Abstract
Purpose
We examined the prognostic impact of specific KRAS mutations in stage III colon adenocarcinoma patients receiving adjuvant FOLFOX alone or combined with cetuximab in a phase III trial (N0147). Analysis was restricted to BRAF-wild type tumors, since BRAF mutation was associated with poor prognosis, and BRAF and KRAS mutations are mutually exclusive.
Experimental Design
The seven most common KRAS mutations in codon 12 and codon 13 were examined in 2,478 BRAF-wild type tumors. Because KRAS mutations in codon 12 (n=779) or 13 (n=220) were not predictive of adjuvant cetuximab benefit, study arms were pooled for analysis. Disease-free survival (DFS) was evaluated by hazard ratios (HR) using Cox models.
Results
KRAS mutations in codon 12 (multivariate HR 1.52; 95% confidence interval [CI] 1.28–1.80; P<.0001) or codon 13 (multivariate HR 1.36; 95% CI 1.04–1.77; P=.0248) were significantly associated with shorter DFS compared to patients with wild type KRAS/BRAF tumors, independent of covariates. KRAS codon 12 mutations were independently associated with proficient mismatch repair (P<.0001), proximal tumor site (P<.0001), low grade, age, and sex, whereas codon 13 mutations were associated with proximal site (P<.0001).
Conclusion
KRAS mutations in either codon 12 or 13 are associated with inferior survival in patients with resected stage III colon cancer. These data highlight the importance of accurate molecular characterization and the significant role of KRAS mutations in both codons in the progression of this malignancy in the adjuvant setting.
Keywords: KRAS, colon cancer, prognosis, BRAF, survival
INTRODUCTION
KRAS is a small G protein that acts as a transducer in the epidermal growth factor receptor (EGFR) signaling pathway (1). Approximately 40% of colorectal cancers (CRCs) harbor activating mutations in KRAS, making it the most commonly mutated gene in the RAS/RAF/MAPK pathway. KRAS mutations are believed to be an early event in colorectal tumorigenesis and lead to constitutive signaling and downstream activation of MAPK- and PI3K-dependent pathways. Most (90%) KRAS mutations occur in codons 12 and 13 in the phosphate-binding loop of KRAS (1), and mutations in either codon possess transforming capacity (2, 3). In vitro evidence indicates that KRAS codon 12 mutations have greater transforming ability characterized by inhibition of apoptosis, enhanced loss of contact inhibition, and increased predisposition to anchorage-independent growth when compared with codon 13 mutations (2-4). The glycine-to-aspartate transition (p.G13D) is the most frequent codon 13 mutation in CRC. In vitro and mouse model data have showed that, although p.G12V-mutated CRC were insensitive to cetuximab, p.G13D-mutated cells were sensitive, as were KRAS wild type cells (5).
Whereas the ability of most KRAS mutations to predict resistance to anti-EGFR therapy in patients with metastatic colorectal cancer is widely accepted, including recommendations for KRAS testing in metastatic disease (6), the prognostic impact of KRAS mutations including in stage III disease is uncertain (7-10). Codon 12 mutations have been associated with adverse prognosis in aggregate colorectal cancer populations of diverse disease stages (11, 12). However, recent data suggest that KRAS codon 13 mutations may not represent an aggressive phenotype or confer resistance to anti-EGFR therapy compared to wild type. In metastatic CRC, codon 13 (p.G13D) mutation, in contrast to those in codon 12, was associated with sensitivity to anti-EGFR therapy that was similar to wild type (5, 13), though the literature is inconsistent (14). Furthermore, recent population-based data suggest that patients with KRAS codon 13 mutations may have similarly favorable prognosis as those with KRAS wild type (11). No study to date has demonstrated that KRAS codon 13 mutations are significantly associated with worse patient survival in patients with non-metastatic colon cancer (5, 11-19). Data from randomized clinical trials are summarized in Table 1. These findings suggest that KRAS codon 13 mutations may not be biologically important in the progression of CRC and question the clinical relevance of analyzing these mutations routinely.
Table 1.
Randomized clinical trials examining the prognostic impact of KRAS codon 12 and 13 mutations in colorectal cancer
| Cohort | No. of Tumors Total (Codon 12 / 13) |
% of Total Cohort |
Tumor, Stage |
Treatment | Findings | ||
|---|---|---|---|---|---|---|---|
| Multivariate HRs for KRAS mutations |
Reference Group a |
||||||
| Codon 12 | Codon 13 | ||||||
| Co.17, BOND, MABEL, EMR202600, EVEREST, BABEL, SALVAGE (5) |
579 (~260 / 45) | CRC IV |
BSC +/− cetuximab; Cetuximab +/− chemotherapy |
c.38G>A HR 1.82 (p =.053) for overall survival b |
BRAF/KRAS wild type or BRAF mutated |
||
| OPUS, CRYSTAL (13) |
1378 (125 / 83) | 90% | CRC IV |
FOLFIRI or FOLFOX +/− cetuximab |
c.35G>T, HR 1.11 (p =.53) for overall survival c |
c.38G>A, HR 1.39 (p=.079) for overall survival c |
BRAF/KRAS wild type or BRAF mutated |
| NSABP C07, C08 (9) |
2299 ( - / - ) | 48% | Colon II—III |
5FU +/− oxaliplatin, FOLFOX +/− bevacizumab |
c.35G>T, HR 1.22 (p=.16) for time to recurrence d |
BRAF/KRAS wild type or BRAF mutated |
|
| PETACC-3 (18) | 1321 (368 / 102) | 40% | Colon II—III |
5FU +/− irinotecan |
c.35G>A, HR 0.98 (p =.91) c.35G>C, HR 0.97 (p =.92) c.35G>T, HR 1.09 (p =.64) c.34G>T, HR 1.40 (p =.15) c.34G>A, HR 0.99 (p =.97) for relapse-free survival d |
c.38G>A, HR 0.99 (p =.97) for relapse-free survival d |
BRAF/KRAS wild type or BRAF mutated |
| CALGB 89803 (21) |
506 (123 / 53) | 40% | Colon III |
5FU +/− irinotecan |
Any Codon 12, HR 1.09 (NS) for disease-free survival d |
c.38G>A, HR 0.82 (NS) for disease-free survival d |
BRAF/KRAS wild type or BRAF mutated |
|
NCCTG N0147
(Alliance); Current Study |
2478 (779 / 220) BRAF wild type only |
82% | Colon III |
FOLFOX +/− cetuximab |
Any Codon 12, HR 1.52 (p <.0001) for disease- free survival d |
c.38G>A, HR 1.36 (p =.025) for disease-free survival d |
BRAF/KRAS
wild type only |
BSC, best supportive care; CRC, colorectal; HR, hazard ratio; 5FU, fluorouracil; NS, not statistically significant
Refers to the patient reference group used for prognostic analysis.
