Abstract
EHD1 has been implicated in the recycling of internalized proteins to the plasma membrane. However, the mechanism by which EHD1 mediates recycling and its relationship to Rab-family–controlled events has yet to be established. To investigate further the mode of EHD1 action, we sought to identify novel interacting partners. GST-EHD1 was used as bait to isolate a ∼120-kDa species from bovine and murine brain cytosol, which was identified by mass spectrometry as the divalent Rab4/Rab5 effector Rabenosyn-5. We mapped the sites of interaction to the EH domain of EHD1, and the first two of five NPF motifs of Rabenosyn-5. Immunofluorescence microscopy studies revealed that EHD1 and Rabenosyn-5 partially colocalize to vesicular and tubular structures in vivo. To address the functional roles of EHD1 and Rabenosyn-5, we first demonstrated that RNA interference (RNAi) dramatically reduced the level of expression of each protein, either individually or in combination. Depletion of either EHD1 or Rabenosyn-5 delayed the recycling of transferrin and major histocompatibility complex class I to the plasma membrane. However, whereas depletion of EHD1 caused the accumulation of internalized cargo in a compact juxtanuclear compartment, Rabenosyn-5-RNAi caused its retention within a dispersed peripheral compartment. Simultaneous RNAi depletion of both proteins resulted in a similar phenotype to that observed with Rabenosyn-5-RNAi alone, suggesting that Rabenosyn-5 acts before EHD1 in the regulation of endocytic recycling. Our studies suggest that Rabenosyn-5 and EHD1 act sequentially in the transport of proteins from early endosomes to the endosomal recycling compartment and back to the plasma membrane.
INTRODUCTION
The ability of cells to internalize plasma membrane proteins is key to many essential physiological processes such as nutrient uptake, retrieval of exocytosed synaptic vesicle components, and the regulated expression of signaling receptors, transmembrane ligands, membrane transporters, and adhesion molecules (Mellman, 1996; Conner and Schmid, 2003). Plasma membrane proteins can be internalized by either clathrin-mediated or nonclathrin-mediated pathways, in both cases resulting in the delivery of the proteins into the endosomal system (Nichols and Lippincott-Schwartz, 2001; Johannes and Lamaze, 2002; Conner and Schmid, 2003). Just as critical is the ability to return a subset of the internalized proteins, in particular endocytic receptors, from endosomes to the plasma membrane, so that they can participate in additional rounds of endocytic uptake. Two distinct recycling pathways have been described. A fast recycling pathway takes proteins directly from early endosomes to the plasma membrane (Sheff et al., 1999; Hao and Maxfield, 2000; Sheff et al., 2002; van Dam et al., 2002), whereas a second, slow pathway involves passage through a pericentriolar endosomal compartment known as the endocytic recycling compartment (ERC) (reviewed by Gruenberg and Maxfield, 1995). The ERC has been described as a dense collection of tubular structures that radiate from the microtubule-organizing center (Hopkins and Trowbridge, 1983; Yamashiro et al., 1984).
Whereas the molecular machinery involved in endocytosis has been characterized in great detail (Mellman, 1996; Conner and Schmid, 2003; Gruenberg, 2003), the mechanisms responsible for recycling to the cell surface are less well understood. Rab GTPases such as Rab4 (van der Sluijs et al., 1992; Daro et al., 1996; Sheff et al., 1999) and Rab11 (Ullrich et al., 1996; Ren et al., 1998; Sheff et al., 1999; Zeng et al., 1999), as well as their corresponding effectors (Mammoto et al., 1999; Zeng et al., 1999; Nagelkerken et al., 2000; Prekeris, et al., 2000; Sonnichsen et al., 2000; Cormont et al., 2001, 2003; Hales et al., 2001; de Renzis et al., 2002; Lindsay et al., 2002; Wallace et al., 2002a,b; Deneka et al., 2003; Fouraux et al., 2004), have been implicated in recycling of internalized receptors. In addition, a genetic screen for Caenorhabditis elegans mutants defective in receptor-mediated uptake of yolk protein resulted in the identification of another component of the recycling machinery, the protein RME-1 (Grant et al., 2001). C. elegans RME-1 is homologous to the family of human proteins named EHD1, EHD2, EHD3, and EHD4 (Mintz et al., 1999; Pohl et al., 2000). EHD family members are characterized by an N-terminal P-loop domain with nucleotide binding motifs, a central coiled-coil region, and a C-terminal Eps15-homology (EH) domain (Mintz et al., 1999; Pohl et al., 2000) (Figure 1A). Interestingly, the inhibition of yolk protein endocytosis upon RNA interference (RNAi) of RME-1 in C. elegans seems to be secondary to impaired recycling of the yolk protein receptor (Grant et al., 2001). In mammalian cells, EHD1 has been shown to control ERC morphology (Lin et al., 2001) and to regulate the recycling of the transferrin receptor (Lin et al., 2001) and major histocompatibility complex class I molecules (MHC-I) (Caplan et al., 2002) to the plasma membrane, with little or no direct effect on internalization.
Figure 1.
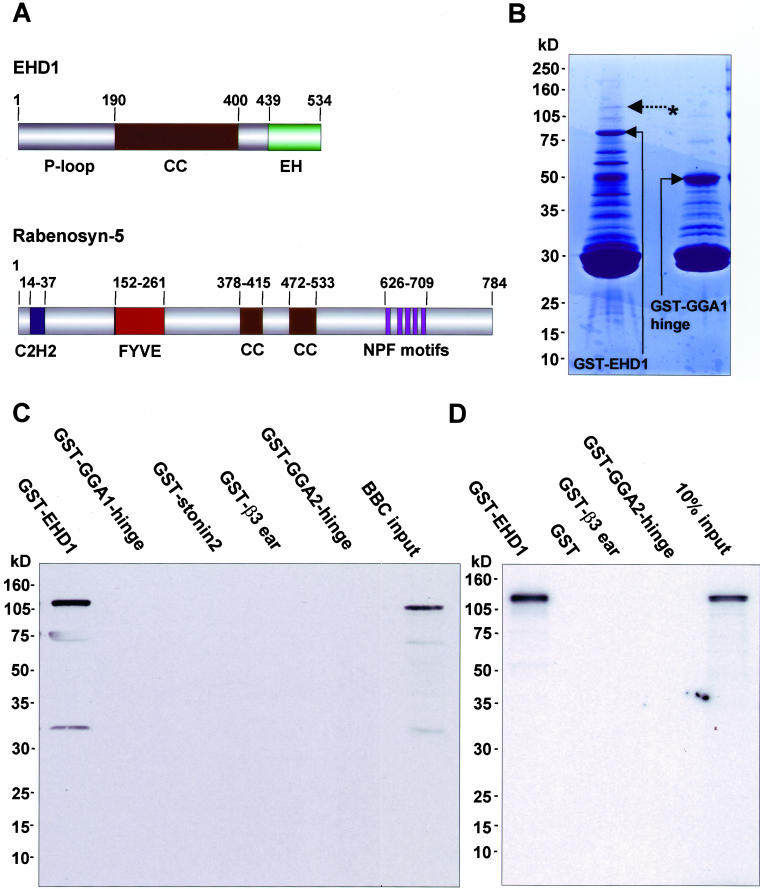
Identification of Rabenosyn-5 as a binding partner for EHD1. (A) Schematic representation of EHD1 and Rabenosyn-5 domain organization. EHD1 comprises an N-terminal P-loop, a central coiled coil (CC) and a C-terminal EH domain. Rabenosyn-5 comprises an N-terminal C2H2 ring, a Fab1p, YOTB, Vac1p, EEA1 (FYVE) domain, two central CC, and five C-terminal NPF motifs. (B) Bacterially expressed, recombinant GST-EHD1 and a GST-fusion protein control (GST-GGA1) were purified and subjected to serial rounds of incubation with bovine brain cytosol. Bound proteins were eluted, separated by SDS-PAGE and visualized by Coomassie Blue staining. The asterisk and dashed arrow indicate a band at ∼120 kDa that was subsequently identified by mass spectrometry as Rabenosyn-5. (C and D) Confirmation of binding between EHD1 and Rabenosyn-5. GST-EHD1 or GST control proteins were incubated with either bovine brain cytosol (C) or 35S-labeled Rabenosyn-5 transcription/translation products (D). Interacting proteins were eluted and subjected to 4–20% gradient SDS-PAGE and analyzed by immunoblotting with antibody to Rabenosyn-5 (C) or by autoradiography (D).
To date, the relationship between the Rab-regulated steps and EHD1 has not been established. In this study, we identify the divalent Rab4/Rab5 effector Rabenosyn-5 (Nielsen et al., 2000; de Renzis et al., 2002) as an interaction partner for EHD1 and present evidence that this interaction regulates recycling by mediating transport of receptors from early endosomes to the ERC.
