again consistent with the GPR56-CD81 association being independent of the presence of Gαq/11. In control experiments, M38 treatment of intact cells had little or no effect on CD81 association with other cell surface partners, EWI-2, EWI-F, and α3β1 integrin (unpublished data). Thus, M38's effects on CD81 interactions appear to be relatively specific and provide a rare example of dynamic regulation of a tetraspanin complex. They also provide independent confirmation of the central role of CD81 in supporting GPR56-Gαq/11 association.
Dynamic Regulation of the GPR56-CD81-Gαq/11 Complex: Phorbol Ester Stimulation
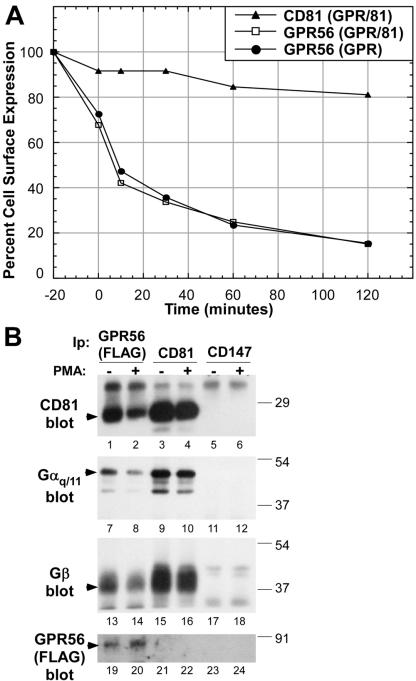
Phorbol ester (PMA) treatment triggers internalization and desensitization of several GPCRs (Diviani et al., 1997; Liang et al., 1998; Tang et al., 1998; Orsini et al., 1999; Xiang et al., 2001; Rochdi and Parent, 2003). We therefore tested whether PMA stimulation mobilized cell surface GPR56, and, if so, what effect this had on CD81 and Gαq/11 association. As shown in Figure 7A, PMA triggered a rapid loss of GPR56-FLAG epitope from the surface of U937 cells such that ∼30% of the GPR56 was lost by the end of the 20-min PMA stimulation, and ∼85% was lost within the following 2 h. In contrast, PMA treatment caused only a modest decrease in cell surface CD81 over a subsequent 2-h interval. The kinetics of the loss of GPR56 were virtually identical in CD81– and CD81+ cells, indicating that CD81 is not required for this type of GPR56 sequestration.
Figure 7.
PMA stimulation causes dissociation of GPR56 from CD81-Gαq/11. (A) U937 cells expressing NFLGPR56 alone (GPR), or together with CD81 (81/GPR), were treated with 100 nM PMA for 20 min at 37°C and then rinsed and returned to 37°C. GPR56 and CD81 cell surface levels were monitored by staining nonpermeabilized cells with M2 anti-FLAG mAb or M38 anti-CD81 mAb, followed by FITC-goat anti-mouse 2o antibody and flow cytometry. The y-axis is the mean fluorescence intensity as a percentage of the values for untreated cells. The 0-min time point corresponds to the end of the PMA treatment. (▴) U937 CD81/GPR56 cells stained for CD81; (□), U937 CD81/GPR56 cells stained for GPR56; (•), U937 GPR56 cells stained for GPR56. (B) 6 × 107 U937 cells expressing both CD81 and NFLGPR56 were divided into two equal pools and maintained at 37°C while one pool was treated for 20 min with 200 nM PMA. Next, cells were extracted with 1% Brij 99, and equal volumes of lysates were immunoprecipitated with M38 anti-CD81 mAb, M2 anti-FLAG agarose, or 8G6 anti-CD147 mAb. The immunoprecipitates were blotted with M38 anti-CD81, anti-Gαq/11 clonal antibody, anti-Gβ polyclonal antibody, or biotinylated M2 anti-FLAG antibody.
