Abstract
Pma1-7 is a mutant plasma membrane ATPase that is impaired in targeting to the cell surface at 37°C and is delivered instead to the endosomal/vacuolar pathway for degradation. We have proposed that Pma1-7 is a substrate for a Golgibased quality control mechanism. By contrast with wild-type Pma1, Pma1-7 is ubiquitinated. Ubiquitination and endosomal targeting of Pma1-7 is dependent on the Rsp5-Bul1-Bul2 ubiquitin ligase protein complex but not the transmembrane ubiquitin ligase Tul1. Analysis of Pma1-7 ubiquitination in mutants blocked in protein transport at various steps of the secretory pathway suggests that ubiquitination occurs after ER exit but before endosomal entry. In the absence of ubiquitination in rsp5-1 cells, Pma1-7 is delivered to the cell surface and remains stable. Nevertheless, Pma1-7 remains impaired in association with detergent-insoluble glycolipid-enriched complexes in rsp5-1 cells, suggesting that ubiquitination is not the cause of Pma1-7 exclusion from rafts. In vps1 cells in which protein transport into the endosomal pathway is blocked, Pma1-7 is routed to the cell surface. On arrival at the plasma membrane in vps1 cells, Pma1-7 remains stable and its ubiquitination disappears, suggesting deubiquitination activity at the cell surface. We suggest that Pma1-7 sorting and fate are regulated by ubiquitination.
INTRODUCTION
Quality control describes the mechanism by which newly synthesized proteins in the endoplasmic reticulum (ER) are monitored and assisted to ensure that proper folding and assembly occur (Ellgard et al., 1999). Quality control also detects misfolded or aberrantly assembled proteins and removes them from the secretory pathway. A major mechanism for destruction of abnormal proteins is by ER-associated degradation that prevents ER export, and targets proteins for retrotranslocation from ER to cytosol followed by degradation via the ubiquitin-proteasome system (Bonifacino and Weissman, 1998; Tsai et al., 2002). A second mechanism for removing abnormal proteins from the secretory pathway has been described involving routing from Golgi to the endosomal/vacuolar system for degradation (Arvan et al., 2002). Although delivery of incorrectly assembled membrane complexes for lysosomal degradation has been described in mammalian cells (Minami et al., 1987; Armstrong et al., 1990), it seems that a Golgi-based quality control mechanism may play a more prominent role in yeast for removing both soluble and membrane proteins (Hong et al., 1996; Jenness et al., 1997; Zhang et al., 2001; Reggiori and Pelham, 2002).
The yeast plasma membrane ATPase mutant Pma1-7 is a well-characterized example of a membrane protein impaired in plasma membrane targeting and routed instead from Golgi to the endosomal/vacuolar system for degradation (Chang and Fink, 1995). pma1-7 cells are temperature sensitive and cannot grow at 37°C because proton pumping activity of the plasma membrane ATPase at the cell surface is essential to generate a membrane potential and to regulate intracellular pH. Defective targeting to the plasma membrane is accompanied by failure of Pma1-7 to associate with sphingolipid and ergosterol-rich membrane microdomains called lipid rafts (Bagnat et al., 2001). Because wild-type Pma1 association with lipid rafts occurs before cell surface delivery, the possibility has been suggested that lipid raft association mediates plasma membrane targeting (Bagnat et al., 2001; Lee et al., 2002). Nevertheless, it is clear that raft association is not the only determinant of cell surface targeting of Pma1 (Gong and Chang, 2001).
Ubiquitination is one regulatory mechanism that serves to target proteins to different pathways (Bonifacino and Weissman, 1998). It is well-established that polyubiquitination targets cytosolic and nuclear proteins for degradation by the 26S proteasome, and it also mediates removal of defective proteins from the secretory pathway by ER-associated degradation. Monoubiquitination of some cell surface proteins has been shown to act as a sorting determinant for internalization from the plasma membrane (Hicke, 2001). Ubiquitination also signals internalization of membrane proteins into multivesicular bodies (Katzmann et al., 2002). Finally, ubiquitination mediates sorting of select proteins from the secretory pathway to the endosomal/vacuolar pathway in response to nutritional signals (Magasanik and Kaiser, 2002; Umebayashi and Nakano, 2003).
Covalent attachment of ubiquitin to protein substrates requires the sequential activities of a ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). The E3 enzyme is thought to mediate substrate recognition by binding the substrate ubiquitination signal (Pickart, 2001). Of the many E3s in yeast, Rsp5 is the best-characterized HECT-family member that is involved in multiple sorting events at the ER (Rape et al., 2001) as well as the plasma membrane (Rotin et al., 2000). Rsp5 and its associated proteins Bul1 and Bul2 are also required for nutritionally regulated sorting of the plasma membrane permeases, Gap1 and Tat2 from Golgi to the endosomal pathway (Helliwell et al., 2001; Umebayashi and Nakano, 2003). A novel transmembrane ubiquitin ligase, Tul1, has been characterized recently and suggested to play a role in Golgi quality control involved in sorting defective proteins into the endosomal pathway for vacuolar degradation (Reggiori and Pelham, 2002).
We have examined whether ubiquitination is involved in sorting of Pma1-7 into the endosomal pathway. By contrast with wild-type Pma1 that has remarkable cell surface stability and a half-life >11 h (Benito et al., 1991), and that is not ubiquitinated (Kolling and Losko, 1997), we now show that targeting-defective Pma1-7 is ubiquitinated. Pma1-7 ubiquitination and its transport into the endosomal system is dependent on the Rsp5–Bul1–Bul2 protein complex. In the absence of ubiquitination in rsp5-1 and bul1 bul2 cells, newly synthesized Pma1-7 is delivered to the cell surface and stabilized. When both transport to the endosomal system and delivery to the cell surface are prevented in vps1 sec4 cells, Pma1-7 ubiquitination was still observed, suggesting that Pma1-7 ubiquitination occurs before entry into the endosomal system. Strikingly, in vps1 cells, Pma1-7 is rerouted to the plasma membrane, and upon arrival, Pma1-7 remains stable and its ubiquitination disappears, suggesting deubiquitination activity at the cell surface. We propose that ubiquitination of Pma1-7 is a regulated process that signals its destination in the secretory pathway.