MATERIALS AND METHODS
Recombinant DNA Constructs
Full-length EHD1 and Rabenosyn-5 constructs, as well as Rabenosyn-5 truncations in pGEM used for in vitro binding assays were described previously (Nielsen et al., 2000; Caplan et al., 2002; de Renzis et al., 2002). Mutations in full-length Rabenosyn-5 to change individual NPF motifs to APA were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), as per the manufacturer's instructions. GST-EHD1 was obtained by subcloning EHD1 into the pGEX 4T-1 vector, and GST-β3ear, GST-GGA1-hinge, and GST-GGA2-hinge have been described previously (Mattera et al., 2003). Two-hybrid constructs for full-length Rabenosyn-5, full-length EHD1, and the EH domain of EHD1 were generated by subcloning into the pGBKT7 and pGADT7 two-hybrid vectors. For both full-length EHD1 and the EH domain of EHD1, tryptophan 485 was mutated to alanine by using the QuikChange site-directed mutagenesis kit. EHD2 and EHD3 were obtained from HeLa mRNA by reverse transcription-polymerase chain reaction and subcloned into pGBKT7. The sequences of all constructs were verified by DNA sequencing.
Antibodies
Purified rabbit polyclonal antiserum against Rabenosyn-5 has been described previously (Nielsen et al., 2000). Rabbit antibodies were prepared against an N-terminal peptide (MFSWVSKDARRKKEP) of EHD1 (AnaSpec, San Jose, CA). Monoclonal antibodies to EEA1 and Rab5 were purchased from BD Transduction Laboratories (Lexington, KY), monoclonal antibody to α-tubulin was purchased from Sigma-Aldrich (St. Louis, MO), W6/32 antibody to MHC-I (peptide-bound) was obtained from the American Type Culture Collection (Manassas, VA), and the 9E10 antibody to the Myc epitope was from Covance (Princeton, NJ). Cy-3-conjugated anti-mouse and anti-rabbit IgG, as well as Alexa-488–conjugated antibody to mouse and rabbit IgG were obtained from Molecular Probes (Eugene, OR).
Identification of Rabenosyn-5 as an EHD1 Binding Partner
GST-EHD1 (50 μg protein) was bound to 30 μl of glutathione-Sepharose 4B slurry in a volume of 1 ml of phosphate-buffered saline (PBS) with 1% (wt/vol) Triton X-100 and 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (GST-binding-buffer), washed four times with GST-binding-buffer, and incubated in the presence of either 2 ml of bovine brain cytosol or 2 ml of murine brain cytosol for 3 h at 4°C. Beads were sedimented, and new brain cytosol was added. This step was repeated three times. Precipitates were washed four times with GST-binding-buffer, twice with the same buffer lacking Triton X-100, eluted, and separated by 4–20% gradient SDS-PAGE. The ∼120-kDa band visualized by Coomassie Blue specifically after pull-down with GST-EHD1, as well as the corresponding region of the gel for GST-control fusion proteins were excised, destained, and in-gel digested using modified porcine trypsin (Promega, Madison, WI). The resulting peptides were extracted and subjected to analysis by liquid chromatographytandem mass spectrometry (LC-MS/MS). Aliquots of the sample were injected onto a capillary gradient MAGIC LC system (Michrom Bioresources, Auburn, CA) operated at 400 nl/min by using a constant pressure splitter and separated using a 10 min linear gradient. A 75-mm i.d. fused silica capillary PicoFritÆ (New Objective, Woburn, MA) column packed with a Vydac C18 resin (5-mm particle, ∼300 pore) of ∼5 cm in length was used for the liquid chromatography. The LC effluent was electrosprayed directly into the sampling orifice of an LCQ DECA (Thermo Finnigan, San Jose, CA) by using an adaptation of the microscale electrospray interface method (Davis et al., 1995). The LCQ DECA was operated in a mode that automatically generated MS/MS spectra of the three most intense peaks present in the full MS scan of the ion trap that exceeded a preset threshold. Peptide partial internal sequence data produced by the LC-MS/MS experiments were subjected to analysis using both MASCOT (Perkins et al., 1999) and SEQUEST (Eng et al., 1994) programs, using the mammalian portion of the NCBI nonredundant data base for the database searches. Sequences of bovine peptides identified that specifically bound to GST-EHD1 were SHLSDFKK, TLQENLR, TLQENLRQLQDEYDQQQTEK, and QLQDEYDQQQTEK.
Other Biochemical Assays
Confirmation of Rabenosyn-5 binding to EHD1 was achieved by pull-down assays performed as described above, either using bovine brain cytosol as input, or 35S-labeled in vitro transcription/translation products (Promega). For the former, proteins separated by SDS-PAGE gels were transferred to nitrocellulose, and immunoblotting was performed using anti-Rabenosyn-5 antiserum. Densitometric analysis was performed using NIH Image (version 1.62).
Immunofluorescence, Fluorescence Recycling Assays, and Quantitation
HeLa cells were grown on cover glasses, transfected using FuGENE-6 (Roche Molecular Biochemicals, Indianapolis, IN), and fixed with 4% (vol/vol) paraformaldehyde in PBS. Fixed cells were then incubated with primary antibodies containing 0.1% (wt/vol) saponin and 0.1% (wt/vol) bovine serum albumin (BSA) for 1 h at room temperature. After washes in PBS, the cells were incubated with the appropriate fluorochrome-conjugated secondary antibody mixture containing 0.1% (wt/vol) saponin and 0.1% (wt/vol) BSA for 30 min at room temperature. To specifically detect proteins on the plasma membrane, antibody binding (primary and secondary) was done in the absence of saponin. Images were acquired on an LSM 410 or LSM 5 Pascal confocal microscope (Carl Zeiss, Thornwood, NY) by using a 63× 1.4 numerical aperture objective with appropriate filters. Transferrin recycling was measured by first starving cells in DMEM lacking serum (but containing 0.5% BSA) for 23 min and then applying a 15-min pulse with Transferrin-Alexa-Fluor 568 (Tf-568; Molecular Probes). The labeled transferrin was then chased for the indicated times by incubating the cells in full medium supplemented with 2 mg/ml unlabeled transferrin at 37°C, and cells were fixed and processed as described above. For quantitative analysis, 30–50 cells were randomly chosen and scanned with an open pinhole, allowing fluorescence to be collected from the entire depth of the cell. Differential interference contrast imaging was used to determine the boundaries of each cell, and LSM software was used to obtain mean fluorescence values. MHC-I recycling was analyzed by performing a 20-min pulse with W6/32 anti-MHC-I, followed by a 20-s acid strip (0.5% acetic acid, 500 mM NaCl) to remove residual surface-bound (noninternalized) antibody (Le Gall et al., 2000; Means et al., 2002; Naslavsky et al., 2003). Cells were returned to 37°C for 30-min chase in full media, fixed and processed as described above. In this case, a secondary Cy3-conjugated anti-mouse antibody was used to label the remaining (nonrecycled) intracellular pool of MHC-I. Quantitation of cell surface transferrin receptor levels was achieved by flow cytometry (BD Transduction Laboratories) by using 10,000 HeLa cells that were released into suspension by a 30-s incubation with either trypsin or EDTA (100 mM) at 37°C. Cells were then fixed with 4% paraformaldehyde, stained first with anti-transferrin receptor antibodies (Zymed Laboratories, South San Francisco, CA), and then with an anti-mouse 488-Alexa secondary antibody (Molecular Probes). Quantitation was also achieved by immunofluorescence microscopy, as described above. In the latter case, the statistical significance of the differences observed for the various treatments was calculated using the one-way analysis of variance for independent or correlated samples. Quantitation of colocalization was done using LSM software, setting thresholds levels by extracting the mean background signal.
Gene Knockdown by RNAi
RNAi duplexes (synthesized by Dharmacon, Lafayette, CO) were transfected using Oligofectamine (Invitrogen, Carlsbad, CA) essentially by the method of Elbashir et al. (2001). Calibration experiments demonstrated that either 48 or 72 h of treatment was sufficient to lower the expression of EHD1 and Rabenosyn-5 to <10% of mock-treated cells. The sequence used for Rabenosyn-5 (base pairs 1559–1579) was cagctgcagatgttgcgtg, and the sequence used for EHD1 (base pairs 943–963) was gaaagagatgcccaatgtc.
Other Procedures
Yeast two-hybrid analysis using the strain AH109 (BD Biosciences Clontech, Palo Alto, CA) was performed as described previously (Caplan et al., 2001).