We next examined the effect of PMA-induced GPR56 sequestration on GPR56 association with CD81, Gαq/11, and Gβ. We chose a time point immediately after PMA stimulation, when cell surface GPR56 was still at ∼70% of untreated levels. Even at this early time point, a dramatic 78–87% decrease in the amount of CD81 associated with GPR56 had already occurred (Figure 7B, compare lanes 1 and 2). Concurrently, the amount of Gαq/11 and Gβ subunit associated with GPR56 also decreased by 67–70% (compare lanes 7 and 8 and lanes 13 and 14), despite the fact that the amount of GPR56 recovered was not diminished and in fact was elevated (compare lanes 19 and 20). On PMA stimulation, the amount of Gαq/11 (lanes 9, 10) and Gβ (lanes 15, 16) associated with CD81 did not decrease. Small apparent diminutions in Gαq/11 and Gβ levels (lanes 10 and 16) can be almost entirely explained by the reduced recovery of CD81 (compare lanes 3 and 4). Thus, PMA stimulation caused a dramatic loss of cell surface GPR56, likely by GPR56 internalization, with the concomitant dissociation of GPR56 from CD81-Gαq/11-Gβ. As a specificity control, no CD81, Gαq/11, Gβ, or GPR56 coprecipitated with CD147 (lanes 5 and 6, all panels). The lack of GPR56 detected by anti-FLAG blotting of CD81 immunoprecipitates reflects the limited sensitivity of the anti-FLAG antibody in immunoblotting, as discussed in Figure 1D. Altogether, these results reveal a second mechanism whereby the GPR56-CD81-Gαq/11-Gβ complex can be dynamically regulated, and they provide yet more evidence for the central role of CD81 in facilitating the interaction of GPR56 with heterotrimeric G protein subunits.
DISCUSSION
Highly Specific Complexes of GPR56 and Gαq/11 with CD9 and CD81
In the present study, we identified the heptahelical protein GPR56 as well as heterotrimeric G protein subunits Gαq, Gα11, and Gβ as novel CD9 and CD81 partners. These interactions are specific at three levels: i) GPR56 but not CD97, a GPCR from the same subfamily, associated with CD9 and CD81. Hence not all GPCRs will form complexes with CD9 and CD81 to the same extent. ii) CD9 and CD81, but not tetraspanins CD151 or CD63, associated with GPR56 and/or Gαq/11 and Gβ. iii) Heterotrimeric G protein subunit Gαq/11, but not Gαi, associated with CD81 and GPR56. Also, GPR56 associated with CD9 and CD81 to a greater extent than did α6 integrin, even though α6 integrin can associate with CD9 and CD81 in some cell types (Berditchevski et al., 1996). We relied on endogenous expression of CD81 (in most cases) and Gαq/11 (in all cases), and the apparent expression of FLAG-tagged GPR56 was similar to that of the endogenous GPCR, CD97 in U937 cells; thus, the interactions are relevant at physiological protein levels. Although other GPCR and heterotrimeric G proteins were absent from our initial mass spectrometry screening results, preliminary data from other mass spectrometry experiments suggest that additional tetraspanin-GPCR complexes exist, with different tetraspanins perhaps forming specific complexes with different GPCR partners in a cell type–specific manner (T. Kolesnikova, X. Yang, C. Stipp, and M. Hemler, unpublished data).
Importantly, GPR56 complexes with CD9 and CD81 appeared in the dense fractions rather than in the light membrane fractions of sucrose gradients, were not disrupted by cholesterol depletion, and did not pellet on centrifugation at 100,000 × g. Hence GPR56-CD9/CD81 complexes are not simply the product of incomplete detergent solubilization, but rather represent discrete, solubilized biochemical entities. It remains to be determined whether CD9 and CD81 complexes with GPR56, Gαq/11, and Gβ will overlap with other CD9 and CD81 complexes containing molecules such as EWI-2 (Clark et al., 2001; Stipp et al., 2001a), EWI-F (Charrin et al., 2001; Stipp et al., 2001b), TGF-α (Shi et al., 2000), HB-EGF (Iwamoto et al., 1994; Nakamura et al., 1995), and CD19/CD21 (Levy et al., 1998).