METHODS
Strains and Media
Standard yeast media and genetic manipulations were as described (Sherman et al., 1986). Strains isogenic with L3852 (MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2) (Chang and Fink, 1995) are ACX58-3C (MATα vps1::LEU2), WLY74 (MATα pep12::HIS3), ACY17 (MATα pep4::URA3), ACX102-3A (MATa sec18-1), and WLX16-1A (MATα vps8::LEU2) (Luo and Chang, 1997). ACX58-3C and WLY74 were made by transformation with disruption cassettes pCRK3A (Rothman et al., 1990) and pCB34 (from C. Burd, University of Pennsylvania), respectively. ACX102-3A was transformed with pWL9 (Luo and Chang, 2000) to generate MPY36 (MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 sec18-1 ura3-52::MET25-HA-pma1-7). WLY103 is MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 ura3-52::MET25-HA-pma1-7 (Luo and Chang, 2000). YHY009K is MATa his3 leu2 ura3 trp1 bul1::TRP1 bul2::ura3::LEU2 (Yashiroda et al., 1996). FW1810 (KY314) is MATa GAL2+ his4-912Δ lys2-128Δ leu2Δ1 rsp5-1 from Fred Winston, Harvard University, Cambridge, MA. via D. Kornitzer. vps45 and vps4 are from a collection of original vps mutants (from Tom Stevens, University of Oregon, Portland, OR and Scott Emr, University of California, San Diego, CA), and isogenic with SF838-9DR2L1 (MATa his4-519 ura3-52 leu2-3, 112 lys2 pep4-3) and SEY6211 (MATα leu2-31 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9). CMY119 is MATa gga1Δ1::TRP1 gga2Δ1::HIS3 ura3-52 leu2-312 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 (from J. Bonifacino, National Institutes of Health, Besthesda, MD). ACX119 is a cross between ACY15 (MATa his3Δ200 lys2Δ201 leu2-3112 ura3-52 ade2 pma1-7) with MPY28 (MATa gga1Δ1::trp1::URA3 gga2Δ1::HIS3 ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9). MPY28 was generated by transforming CMY119 with pTV10, a plasmid to swap the TRP1 marker with URA3 (Cross, 1997). MPY33 (MATa gga1Δ1::trp1::ura3::TRP1 gga2Δ1::HIS3 ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 pma1-7) was generated by transforming ACX119-4A with pUT11 to swap the URA3 marker with TRP1. tul1Δ is from the Euroscarf collection. SNY24 and SNY31 are MATα ura3-52 leu2-3,112 his4-519 ade6 pho8-ΔX sec4-8 and MATα ura3-52 leu2-3,112 his4-519 ade6 pho8-ΔX sec4-8 vps1Δ::LEU2, respectively (Nothwehr et al., 1995); these strains were transformed with pWL9 to generate MPY39 and MPY35, respectively.
Molecular Biology
pS3 and pMP4 are centromeric plasmids marked with HIS3 and LEU2, (Sikorski and Hieter, 1989), respectively, bearing MET-HA-pma1-7 (Luo and Chang, 2000); they were made by moving the SacI-XhoI 4.6-kb insert from pWL9 into pRS313 and pRS315, respectively. pWQ13 is a URA3-marked centromeric plasmid bearing MET-HA-PMA1. It was constructed by first amplifying an amino terminal fragment from pJC016 (bearing HA-PMA1 from M. Ziman and R. Schekman) by using oligos CCTTCACCTCTCTTAACA and TCCCCCGGGAGCTAGTTAAAGAAAATC; the fragment was used to replace the SmaI-BstEII fragment from pWL4 (Luo and Chang, 2000).
Protein Induction, Immunoprecipitation, and Western Blot
Cells with plasmids bearing wild-type or mutant tagged Pma1 under the control of the MET25 promoter were grown overnight at 25°C in minimal medium supplemented with 600 μM methionine (Luo and Chang, 2000). Mid-log cultures were harvested, washed once with water, and resuspended at 0.5 OD600/ml in minimal medium without methionine to induce synthesis of hemagglutinin (HA)-tagged Pma1. Cells were shifted to 37°C for 2h and harvested for lysis after addition of 10 mM Na azide.
Lysates were prepared by vortexing with glass beads in the presence of a protease inhibitor cocktail including 1 mM phenylmethylsulfonyl fluoride, as described previously (Chang and Slayman, 1991). For detection of ubiquitinated protein, 5 mM NEM was included during lysis. Immunoprecipitations with anti-HA antibody (Covance, Princeton, NJ) were with 200 μg of lysate protein in RIPA buffer. Immunoprecipitations (IPs) were analyzed by SDS-PAGE and Western blot with anti-ubiquitin (1 μg/ml; Zymed Laboratories, South San Francisco, CA), processed as described previously (Swerdlow et al., 1986). After chemiluminescence detection, blots were stripped and reprobed with anti-HA antibody.
Metabolic Labeling and Cell Fractionation
Cells were induced to synthesize HA-tagged Pma1 for 1 h at 25°C and shifted to 37°C for 15 min before pulse-labeling with Expre35S35S for 10 min. Cells were chased for various times. Cells were lysed and used for anti-HA IP directly or after fractionation on Renografin density gradients, as described previously (Chang, 2002).
Association with Detergent-resistant Membranes
Detergent-insoluble glycolipid (DIG)-enriched complexes were isolated as described previously (Bagnat et al., 2000). Cells were lysed by vortexing with glass beads in TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA) in the presence of protease inhibitors. After centrifugation at 2500 × g for 5 min, lysate (100 μg of protein) was incubated in 250 μl of cold 1% Triton X-100 in TNE buffer for 30 min on ice, and then adjusted to 750 μl by mixing with Optiprep to 40% (Greiner Bio-One, Longwood, FL). The sample was placed at the bottom of a centrifuge tube and overlaid with 1.2 ml of 30% Optiprep, 0.1% Triton X-100 in TNE buffer followed by 200 μl of 0.1% Triton X-100 in TNE buffer. Samples were centrifuged in a Sorvall RP55-S at 52,000 rpm for 2.5 h. Six fractions were collected from the top, and protein was trichloroacetic acid precipitated.
RESULTS
Ubiquitination of Pma1-7 Is Dependent on Rsp5
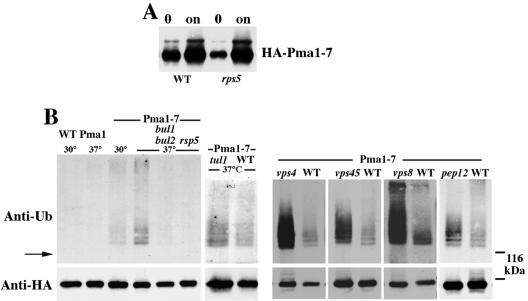
Because ubiquitination is one mechanism for sorting proteins in the secretory pathway (Bonifacino and Weissman, 1998; Katzmann et al., 2002), we asked whether ubiquitination accompanies targeting of Pma1-7 into the endosomal/vacuolar pathway. For these studies, we used a construct in which Pma1-7 is tagged with an HA epitope and its expression is under the control of a MET25 promoter, as described previously (Luo and Chang, 2000). In the presence of methionine, HA-Pma1-7 is synthesized at a low level (Figure 1A). By removing methionine from the medium, there is induction (derepression) of HA-Pma1-7 expression (Figure 1A). After 2-h induction at 37°C, newly synthesized HA-Pma1 was immunoprecipitated and assayed by Western blot with anti-ubiquitin. Figure 1B shows no signal was detected from immunoprecipitates of wild-type Pma1, consistent with reports that wild-type Pma1 is not ubiquitinated (Kolling and Losko, 1997; Wang and Chang, 2002). By contrast, a series of high-molecular-weight bands migrating above the 100-kDa position of Pma1 (Figure 1B, arrow) were observed in immunoprecipitates of Pma1-7 synthesized at 37°C, indicating ubiquitination (Figure 1B). The estimated molecular mass of the high-molecular-weight bands are consistent with addition of one to four ubiquitin moieties to Pma1. At the permissive temperature 30°C, because a larger fraction of newly synthesized Pma1-7 is permitted to move to the plasma membrane (Chang and Fink, 1995), ubiquitination of Pma1-7 is decreased (Figure 1B). These results suggest the modification is associated with increased delivery to the endosomal/vacuolar pathway.