BSC-alone arm
Chemotherapy-alone arms across both trials
Data pooled across both arms
Few studies examining the prognostic impact of specific KRAS mutations in CRC have controlled for BRAF mutation as a confounder. However, the most rigorous approach to isolate the prognostic impact of KRAS is to restrict analysis to BRAF-wild type tumors, given that BRAF and KRAS mutations are mutually exclusive (6) and that BRAF mutations are associated with adverse prognosis (7, 18, 20-24). It is also important to account for DNA mismatch repair (MMR) status, since the subset of CRCs with deficient MMR (dMMR) and microsatellite instability (MSI) have a relatively low rate of KRAS mutations as compared to proficient MMR (pMMR) and microsatellite stable tumors (25).
In this report, we determined the association of the seven most common KRAS mutations in codon 12 and 13 with disease-free survival (DFS) in prospectively collected, stage III colon adenocarcinomas from participants of a phase III trial (N0147). Patients were randomized to adjuvant 5-fluorouracil, oxaliplatin, and leucovorin (mFOLFOX6) alone or combined with cetuximab, and the addition of cetuximab to FOLFOX failed to improve DFS overall or in patients with wild type KRAS tumors (26). The current prognostic analysis was restricted to patients whose tumors were wild type for BRAF. In this cohort, we previously reported that KRAS (all codons combined) or BRAF mutations were each associated with shorter DFS (25). In the current study, we examined KRAS mutations in codons 12 and 13 separately, with a focus on determining whether codon 13 mutations are prognostic. Our findings indicate that KRAS mutations in both codon 12 and 13 confer a worse prognosis in stage III colon cancers.
METHODS
Study Population
Subjects with completely resected, stage III colon adenocarcinoma (TanyN1-2M0) participated in a phase III randomized trial (North Central Cancer Treatment Group [NCCTG] N0147; 2004 to 2009) of adjuvant mFOLFOX6 alone or combined with cetuximab, which was previously described (26). Prospective and centralized KRAS mutation testing was required, although randomization was done irrespective of KRAS status in the original trial design. In August 2008, the trial was amended to restrict randomization to patients with KRAS-wild type tumors based upon data demonstrating the predictive utility of KRAS for anti-EGFR antibody therapy (26). Post-amendment, eligible patients with KRAS-mutated tumors (n=332) were treated at investigator discretion (97% received FOLFOX) and followed for disease recurrence. To avoid selection bias, the current analysis includes all randomized study patients and those with KRAS-mutated tumors who enrolled post-amendment (n=3,018 total). Tissues were prospectively collected and required for study participation. Central pathology review was performed. Proximal tumor site included the cecum, ascending and transverse colon; distal site included the splenic flexure, descending and sigmoid colon.
Patients initiated chemotherapy within 10 weeks of surgery. After completing protocol-specified treatment, disease recurrence was assessed every 6 months until 5 years post-randomization with a physical examination, computed tomographic scan, and laboratory assessment. Follow-up colonoscopy was recommended at years 1 and 4 post-resection.
The study was approved by the Mayo Clinic Institutional Review Board (IRB) and the NCCTG (now part of Alliance for Clinical Trials in Oncology). Patients signed an IRB-approved consent.
KRAS and BRAF mutation
Assessment of KRAS and BRAF (NCBI Entrez Gene 673) mutational status was performed centrally at the Mayo Clinic in a Clinical Laboratory Improvement Amendments (CLIA)-compliant laboratory, using appropriate quality control procedures. Both KRAS and BRAF mutation status was determined using DNA extracted from macrodissected formalin-fixed, paraffin-embedded tumor tissue.
For KRAS, testing was performed with the DxS mutation test kit KR-03/04 (DxS), together with the Light-Cycler 480 (Roche Applied Sciences), which assesses for 7 missense point mutations: six mutations in codons 12 (c.35G>C [p.G12A, GGT>GCT], c.34G>C [p.G12R, GGT>CGT], c.35G>A [p.G12D, GGT>GAT], c.34G>T [p.G12C, GGT>TGT], c.34G>A [p.G12S, GGT>AGT], and c.35G>T [p.G12V, GGT>GTT] and one mutation in codon 13 (c.38G>A [p.G13D, GGC>GAC]). The level of detection was set at 5%. Assessment for the BRAF c.1799T>A (p.V600E) mutation was performed using a multiplex allele specific polymerase chain reaction (PCR)–based assay. The polymerase chain reaction primers used for this assay were fluorescently labeled and included the following (wild type forward NEDTGATTTTGGTCATGCTACAGT]; mutant forward [6-Fam-CAGTGATTTTGGTCTAGCTTCAGA]; and reverse [GTTTCTTTCTAGTAACTCAGCAGC]). Following amplification, PCR products were analyzed on an ABI 3130×l instrument (Life Technologies, Applied Biosystems) and scored for the presence or absence of the V600E variant only.
DNA Mismatch Repair Proteins
MMR protein (MLH1, MSH2 and MSH6) expression was analyzed in formalin-fixed, paraffin-embedded tumor sections using an immunoperoxidase method (27). Monoclonal antibodies included mouse anti-human MLH1 (clone G168-15, Biocare Medical, Concord, CA), anti-human MSH2 (clone FE11, Biocare Medical, Concord, CA), and anti-human MSH6 (clone BC/44; Biocare Medical, Concord, CA). MMR protein loss was defined as the absence of nuclear staining in tumor cells in the presence of positive nuclear staining in normal colonic epithelium and lymphocytes. Tumors were classified as MMR-deficient (vs MMR-proficient) if loss of one or more MMR proteins was detected.