A Central Role for CD81 in Facilitating GPR56-Gαq/11 Interactions
Several results strongly support the existence of GPR56-CD81-Gαq/11 complexes. Immunodepletion of CD81 essentially abolished Gαq/11 association with GPR56. Conversely, reexpressing CD81 in CD81– cells dramatically enhanced GPR56-Gαq/11 complex formation. Furthermore, anti-CD81 antibody treatment (or harsh detergent) led to separation of GPR56-CD81 from Gαq/11, whereas PMA treatment separated GPR56 from CD81-Gαq/11. Under no conditions did we observe separation of CD81 from GPR56-Gαq/11. Hence CD81 clearly plays a central role in the GPR56-CD81-Gαq/11 complex. The physical arrangement of the molecules within the GPR56-CD81-Gαq/11 complexes remains to be more precisely defined. Because CD81 may typically exist as a dimer (Kovalenko et al., 2004) and can form an expanded network of interactions with other molecules (Kolesnikova et al., 2004), we suspect that GPR56-CD81-Gαq/11 complexes also exist as part of an expanded network of interactions, rather than as a simple 1:1:1 linear complex. The presence of the Gβ subunit in GPR56-CD81-Gαq/11 complexes is consistent with CD81 promoting GPR56 association with an intact Gq/11 heterotrimeric G protein. Because immunodepletion experiments indicate that the pool of GPR56 associated with Gq/11 and the pool associated with CD81 are one and the same, CD81 may be perfectly positioned to influence the extent of GPR56 coupling to Gq/11 upon activation.
Dynamic Regulation of the GPR56-CD81-Gαq/11 Complex on Intact Cells
For most tetraspanin complexes, there is little evidence suggesting that they are rapidly altered upon cell stimulation or activation. Our data revealed two different types of dynamic regulation of the GPR56-CD81-Gq/11 complex (Figure 8). First, engaging the complex on intact cells with the anti-CD81 antibody, M38, triggered the simultaneous loss of Gαq/11 and Gβ from CD81 and GPR56, while maintaining the CD81-GPR56 association. Conceivably, the M38 antibody could be acting as a surrogate ligand, mimicking some aspects of GPCR ligand binding, such as G protein mobilization (Rodbell, 1997) or translocation of GPCR into or out of different cell surface microdomains (Ostrom et al., 2000). Ligand binding frequently triggers GPCR phosphorylation (Pierce et al., 2002); however, we failed to detect any phosphorylation of GPR56 upon M38 treatment of 32PO -labeled cells (unpublished data). Furthermore, M38 did not 4trigger a redistribution of Gαq/11 into or out of detergent insoluble membrane microdomains, and, using cells labeled with [3H]inositol, we did not observe an increase in total cellular IP3 upon M38 treatment (unpublished data). Collectively, these data suggest that the loss of Gαq/11 from CD81-GPR56 complexes does not correspond to activation of the Gαq/11-phospholipase Cβ (PLCβ) signaling cascade. However, local activation of Gq/11 or PLCβ might be overlooked in whole cell lysates.
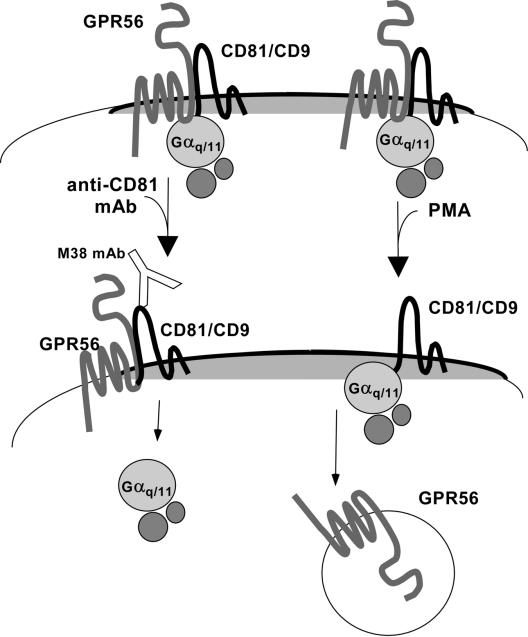
Figure 8.
Dynamic regulation of GPR56-CD81-Gαq/11 complexes. Left: An anti-CD81 mAb triggers dissociation of Gαq/11 from CD81-GPR56. This might involve mimicking some aspects of events that occur downstream of GPCR ligand binding, such as a mobilization of G proteins or transit of GPCRs into or out of different cell surface microdomains. Right: By a distinct mechanism, which resembles heterologous GPCR desensitization, PMA treatment triggers the apparent loss of GPR56 from the cell surface, resulting in a loss of GPR56 from CD81-Gαq/11.