Figure 1.
Pma1-7 is ubiquitinated. Cells bearing pMET-HA-pma1-7 (pMP4 or pS3) or pMET-HA-PMA1 (pWQ13) were induced for 2 h at 30 or 37°C to express tagged Pma1. (A) Pma1-7 induction in RSP5+ and rsp5-1 cells. Lysate was analyzed by anti-HA Western before (“0”) and after 2-h induction at 37°C (“on”). (B) Western blot with anti-ubiquitin (top) followed by anti-HA (bottom). HA-Pma1-7 was immunoprecipitated with anti-HA in bul1 bul2 (YHY009K), rsp5-1 (FW1810), tul1 (BY4742 background), vps45 (T. Stevens collection), vps4 (SEY6211 background), vps8 (WLX16-1A), pep12 (WLY74), and a variety of corresponding wild-type strain backgrounds. The position of Pma1 and the 116-kDa molecular weight marker are indicated by an arrow and dashed lines, respectively.
HA-Pma1-7 ubiquitination was studied after blocking Pma1-7 transport at various steps through the endosomal pathway. Previously, we used indirect immunofluorescence to visualize localization of HA-Pma1-7 after induction at 37°C. In vps8 cells in which transport from early to late endosomes is impaired, newly synthesized HA-Pma1-7 was localized in endosomal/Golgi-like compartments (Luo and Chang, 2000). Figure 1B shows Pma1-7 ubiquitination is increased in vps8 by comparison with that seen in VPS8+ cells: more ubiquitinated Pma1-7 molecules were detected and the extent of modification of Pma1-7 by ubiquitin seemed increased. Pma1-7 ubiquitination also is increased in pep12 cells, which, like vps8, are impaired in transport from early to late endosomes, and in vps45 in which Golgi-derived vesicles accumulate before arrival at the endosome (Cowles et al., 1994). In class E vps mutants, Pma1-7 has been shown to accumulate in an aberrant prevacuolar compartment (Luo and Chang, 2000); in the class E mutant, vps4, there is defective protein sorting to multivesicular bodies (Babst et al., 2002; Katzmann et al., 2002), and ubiquitination of Pma1-7 is also increased by comparison with that in VPS4+ cells.
The transmembrane ubiquitin ligase Tul1 has been proposed to participate in a quality control mechanism that regulates retention of Golgi membrane proteins (Reggiori and Pelham, 2002). Because we have suggested that Pma1-7 is recognized by Golgi quality control (Arvan et al., 2002; Chang and Fink, 1995), we tested whether Pma1-7 ubiquitination is mediated by Tul1. Figure 1B shows Pma1-7 ubiquitination is not impaired in tul1Δ cells. However, Pma1-7 ubiquitination is dependent on the ubiquitin ligase Rsp5 because ubiquitination was not detectable in rsp5-1 cells, defective in E3 activity at 37°C (Andoh et al., 2000; Rotin et al., 2000). Similarly, no ubiquitination of Pma1-7 was detected in bul1 bul2 cells (Figure 1B). These observations are consistent with reports that Bul1 and Bul2 bind to Rsp5, and a bul1 bul2 mutant has phenotypes similar to that of rps5 (Andoh et al., 2000).
Relationship between Ubiquitination, Endosomal Delivery, and Raft Association
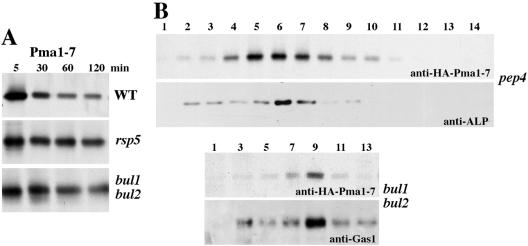
The fate of Pma1-7 was examined in the absence of ubiquitination in rsp5-1 and bul1 bul2 cells. In Figure 2A, cells were pulse labeled with Expre35S35S, chased for various times, and HA-Pma1-7 was immunoprecipitated. In agreement with previous reports (Luo and Chang, 2000), HA-Pma1-7 undergoes degradation in wild-type cells at 37°C; however, stabilization of newly synthesized HA-Pma1-7 was observed in rsp5-1 as well as bul1 bul2 cells, suggesting Pma1-7 was deterred from vacuolar degradation.
Figure 2.
Pma1-7 is routed to the plasma membrane in the absence of ubiquitination in rsp5 and bul1 bul2 cells. Cells were induced to express HA-Pma1-7 in the absence of methionine for 1 h at 25°C. Cells were shifted to 37°C for 15 min, pulse labeled 10 min with Expre35S35S, and chased for various times. (A) Stabilization of Pma1-7. Wild-type (L3852), rsp5-1 (FW1810), and bul1 bul2 (YHY009K) cells bearing MET-HA-pma1-7 (pMP4). HA-Pma1-7 was immunoprecipitated from cells lysed at various times of chase and analyzed by SDS-PAGE and fluorography. (B) Cell fractionation on Renografin density gradients. HA-Pma1-7 was induced in pep4 (ACY17) and bul1 bul2 cells followed by pulse labeling and chase for 1 h at 37°C. After gradient centrifugation, HA-Pma1-7 was immunoprecipitated from each fraction. Plasma membrane (Gas1) and vacuolar membrane (alkaline phosphatase, ALP) markers were assayed by Western blot after pelleting membranes from each fraction.
Localization of HA-Pma1-7 was determined by cell fractionation on Renografin density gradients. It has been well established that Renografin density gradient fractionation leads to efficient separation of intracellular membranes from plasma membrane, migrating in low-density and high-density (9–11) fractions, respectively (Chang, 2002). Fractionation of HA-Pma1-7 was examined after metabolically labeling cells followed by chase for 1 h. Newly synthesized HA-Pma1-7 was immunoprecipitated from gradient fractions and analyzed by SDS-PAGE and fluorography. In pep4 cells, defective in vacuolar proteolysis (Jones, 1977), newly synthesized HA-Pma1-7 is stabilized; fractionation revealed HA-Pma1-7 is predominantly colocalized with the vacuolar membrane marker alkaline phosphatase (Figure 2B), in agreement with localization to the vacuolar membrane and lumen determined previously by indirect immunofluorescence (Luo and Chang, 2000). By contrast, newly synthesized wild-type Pma1 is delivered to the cell surface by 30-min chase (Gong and Chang, 2001). In bul1 bul2 cells, HA-Pma1-7 was found predominantly in high-density fractions 9 and 10, coincident with the plasma membrane marker Gas1 (Figure 2B). These results indicate that in the absence of ubiquitination vacuolar targeting of Pma1-7 is impaired, and Pma1-7 is delivered to the plasma membrane. Previously, we have shown that routing of Pma1-7 to the plasma membrane results in suppression of temperaturesensitive growth (Chang and Fink, 1995; Luo and Chang, 1997). Consistently, bul1 bul2 pma1-7 cells grow at 37°C (our unpublished data).