Statistical Methods
Our primary objective was to compare survival among patients carrying any mutation in codon 12, mutated codon 13, and wild type KRAS. The primary clinical endpoint was DFS, and a secondary endpoint was time to recurrence (TTR). DFS was defined as the time from randomization to first documented recurrence or any-cause death, whichever occurred first. TTR was defined as the time from randomization to first documented recurrence. Survival was evaluated by hazard ratios (HR) using Cox models. Kaplan-Meier methods were used to describe the distributions of DFS and TTR, which were censored at 5 years after randomization. Multivariable Cox models were adjusted for age, gender, T stage, N stage, number of examined nodes, histologic grade, performance status, primary tumor site, mismatch repair status, and treatment. Analysis of KRAS mutations included analysis of codon 12 mutations grouped together and codon 13, as well as each mutation individually. Interactions between KRAS mutation and treatment were assessed. All analyses were based on the study database frozen on Sept. 4, 2012. Two-sided P values, with values <.05 considered statistically significant, and 95% confidence intervals (CI) are reported. Analyses were performed using SAS version 9.2 (SAS Institute Inc, Cary NC). Data collection and statistical analyses were conducted by the Alliance Statistics and Data Center.
RESULTS
KRAS mutations in colon cancer
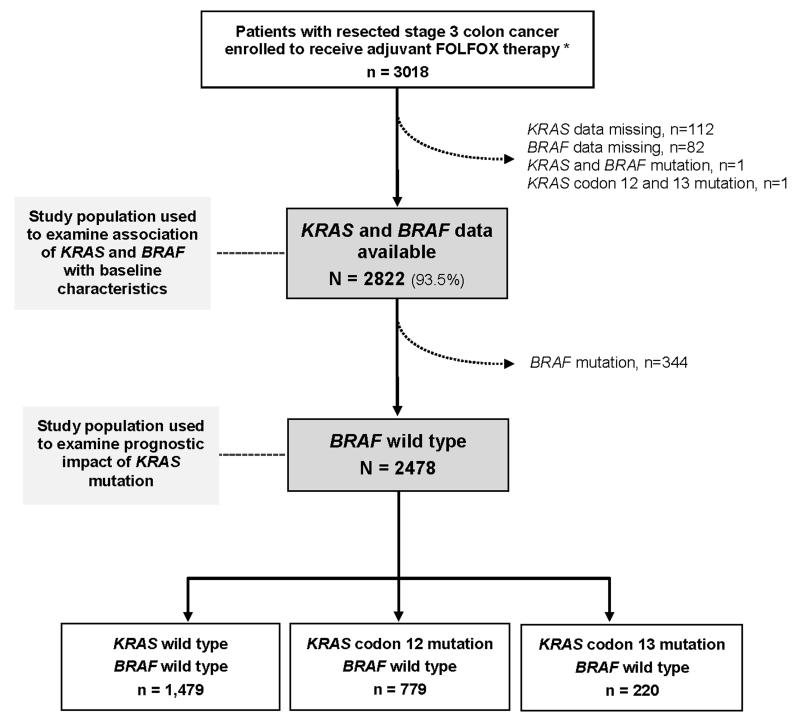
The study population comprises patients with completely resected stage III colon cancer (n=3018) who received adjuvant FOLFOX-based chemotherapy in a North American phase III clinical trial (N0147) (Figure 1). KRAS and BRAF data were available in 93.5% (2822/3018) of patients. Tumors with both KRAS and BRAF mutations (n=1) or KRAS mutation in both codon 12 and 13 (n=1) were excluded.
Figure 1. Study profile.
BRAF-mutated cases were excluded to assess the prognostic role of KRAS mutation in BRAF-wild type tumors. * Includes patients with KRAS-mutated tumors (n=332) enrolled post-study modification (see Methods), of whom 97% received FOLFOX.
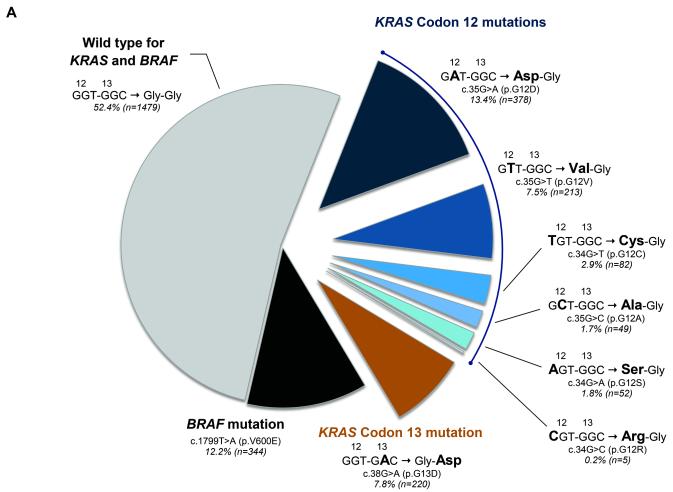
Figure 2a shows the frequencies and types of KRAS mutations, which are consistent with prior reports (28), and the corresponding predicted amino acid sequence alterations. KRAS codon 12 or 13 (c.38G>A [p.G13D]) mutations were detected in 35.4% (999/2822) of tumors, with 27.6% in codon 12 and 7.8% in codon 13. Within codon 12, most (82%) mutations occurred in the second base position, and the frequency of transversions (G>C, G>T) and transitions (G>A) were similar (45% and 55%, respectively). BRAF mutation occurred in 12.2% (344/2822) (Fig 2a).
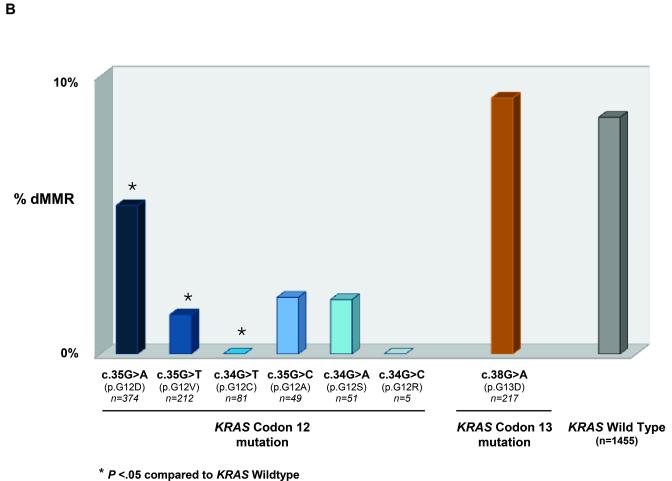
Figure 2. KRAS (codon 12 and 13) and BRAF mutation frequencies in 2,904 stage III colon adenocarcinomas.