The second example of dynamic regulation of the GPR56-CD81-Gq/11 complex resembles the phenomenon of heterologous GPCR desensitization, in which signaling through PKC or PKA pathways can drive desensitization/internalization of non-ligand–bound GPCRs (Ferguson, 2001; Pierce et al., 2002). Indeed we found that PMA stimulation of intact cells resulted in the apparent internalization of GPR56, with a concomitant loss of GPR56 from the GPR56-CD81-Gαq/11-Gβ complex. After PMA treatment, CD81 interactions with both Gαq/11 and Gβ remained intact, and GPR56 did not undergo phosphorylation (unpublished data). Hence, GPR56 dissociation does not appear to yield G protein activation or depend on PKC phosphorylation of GPR56. In one of the few other examples of dynamic regulation of cell surface tetraspanin complexes, CD9 showed a regulated interaction with the integral membrane metalloproteinase, kuzbanian/ADAM 10. That example also involves heterotrimeric G protein signaling pathways (Yan et al., 2002; see below). PMA does not alter the association of CD81 with integrins, or with its partner EWI-2 (T. Kolesnikova, unpublished results), thus further emphasizing the unique aspects of the GPR56-CD81 interaction.
Are CD9 and CD81 GPCR Scaffolding Proteins?
A variety of intracellular scaffolding proteins are emerging as potential answers to longstanding questions about how the vast number of GPCRs in the human genome achieve specificity in signaling through a much more limited number of heterotrimeric G proteins (Hall et al., 1999; Pierce et al., 2002). GPCR-binding proteins such as the β-arrestins, PDZ, SH2, and polyproline binding proteins may help create GPCR signaling specificity by engaging additional signaling pathways or localizing GPCR signaling events to specific subcellular sites. The central role of CD81 in promoting GPR56-Gαq/11 association argues that CD9/CD81 complexes may have analogous GPCR scaffolding functions. The apparently low fraction of GPR56 and Gαq/11 associated with CD9 and CD81 is not inconsistent with the low fractional association of known scaffolding proteins, such as β-arrestin 1 associating with the ligand-bound GPCR, β2 adrenergic receptor (Luttrell et al., 1999).
Association of GPR56 and Gαq/11 with CD81 and CD9 opens up a range of scaffolding possibilities. These tetraspanins have been linked to several other proteins relevant to G protein signaling, including PKC (Zhang et al., 2001a), the integral membrane metalloproteinase kuzbanian/ADAM 10 (Yan et al., 2002), EGF receptor ligands (Iwamoto et al., 1994; Nakamura et al., 1995; Shi et al., 2000), and phosphatidylinositol 4-kinase (Berditchevski et al., 1997b; Yauch et al., 1998). CD9 and CD81 can functionally link conventional PKC isoforms to α3 or α6 integrins, leading to integrin phosphorylation accompanied by altered cell motility (Zhang et al., 2001a, 2001b). Because these PKC isoforms are major downstream effectors of Gq/11, it is tempting to speculate that CD9/CD81 might likewise promote linkage of GPR56 and Gq/11 to PKC. A further suggestion of a GPCR scaffolding function for CD9/CD81 complexes comes from a recent study of the GPCR-mediated transactivation of the EGF receptor (EGFR; Yan et al., 2002). In this study, the GPCR ligand, bombesin, triggered EGFR phosphorylation and signaling and also promoted CD9 association with kuzbanian/ADAM 10, which can cleave cell surface HB-EGF, itself a CD9-associated protein (Iwamoto et al., 1994; Nakamura et al., 1995). The kuzbanian-mediated shedding of HB-EGF appears to be responsible for EGFR transactivation in this study. Thus, complexes organized by CD9 may be important for promoting a GPCR activation–dependent association between kuzbanian and HB-EGF. Finally, the activation of phosphatidylinositol 4-kinase by GPCR ligands such as mastoparan (Gasman et al., 1998) suggests yet another potential scaffolding function for CD9 and CD81.
Possible Physiological Relevance of GPR56-CD9/81-Gq/11 Complexes
The placement of GPR56 within the LNB-TM7 subfamily of GPCRs provides clues to potential functions in vivo. The 30 or more subfamily members share the unusual property of long extracellular N-termini that contain structural motifs from other protein superfamilies (Stacey et al., 2000; Krasnoperov et al., 2002; Fredriksson et al., 2003). These motifs, which include EGF-like, Ig-like, laminin G domain-like, thrombospondin type I-like, and protocadherin-like domains, suggest that many LNB-TM7 family members may bind to extracellular matrix proteins or cellular counterreceptors, rather than small molecule ligands. By analogy, GPR56 and its ligand may be involved in cell-cell or cell-substrate interactions. One function of CD9 and CD81 might be to enhance GPR56 ligand binding, in the same way that CD9 association promotes HB-EGF diphtheria toxin binding (Iwamoto et al., 1994) and potentiates juxtacrine HB-EGF signaling (Higashiyama et al., 1995).