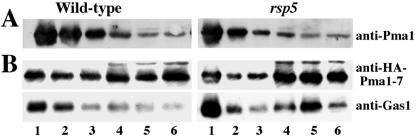
It has been suggested that failure of Pma1-7 to associate with lipid rafts or DIG-enriched complexes may account for its targeting to the endosomal/vacuolar pathway (Bagnat et al., 2001). According to this model, it is possible that ubiquitination of Pma1-7 may occur after, or as a consequence of, its inability to become raft-associated. To examine this possibility, raft association of Pma1-7 was analyzed in the absence of ubiquitination in rsp5-1 cells. Lipid raft association was studied by flotation of Triton X-100–resistant membranes on Optiprep density gradients (Bagnat et al., 2000), and Pma1 distribution within the gradient was assayed by Western blot. Figure 3A shows wild-type Pma1 predominantly in lipid rafts in both RSP5+ and rsp5-1 cells, floating with DIGs at the top of the Optiprep gradient (fractions 1 and 2). Figure 3B examines the ability of Pma1-7 to partition into DIGs after induction of HA-Pma1-7 synthesis for 2h at 37°C. In RSP5+ cells, raft association of HA-Pma1-7 is impaired with ∼30% of total HA-Pma1-7 floating in fractions 1 and 2, in agreement with a previous report (Bagnat et al., 2001). In rsp5-1 cells, DIG association of HA-Pma1-7 remains defective although newly synthesized HA-Pma1-7 is delivered to and stable at the plasma membrane in these cells (Figure 2). Lipid rafts are not perturbed in rsp5-1 cells because the lipid raft-associated marker, Gas1, remains predominantly DIG-associated (Figure 3B, bottom). Thus, ubiquitination does not cause exclusion from rafts. The results are consistent with (but do not prove) ubiquitination and targeting to a degradative pathway occur after, or as a consequence of, failure to associate with rafts.
Figure 3.
Pma1-7 fails to associate with rafts. Cells were shifted to 37°C for 2 h before lysis. After incubation with cold Triton X-100, samples were mixed with Optiprep, overlaid with an Optiprep step gradient, and centrifuged. Gradient fractions were analyzed after trichloroacetic acid precipitation. (A) Pma1 association with DIG-enriched complexes in wild-type and rsp5 (KY314) cells by Western blot with anti-Pma1. (B) DIG association of Pma1-7. Wild-type and rsp5 (KY314) cells bearing pMET-HA-pma1-7 (pMP4) were induced for 2 h at 37°C. Optiprep gradient fractions were analyzed by Western blot with anti-HA and anti-Gas1.
Ubiquitination of Pma1-7 Occurs before Rerouting to the Cell Surface in gga1 gga2 Cells
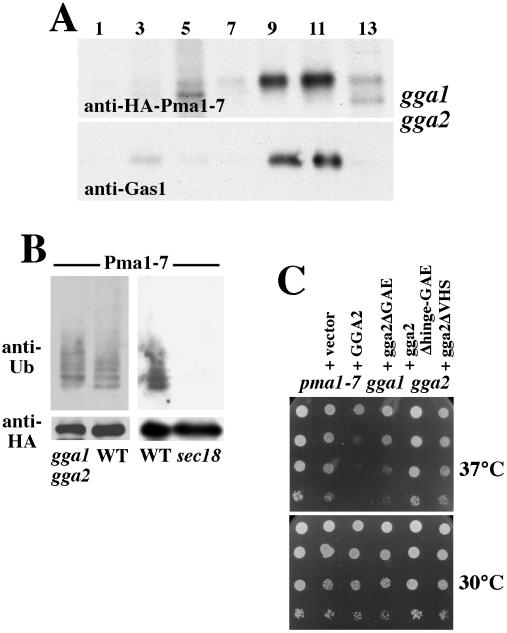
Imposing a general block of protein transport in the endosomal pathway prevents delivery of Pma1-7 for vacuolar degradation and causes Pma1-7 to move to the cell surface (Luo and Chang, 1997; Luo and Chang, 2000). Recent work indicates that the GGA proteins (Golgi-localized, γ-ear-containing, ARF-binding) are conserved, interact with clathrin and the AP-1 adaptor protein complex, and play an important role in protein sorting from Golgi into the endosomal pathway (Mullins and Bonifacino, 2001; Hinners and Tooze, 2003). Therefore, we tested whether the two yeast GGAs, Gga1 and Gga2, play a role in Pma1-7 trafficking. gga1 gga2 cells were pulse labeled with Expre35S35S and chased for 1 h, followed by fractionation on Renografin density gradients and immunoprecipitation of HA-Pma1-7 from each gradient fraction. As one might expect, newly synthesized HAPma1-7 is predominantly localized in dense fractions (9–12) in gga1 gga2 cells, coincident with the plasma membrane marker Gas1 (Figure 4A). These results indicate Pma1-7 is blocked from endosomal delivery and are consistent with reported defects in sorting of the vacuolar protease CPY and recycling of Kex2 in gga1 gga2 (Mullins and Bonifacino, 2001).
Figure 4.
Ubiquitination and cell surface delivery of Pma1-7 in gga1 gga2 cells. HA-Pma1-7 was induced for 2 h at 37°C in gga1 gga2 (CMY119) cells. (A) Localization of newly synthesized HA-Pma1-7. Cells were pulse labeled at 37°C, chased for 1 h, and fractionated on Renografin density gradients. HA-Pma1-7 was immunoprecipitated from each fraction and analyzed by SDS-PAGE and fluorography. Gas1 localization in each fraction was assayed by Western blot. (B) HA-Pma1-7 is ubiquitinated in gga1 gga2 but not sec18 (MPY36). HA-Pma1-7 was immunoprecipitated and then analyzed by Western blot with antiubiquitin followed by anti-HA. (C) gga2 is a suppressor of pma1-7. gga1 gga2 pma1-7 triple mutant (MPY33) was transformed with URA3-marked centromeric vectors bearing no insert or GGA2 (pCM53-3), gga2-ΔGAE (pCM62-2), gga2-Δhinge/GAE (pCM63-1), and gga2-ΔVHS (pCM64-1). Cells were spotted in serial 10-fold dilutions on plates with synthetic complete medium without uracil and incubated at 30 or 37°C.