(a) Frequencies of KRAS mutations and corresponding amino acid sequence alterations are shown. (b) Frequency of deficient mismatch repair (MMR) among KRAS-mutated and BRAF-wild type tumors are shown (numbers differ slightly from [a] due to missing MMR data). Asterisks (*) denote statistically significant differences compared to KRAS/BRAF wild type (P <.05).
KRAS mutations and clinicopathologic characteristics
Table 2 summarizes the baseline clinicopathologic characteristics of study subjects according to KRAS and BRAF mutation status. Compared to wild type, KRAS mutations were significantly associated with older age and female sex, primarily due to mutations in codon 12, and did not differ by T stage or number of positive nodes. Compared to KRAS wild type, codon 12 and 13 mutations were each associated with proximal (vs distal) tumor site within the colon (P<.0001). Codon 12 and 13 mutations were associated with low and high grade histology, respectively, in primary tumors.
Table 2.
KRAS codon 12 and 13 mutations in relation to clinicopathologic and molecular characteristics in stage III colon cancers (N = 2,822)
| Variable | Wild type for KRAS and BRAF (n=1479) |
Any KRAS mutation in Codon 12 or 13 (n=999) |
Specific KRAS Mutation |
BRAF Mutation (n=344) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Codon 12 only (n=779) |
Codon 13 only (n=220) |
||||||||
| N (%) | N (%) | P a | N (%) | P a | N (%) | P a | N (%) | P a | |
| Age, years | |||||||||
| Median (range) | 56 (19-84) | 58 (22-85) | 0.0008 | 58 (22-85) | .0002 | 57 (22-82) | .6052 | 65 (31-86) | <.0001 |
|
| |||||||||
| Gender | |||||||||
| Female (n=1336) | 630 (43) | 484 (48) | 0.0041 | 387 (50) | .0013 | 97 (44) | .6759 | 222 (65) | <.0001 |
| Male (n=1486) | 849 (57) | 515 (52) | 392 (50) | 123 (56) | 122 (35) | ||||
|
| |||||||||
| T stage | |||||||||
| T1-2 (n=423) | 238 (16) | 149 (15) | 0.4346 | 111 (14) | .2545 | 38 (17) | .6578 | 36 (11) | .0085 |
| T3-4 (n=2398) | 1241 (84) | 849 (85) | 667 (86) | 182 (83) | 308 (89) | ||||
| missing | 0 | 1 | 1 | 0 | 0 | ||||
|
| |||||||||
| Grade | |||||||||
| Low (n=2116) | 1145 (77) | 792 (79) | 0.2710 | 639 (82) | .0105 | 153 (70) | .0103 | 179 (52) | <.0001 |
| High (n=706) | 334 (23) | 207 (21) | 140 (18) | 67 (30) | 165 (48) | ||||
|
| |||||||||
| No. positive nodes | |||||||||
| 1-3 (n=1650) | 871 (59) | 610 (61) | 0.2799 | 487 (63) | .0944 | 123 (56) | .4023 | 169 (49) | .0010 |
| 4 or more (n=1172) | 608 (41) | 389 (39) | 292 (37) | 97 (44) | 175 (51) | ||||
|
| |||||||||
| Tumor Site | |||||||||
| Proximal (n=1407) | 545 (37) | 577 (59) | <.0001 | 443 (58) | <.0001 | 134 (62) | <.0001 | 285 (84) | <.0001 |
| Distal (n=1370) | 914 (63) | 402 (41) | 321 (42) | 81 (38) | 54 (16) | ||||
| Missing | 20 | 20 | 15 | 5 | 5 | ||||
|
| |||||||||
| Mismatch Repair | |||||||||
| Deficient (n=318) | 124 (8) | 45 (5) | 0.0001 | 25 (3) | <.0001 | 20 (9) | .7338 | 149 (44) | <.0001 |
| Proficient (n=2464) | 1331 (92) | 944 (95) | 747 (97) | 197 (91) | 189 (56) | ||||
| Missing | 24 | 10 | 7 | 3 | 6 | ||||
Comparison with KRAS/BRAF wild type.
A low frequency of KRAS mutations was detected in dMMR compared to pMMR tumors (14% [45/318] vs 38% [944/2464]; Table 2). Mutations in codon 12 were significantly less frequent in dMMR tumors compared to wild type (3% vs 8%; P<.0001; Table 2), and this low frequency was observed across codon 12 mutations (Figure 2b). Deficient MMR showed a strong inverse association with KRAS codon 12 mutation (OR 0.28; 95% CI 0.18–0.44; P<.0001), independent of covariates (Table S1). However, the frequency of dMMR was similar in KRAS codon 13 mutations and KRAS/BRAF-wild type (9% vs 8%; P=.7338; Table 2).
Proximal tumor site, older age, female sex, and low grade were each significantly associated with KRAS codon 12 mutation independent of covariates (all P values <.030; Table S1). By contrast, only proximal site (P<.0001) showed an independent association with KRAS codon 13 mutation compared to KRAS/BRAF-wild type (Table S1).
Similar to KRAS mutations, BRAF mutation was associated with older age, female sex, proximal site, and dMMR; and unlike KRAS, BRAF mutation was also associated with higher T and N stage, and higher histologic grade (Table 2), as previously reported (25).
KRAS mutation and patient survival in BRAF-wild type cases
To remove the confounding effect of BRAF mutation on the prognostic impact of KRAS mutation, we analyzed BRAF-wild type tumors only (n=2478) when examining patient survival and compared KRAS-mutated/BRAF-wild type cases with KRAS-wild type/BRAF-wild type cases (Fig. 1). Among the 687 DFS events, there were 353 deaths during a median follow-up of 43.2 (interquartile range, 31.0–55.3) months and 616 TTR events during a median follow-up of 42.4 (interquartile range, 30.4–55.0) months for censored cases.