RESULTS
Identification of Rabenosyn-5 as a Binding Partner for EHD1
EHD1 has been shown to play a role in the recycling of internalized proteins from the pericentriolar ERC to the plasma membrane (Lin et al., 2001; Caplan et al., 2002; Picciano et al., 2003). However, little is known about its mechanism of action and, in particular, how it might connect with Rab proteins involved in the recycling pathway. Accordingly, we sought to identify components of the recycling machinery that interact with EHD1. To this end, recombinant, full-length EHD1 was expressed as a GST-fusion protein, and used as bait in pull-down assays with either bovine or murine brain cytosol. Bound and eluted proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining (Figure 1B). Pull-down experiments using bovine brain cytosol resulted in the isolation of a ∼120-kDa species (Figure 1B, asterisk and dashed arrow) that was specific for GST-EHD1, but not GST-GGA1-hinge (Figure 1B) or other GST-fusion proteins (our unpublished data). Other proteins on the gel either bound to all the GST-fusion proteins tested or could not be visualized because of interference by degradation products of the GST-fusion proteins (i.e., those of ∼80 kDa or less). Similar results were obtained using murine brain cytosol as a protein source (our unpublished data). Mass spectrometry analysis (LC-MS/MS) of tryptic peptides derived from the specific ∼120-kDa species from both bovine and murine sources resulted in the identification of four peptides matching the sequence of orthologues of the divalent Rab4 and Rab5 effector Rabenosyn-5 (Nielsen et al., 2000; de Renzis et al., 2002) (Figure 1A).
To confirm the association of EHD1 with Rabenosyn-5, we performed immunoblot analysis of proteins isolated in GST pull-down experiments from bovine brain cytosol (Figure 1C). We observed that an antiserum to Rabenosyn-5 recognized a band of ∼120 kDa from bovine brain cytosol (Figure 1C, BBC input) as well as from a pull-down with GST-EHD1 but not with other GST-fusion proteins (Figure 1C). In addition, radiolabeled, in vitro transcribed-translated Rabenosyn-5 was generated and assayed for binding to GST-EHD1, or to control proteins. As shown in Figure 1D, only GST-EHD1 bound to Rabenosyn-5. These experiments thus identified Rabenosyn-5 as a specific binding partner for EHD1.
The EH Domain of EHD1 Interacts with Rabenosyn-5 NPF Motifs
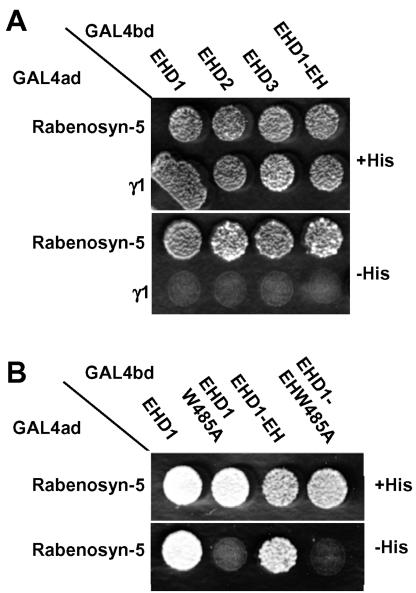
A prominent feature of EHD1 is its C-terminal EH-domain (Figure 1A), which for other proteins has been demonstrated to bind the asparagine-proline-phenylalanine (NPF) motif (de Beer et al., 1998, 2000). We therefore hypothesized that the EH domain of EHD1 could interact with either of five NPF motifs present in Rabenosyn-5 (Figure 1A). To test this hypothesis, we performed yeast two-hybrid analyses. As shown in Figure 2A (bottom, –His), both full-length EHD1 and its isolated EH domain were capable of interacting with Rabenosyn-5. Moreover, other EHD family members (i.e., EHD2 and EHD3), which also contain EH domains, similarly interacted with Rabenosyn-5 (Figure 2A). A conserved tryptophan residue within the EH domain of other EH-containing proteins has been shown to be critical for interactions with NPF motifs (de Beer et al., 2000). Mutation of the homologous tryptophan 485 of EHD1 in the context of the full-length protein or its isolated EH domain abrogated interactions with Rabenosyn-5 (Figure 2B, bottom, –His). These results hinted at EH-domain–NPF motif interactions as the probable basis for the binding of EHD1 to Rabenosyn-5.
Figure 2.
The EHD1 EH domain is responsible for the interaction with Rabenosyn-5. The Saccharomyces cerevisiae yeast strain AH109 was cotransformed with the following GAL4 transcription activation (GAL4ad) fusion constructs: GAL4ad-Rabenosyn-5 and GAL4ad-γ1 (negative control), together with the GAL4 DNA-binding domain (GAL4bd) fusion products GAL4bd-EHD1, GAL4bd-EHD2, GAL4bd-EHD3, GAL4bd-EHD1-EH-domain, GAL4bd-EHD1-W485A, and GAL4bd-EHD1-EH-domain W485A. Cotransformants were assayed for growth on nonselective (+His) and selective (–His) media.
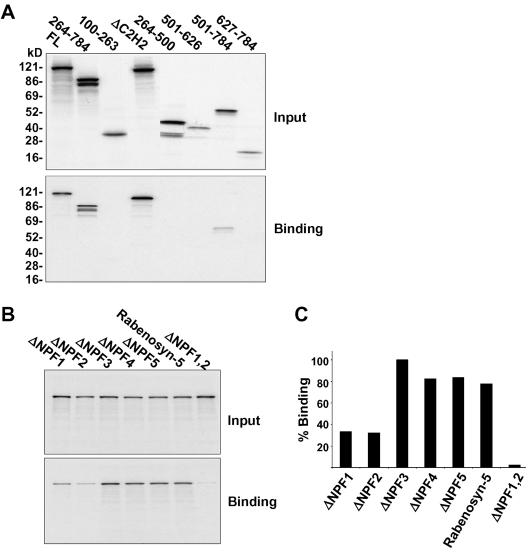
To delineate the region of Rabenosyn-5 that is required for binding to EHD1, a series of deletions and truncations of Rabenosyn-5 produced by in vitro transcription-translation were used for binding assays. As shown in Figure 3A, full-length Rabenosyn-5 (FL) or mutants deleted in the N-terminal region (i.e., 264–784, ΔC2H2) bound efficiently to GST-EHD1. In contrast, proteins truncated in the C-terminal region displayed impaired binding to EHD1 (Figure 3A). Loss of the five NPF motifs (as in the 100–263, 264–500, 501–626 constructs) resulted in complete abrogation of binding, whereas inclusion of all five NPF motifs in a short deletion construct (501–784) displayed significant, albeit reduced, binding. The short 627–784 Rabenosyn-5 deletion, which contains the last four NPF motifs but lacks the first NPF, did not bind to EHD1 (Figure 3A), indicating either that this truncated protein is improperly folded or that the first NPF motif is critical for binding. Overall, these results highlight the importance of the Rabenosyn-5 C-terminal region for binding to EHD1 and further indicate the likelihood of EH-domain–NPF motif interactions.
Figure 3.
The first two of five Rabenosyn-5 NPF motifs are critical for binding to EHD1. (A) Bacterially expressed, recombinant GST-EHD1 was incubated with full-length wild-type Rabenosyn-5 (FL), or various 35S-labeled Rabenosyn-5 transcription/translation products with deletions and/or truncations, and the bound proteins were analyzed by SDS-PAGE and autoradiography. Five percent of the total input is shown in the top panel, and the bound Rabenosyn-5 products are shown in the bottom panel. (B) Individual NPF motifs were mutated from NPF to APA (ΔNPF1–5), or the first two NPF motifs were mutated to APA (ΔNPF1,2). 35S-labeled Rabenosyn-5 transcription/translation products of wild-type Rabenosyn-5 or NPF mutants were pulled down with GST-EHD1. Five percent of input is shown in the top panel, whereas bound Rabenosyn-5 products are shown in the bottom panel. (C) Densitometric analysis of the binding experiment depicted in B is shown in histogram format.
To address this directly, we generated a series of NPF mutants. Each of the individual NPF motifs was mutated to APA, leaving the proline residue intact to avoid structural changes (Martina et al., 2001). In addition, a double-mutant with substitution of the first two NPF motifs was also generated. As shown in Figure 3B, and quantified in Figure 3C, >60% of the binding to EHD1 was lost when either the first or second NPF motifs were mutated. Mutations in both of the first two NPF motifs resulted in very weak binding, at <10% of the wild-type full-length protein. These data are consistent with the EHD1 EH domain interacting with Rabenosyn-5 primarily via its first two NPF motifs.