To determine whether Pma1-7 is ubiquitinated before it undergoes rerouting to the plasma membrane in gga1 gga2 cells, HA-Pma1-7 was immunoprecipitated after induction for 2 h at 37°C, and the IPs were analyzed by Western with anti-ubiquitin. Figure 4B shows that ubiquitination of HA-Pma1-7 in gga1 gga2 cells remains similar to that in GGA+ cells, suggesting ubiquitination occurs before entry into the endosomal pathway. By contrast with Pma1-7, ubiquitination of the vacuolar protein CPS is prevented in gga1 gga2 cells and thus is thought to occur after entry into the endosomal system (Katzmann et al., 2001). Ubiquitination of Pma1-7 was not detected when Pma1-7 synthesis was induced in sec18 cells in which export from the ER is blocked (Figure 4B). These observations suggest Pma1-7 ubiquitination occurs after ER export but before entry into the endosomal system.
Previously, we established that routing of Pma1-7 to the cell surface allows cells to grow because Pma1-7 is enzymatically active (Chang and Fink, 1995; Luo and Chang, 1997). Thus, gga1 gga2 is a suppressor of temperature-sensitive growth of pma1-7 (Figure 4C), as are other trafficking mutants that prevent Golgi-to-endosome traffic and consequently cause Pma1-7 movement to the cell surface. Figure 4C shows the gga1 gga2 pma1-7 triple mutant grows at 37°C. Addition of a centromeric plasmid bearing wild-type GGA2 reverses suppression of pma1-7 so that the cells lose the ability to grow at 37°C, suggesting deletion of both GGA1 and GGA2 is required for pma1-7 suppression. The GGAs have a multidomain structure, including a VHS domain that recognizes acidic cluster-dileucine sorting signals, a hinge domain with putative clathrin-binding motifs, and GAE domain with similarity to gamma-adaptin ear domain; a series of gga2 mutants have been made to test the role of these distinct domains (Mullins and Bonifacino, 2001). We used these mutants to test the role of VHS, hinge, and GAE domains in GGA function by using suppression of pma1-7 as an assay. Figure 4C shows a complementation test of the triple gga1 gga2 pma1-7 mutant with a centromeric plasmid bearing truncations of either VHS, hinge-GAE, or GAE domains of Gga2. Expression of the mutant Gga2 proteins is driven by the strong constitutive triose-phosphate isomerase promoter (Mullins and Bonifacino, 2001). Cells transformed with truncations of VHS and hinge-GAE domains grow well at 37°C, indicating these gga2 mutants fail to reverse pma1-7 suppression. By contrast, gga1 gga2 pma1-7 cells transformed with the ΔGAE gga2 mutant grow almost as poorly at 37°C as cells transformed with wild-type GGA2. Thus, the VHS and hinge regions, but not the GAE domain, play important roles in delivery of Pma1-7 to the endosomal/vacuolar pathway, in agreement with structural requirements for GGA-mediated sorting of pro-CPY and pro-α factor (Mullins and Bonifacino, 2001).
Pma1-7 at the Cell Surface Is Not Ubiquitinated
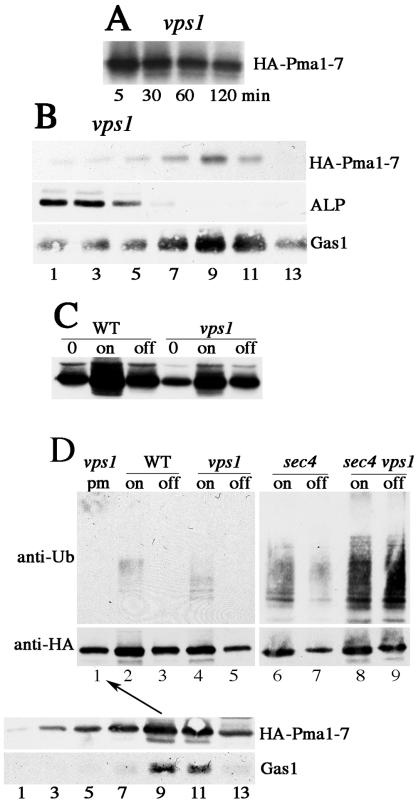
As in gga1 gga2 cells, endosome-directed protein traffic is rerouted to the plasma membrane in vps1 cells (Nothwehr et al., 1995; Wilsbach and Payne, 1993). Therefore, as expected, pulse-chase analysis showed that newly synthesized Pma1-7 is stabilized in vps1 (Figure 5A), and localized to the cell surface by 1 h of chase, coincident with the plasma membrane marker Gas1 on Renografin density gradients (Figure 5B). At the same time, HA-Pma1-7 induced for 2 h at 37°C is ubiquitinated in vps1 (Figure 5D), as in gga1 gga2 cells (Figure 4B). Because newly synthesized Pma1-7 is stabilized in both gga1 gga2 and vps1 cells, it was of interest to determine the localization of ubiquitinated Pma1-7 detected in these cells. Previous indirect immunofluorescence data using pMET-HA-pma1-7 showed after induction in vps1 cells, newly synthesized HA-Pma1-7 is localized in Golgi-like structures as well as the plasma membrane; after chase (by shutting off further synthesis), HA-Pma1-7 was largely found at the plasma membrane (Luo and Chang, 2000). To examine whether ubiquitination in vps1 cells is dependent on HA-Pma1-7 localization, HA-Pma1-7 synthesis was first induced and then its synthesis was shut off for 2h at 37°C. Western blot analysis of cell lysate shows that HA-Pma1-7 is stabilized after chase in vps1 cells (compare “on” and “off”), although induction is less than that in VPS1+ cells (Figure 5C). Ubiquitination was then examined by immunoprecipitating HA-Pma1-7 and analysis by Western blot with anti-HA and anti-ubiquitin. After chase, HA-Pma1-7 ubiquitination disappears in vps1 as well as VPS1+ cells (Figure 5D, top). These results suggest that Pma1-7 undergoes ubiquitination before delivery to the endosomal pathway, and after chase in vps1 cells, ubiquitin is removed upon delivery of HA-Pma1-7 to the cell surface.
Figure 5.
Ubiquitination of cell surface Pma1-7. Cells bearing pMET-HA-pma1-7 were induced to express HA-Pma1-7. (A) Newly synthesized Pma1-7 is stabilized in vps1. After induction for 1 h at 25°C, cells were shifted to 37°C and pulse labeled with Expre35S35S and chased for various times. HA-Pma1-7 was immunoprecipitated and analyzed by SDS-PAGE and fluorography. (B) Pma1-7 is routed to the plasma membrane in vps1. After pulse-labeling and chase for 1 h, cells were lysed and fractionated on Renografin density gradients. The distribution of HA-Pma1-7 on the gradient was determined by immunoprecipitation, whereas Gas1 and alkaline phosphatase were detected by Western blot. (C) Pma1-7 synthesis and degradation in VPS1 and vps1. HA-Pma1-7 level was determined by anti-HA Western blot before induction (“0”) and after 2-h induction at 37°C (“on”). Methionine (2 mM) was then added to terminate further synthesis of HA-Pma1-7 and incubation continued for another 2 h at 37°C (“off”). (D) Ubiquitination of HA-Pma1-7 in vps1 disappears after chase. vps1 (ACX58-3C), vps1 sec4 (MPY 39), sec4 (MPY35), and wild-type (L3852) cells were induced to express HA-Pma1-7 for 2 h at 37°C (on) and chased for another 2 h. HA-Pma1-7 was immunoprecipitated with anti-HA and analyzed by Western blot with anti-ubiquitin followed by anti-HA. Lane 1, Pma1-7 from isolated plasma membranes is not ubiquitinated. After induction of HA-Pma1-7 synthesis for 2 h followed by chase for another 2hat37°C, cell lysate was fractionated on Renografin density gradients in the presence of 5 mM NEM. Fractions 9 and 10 (arrow) containing peak distribution of HA-Pma1-7 (confirmed by Western blot with anti-HA, bottom) were collected for immunoprecipitation with anti-HA and analyzed by anti-ubiquitin Western blot.