As shown in Figure 3a and Table 3 (top panel), patient tumors with KRAS codon 13 mutations experienced shorter DFS (univariate HR 1.46; 95% CI 1.13–1.89; P=.0035; multivariate HR 1.36; 95% CI 1.04–1.77; P=.0248), compared with those that were wild type for KRAS and BRAF, independent of clinicopathologic variables and MMR status. KRAS codon 12 mutation was also significantly associated with worse DFS (univariate HR 1.50; 95% CI 1.28–1.76; P<.0001; multivariate HR 1.52; 95% CI 1.28–1.80; P<.0001), compared with patients whose tumors were wild type for KRAS and BRAF. Results were similar when the full cohort was analyzed adjusting for BRAF mutation (codon 13, multivariate HR 1.334 [95% CI 1.003, 1.773], P=.0474; codon 12, multivariate HR 1.584 [95% CI 1.328, 1.890], P<.0001). When TTR was analyzed as the outcome variable in the BRAF-wild type subgroup (Figure 3b), results were consistent both for codon 13 (univariate HR 1.46; 95% CI 1.11– 1.92; P=.0064; multivariate HR 1.34; 95% CI 1.01–1.78; P=.0446) and for codon 12 (univariate 1.59; 95% CI 1.34–1.88; P<.0001; multivariate HR 1.60; 95% CI 1.34–1.91; P<.0001).
Figure 3. Prognostic impact of specific KRAS mutations in 2,478 patients with BRAF-wild type resected stage III colon cancer.

KRAS mutations in codon 12 and 13, compared to wild type BRAF and KRAS, are shown in relation to (a) disease free-survival and (b) time to recurrence.
Table 3.
Cox proportional hazards models examining association of KRAS mutation status with disease-free survival in 2,478 BRAF-wild type colon cancer patients
| KRAS status | N (Events) | 3-year disease-free survival rate (95% CI) |
Univariate |
Multivariate a |
||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| Model 1 | ||||||
|
| ||||||
| Any codon 12 mutation | 779 (256) | 68% (64%-71%) | 1.50 (1.28, 1.76) | <.0001 | 1.52 (1.28, 1.80) | <.0001 |
|
| ||||||
| Codon 13 mutation | 220 (71) | 67% (60%-73%) | 1.46 (1.13, 1.89) | 0.0035 | 1.36 (1.04, 1.77) | 0.0248 |
|
| ||||||
| Wild type b | 1479 (360) | 77% (75%-80%) | reference | Reference | ||
|
| ||||||
| Model 2 | ||||||
|
| ||||||
| Individual codon 12 mutations |
||||||
| c.35G>A (p.G12D) | 378 (122) | 68% (63%-73%) | 1.51 (1.23, 1.85) | <.0001 | 1.53 (1.23, 1.89) | 0.0001 |
| c.35G>T (p.G12V) | 213 (68) | 70% (63%-76%) | 1.38 (1.07, 1.79) | 0.0145 | 1.40 (1.07, 1.82) | 0.0139 |
| c.34G>T (p.G12C) | 82 (30) | 61% (50%-73%) | 1.66 (1.14, 2.41) | 0.0078 | 1.63 (1.11, 2.41) | 0.0128 |
| c.35G>C (p.G12A) | 49 (19) | 63% (49%-77%) | 1.78 (1.12, 2.82) | 0.0148 | 1.75 (1.10, 2.79) | 0.0178 |
| c.34G>A (p.G12S) | 52 (14) | 72% (59%-85%) | 1.28 (0.75, 2.19) | 0.3624 | 1.37 (0.80, 2.35) | 0.2485 |
| c.34G>C (p.G12R) | 5 (3) | 50% (1%-99%) | 3.81 (1.23, 11.87) | 0.0209 | 5.30 (1.69, 16.64) | 0.0043 |
|
| ||||||
| Codon 13 mutation | ||||||
| c.38G>A (p.G13D) | 220 (71) | 67% (60%-73%) | 1.46 (1.13, 1.89) | 0.0035 | 1.36 (1.04, 1.77) | 0.0246 |
|
| ||||||
| Wild type b | 1479 (360) | 77% (75%-80%) | reference | reference | ||
CI, confidence interval; HR, hazard ratio.
Adjusted for age, gender, T stage, N stage, no. examined nodes, grade, performance status, tumor site, mismatch repair status, treatment.
KRAS and BRAF wild type.
Individual KRAS mutations within codon 12 were also examined in relation to patient survival (Table 3, bottom panel). Each mutation was associated with worse DFS compared to KRAS/BRAF-wild type (all HR point estimates >1). Five of the 6 KRAS codon 12 mutations (c.34G>A [p.G12D], c.35G>T [p.G12V], c.34G>T [p.G12C], c.35G>C [p.G12A], c.34G>C [p.G12R]) demonstrated a statistically significant association with worse DFS in univariate and multivariate analysis. Results were consistent when TTR was analyzed as the outcome (data not shown).
In an exploratory analysis, we examined the prognostic association of KRAS codon 12 or 13 mutations (vs wild type) among BRAF-wild type tumors within various strata, including tumor site, N stage, and MMR status. No significant modifying effect by these variables was observed (all P interaction >.18).
The predictive value of KRAS status for cetuximab benefit was determined among patients that enrolled prior to August 2008, when both KRAS-mutated and -wild type patients were randomized to chemotherapy with or without cetuximab (see Methods). KRAS codon 12 or 13 mutations were not associated with differential DFS among treatment arms (any KRAS mutation vs wild type, Pinteraction =.988; codon 12 vs codon 13 KRAS mutations vs wild type, Pinteraction =.628; Figure S1). Individual mutations within codon 12 were also not predictive of cetuximab benefit (Figure S1).