Partial Colocalization of EHD1 and Rabenosyn-5
We and others have previously shown that EHD1 and other EHD family proteins localize to membrane-bound vesicular and tubular structures (Lin et al., 2001; Rotem-Yehudar et al., 2001; Caplan et al., 2002; Galperin et al., 2002; Shao et al., 2002). Rabenosyn-5 has also been observed in association with vesicular structures (Nielsen et al., 2000). To determine whether EHD1 and Rabenosyn-5 colocalize, Myc-EHD1 was transfected into HeLa cells (Figure 4, A–C). This transfection approach was necessary because antibodies to EHD1 gave very weak staining for the endogenous protein. EHD1 was found to colocalize with endogenous Rabenosyn-5 on cytoplasmic vesicles and tubules (Figure 4, A and B, yellow arrows). The overlap, however, was partial because EHD1 tended to be on tubular structures and Rabenosyn-5 on vesicular structures. Quantitation by confocal microscopy indicates that 5–10% of Myc-EHD1 or GFP-EHD1 colocalizes with Rabenosyn-5, whereas ∼3–5% of Rabenosyn-5 is found on common structures with GFP-EHD1. Together with the binding of EHD1 to Rabenosyn-5, these data indicate that EHD1 and Rabenosyn-5 might functionally interact in endocytic/recycling pathways.
Figure 4.
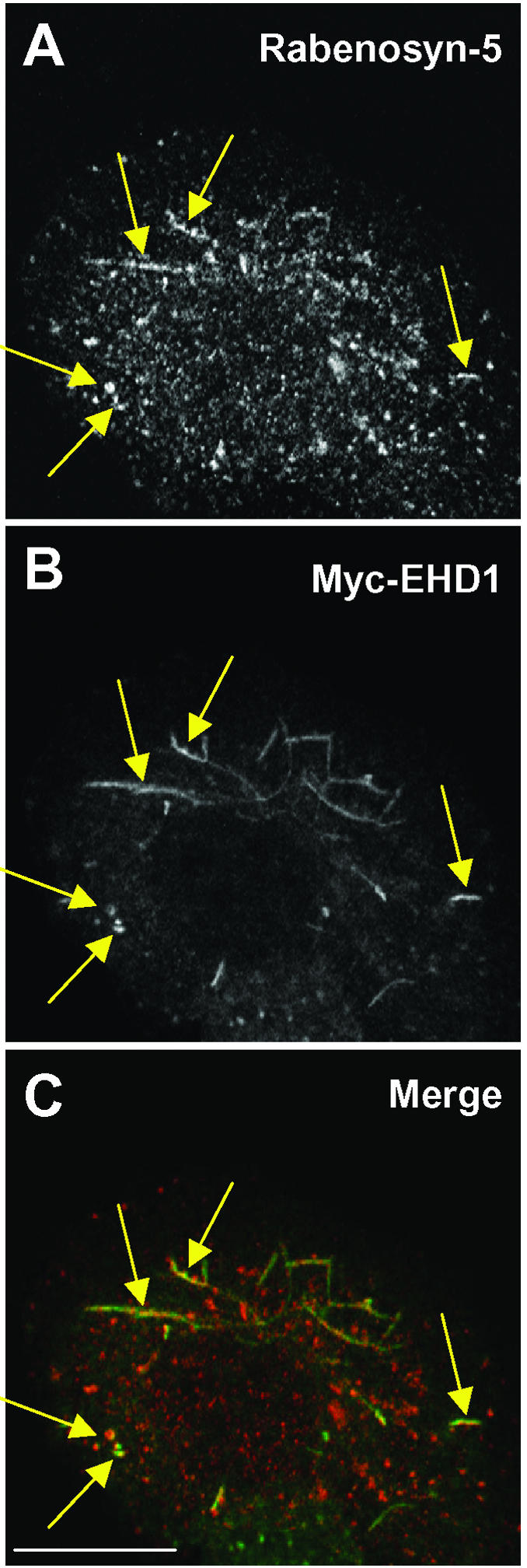
Partial colocalization of Rabenosyn-5 and EHD1. HeLa cells were transfected with Myc-EHD1, and 24 h later the cells were fixed, permeabilized, and incubated with rabbit polyclonal antibodies to Rabenosyn-5 (A) and mouse monoclonal antibody to the Myc epitope (B). Cells were then washed and incubated with Alexa-488–conjugated donkey anti-mouse antibody (green channel) and Cy3-conjugated donkey anti-rabbit IgG (red channel). Yellow arrows mark the colocalization of Rabenosyn-5 and EHD1 to tubular and vesicular structures. Levels of colocalization were assessed using LSM software. Bar, 10 μm.
Impairment of Protein Recycling to the Plasma Membrane in the Absence of Either Rabenosyn-5 or EHD1
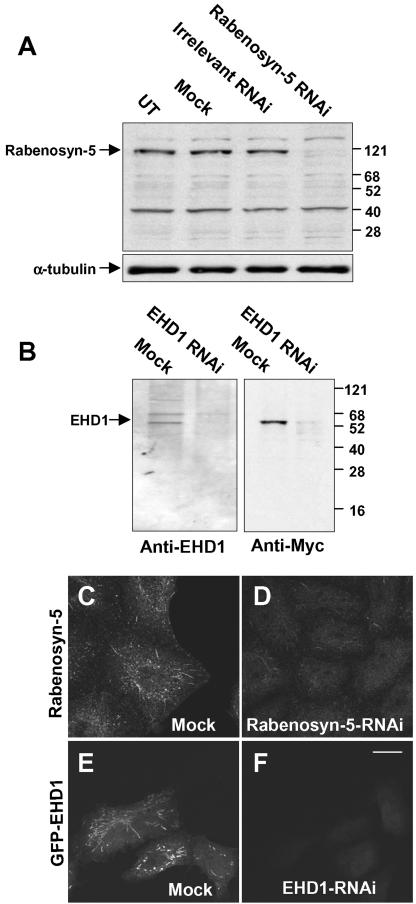
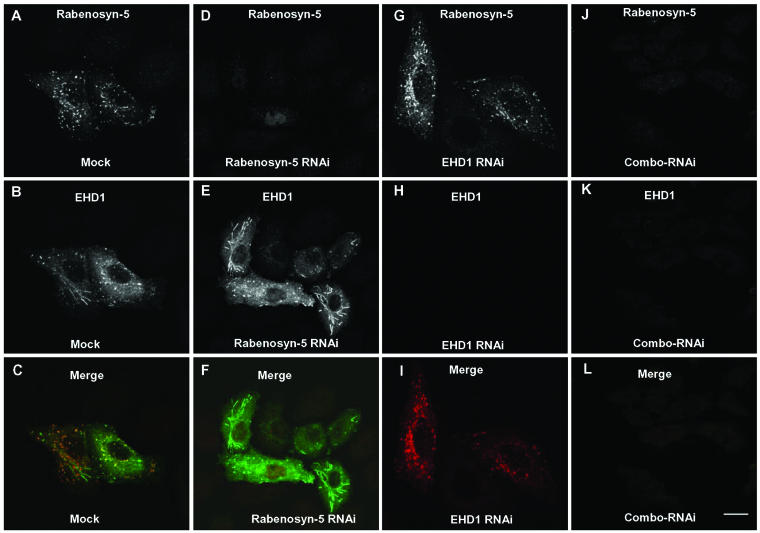
Previous studies using overexpression and dominant-negative approaches have implicated both EHD1 (Lin et al., 2001; Caplan et al., 2002) and Rabenosyn-5 (de Renzis et al., 2002) in endocytic recycling events. We hypothesized that the absence of expression of either of these proteins should similarly result in impaired recycling to the plasma membrane. To this end, we undertook an RNAi approach to block expression of EHD1 and Rabenosyn-5 in HeLa cells. The effects of RNAi were assessed by immunoblot analysis and immunofluorescence microscopy (Figure 5). Immunoblotting of HeLa cell lysates with antibodies to Rabenosyn-5 revealed a specific ∼120-kDa band in untreated cells (UT), cells treated with mock-RNAi (Mock), and cells treated with irrelevant RNAi (Figure 5A, top). This band was not detected in cells treated with RNAi specific for Rabenosyn-5 (Figure 5A, top). As a control, nitrocellulose blots were stripped and reprobed with antibodies to α-tubulin, demonstrating similar levels of protein in all lysates (Figure 5A, bottom). Immunofluorescent staining of mock-treated cells with antibody to Rabenosyn-5 revealed a pattern of endogenous cytoplasmic vesicles and tubules (Figure 5C), as described previously (Nielsen et al., 2000; de Renzis et al., 2002). In agreement with the immunoblot analysis, treatment with Rabenosyn-5-RNAi resulted in a dramatic reduction in the intensity of immunofluorescence staining (Figure 5D). Immunoblotting with an antibody to endogenous EHD1 in mock-treated cell lysates revealed a band of ∼55 kDa and a second band of ∼68 kDa (Figure 5B, left), as reported previously (Rotem-Yehudar et al., 2001; Caplan et al., 2002). RNAi for EHD1 almost completely eliminated the ∼55-kDa band, indicating that this corresponds to endogenous EHD1. Mock- and EHD1-RNAi–treated cells were also transfected with Myc-EHD1 and subjected to the same immunoblotting analysis by using anti-Myc antibodies (Figure 5B, right). A single band at ∼55 kDa was visualized in the mock-treated lysates but not in the EHD1-RNAi lysate, thus confirming the efficacy of the RNAi approach. To analyze EHD1-RNAi efficacy by immunofluorescence microscopy, it was necessary to transfect the cells with a tagged construct (i.e., GFP-EHD1; Figure 5, E and F) because our antibodies to EHD1 do not work well for immunofluorescent staining of the endogenous protein. As shown in Figure 5, E and F, mock-treated cells displayed typical EHD1 vesicles and tubules, whereas EHD1-RNAi cells showed no staining. These results indicate that the expression of both Rabenosyn-5 and EHD1 can be specifically abolished with RNAi treatment.