Ubiquitination of Pma1-7 was further analyzed in vps1 sec4 cells in which protein diverted from entry into the endosomal pathway should accumulate in post-Golgi secretory vesicles (Nothwehr et al., 1995). In sec4 cells, in the absence of vps1 mutation, accumulation of post-Golgi secretory vesicles should have no effect on vacuolar delivery of Pma1-7. Ubiquitination of Pma1-7 was examined after induction and chase in vps1 sec4 and sec4 cells at 37°C. As expected, upon induction, HA-Pma1-7 is ubiquitinated in sec4 cells, and after chase, ubiquitination disappears as HA-Pma1-7 is degraded (Figure 5D, lanes 6 and 7). In vps1 sec4 cells, ubiquitination of Pma1-7 was seen upon induction (lane 8). Strikingly, after chase in vps1 sec4 cells, Pma1-7 ubiquitination does not disappear but seems even somewhat increased (Figure 5D, top, lanes 8 and 9). These observations support the idea that ubiquitination occurs before entry into the endosomal pathway but disappears upon cell surface arrival when Pma1-7 is routed to the plasma membrane (Figure 5D, compare lanes 5 and 9).
Further evidence in support of deubiquitination of Pma1-7 at the plasma membrane comes from isolating plasma membranes after fractionation of cell lysate on Renografin gradients. NEM was included to prevent deubiquitination occurring during cell fractionation. Figure 5D, bottom, shows the fractionation pattern of HA-Pma1-7 after 2-h induction followed by 2-h chase in vps1 cells at 37°C. HA-Pma1-7 is predominantly present in dense fractions containing plasma membrane (coincident with the plasma membrane marker Gas1 (Figure 5D, bottom). HA-Pma1-7 was immunoprecipitated from plasma membrane collected from fractions 9 and 10; the ubiquitination status of HA-Pma1-7 was assessed by anti-ubiquitin Western blot (lane 1, arrow). In agreement with the “pulse-chase” experiment in lanes 4 and 5, ubiquitination of plasma membrane HA-Pma1-7 was not detectable (Figure 5D, top, lane 1). These data suggest deubiquitination of Pma1-7 occurring upon arrival at the plasma membrane.
DISCUSSION
Ubiquitination is a mechanism used to target proteins for internalization into multivesicular bodies before vacuolar degradation (Katzmann et al., 2002). Because Pma1-7 undergoes vacuolar degradation (Chang and Fink, 1995), it is likely that ubiquitination signals its internalization into multivesicular bodies. However, our evidence suggests ubiquitination of Pma1-7 occurs before its entry into the endosomal pathway. Ubiquitination was analyzed by inducing synthesis of HA-Pma1-7 for 2 h at 37°C. On induction in vps1 and gga1 gga2 mutants in which protein transport from Golgi to endosome is blocked, ubiquitination of HA-Pma1-7 was observed even though Pma1-7 is prevented from endosomal entry (Figures 4B and 5B). In sec18 cells blocked in ER exit, no Pma1-7 ubiquitination was detected (Figure 4B), suggesting ubiquitination occurs after ER export.
The Pma1-7 ubiquitination pattern is consistent with modification by one to four ubiquitin molecules (Figure 1). It is unknown which of 48 lysines, all predicted to reside in the cytoplasmic domains of Pma1, are recognized for ubiquitination. Analysis of Pma1-7 in mutants blocked in various steps along the endosomal pathway revealed an increase in number of ubiquitinated Pma1-7 molecules as well as an increase in the extent of ubiquitination in vps8, pep12, vps45 cells (Figure 1B). Pma1-7 ubiquitination also seems increased in vps4 cells (Figure 1B), impaired in formation of multivesicular bodies (Katzmann et al., 2002). Increased ubiquitination may reflect Pma1-7 accumulating before deubiquitination in the vacuole. Similarly, ubiquitination of the vacuolar hydrolase carboxypeptidase S (which undergoes internalization into multivesicular bodies) is increased in mutants blocked in endosomal transport (Katzmann et al., 2001; Shih et al., 2002). By contrast, Pma1-7 ubiquitination does not seem substantially increased when vacuolar degradation is prevented in pep4 cells (our unpublished data), presumably because deubiquitination is unimpaired in these cells. The idea that Pma1-7 may undergo ubiquitination before and then again after entry into the endosomal pathway is also consistent with increased Pma1-7 ubiquitination when the endosomal transport pathway is blocked is (see below).
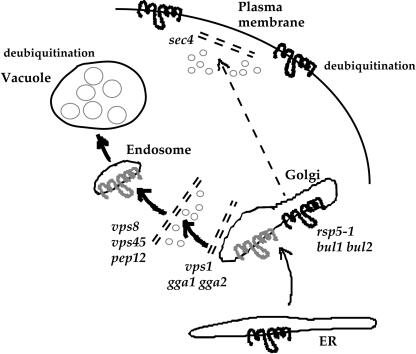
Pma1-7 ubiquitination is dependent on Rsp5 and its associated proteins Bul1 and Bul2 (Figure 1B). In the absence of ubiquitination, Pma1-7 is stabilized in rps5-1 cells, as revealed by pulse-chase analysis; it is prevented from entering the endosomal pathway and is routed instead to the cell surface in bul1 bul2 cells, as revealed by cell fractionation (Figure 2). Thus, we propose that Pma1-7 ubiquitination mediates its delivery into the endosomal pathway (see model in Figure 6).
Figure 6.
Model for ubiquitination and intracellular transport of Pma1-7. Pma1-7 acquires ubiquitination (in grey) after ER export and before entry into the endosomal pathway. In mutants blocked in various steps of the endosomal pathway (vps8, vps45, pep12) before deubiquitination at the vacuole, Pma1-7 ubiquitination is increased. In the absence of ubiquitination in rsp5-1 and bul1 bul2, Pma1-7 (in black) is delivered to the plasma membrane. In vps1 and gga1 gga2 cells, ubiquitinated Pma1-7 is diverted to the plasma membrane where it undergoes deubiquitination (in black).