DISCUSSION
We analyzed the frequency of KRAS codon 12 and 13 mutations in prospectively collected stage III colon cancers from a clinical trial of adjuvant chemotherapy. KRAS mutations were detected in 35.4% (999/2822) of tumors, with 27.6% detected in codon 12 and 7.8% in codon 13 (c.38G>A [p.G13D]). The specific KRAS mutations identified and their relative frequencies are consistent with other studies across tumor stages (28). We also determined the association of KRAS codon 12 and 13 mutations with clinicopathologic variables and survival. The study arms were combined for analysis since the addition of cetuximab to FOLFOX trial did not improve outcome in the parent trial, and no interaction between treatment and KRAS mutation status was observed. We restricted prognostic analysis to BRAF-wild type tumors so as to control for the confounding effect of BRAF c.1799T>A mutations. We found that KRAS mutations in codons 12 or 13 (c.38G>A) were each significantly associated with worse DFS compared with KRAS-wild type/BRAF-wild type cases. Specifically, patients whose tumors carried KRAS codon 12 or 13 mutations experienced a 52% or 36% higher relative risk, respectively, of colon cancer recurrence or any-cause death that was independent of clinicopathological variables or MMR status. Results were similar when TTR was used as the outcome variable. We emphasize that only the c.38G>A mutation was analyzed in codon 13, whereas multiple mutations within codon 12 were found that showed a consistent association with adverse outcome.
To our knowledge, our data are the first to demonstrate that KRAS codon 13 (c.38G>A) mutations adversely impact survival in non-metastatic colon cancer. In both a population-based cohort and a meta-analysis using individual patient data of stage I to IV CRCs, codon 13 mutations were not prognostic, in contrast to codon 12 mutations (11, 12). In smaller studies examining CRCs of metastatic or mixed stage, non-significant trends were reported between codon 13 mutations and worse prognosis (13, 15, 17, 29). Furthermore, a study of 160 CRCs of varying tumor stages and treatments found that KRAS codon 13, but not codon 12, mutations were associated with higher S-phase fractions, increased nodal metastases, and adverse outcome compared to wild type (16). A Swedish population-based study of 525 CRCs reported that individuals with KRAS codon 13 (but not codon 12) mutations experienced shorter cancer-specific survival in unadjusted, but not adjusted, analysis (30). Limitations of prior studies include the inconsistent incorporation of patients with BRAF mutations (in the comparison group) and variable patient therapies, which can confound the interpretation of the KRAS prognostic data (31-33). Most prior studies included stage IV patients and had fewer codon 13 mutation patients. Of note, the adverse impact of KRAS codon 13 mutations on survival in our study appeared to be attenuated compared to codon 12 mutations (36% vs 52%, respectively, higher risk of DFS). Consistent with this finding are laboratory data showing that KRAS codon 12 mutations display greater transforming ability, enhanced anchorage-independent growth, and an increased ability to suppress apoptosis when compared with codon 13 mutants (2-4). Computational analysis of the structural implications of KRAS mutations suggests that codon 12 mutation (c.35G>A, p.G12D) may impair hydrolysis of GTP, leaving KRAS in an active GTP-bound state, to a greater degree than codon 13 mutation (c.38G>A, p.G13D) or wild type KRAS (34). In metastatic CRCs codon 13 mutations (p.G13D), but not codon 12 mutations, were associated with sensitivity to anti-EGFR therapy that was similar to wild type tumors (5, 13), However, cetuximab was ineffective in our study and, therefore, KRAS mutations including those in codon 13 did not predict outcomes from adjuvant cetuximab treatment.
Within KRAS codon 12, each of the six individual mutations showed an association with shorter DFS compared to wild type KRAS/BRAF. Although c.35G>A (p.G12D) was most common, four other mutations (c.35G>T [p.G12V], c.34G>C [p.G12R], c.34G>T [p.G12C], c.35G>C [p.G12A]) also demonstrated a significant association with adverse outcome that was independent of covariates and sometimes appeared to be stronger. The c.34G>A [p.G12S] mutation showed the weakest association. Codon 12 RAS mutations encoding valine (p.G12V) or arginine (p.G12R) have been reported to demonstrate stronger transforming ability and a more aggressive tumorigenic phenotype than other codon 12 mutations (35-37) and to be associated with shorter patient survival compared to wild type (11, 12). Interestingly, c.34G>C [p.G12R] demonstrated the strongest association with poor survival in both our study (HR >5 for DFS) and in a population-based cohort (HR > 3 for cancer-specific death), suggesting that this codon 12 mutation is particularly aggressive despite being rare (<1%). Our findings confirm the adverse prognostic impact of c.35G>T (p.G12V) and, consistent with prior studies, suggest that c.34G>C (p.G12R) mutations are also adverse. In addition, our findings suggest the adverse impact of lower frequency mutations within codon 12 (c.34G>T [p.G12C], c.35G>C [p.G12A]) and c.35G>A (p.G12D) that has not been previously reported in non-metastatic colon cancers.
In our study, tumors with KRAS codon 12 mutations had a lower frequency of deficient MMR compared to tumors with codon 13 mutation or wild type, consistent with findings from a smaller report (38). Admittedly, this difference may be related to smaller size of the codon 13 subgroup, yet the frequency of deficient MMR was consistently low across all KRAS codon 12 mutations. In addition, codon 12 mutations were associated with low-grade histology whereas cancers with codon 13 mutations were more likely to show high-grade histology. These findings are consistent with evidence indicating that KRAS mutations may arise in unique molecular and clinical contexts, as the mutational spectrum can depend on the nature of the underlying genetic instability (38, 39). Epidemiologically, colorectal cancers with codon 12 and 13 mutations have been associated with different dietary intake patterns (40, 41). Furthermore, laboratory studies have shown that codon 12 mutations demonstrate increased PI3K pathway activation (2) and a distinct metabolic phenotype that promotes resistance to apoptosis (42) compared to codon 13 mutations. We found that KRAS mutations showed a higher frequency in proximal (vs distal) colon tumors, independent of other variables (43, 44). The distribution of KRAS codon 12 vs 13 mutations did not differ considerably by tumor subsite (data not shown). Proximal colon tumors are more likely than distal tumors to be KRAS-, BRAF-, and hypermutated, hypermethylated, and MMR-deficient (45). The explanation for why KRAS mutations show a predilection for the proximal tumor is unknown except to invoke molecular differences based on midgut and hindgut embryology. As expected, BRAF c.1799T>A mutations were enriched in tumors with dMMR and showed clinicopathologic features in common that included proximal tumor predominance, high-grade histology, older age, and female sex (46). In the N0147 study cohort and other reports, BRAF mutations are associated with shorter patient survival rates (9, 18, 21, 25).