Figure 5.
RNAi reduces expression of Rabenosyn-5 and EHD1. Plates of cells transfected with either 10 μM Rabenosyn-5-RNAi (A) 10 μM EHD1-RNAi (B) or mock-RNAi and controls (A and B) were harvested after 72 h, and lysed in detergent. Detergent-solubilized proteins were separated by SDS-PAGE, transferred to nitrocellulose, and subjected to immunoblotting with antibodies to Rabenosyn-5 (A, top), α-tubulin (A, bottom), EHD1 (B, left) or the Myc epitope (B, right). For analysis by immunofluorescence microscopy, cells were mock-treated (C and E) or treated with either 10 μM Rabenosyn-5-RNAi (D) or 10 μM EHD1-RNAi (F). Forty-eight hours later, GFP-EHD1 was transfected to EHD1-RNAi–treated cells. After a total of 72 h, cells were fixed in 4% paraformaldehyde, permeabilized, incubated with rabbit polyclonal antibodies to Rabenosyn-5, and then incubated with Cy3-conjugated donkey anti-rabbit IgG antibodies (C and D), or mounted directly on cover glasses (E and F). Bar, 10 μm.
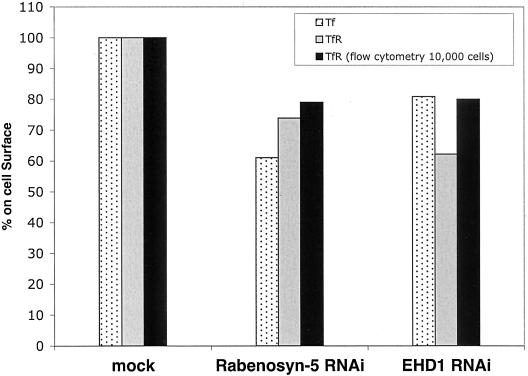
It has been demonstrated that loss of RME-1/EHD1 expression in C. elegans inhibits yolk protein endocytosis, primarily due to retention of the yolk protein receptor in an expanded endocytic compartment, and it has been proposed that delayed recycling decreases yolk receptor expression on the plasma membrane (Grant et al., 2001). Because overexpression of dominant-negative mammalian EHD1 proteins interferes with the recycling of transferrin receptor and MHC-I to the plasma membrane (Lin et al., 2001; Caplan et al., 2002), we hypothesized that RNAi-based knock-down of EHD1 and/or Rabenosyn-5 in human HeLa cells might reduce the levels of transferrin receptor localized to the plasma membrane. As shown in Figure 6, Rabenosyn-5-RNAi treatment led to a ∼27% (p < 0.01) reduction of transferrin receptor on the plasma membrane at steady state compared with mock-treated cells as calculated by quantitation of fluorescence on the cell surface. EHD1-RNAi treatment led to a 38% (p < 0.01) decrease in plasma membrane levels of transferrin receptor compared with mock-treated cells. In agreement with these results, experiments performed using Transferrin-Alexa-Fluor 568 (Tf-568) as a ligand for the cell-surface expressed transferrin receptor also indicated that there are decreased levels of transferrin receptor localized to the plasma membrane. About a 40% (p < 0.01) and 20% (p < 0.01) decrease in bound ligand was observed after Rabenosyn-5-RNAi and EHD1-RNAi treatments, respectively, compared with mock-treated cells. No statistically significant difference in plasma membrane levels of transferrin receptor was observed when Rabenosyn-5-RNAi and EHD1-RNAi treatments were compared directly. To verify these findings using a larger sample population, we performed flow cytometry analysis, comparing the levels of transferrin receptor expressed on 10,000 cells from each treatment (Figure 6, black columns). By this assay, the level of transferrin receptor on the plasma membrane at steady state in mock-treated cells was 22% higher than in Rabenosyn-5- and EHD1-RNAi–treated cells. These data are consistent with both Rabenosyn-5 and EHD1 regulating recycling of receptors to the plasma membrane.
Figure 6.
Decreased plasma membrane levels of transferrin receptor in the absence of Rabenosyn-5 or EHD1. HeLa cells on cover glasses were mock-treated, treated with Rabenosyn-5-RNAi, or with EHD1-RNAi. After 48 h, the levels of transferrin receptor and/or bound transferrin on the plasma membrane were determined either by flow cytometry analysis or by confocal microscopy. For flow cytometry analysis, adherent HeLa cells were starved in serum free media for 30 min, released into suspension by a 30-s incubation with trypsin or 100 mM EDTA, and fixed with 4% paraformaldehyde. Suspended cells were incubated with mouse anti-transferrin receptor antibodies and goat anti-mouse Alexa 488 antibody. Data from 10,000 cells (for each treatment) were analyzed by flow cytometry (BD Transduction Laboratories). The bar graph depicts the percentage of mean fluorescence counts for RNAi-treated cells compared with mock-treated cells in a representative experiment. For confocal microscopic analysis, HeLa cells were incubated on ice with Transferrin-Alexa-Fluor (Tf-568) extensively rinsed and fixed in 4% paraformaldehyde, or fixed and then incubated with mouse anti-transferrin antibodies followed by a secondary goat anti-mouse Alexa 488 antibody. Thirty to 50 cells were randomly chosen, scanned by confocal microscopy, and their mean fluorescence intensity calculated by LSM software. Differential interference contrast scanning was used to detect the precise area of the cells, and an open pinhole allowed the collection of fluorescence throughout the depth of the cells. Mock values were normalized to 100% to allow comparison with Rabenosyn-5-RNAi– and EHD1-RNAi–treated cells. One-way analysis of variance for independent or correlated samples was used to test statistical significance of the data. The p values for the Tukey HSD test were as follows: Mock versus Rabenosyn-5-RNAi, p < 0.01; Mock versus EHD1-RNAi, p < 0.01; and Rabenosyn-5-RNAi versus EHD1-RNAi, p is insignificant. The graph portrayed is a representative one from four individual experiments.
Rabenosyn-5 and EHD1 Sequentially Regulate Recycling to the Plasma Membrane
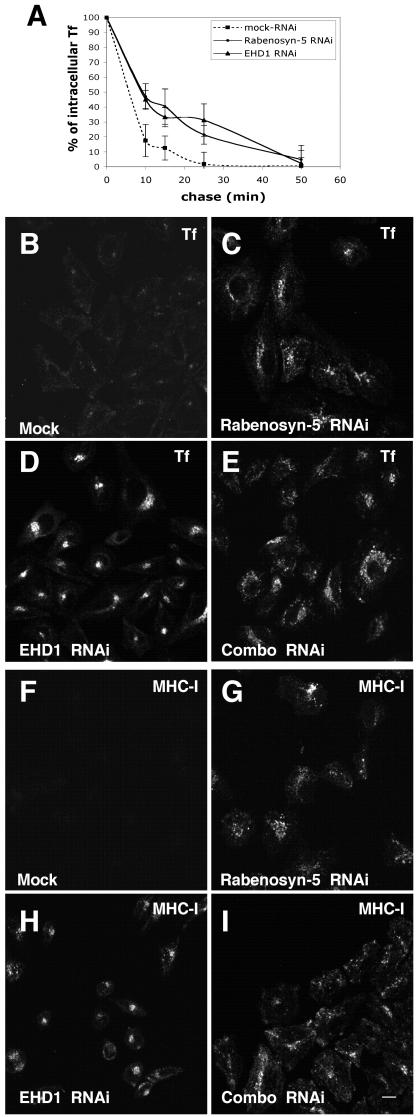
Based on the observations shown in Figure 6, as well as previous evidence for a role of Rabenosyn-5 (de Renzis et al., 2002) and EHD1 (Grant et al., 2001; Lin et al., 2001; Caplan et al., 2002) in recycling, we hypothesized that Rabenosyn-5 and EHD1 regulate recycling events en route to the plasma membrane. To address this hypothesis, we obtained quantitative data comparing the kinetics of recycling in mock-, Rabenosyn-5-, and EHD1-RNAi–treated HeLa cells (Figure 7A). In these experiments, recycling was measured by pulsing the cells with fluorochrome-labeled transferrin (Tf-568), and analyzing the loss of intracellular Tf-568 over time (during a “chase” in the presence of excess nonlabeled transferrin). Because the steady-state levels of transferrin receptor on the plasma membrane are ∼20% lower in cells treated with either Rabenosyn-5- or EHD1-RNAi (Figure 6), it was necessary to normalize the initial levels of internalized (“pulsed”) transferrin for each treatment, so that the rates of recycling could be compared. As demonstrated in Figure 7A, after 10, 15, and 25 min of chase, there was a significant decrease in the rate of transferrin recycling in HeLa cells treated with Rabenosyn-5- or EHD1-RNAi (compared with mock-treated cells). Thus, the amount of recycled transferrin is impacted in two ways by the loss of either Rabenosyn-5 or EHD1, as a result of 1) less transferrin receptor on the plasma membrane at steady state (and subsequently, less binding and internalization of transferrin), and 2) a slower rate of recycling of transferrin and its receptor to the plasma membrane.