The presence of GPR56 mRNA in hematopoietic and neural precursor cell populations (Terskikh et al., 2001) suggests possible functions in stem cell proliferation or differentiation, and indeed GPR56 expression is selectively high in ventricular zones in the developing CNS (Terskikh et al., 2001), where neural cells are rapidly proliferating. This is potentially interesting in light of dramatic overproduction of glial cells during brain development in CD81-null mice (Geisert et al., 2002). GPR56 expression also persists in several regions of the adult brain (Liu et al., 1999), suggesting possible functions in differentiated neurons or glia as well. GPR56 expression is downregulated on highly metastatic melanoma cells (Zendman et al., 1999), whereas the loss of CD9 also correlates with increased metastases in many cases (Hemler et al., 1996; Boucheix and Rubinstein, 2001). It remains to be determined whether GPR56 will functionally associate with CD9 or CD81 on metastatic tumor cells or with CD9 during sperm-egg fusion (Le Naour et al., 2000; Miyado et al., 2000) or with CD81 while it is regulating Th2 immune responses (Deng et al., 2002).
In conclusion, we have presented evidence that the LNB-7TM protein, GPR56, and heterotrimeric G protein subunits, Gαq, Gα11, and Gβ, associate specifically with CD9 and CD81. The central role of CD81 (and CD9 when present) suggests possible scaffolding functions for CD81 and CD9 in GPR56 signaling. Further, we demonstrated two different ways in which these GPR56-CD81-Gαq/11 complexes can be dynamically regulated: triggering with an anti-CD81 antibody or stimulating with PMA. Several major questions remain to be answered: can functional coupling of GPR56 to Gq/11 be established? What role might CD9 and CD81 play in GPR56-mediated signaling events? Are other CD9/CD81 partners, such as α3 integrin or EWI proteins, affected by GPR56 activation? The answers to these questions await the identification either of the GPR56 ligand or of other CD9/CD81-associated GPCRs whose ligands are already known.
Acknowledgments
The authors are grateful to S. Levy, Stanford University, for providing CD81-negative U937 cells and to W.S. Lane, R. Robinson, and K. Pierce (Harvard Microchemistry Facility) for HPLC, mass spectrometry, and peptide sequencing. We also thank Dr. Fabio Re for a critical reading of the manuscript. This work was supported by grants from the Claudia Adams Barr Foundation and the Medical Foundation (Charles A. King Trust Fellowship; to C.S.S.), and National Institutes of Health Grant CA86712 (to M.E.H.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-12-0886. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–12–0886.
References
- Berditchevski, F. (2001). Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114, 4143–4151. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., Bazzoni, G., and Hemler, M.E. (1995). Specific association of CD63 with the VLA-3 and VLA-6 integrins. J. Biol. Chem. 270, 17784–17790. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., Chang, S., Bodorova, J., and Hemler, M.E. (1997a). Generation of monoclonal antibodies to integrin-associated proteins. Evidence that alpha3beta1 complexes with EMMPRIN/basigin/OX47/M6. J. Biol. Chem. 272, 29174–29180. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., Tolias, K.F., Wong, K., Carpenter, C.L., and Hemler, M.E. (1997b). A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272, 2595–2598. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., Zutter, M.M., and Hemler, M.E. (1996). Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol. Biol. Cell 7, 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J.M., St. John, N.F., Kawaguchi, S., Pasqualini, R., Berdichevsky, F., Hemler, M.E., and Finberg, R.W. (1994). The I domain is essential for echovirus 1 interaction with VLA-2. Cell Adhes. Commun. 2, 455–464. [DOI] [PubMed] [Google Scholar]