There are several similarities between Pma1-7 delivery for degradation and nutrient-dependent regulation of plasma membrane transporters by targeting to the endosomal/vacuolar pathway. Like Pma1-7, the tryptophan permease Tat2, the general amino acid permease Gap1, and the metal transporters Smf1 and Smf2 are sorted at the Golgi or early endosome (Beck et al., 1999; Liu and Culotta, 1999; Helliwell et al., 2001; Umebayashi and Nakano, 2003). Tat2 and Gap1 sorting depend on ubiquitination via the Rsp5–Bul1–Bul2 ubiquitin ligase complex. Tat2 and Gap1 are inversely regulated in response to nitrogen source: when cells are starved for nitrogen Gap1 is sorted to the plasma membrane, whereas Tat2 is sent for vacuolar degradation; moreover, the TOR signaling pathway regulates Tat2 sorting (but not Gap1) so that rapamycin treatment to inactivate TOR induces endosomal sorting of Tat2. By contrast, Pma1-7 delivery to the endosomal pathway seems constitutive. In addition to regulating protein sorting at the Golgi complex, Rsp5 is reported to play roles in down-regulation of plasma membrane proteins (Rotin et al., 2000), mitochondrial inheritance (Fisk and Yaffe, 1999), and proteolytic processing and activation of ER membrane-bound transcription factors (Hoppe et al., 2000). Clearly, critical issues are how Rsp5 recognizes substrates in multiple cellular compartments and how this recognition is regulated. The WW domains of Rsp5 mediate interactions with some substrates (Rotin et al., 2000). It has also been suggested that the C2 domain of the mammalian Rsp5 homolog, Nedd4, regulates its membrane and lipid raft association, and in this way perhaps also its substrate selection (Gormley et al., 2003).
We suggest the possibility that Rsp5 modification of Pma1-7 is a response to activation of a quality control mechanism which detects Pma1-7 as a misfolded protein. Pma1-7 is more sensitive than wild-type Pma1 to limited trypsinolysis (Chang, unpublished data), suggesting conformational differences. It remains an unresolved question whether interaction of the Rsp5-Bul1-Bul2 complex with Pma1-7 is direct or whether Pma1-7 recognition is indirect and mediated by other factors; for instance, ubiquitination of some substrates is promoted by phosphorylation (Hicke, 1999).
The relationship was examined between Pma1 ubiquitination, sorting, and association with lipid rafts because it has been proposed that Pma1 targeting requires lipid raft association (Bagnat et al., 2001). Wild-type Pma1 and the GPI-anchored Gas1 are DIG-associated (Bagnat et al., 2000), but Pma1-7 is not present in a Triton-insoluble glycolipid-enriched complex (DIG) (Figure 3; Bagnat et al., 2001). In the absence of ubiquitination, Pma1-7 remains excluded from DIGs, even though it is stabilized and delivered to the plasma membrane in rsp5 and bul1 bul2 cells (Figure 2). Rafts are not perturbed in rsp5 cells as Gas1 is DIG-associated in these cells (Figure 2). Thus, ubiquitination is not the cause of Pma1-7 exclusion from DIGs.
As in rsp5 cells, DIG association of Pma1-7 remains impaired in vps1 cells, even though Pma1-7 is stabilized and routed to the plasma membrane (Bagnat et al., 2001), suggesting that cell surface delivery and stabilization are not sufficient to induce lipid raft association. Although we have suggested that perturbation of lipid raft integrity may promote instability of wild-type Pma1 at the plasma membrane (Wang and Chang, 2002), it is clearly not the only determinant of Pma1 stability. Analogous with Pma1-7, Tat2 is a raft associated protein, and inhibition of Tat2 ubiquitination in bul1 cells has been reported to lead to cell surface delivery without causing recovery of DIG association (Umebayashi and Nakano, 2003).
Although failure of Pma1-7 to associate with lipid rafts may act as a signal for ubiquitination and endosomal delivery, we cannot at present exclude the possibility that impaired raft association is simply one of several phenotypes that reflects its conformational defect. In this regard, it is relevant that when Tat2 association with rafts is perturbed in erg6 cells, Tat2 undergoes Rsp5–Bul1–Bul2-mediated ubiquitination and delivery to the endosomal/vacuolar pathway even under nutritional conditions that signal plasma membrane targeting (Umebayashi and Nakano, 2003). Under these circumstances, Tat2 ubiquitination occurs on anomalous sites distinct from those modified in response to nutritional signals (Umebayashi and Nakano, 2003). According to the quality control model we have suggested for Pma1-7, it is possible that Tat2 is misfolded as a result of impaired raft association and recognition of the conformational defect leads to endosomal targeting.
Ubiquitination is a reversible modification and the activity of deubiquitinating enzymes represents one means of regulation. In yeast, the family of deubiquitinating enzymes is large but less well-characterized by comparison with enzymes involved in ubiquitin attachment (Hochstrasser, 1996). Of the 17 yeast deubiquitinating enzymes, Doa4 is perhaps the best characterized, playing a general role in deubiquitination of membrane proteins at the vacuole, recovery of ubiquitin, and maintenance of cellular free ubiquitin levels (Katzmann et al., 2002). Accordingly, ubiquitination of newly synthesized Pma1-7 is markedly reduced in doa4 cells (Pizzirusso, unpublished data). Evidence suggesting deubiquitination of Pma1-7 at the plasma membrane comes from analysis of vps1 cells (Figure 5D). After induction and chase in vps1 cells, HA-Pma1-7 is prevented from endosomal entry and routed instead to the cell surface (Luo and Chang, 2000; Figure 5). Under these conditions, Pma1-7 ubiquitination disappears (Figure 5D), suggesting ubiquitination is removed at the plasma membrane. Indeed, in vps1 sec4 cells in which HA-Pma1-7 is barred from endosomal entry as well as plasma membrane arrival, ubiquitination seemed increased (Figure 5D). Consistently, Pma1-7 ubiquitination was not detectable in plasma membrane isolated after fractionation on Renografin density gradients (Figure 5D). Together, these observations support the hypothesis that Pma1-7 becomes deubiquitinated at the plasma membrane. Deubiquitination of Pma1-7 is consistent with its stabilization at the cell surface in vps1 (Figure 5A), as well as in multiple suppressor strains that cause Pma1-7 routing to the plasma membrane (Luo and Chang, 1997). By contrast with Pma1-7, ubiquitination of a number of plasma membrane proteins induces their rapid turnover (Rotin et al., 2000), and fusion of ubiquitin or ubiquitination motifs to wild-type Pma1 has been shown to destabilize and cause rapid cell surface internalization of the chimera (Kolling and Losko, 1997; Roth et al., 1998; Shih et al., 2000). Although the mechanism by which plasma membrane proteins are deubiquitinated will require further investigation, it is clear that multiple mechanisms regulating ubiquitination can control protein fate in the secretory pathway.