Figure 7.
Impaired transferrin and MHC-I recycling in the absence of Rabenosyn-5 and EHD1. Cells treated with mock-RNAi (A, B, and F), RNAi specific for Rabenosyn-5 (A, C, and G), RNAi specific for EHD1 (A, D, and H), or combinations with RNAi for both proteins (E and I) were subjected to pulse-chase analysis with either Transferrin-Alexa-Fluor (Tf-568) (A and B–E) or with antibodies to MHC-I (F–I). In A–I, cells were pulsed for 15 min and chased in full media (with excess unlabeled transferrin in A–I) for the indicated times. Cells were then fixed and analyzed by confocal microscopy (A–E), or first permeabilized and incubated with Alexa-488–conjugated donkey anti-mouse antibody (F–I), before analysis. In A, differential interference contrast imaging was used to identify the cell boundaries, and LSM software was used to quantitate mean fluorescence values for the cells; 100% represents the initial level of mean fluorescence for each treatment before the chase. Bar, 10 μm.
To visualize cargo-recycling, we analyzed the distribution of internalized Tf-568 (Figure 7, B–E) or antibody to MHC-I (Figure 7, F–I) in pulse-chase experiments by confocal microscopy comparing cells treated with mock-, Rabenosyn-5-, or EHD1-RNAi. After 15-min internalization of Tf-568 or anti-MHC-I, followed by a 15-min chase, cells treated with mock-RNAi emptied of Tf-568 (Figure 7B) and anti-MHC-I (Figure 7F), as a result of recycling to the plasma membrane. Cells treated with Rabenosyn-5-RNAi displayed a decreased rate of recycling of Tf-568 (Figure 7C) and MHC-I (Figure 7G), with the cargo being retained longer in a dispersed, peripheral compartment. Treatment with EHD1-RNAi also slowed recycling of Tf-568 and anti-MHC-I, but these cargo proteins accumulated in a compact, pericentriolar compartment (Figure 7, D and H) reminiscent of the compartment described upon overexpression of a dominant-negative EHD1 mutant (Lin et al., 2001). These data collectively indicate that Rabenosyn-5 and EHD1 regulate recycling at distinct but partially overlapping stages of recycling.
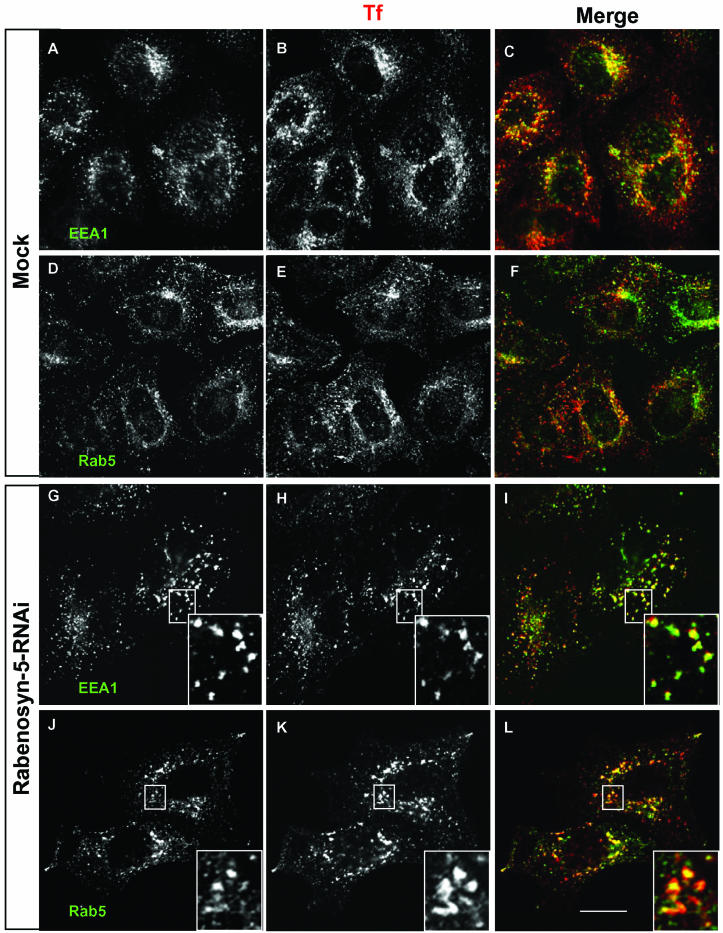
Previous studies provide evidence that loss of EHD1 function causes a delay in recycling and accumulation of transferrin (Lin et al., 2001), MHC-I (Caplan et al., 2002), and cholesterol (Hao et al., 2002) at the endocytic recycling compartment. Overexpression of EHD1, on the other hand, does not significantly alter the morphology of transferrin-containing compartments (Supplemental Figure 1). To characterize the identity of the compartment within which transferrin accumulates upon Rabenosyn-5-RNAi treatment, mock-treated cells or cells treated with Rabenosyn-5-RNAi were subjected to pulse-chase recycling assays with Tf-568, and then fixed and immunostained with antibodies for the endocytic marker proteins EEA1 and Rab5 (Figure 8). To view the colocalization of transferrin with EEA1 and Rab5 in the Mock-treated cells, the cells were pulsed without a chase, preventing the recycling and exit of Tf-568 from the cells (Figure 8, A–F). On the other hand, cells treated with RNAi for Rabenosyn-5 were chased for 10 min, to allow accumulation of the Tf-568 within the intracellular compartments (Figure 8, G–L). As expected, in mock-treated cells internalized transferrin was distributed primarily in small, dispersed, punctate compartments (Figure 8, B and E), showing substantial colocalization with EEA1 and Rab5 (Figure 8, A–F). In cells treated with Rabenosyn-5-RNAi, internalized transferrin accumulated in an intracellular vesicular compartment (Figure 8, H and K). These transferrin-containing vesicles seemed somewhat larger than those in the mock-treated cells, but they also substantially colocalized with both EEA1 and Rab5 (Figure 8, G–L), suggesting that Rabenosyn-5 affects transport through early endosomes. These data raise the question as to where endogenous Rabenosyn-5 and EHD1 act in the endocytic pathway with respect to one another.
Figure 8.
Transferrin accumulates in an early endocytic compartment containing Rab5 and EEA1 in cells treated with Rabenosyn-5-RNAi. HeLa cells on cover glasses were treated with Mock-RNAi or Rabenosyn-5-RNAi for 48 h and then pulsed with Tf-568 for 15 min. Mock-treated cells (A–F) were fixed in paraformaldehyde, whereas cells treated with Rabenosyn-5-RNAi were subjected to a 10-min chase in full media (containing excess unlabeled transferrin) before fixation. Fixed permeabilized cells were incubated first with mouse antibodies against EEA1 (A–C, G–I) or Rab5 (D–F, J–L), and then with secondary Alexa-488–conjugated donkey anti-mouse antibody (A–L). Images were obtained by confocal microscopy. Insets depict the colocalization of Tf-568 with EEA1 (G–I) and Rab5 (J–L) on vesicles in Rabenosyn-5-RNAi–treated cells. Bar, 10 μm.