- Bockaert, J., and Pin, J.P. (1999). Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 18, 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucheix, C., and Rubinstein, E. (2001). Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S., Le Naour, F., Oualid, M., Billard, M., Faure, G., Hanash, S.M., Boucheix, C., and Rubinstein, E. (2001). The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276, 14329–14337. [DOI] [PubMed] [Google Scholar]
- Chittum, H.S., Lane, W.S., Carlson, B.A., Roller, P.P., Lung, F.D., Lee, B.J., and Hatfield, D.L. (1998). Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- Claas, C., Stipp, C.S., and Hemler, M.E. (2001). Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276, 7974–7984. [DOI] [PubMed] [Google Scholar]
- Clark, K.L., Zeng, Z., Langford, A.L., Bowen, S.M., and Todd, S.C. (2001). PGRL is a major CD81-associated protein on lymphocytes and distinguishes a new family of cell surface proteins. J. Immunol. 167, 5115–5121. [DOI] [PubMed] [Google Scholar]
- Deng, J., Dekruyff, R.H., Freeman, G.J., Umetsu, D.T., and Levy, S. (2002). Critical role of CD81 in cognate T-B cell interactions leading to Th2 responses. Int. Immunol. 14, 513–523. [DOI] [PubMed] [Google Scholar]
- Diviani, D., Lattion, A.L., and Cotecchia, S. (1997). Characterization of the phosphorylation sites involved in G protein-coupled receptor kinase- and protein kinase C-mediated desensitization of the alpha1B-adrenergic receptor. J. Biol. Chem. 272, 28712–28719. [DOI] [PubMed] [Google Scholar]
- Eng, J.K., McCormick, A.L., and Yates, J.R.I. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Ferguson, S.S. (2001). Evolving concepts in G protein-coupled receptor endocytosis: the role in receptor desensitization and signaling. Pharmacol. Rev. 53, 1–24. [PubMed] [Google Scholar]
- Fredriksson, R., Gloriam, D.E., Hoglund, P.J., Lagerstrom, M.C., and Schioth, H.B. (2003). There exist at least 30 human G-protein-coupled receptors with long Ser/Thr-rich N-termini. Biochem. Biophys. Res. Commun. 301, 725–734. [DOI] [PubMed] [Google Scholar]
- Fukudome, K., Furuse, M., Imai, T., Nishimura, M., Takagi, S., Hinuma, Y., and Yoshie, O. (1992). Identification of membrane antigen C33 recognized by monoclonal antibodies inhibitory to human T-cell leukemia virus type 1 (HTLV-1)-induced syncytium formation: altered glycosylation of C33 antigen in HTLV-1-positive T cells. J. Virol. 66, 1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasman, S., Chasserot-Golaz, S., Hubert, P., Aunis, D., and Bader, M.F. (1998). Identification of potential effector pathway for the trimeric Go protein associated with secretory granules.Gostimulatesagranule-boundphosphatidylinositol 4-kinase by activating RhoA in chromaffin cells. J. Biol. Chem. 273, 16913–16920. [DOI] [PubMed] [Google Scholar]
- Geisert, E.E., Jr., Williams, R.W., Geisert, G.R., Fan, L., Asbury, A.M., Maecker, H.T., Deng, J., and Levy, S. (2002). Increased brain size and glial cell number in CD81-null mice. J. Comp. Neurol. 453, 22–32. [DOI] [PubMed] [Google Scholar]
- George, S.R., O'Dowd, B.F., and Lee, S.P. (2002). G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discov. 1, 808–820. [DOI] [PubMed] [Google Scholar]
- Hall, R.A., Premont, R.T., and Lefkowitz, R.J. (1999). Heptahelical receptor signaling: beyond the G protein paradigm. J. Cell Biol. 145, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaia, S., Li, C., and Allain, J.P. (2001). The dynamics of hepatitis C virus binding to platelets and 2 mononuclear cell lines. Blood 98, 2293–2300. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. (2003). Tetraspanin proteins mediate cellular penetration, invasion and fusion events, and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E., Mannion, B.A., and Berditchevski, F. (1996). Association of TM4SF proteins with integrins: relevance to cancer. Biochim. Biophys. Acta 1287, 67–71. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E., Sanchez-Madrid, F., Flotte, T.J., Krensky, A.M., Burakoff, S.J., Bhan, A.K., Springer, T.A., and Strominger, J.L. (1984). Glycoproteins of 210,000 and 130,000 m.w. on activated T cells: cell distribution and antigenic relation to components on resting cells and T cell lines. J. Immunol. 132, 3011–3018. [PubMed] [Google Scholar]