This study is the largest to evaluate the prognostic impact of specific KRAS codons 12 and 13 in stage III colon cancer. Other strengths of this study include prospective collection of tissue specimens from a large clinical trial with meticulous collection of survival data. Systemic treatment consisted of a modern chemotherapy regimen (FOLFOX) generalizable to most stage III patients in the world. KRAS and BRAF mutation status was determined in a CLIA-certified laboratory. Limitations of the study include the fact that overall survival data have not yet matured; however, the reliability of DFS as a surrogate for OS in a stage III colon cancer population has been demonstrated by our group and others (47). We await biomarker results from PETACC-8, a phase 3 trial of colon cancer patients in which the addition of cetuximab to FOLFOX did not improve DFS or OS (48). We did not examine other less common mutations in KRAS, NRAS, or HRAS; recent data suggest that 17%-18% of patients with metastatic CRC that are wild type for KRAS codon 12 or 13 harbor additional RAS activating mutations that predict a lack of response to panitumumab (49, 50).
In conclusion, we found that KRAS mutations in codon 12 and 13 were each significantly associated with shorter DFS, compared to tumors with wild type KRAS/BRAF. In contrast to prior reports, our data establish codon 13 mutations as being adversely associated with outcome in stage III colon cancers. KRAS mutations were significantly more frequent in proximal tumors, and codon 12 mutations were less frequent in tumors with deficient vs proficient MMR. Our findings support testing for KRAS mutations in codons 12 and 13 in stage III colon cancers as these results provide important prognostic information.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
The most common mutations in the EGFR pathway in colorectal cancers occur in KRAS codons 12 and 13. However, recent data suggests that codon 13 mutations may not represent an aggressive phenotype. We examined the prognostic impact of the seven most common KRAS mutations in codon 12 and 13 in stage III colon adenocarcinomas from a phase III adjuvant trial of FOLFOX with or without cetuximab. To minimize confounding, analysis was restricted to 2,478 BRAF-wild type tumors. KRAS mutations, including those in codon 13 only, were prognostic, showing a significant association with shorter disease-free survival compared to wild type KRAS/BRAF. These data demonstrate for the first time that KRAS codon 13 mutations are associated with inferior survival in patients with non-metastatic colon cancer, and highlight the important role of both codon 12 and 13 mutations in the progression of this malignancy in the adjuvant setting.
Acknowledgments
FINANCIAL SUPPORT: Supported by a National Cancer Institute Senior Scientist Award (K05CA-142885 to F.A.S) and the North Central Cancer Treatment Group Biospecimen Resource Grant (CA-114740) from the National Institutes of Health. Support for correlative studies was also provided by unrestricted funds from Bristol-Myers Squibb, ImClone Systems, Sanofi-Aventis, and Pfizer. The study was conducted as a collaborative trial of the North Central Cancer Treatment Group, Mayo Clinic and was supported in part by Public Health Service grants CA-25224 and CA37404 from the National Cancer Institute, Department of Health and Human Services. The study was also supported, in part, by grants from the National Cancer Institute (CA31946) to the Alliance for Clinical Trials in Oncology and to the Alliance Statistics and Data Center (CA33601). The content is solely the responsibility of the authors and does not necessarily represent the views of the National Cancer Institute or the National Institute of Health.
Footnotes
CONFLICT OF INTEREST: none
REFERENCES
- 1.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–85. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750–6. [PubMed] [Google Scholar]
- 3.Smith G, Bounds R, Wolf H, Steele RJC, Carey FA, Wolf CR. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerrero S, Figueras A, Casanova I, Farre L, Lloveras B, Capella G, et al. Codon 12 and codon 13 mutations at the K-ras gene induce different soft tissue sarcoma types in nude mice. FASEB J. 2002;16:1642–4. doi: 10.1096/fj.02-0050fje. [DOI] [PubMed] [Google Scholar]
- 5.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA : the journal of the American Medical Association. 2010;304:1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 6.Febbo PG, Ladanyi M, Aldape KD, De Marzo AM, Hammond ME, Hayes DF, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. quiz S3. [DOI] [PubMed] [Google Scholar]
- 7.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF Mutations in Advanced Colorectal Cancer Are Associated With Poor Prognosis but Do Not Preclude Benefit From Oxaliplatin or Irinotecan: Results From the MRC FOCUS Trial. J Clin Oncol. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 8.Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, et al. KRAS Mutation in Stage III Colon Cancer and Clinical Outcome Following Intergroup Trial CALGB 89803. Clin Cancer Res. 2009;15:7322–9. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavin PG, Colangelo LH, Fumagalli D, Tanaka N, Remillard MY, Yothers G, et al. Mutation profiling and microsatellite instability in stage II and III colon cancer: an assessment of their prognostic and oxaliplatin predictive value. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6531–41. doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, et al. Value of Mismatch Repair, KRAS, and BRAF Mutations in Predicting Recurrence and Benefits From Chemotherapy in Colorectal Cancer. J Clin Oncol. 2011;29:1261–70. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 11.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:4753–63. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreyev HJN, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D Tumor Mutations With Outcome in Patients With Metastatic Colorectal Cancer Treated With First-Line Chemotherapy With or Without Cetuximab. J Clin Oncol. 2012;30:3570–7. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 14.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, et al. Mutant KRAS Codon 12 and 13 Alleles in Patients With Metastatic Colorectal Cancer: Assessment As Prognostic and Predictive Biomarkers of Response to Panitumumab. J Clin Oncol. 2013;31:759–65. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 15.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: A population-based study. Cancer Epidemiology Biomarkers & Prevention. 2000;9:1193–7. [PubMed] [Google Scholar]