We hypothesized that these two proteins may act sequentially to control recycling to the plasma membrane, in which case the simultaneous absence of Rabenosyn-5 and EHD1 should result in the accumulation of recycling cargo within the first of the two affected compartments. To test this hypothesis, we first demonstrated that simultaneous treatment with RNAi for both Rabenosyn-5 and EHD1 reduces expression of both proteins in single cells (Figure 9). To this end, mock-treated HeLa cells were cotransfected with Rabenosyn-5 and GFP-EHD1 and displayed good coexpression (>95%) and partial colocalization (Figure 9, A–C). When cotransfected cells were treated only with Rabenosyn-5-RNAi, expression of Rabenosyn-5, but not GFP-EHD1, was abolished (Figure 9, D–F). The reciprocal experiment, using only EHD1-RNAi, demonstrated that GFP-EHD1 expression was abrogated, whereas Rabenosyn-5 expression remained intact (Figure 9, G–I). Finally, simultaneous treatment of HeLa cells with RNAi for both Rabenosyn-5 and EHD1 (combo-RNAi) resulted in loss of expression of both proteins (Figure 9, J–L), indicating the feasibility of this approach. Cells treated with combo-RNAi were then subjected to microscopic pulse-chase analysis using either Tf-568 (Figure 7E) or antibody to MHC-I (Figure 7I). Simultaneous reduction in expression of both Rabenosyn-5 and EHD1 resulted in a distribution pattern of Tf-568 (Figure 7E) and anti-MHC-I (Figure 7I) resembling that observed with only Rabenosyn-5-RNAi (Figure 7, C and G). These results support a model in which Rabenosyn-5 acts upstream of EHD1, probably by mediating transport from early endosomes to the ERC (see model in Figure 10).
Figure 9.
Simultaneous RNAi suppression of Rabenosyn-5 and EHD1 in single cells. HeLa cells were mock-treated (A–C), treated with RNAi specific for Rabenosyn-5 (D–F), or RNAi specific for EHD1 (G–I), or treated simultaneously with RNAi specific for both Rabenosyn-5 and EHD1 (J–L). Forty-eight hours later, cells were cotransfected with Rabenosyn-5 and GFP-EHD1. Seventy-two hours after the start of the RNAi treatment, cells were fixed, permeabilized, and incubated with rabbit polyclonal antibodies to Rabenosyn-5, followed by Cy3-conjugated donkey anti-rabbit IgG antibody (red channel). More than 300 cells were examined on four cover glasses for each RNAi treatment to examine expression of GFP-EHD1 (B, E, H, and K; green channel) and Rabenosyn-5 (A, D, G, and J; red channel). The images shown are representative, with the efficacy for either individual or combination RNAi (Combo-RNAi) >95%.
Figure 10.
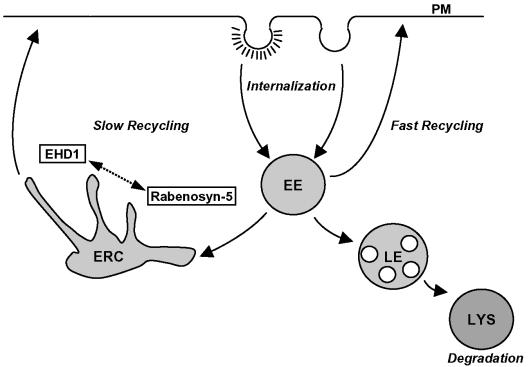
Schematic illustration of endocytic recycling pathways. Proteins internalized into vesicles either via clathrin-coated pits or independently of clathrin quickly fuse with early endosomes (EE), which play a role in determining whether proteins are delivered to late endosomes (LE) and lysosomes (LYS) for degradation, or recycled to the plasma membrane. At least two partially distinct recycling pathways have been described. The first is a “Fast Recycling” pathway that is thought to return cargo to the plasma membrane directly from the EE. The second recycling route is a slower pathway, by which receptors are recycled via the pericentriolar ERC. EHD1 seems to be involved primarily in the latter pathway, regulating the exit of receptors from the ERC. Rabenosyn-5 seems to be critical for transport from the EE to the ERC, but a role cannot be ruled out in direct recycling from the EE to the plasma membrane.
DISCUSSION
Using a combination of biochemical approaches, we have identified Rabenosyn-5 as a novel binding partner for EHD1. The interactions between these two proteins are mediated by the first two of five NPF motifs in Rabenosyn-5 and the EH domain of EHD1. The role of the other three NPF motifs in Rabenosyn-5 remains to be established; one possibility is that they are involved in binding other EH-domain–containing proteins. Rabenosyn-5 and EHD1 colocalize to cytoplasmic vesicles and tubules, but this colocalization is partial. By quantitation, we estimate that 5–10% of EHD1 colocalizes with Rabenosyn-5, whereas 3–5% of Rabenosyn-5 colocalizes with EHD1. Rabenosyn-5 tends to give a more peripheral vesicular pattern, as reported previously (Nielsen et al., 2000), whereas EHD1 is also concentrated in tubules that radiate from the ERC (Lin et al., 2001; Rotem-Yehudar et al., 2001; Caplan et al., 2002) (Figure 4). These observations are consistent with Rabenosyn-5 and EHD1 being associated with successive compartments of the same pathway, with the overlap representing sites of interaction or transition between the two compartments.
RNAi experiments with HeLa cells support the notion that Rabenosyn-5 and EHD1 function in the same membrane trafficking pathway. Consistent with previous studies involving overexpression of normal and mutant forms of these proteins (Nielsen et al., 2000; Lin et al., 2001; de Renzis et al., 2002; Caplan et al., 2002), our RNAi experiments demonstrated that both proteins are required for efficient recycling of internalized proteins to the plasma membrane. Internalized Tf-568 and anti-MHC-I in cells lacking Rabenosyn-5 were delayed in their ability to reach the ERC and hence the cell surface. Similarly, EHD1-depleted cells exhibited decreased recycling of both internalized proteins to the cell surface. Our kinetic analysis indicates that the rate of recycling is slower in cells treated with Rabenosyn-5- or EHD1-RNAi; however, over long periods of chase the internalized cargo is eventually recycled in these cells. This is consistent with the modest but measurable (∼20%) decrease in the level of transferrin receptor localized to the plasma membrane at steady state for Rabenosyn-5– and EHD1-RNAi–treated cells. These findings are also reminiscent of the changes observed in yolk receptor distribution in C. elegans upon loss of the EHD1 homolog (Grant et al., 2001). This reduced rate of transferrin and anti-MHC-I recycling caused by EHD1-RNAi occurred despite the existence of three other proteins closely related to EHD1, namely EHD2, EHD3, and EHD4 (Mintz et al., 1999; Pohl et al., 2000). This indicates that at least in HeLa cells, EHD1 is a key member of this family for the recycling of internalized proteins to the plasma membrane. The other family members may not be sufficiently active or abundant in HeLa cells to compensate for the loss of EHD1 and may be active in other cell types, consistent with a role described for EHD4/Pincher in nerve cells (Shao et al., 2002). Indeed, it has been recently demonstrated that EHD2 plays a critical role in the internalization of transferrin into early endosomes in cultured adipocytes (Guilherme et al., 2003). Because transferrin and anti-MHC-I are internalized by clathrin-dependent and -independent processes, respectively (Naslavsky et al., 2003), our observations indicate that Rabenosyn-5 and EHD1 function after the merging of the two endocytic pathways.
Our data demonstrate a similar decrease in the rate of recycling caused by the absence of either Rabenosyn-5 or EHD1. However, RNAi specific for either EHD1 or Rabenosyn-5 resulted in a different subcellular accumulation pattern of internalized cargo. Rabenosyn-5-RNAi caused the internalized cargo to accumulate in a dispersed, slightly enlarged vesicular compartment extending to the periphery of the cell (Figure 7, B and F); this compartment largely colocalized with the endosomal markers EEA1 and Rab5 (Figure 8, A–F). Compartments containing accumulated transferrin in cells treated simultaneously with Rabenosyn-5- and EHD1-RNAi (Combo-RNAi) were similarly positive for early endosomal markers (our unpublished data). On the other hand, EHD1-RNAi caused delayed cargo recycling and retention in a tight, compact pericentriolar compartment characteristic of the ERC (Figure 7, D and H), and similar to the compartment described upon utilization of a dominant-negative EHD1 (Lin et al., 2001; Hao et al., 2002; Picciano et al., 2003).