- Higashiyama, S., Iwamoto, R., Goishi, K., Raab, G., Taniguchi, N., Klagsbrun, M., and Mekada, E. (1995). The membrane protein CD9/DRAP 27 potentiates the juxtacrine growth factor activity of the membrane-anchored heparin-binding EGF-like growth factor. J. Cell Biol. 128, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto, R., Higashiyama, S., Mitamura, T., Taniguchi, N., Klagsbrun, M., and Mekada, E. (1994). Heparin-binding EGF-like growth factor, which acts as the diphtheria toxin receptor, forms a complex with membrane protein DRAP27/CD9, which up-regulates functional receptors and diphtheria toxin sensitivity. EMBO J. 13, 2322–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko, O.V., Yang, X., Kolesnikova, T.V., and Hemler, M.E. (2004). Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteines available for crosslinking. Biochem. J. 377, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova, T.V., Stipp, C.S., Rao, R.M., Lane, W.S., Luscinskas, F.W., and Hemler, M.E. (2004). EWI-2 modulates lymphocyte integrin α4β1 functions. Blood (Epub, ahead of print). [DOI] [PubMed]
- Krasnoperov, V., Lu, Y., Buryanovsky, L., Neubert, T.A., Ichtchenko, K., and Petrenko, A.G. (2002). Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J. Biol. Chem. 277, 46518–46526. [DOI] [PubMed] [Google Scholar]
- Le Naour, F., Rubinstein, E., Jasmin, C., Prenant, M., and Boucheix, C. (2000). Severely reduced female fertility in CD9-deficient mice. Science 287, 319–321. [DOI] [PubMed] [Google Scholar]
- Lee, R.T., Berditchevski, F., Cheng, G.C., and Hemler, M.E. (1995). Integrin-mediated collagen matrix reorganization by cultured human vascular smooth muscle cells. Circ. Res. 76, 209–214. [DOI] [PubMed] [Google Scholar]
- Levy, S., Todd, S.C., and Maecker, H.T. (1998). CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16, 89–109. [DOI] [PubMed] [Google Scholar]
- Liang, M., Eason, M.G., Jewell-Motz, E.A., Williams, M.A., Theiss, C.T., Dorn, G.W., 2nd, and Liggett, S.B. (1998). Phosphorylation and functional desensitization of the alpha2A-adrenergic receptor by protein kinase C. Mol. Pharmacol. 54, 44–49. [DOI] [PubMed] [Google Scholar]
- Liu, M. et al. (1999). GPR56, a novel secretin-like human G-protein-coupled receptor gene. Genomics 55, 296–305. [DOI] [PubMed] [Google Scholar]
- Luttrell, L.M. et al. (1999). Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science 283, 655–661. [DOI] [PubMed] [Google Scholar]
- Marinissen, M.J., and Gutkind, J.S. (2001). G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol. Sci. 22, 368–376. [DOI] [PubMed] [Google Scholar]
- Miyado, K. et al. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321–324. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Iwamoto, R., and Mekada, E. (1995). Membrane-anchored heparin-binding EGF-like growth factor (HB-EGF) and diphtheria toxin receptor-associated protein (DRAP27)/CD9 form a complex with integrin alpha 3 beta 1 at cell-cell contact sites. J. Cell Biol. 129, 1691–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini, M.J., Parent, J.L., Mundell, S.J., Benovic, J.L., and Marchese, A. (1999). Trafficking of the HIV coreceptor CXCR4. Role of arrestins and identification of residues in the c-terminal tail that mediate receptor internalization. J. Biol. Chem. 274, 31076–31086. [DOI] [PubMed] [Google Scholar]
- Ostrom, R.S., Post, S.R., and Insel, P.A. (2000). Stoichiometry and compartmentation in G protein-coupled receptor signaling: implications for therapeutic interventions involving G(s). J. Pharmacol. Exp. Ther. 294, 407–412. [PubMed] [Google Scholar]
- Pesando, J.M., Hoffman, P., and Conrad, T. (1986). Malignant human B cells express two populations of p24 surface antigens. J. Immunol. 136, 2709–2714. [PubMed] [Google Scholar]
- Pierce, K.L., Premont, R.T., and Lefkowitz, R.J. (2002). Seven-transmembrane receptors. Nat. Rev. Mol. Cell. Biol. 3, 639–650. [DOI] [PubMed] [Google Scholar]