Acknowledgments
We thank Juan Bonifacino for strains and the GGA plasmids; Howard Riezman for anti-Gas1; and Daniel Kornitzer, Steve Nothwehr, and Yoshiko Kikuchi for strains. We thank Qiongqing Wang for assisting with the lipid raft assay. This work was supported by grant GM-58212 from the National Institutes of Health.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0727. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–10–0727.
References
- Andoh, T., Hirata, Y., and Kikuchi, A. (2000). Yeast glycogen synthase kinase 3 is involved in protein degradation in cooperation with Bul1, Bul2, and Rsp5. Mol. Cell. Biol. 20, 6712–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, J., Patel, S., and Riddle, P. (1990). Lysosomal sorting mutants of coronavirus E1 protein, a Golgi membrane protein. J. Cell Sci. 95, 191–197. [DOI] [PubMed] [Google Scholar]
- Arvan, P., Zhao, X., Ramos-Castaneda, J., and Chang, A. (2002). Secretory pathway quality control in the Golgi, plasmalemmal, and endosomal systems. Traffic 3, 771-780. [DOI] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., Estepa-Sabal, E.J., Meerloo, T., and Emr, S.D. (2002). ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., Chang, A., and Simons, K. (2001). Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 12, 4129–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., Keranen, S., Shevchenko, A., and Simons, K. (2000). Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97, 3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., Schmidt, A., and Hall, M.N. (1999). Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146, 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, B., Moreno, E., and Lagunas, R. (1991). Half-life of plasma membrane ATPase and its activating system in resting yeast cells. Biochim. Biophys. Acta 1063, 265–268. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Weissman, A.M. (1998). Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell Dev. Biol. 14, 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A. (2002). Plasma membrane biogenesis. Methods Enzymol. 351, 339–350. [DOI] [PubMed] [Google Scholar]
- Chang, A., and Fink, G.R. (1995). Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J. Cell Biol. 128, 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A., and Slayman, C.W. (1991). Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 115, 289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C.R., Emr, S.D., and Horazdovsky, B.F. (1994). Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 107, 3449–3459. [DOI] [PubMed] [Google Scholar]
- Cross, F. (1997). `Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13, 647–653. [DOI] [PubMed] [Google Scholar]
- Ellgard, L., Molinari, M., and Helenius, A. (1999). Setting the standards: quality control in the secretory pathway. Science 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Fisk, H. A., and Yaffe, M. P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, X., and Chang, A. (2001). A mutant plasma membrane ATPase, Pma1–10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. USA 98, 9104–9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley, K., Dong, Y., and Sagnella, G.A. (2003). Regulation of the epithelial sodium channel by accessory proteins. Biochem. J. 371, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, S.B., Losko, S., and Kaiser, C.A. (2001). Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L. (1999). Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 9, 107–112. [DOI] [PubMed] [Google Scholar]
- Hicke, L. (2001). Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201. [DOI] [PubMed] [Google Scholar]
- Hinners, I., and Tooze, S.A. (2003). Changing directions: clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 116, 763–771. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (1996). Ubiquitin dependent protein degradation. Annu. Rev. Genet. 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hong, E., Davidson, A.R., and Kaiser, C.A. (1996). A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 135, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe, T., Matuschewski, K., Rape, M., Schlenker, S., Ulrich, H.D., and Jentsch, S. (2000). Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577–586. [DOI] [PubMed] [Google Scholar]
- Jenness, D.D., Li, Y., Tipper, C., and Spatrick, P. (1997). Elimination of defective α-factor pheromone receptors. Mol. Cell Biol. 17, 6236–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E.W. (1977). Proteinase mutants of Saccharomyces cerevisiae. Genetics 35, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst, M., and Emr, S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-1. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. 3, 893–905. [DOI] [PubMed] [Google Scholar]
- Kolling, R., and Losko, S. (1997). The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J. 16, 2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.C.S., Hamamoto, S., and Schekman, R. (2002). Ceramide biosynthesis is required for the formation of oligomeric H+-ATPase, Pma1p, in the yeast endoplasmic reticulum. J. Biol. Chem. 277, 22395–22401. [DOI] [PubMed] [Google Scholar]
- Liu, X.F., and Culotta, V.C. (1999). Post-translation control of Nramp metal transport in yeast. J. Biol. Chem. 274, 4863–4868. [DOI] [PubMed] [Google Scholar]
- Luo, W.-J., and Chang, A. (2000). An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast. Mol. Biol. Cell 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W.-J., and Chang, A. (1997). Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J. Cell Biol. 138, 731-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik, B., and Kaiser, C.A. (2002). Nitrogen regulation in Saccharomyces cerevisiae. Gene 290, 1–18. [DOI] [PubMed] [Google Scholar]
- Minami, Y., Weissman, A.M., Samelson, L.E., and Klausner, R.D. (1987). Building a multichain receptor: synthesis, degradation, and assembly of the T-cell antigen receptor. Proc. Natl. Acad. Sci. USA 84, 2688–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, C., and Bonifacino, J.S. (2001). Structural requirements for function of yeast GGAs in vacuolar protein sorting, α-factor maturation, and interactions with clathrin. Mol. Cell Biol. 21, 7981-7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr, S.F., Conibear, E., and Stevens, T.H. (1995). Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J. Cell Biol. 129, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart, C.M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–33. [DOI] [PubMed] [Google Scholar]
- Rape, M., Hoppe, T., Gorr, I., Kalocay, M., Richly, H., and Jentsch, S. (2001). Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin-selective chaperone. Cell 107, 667–677. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and Pelham, H.R.B. (2002). A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4, 117–123. [DOI] [PubMed] [Google Scholar]
- Roth, A.F., Sullivan, D.M., and Davis, N.G. (1998). A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J. Cell Biol. 142, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, J.H., Raymond, C.K., Gilbert, T., O'Hara, P.J., and Stevens, T.H. (1990). A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell 61, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Rotin, D., Staub, O., and Haguenauer-Tsapis, R. (2000). Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176, 1–17. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Hicks, J.B., and Fink, G.R. (1986). Methods in Yeast Genetics: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shih, S.C., Katzmann, D.J., Schnell, J.D., Sutanto, M., Emr, S.D., and Hicke, L. (2002). Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389–393. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., Sloper-Mould, K.E., and Hicke, L. (2000). Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 19, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow, P.S., Finley, D., and Varshavsky, A. (1986). Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 156, 147–153. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. 3, 246–255. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., and Nakano, A. (2003). Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161, 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., and Chang, A. (2002). Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc. Natl. Acad. Sci., USA 99, 12853–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach, K., and Payne, G.S. (1993). Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 12, 3049–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda, H., Oguchi, T., Yasuda, Y., Toh-e, A., and Kikuchi, Y. (1996). Bul1, a new protein that binds to the Rps5 ubiquitin ligase in Saccharomyces cerevisiae. Mol. Cell Biol. 16, 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B.-Y., Chang, A., Kjeldsen, T.B., and Arvan, P. (2001). Intracellular retention of newly-synthesized insulin in yeast is caused by endoproteolytic processing in the Golgi complex. J. Cell Biol. 153, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]