- 16.Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13:1438–46. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 17.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–62. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic Role of KRAS and BRAF in Stage II and III Resected Colon Cancer: Results of the Translational Study on the PETACC-3, EORTC 40993, SAKK 60-00 Trial. J Clin Oncol. 2010;28:466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 19.Zlobec I, Kovac M, Erzberger P, Molinari F, Bihl MP, Rufle A, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. 2010;127:2569–75. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- 20.Phipps AI, Buchanan DD, Makar KW, Burnett-Hartman AN, Coghill AE, Passarelli MN, et al. BRAF Mutation Status and Survival after Colorectal Cancer Diagnosis According to Patient and Tumor Characteristics. Cancer Epidemiology Biomarkers & Prevention. 2012;21:1792–8. doi: 10.1158/1055-9965.EPI-12-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogino S, Shima K, Meyerhardt JA, McCleary NJ, Ng K, Hollis D, et al. Predictive and Prognostic Roles of BRAF Mutation in Stage III Colon Cancer: Results from Intergroup Trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 23.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab Plus Irinotecan, Fluorouracil, and Leucovorin As First-Line Treatment for Metastatic Colorectal Cancer: Updated Analysis of Overall Survival According to Tumor KRAS and BRAF Mutation Status. J Clin Oncol. 2011;29:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 24.Price TJ, Hardingham JE, Lee CK, Weickhardt A, Townsend AR, Wrin JW, et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29:2675–82. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 25.Sinicrope FA, Mahoney MR, Smyrk TC, Thibodeau SN, Warren RS, Bertagnolli MM, et al. Prognostic Impact of Deficient DNA Mismatch Repair in Patients With Stage III Colon Cancer From a Randomized Trial of FOLFOX-Based Adjuvant Chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3664–72. doi: 10.1200/JCO.2013.48.9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alberts SR, Sargent DJ, Nair S, Mahoney MR, Mooney M, Thibodeau SN, et al. Effect of oxaliplatin, fluorouracil, and leucovorin with or without cetuximab on survival among patients with resected stage III colon cancer: a randomized trial. JAMA : the journal of the American Medical Association. 2012;307:1383–93. doi: 10.1001/jama.2012.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinicrope FA, Rego RL, Garrity-Park MM, Foster NR, Sargent DJ, Goldberg RM, et al. Alterations in cell proliferation and apoptosis in colon cancers with microsatellite instability. Int J Cancer. 2007;120:1232–8. doi: 10.1002/ijc.22429. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Ye Y, Sun HZ, Shi GM. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother Pharmacol. 2013;71:265–72. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 29.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The lancet oncology. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 30.Wangefjord S, Sundstrom M, Zendehrokh N, Lindquist KE, Nodin B, Jirstrom K, et al. Sex differences in the prognostic significance of KRAS codons 12 and 13, and BRAF mutations in colorectal cancer: a cohort study. Biol Sex Differ. 2013;4:17. doi: 10.1186/2042-6410-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smakman N, van den Wollenberg DJ, Elias SG, Sasazuki T, Shirasawa S, Hoeben RC, et al. KRAS(D13) Promotes apoptosis of human colorectal tumor cells by ReovirusT3D and oxaliplatin but not by tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2006;66:5403–8. doi: 10.1158/0008-5472.CAN-05-4108. [DOI] [PubMed] [Google Scholar]
- 32.Basso M, Strippoli A, Orlandi A, Martini M, Calegari MA, Schinzari G, et al. KRAS mutational status affects oxaliplatin-based chemotherapy independently from basal mRNA ERCC-1 expression in metastatic colorectal cancer patients. Br J Cancer. 2013;108:115–20. doi: 10.1038/bjc.2012.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YL, Liau JY, Yu SC, Ou DL, Lin LI, Tseng LH, et al. KRAS mutation is a predictor of oxaliplatin sensitivity in colon cancer cells. PLoS One. 2012;7:e50701. doi: 10.1371/journal.pone.0050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CC, Er TK, Liu YY, Hwang JK, Barrio MJ, Rodrigo M, et al. Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations. PLoS One. 2013;8:e55793. doi: 10.1371/journal.pone.0055793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fasano O, Aldrich T, Tamanoi F, Taparowsky E, Furth M, Wigler M. Analysis of the transforming potential of the human H-ras gene by random mutagenesis. Proc Natl Acad Sci U S A. 1984;81:4008–12. doi: 10.1073/pnas.81.13.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cespedes MV, Sancho FJ, Guerrero S, Parreno M, Casanova I, Pavon MA, et al. K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis. 2006;27:2190–200. doi: 10.1093/carcin/bgl063. [DOI] [PubMed] [Google Scholar]
- 37.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–5. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–11. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 40.Slattery ML, Curtin K, Anderson K, Ma KN, Edwards S, Leppert M, et al. Associations between dietary intake and Ki-ras mutations in colon tumors: A population-based study. Cancer Res. 2000;60:6935–41. [PubMed] [Google Scholar]
- 41.Kampman E, Voskuil DW, van Kraats AA, Balder HF, van Muijen GNP, Goldbohm RA, et al. Animal products and K-ras codon 12 and 13 mutations in colon carcinomas. Carcinogenesis. 2000;21:307–9. doi: 10.1093/carcin/21.2.307. [DOI] [PubMed] [Google Scholar]
- 42.Vizan P, Boros LG, Figueras A, Capella G, Mangues R, Bassilian S, et al. K-ras codon-specific mutations produce distinctive metabolic phenotypes in NIH3T3 mice [corrected] fibroblasts. Cancer Res. 2005;65:5512–5. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- 43.Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013 doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–54. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinicrope FA. DNA mismatch repair and adjuvant chemotherapy in sporadic colon cancer. Nature reviews Clinical oncology. 2010;7:174–7. doi: 10.1038/nrclinonc.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sargent DJ, Wieand HS, Haller DG, Gray R, Benedetti JK, Buyse M, et al. Disease-free survival versus overall survival as a primary end point for adjuvant colon cancer studies: Individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2005;23:8664–70. doi: 10.1200/JCO.2005.01.6071. [DOI] [PubMed] [Google Scholar]
- 48.Taïeb J, Tabernero J, Mini E, Subtil F, Folprecht G, Van Laethem J-L, et al. Subgroup analyses results of the PETACC8 phase III trial comparing adjuvant FOLFOX4 with or without cetuximab (CTX) in resected stage III colon cancer (CC) J Clin Oncol. 2013;31(suppl) abstr 3525. [Google Scholar]
- 49.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 50.Peeters M, Oliner KS, Price TJ, Cervantes A, Sobrero AF, Ducreux M, et al. Analysis of KRAS/NRAS mutations in phase 3 study 20050181 of panitumumab (pmab) plus FOLFIRI versus FOLFIRI for second-line treatment (tx) of metastatic colorectal cancer (mCRC) J Clin Oncol. 2014;32 abstr LBA387. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.