- Pleasure, S.J., Page, C., and Lee, V.M. (1992). Pure, postmitotic, polarized human neurons derived from NTera 2 cells provide a system for expressing exogenous proteins in terminally differentiated neurons. J. Neurosci. 12, 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochdi, M.D., and Parent, J.L. (2003). Galpha q-coupled receptor internalization specifically induced by Galpha q signaling: regulation by EBP50. J. Biol. Chem. 278, 17827–17837. [DOI] [PubMed] [Google Scholar]
- Rodbell, M. (1997). The complex regulation of receptor-coupled G-proteins. Adv. Enzyme Regul. 37, 427–435. [DOI] [PubMed] [Google Scholar]
- Rohrer, D.K., and Kobilka, B.K. (1998). G protein-coupled receptors: functional and mechanistic insights through altered gene expression. Physiol. Rev. 78, 35–52. [DOI] [PubMed] [Google Scholar]
- Shi, W., Fan, H., Shum, L., and Derynck, R. (2000). The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced EGF receptor activation and cell proliferation. J. Cell Biol. 148, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubitz, K.M., Campbell, K.D., and Skubitz, A.P. (2000). CD63 associates with CD11/CD18 in large detergent-resistant complexes after translocation to the cell surface in human neutrophils. FEBS Lett. 469, 52–56. [DOI] [PubMed] [Google Scholar]
- Stacey, M., Lin, H.H., Gordon, S., and McKnight, A.J. (2000). LNB-TM7, a group of seven-transmembrane proteins related to family-B G-protein-coupled receptors. Trends Biochem. Sci. 25, 284–289. [DOI] [PubMed] [Google Scholar]
- Stadel, J.M., Wilson, S., and Bergsma, D.J. (1997). Orphan G protein-coupled receptors: a neglected opportunity for pioneer drug discovery. Trends Pharmacol. Sci. 18, 430–437. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., Kolesnikova, T.V., and Hemler, M.E. (2001a). EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276, 40545–40554. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., Orlicky, D., and Hemler, M.E. (2001b). FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 276, 4853–4862. [DOI] [PubMed] [Google Scholar]
- Tang, H., Guo, D.F., Porter, J.P., Wanaka, Y., and Inagami, T. (1998). Role of cytoplasmic tail of the type 1A angiotensin II receptor in agonist- and phorbol ester-induced desensitization. Circ. Res. 82, 523–531. [DOI] [PubMed] [Google Scholar]
- Terskikh, A.V., Easterday, M.C., Li, L., Hood, L., Kornblum, H.I., Geschwind, D.H., and Weissman, I.L. (2001). From hematopoiesis to neuropoiesis: evidence of overlapping genetic programs. Proc. Natl. Acad. Sci. USA 98, 7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, B., Yu, G.H., Guo, J., Chen, L., Hu, W., Pei, G., and Ma, L. (2001). Heterologous activation of protein kinase C stimulates phosphorylation of delta-opioid receptor at serine 344, resulting in beta-arrestin- and clathrin-mediated receptor internalization. J. Biol. Chem. 276, 4709–4716. [DOI] [PubMed] [Google Scholar]
- Yan, Y., Shirakabe, K., and Werb, Z. (2002). The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J. Cell Biol. 158, 221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey, P.G., Rodrigueza, W.V., Kilsdonk, E.P., Stoudt, G.W., Johnson, W.J., Phillips, M.C., and Rothblat, G.H. (1996). Cellular cholesterol efflux mediated by cyclodextrins. Demonstration of kinetic pools and mechanism of efflux. J. Biol. Chem. 271, 16026–16034. [DOI] [PubMed] [Google Scholar]
- Yauch, R.L., Berditchevski, F., Harler, M.B., Reichner, J., and Hemler, M.E. (1998). Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell 9, 2751–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendman, A.J., Cornelissen, I.M., Weidle, U.H., Ruiter, D.J., and van Muijen, G.N. (1999). TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett. 446, 292–298. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A., Bontrager, A.L., and Hemler, M.E. (2001a). Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta 1 integrins. J. Biol. Chem. 276, 25005–25013. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A., Bontrager, A.L., Stipp, C.S., Kraeft, S.K., Bazzoni, G., Chen, L.B., and Hemler, M.E. (2001b). Phosphorylation of a conserved integrin alpha 3 QPSXXE motif regulates signaling, motility, and cytoskeletal engagement. Mol. Biol. Cell 12, 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]