Based on the phenotypic differences observed after treatment with either Rabenosyn-5-RNAi or EHD1-RNAi, we hypothesize that these proteins act at consecutive stages of the recycling pathway. We cannot rule out the possible involvement of EHD1 in the regulation of the fast recycling pathway, directly from early endosomes to the plasma membrane (Figure 10); indeed, little is known about the regulation of this pathway. However, the accumulation of internalized transferrin (and MHC-I) in a compact perinuclear region in the presence of EHD1-RNAi or a dominant-negative EHD1 (Lin et al., 2001; Hao et al., 2002; Picciano et al., 2003) fosters the notion that the primary site of EHD1 action is at the ERC. Moreover, our experiments simultaneously using RNAi for both EHD1 and Rabenosyn-5 support a model in which Rabenosyn-5 is specifically involved in the transport of cargo from early endosomes to the ERC, whereas EHD1 participates in transport from the ERC to the plasma membrane (Figure 10). Rabenosyn-5 might thus serve as a relay, steering cargo from early endosomes to the ERC, where contact is made with EHD1-containing structures before returning to peripheral endosomes.
As a result of its location downstream from Rab5 and Rab4 but upstream from EHD1, Rabenosyn-5 may connect these two types of proteins involved in endosomal recycling. Rab4, in particular, is localized to a recycling domain of early endosomes (Daro et al., 1996; Sonnichsen et al., 2000) and regulates recycling to the plasma membrane (Van der Sluijs et al., 1992). Rab4 could recruit Rabenosyn-5 (de Renzis et al., 2002), after which Rabenosyn-5 could become incorporated into vesicular carriers that mediate transport from early to recycling endosomes. Rabenosyn-5 would then interact with EHD1 in the process of relaying cargo to the ERC. After dissociation from Rabenosyn-5, EHD1 would assume its role in recycling cargo to the plasma membrane via tubular and/or vesicular structures, possibly in conjunction with Rab11 and other recycling regulatory proteins.
In conclusion, our observations indicate that Rabenosyn-5 may serve as a nexus between Rab4 on early endosomes, and EHD1 on the ERC, thus linking consecutive molecular machineries involved in recycling internalized proteins to the plasma membrane.
Supplementary Material
Acknowledgments
We are indebted to J.S. Bonifacino for ideas, discussions, and involvement throughout this project, as well as the critical reading of this manuscript. We thank M. Zerial for very generous gifts of reagents, and R. Weigert for sharing expertise in extremely helpful discussions. We also thank X. Zhu and A. San Miguel for excellent technical assistance.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-10-0733. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–10–0733.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Caplan, S., Hartnell, L.M., Aguilar, R.C., Naslavsky, N., and Bonifacino, J.S. (2001). Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 154, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, S., Naslavsky, N., Hartnell, L.M., Lodge, R., Polishchuk, R.S., Donaldson, J.G., and Bonifacino, J.S. (2002). A tubular EHD1-containing compartment involved in the recycling of Major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, S.D., and Schmid, S.L. (2003). Regulated portals of entry into the cell. Nature 422, 37–44. [DOI] [PubMed] [Google Scholar]
- Cormont, M., Mari, M., Galmiche, A., Hofman, P., and Le Marchand-Brustel, Y. (2001). A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. USA 98, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormont, M., Meton, I., Mari, M., Monzo, P., Keslair, F., Gaskin, C., McGraw, T.E., and Le Marchand-Brustel, Y. (2003). CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic 4, 97–112. [DOI] [PubMed] [Google Scholar]
- Daro, E., van der Sluijs, P., Galli, T., and Mellman, I. (1996). Rab4 and cellulobrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA 93, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M.T., Stahl, D.C., Hefta, S.A., and Lee, T.D. (1995). A microscale electrospray interface for on-line, capillary liquid chromatography/tandem mass spectrometry of complex peptide mixtures. Anal. Chem. 67, 4549–4556. [DOI] [PubMed] [Google Scholar]
- de Beer, T., Carter, R.E., Lobel-Rice, K.E., Sorkin, A., and Overduin, M. (1998). Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science 281, 1357–1360. [DOI] [PubMed] [Google Scholar]
- de Beer, T., Hoofnagle, A.N., Enmon, J.L., Bowers, R.C., Yamabhai, M., Kay, B.K., and Overduin, M. (2000). Molecular mechanism of NPF recognition by EH domains. Nat. Struct. Biol. 7, 1018–1022. [DOI] [PubMed] [Google Scholar]
- Deneka, M., Neeft, M., Popa, I., van Oort, M., Sprong, H., Oorschot, V., Klumperman, J., Schu, P., and van der Sluijs, P. (2003). Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes. EMBO J. 22, 2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis, S., Sonnichsen, B., and Zerial, M. (2002). Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4, 124–133. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Eng, J. K., McCormack, A. L. and Yates, J. R., III. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- Fouraux, M.A., Deneka, M., Ivan, V., van der Heijden, A., Raymackers, J., van Suylekom, D., van Venrooij, W.J., van der Sluijs, P., and Pruijn, G.J.M. (2004). rab4ip' is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol. Biol. Cell 15, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin, E., Benjamin, S., Rapaport, D., Rotem-Yehudar, R., Tolchinsky, S., and Horowitz, M. (2002). EHD 3, A protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic 3, 575–589. [DOI] [PubMed] [Google Scholar]
- Grant, B., Zhang, Y., Paupard, M.C., Lin, S.X., Hall, D.H., and Hirsh, D. (2001). Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J., and Maxfield, F.R. (1995). Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 7, 552–563. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. (2003). Lipids in endocytic membrane transport and sorting. Curr. Opin. Cell Biol. 15, 382–388. [DOI] [PubMed] [Google Scholar]
- Guilherme, A., Soriano, N.A., Bose, S., Holik, J., Bose, A., Pomerleau, D.P., Furcinitti, P., Leszyk, J., Corvera, S., and Czech, M.P. (2003). EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. J. Biol. Chem. 279, 10593–10605. [DOI] [PubMed] [Google Scholar]
- Hales, C.M., Griner, R., Hobdy-Henderson, K.C., Dorn, M.C., Hardy, D., Kumar, R., Navarre, J., Chan, E.K., Lapierre, L.A., and Goldenring, J.R. (2001). Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276, 39067–39075. [DOI] [PubMed] [Google Scholar]
- Hao, M., and Maxfield, F.R. (2000). Characterization of rapid membrane internalization and recycling. J. Biol. Chem. 275, 15279–15286. [DOI] [PubMed] [Google Scholar]
- Hao, M., Lin, S.X., Karylowski, O.J., Wustner, D., McGraw, T.E., and Maxfield, F.R. (2002). Vesicular and non-vesicular sterol transport in living cells. The endocytic recycling compartment is a major sterol storage organelle. J. Biol. Chem. 277, 609–617. [DOI] [PubMed] [Google Scholar]
- Hopkins, C.R., and Trowbridge, I.S. (1983). Internalization and processing of transferrin and the transferrin receptor in human carcinoma A431 cells. J. Cell Biol. 97, 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, L., and Lamaze, C. (2002). Clathrin dependent or not: is it still the question? Traffic 3, 443–451. [DOI] [PubMed] [Google Scholar]
- Le Gall, S., Buseyne, F., Trocha, A., Walker, B.D., Heard, J.-M., and Schwartz, O. (2000). Distinct trafficking pathways mediate Nef-induced and clathrin-dependent major histocompatibility complex class I down-regulation. J. Virol. 74, 9256–9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.X., Grant, B., Hirsh, D., and Maxfield, F.R. (2001). RME-1 regulates distribution and function of the endocytic recycling compartment in mammalian cells. Nat. Cell Biol. 3, 567–572. [DOI] [PubMed] [Google Scholar]
- Lindsay, A.J., Hendrick, A.G., Cantalupo, G., Senic-Matuglia, F., Goud, B., Bucci, C., and McCaffrey, M.W. (2002). Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 277, 12190–12199. [DOI] [PubMed] [Google Scholar]
- Mammoto, A., Ohtsuka, T., Hotta, I., Sasaki, T., and Takai, Y. (1999). Tab11BP/Rabphilin-11, a downstream target of Rab11 small G protein implicated in vesicle recycling. J. Biol. Chem. 274, 25517–25524. [DOI] [PubMed] [Google Scholar]
- Martina, J.A., Bonangelino, C.J., Aguilar, R.C., and Bonifacino, J.S. (2001). Stonin-2, an adaptor-like protein that interacts with components of the endocytic machinery. J. Cell Biol. 153, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera, R., Arighi, C.N., Lodge, R., Zerial, M., and Bonifacino, J.S. (2003). Divalent interaction of the GGAs with the Rabaptin-5–Rabex-5 complex. EMBO J. 22, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means, R.E., Ishido, S., Alvarez, X., and Jung, J.U. (2002). Multiple endocytic trafficking pathways of MHC class I molecules induced by Herpesvirus protein. EMBO J. 21, 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I. (1996). Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12, 575–625. [DOI] [PubMed] [Google Scholar]
- Mintz, L., Galperin, E., Pasmanik-Chor, M., Tulzinsky, S., Bromberg, Y., Kozak, C.A., Joyner, A., Fein, A., and Horowitz, M. (1999). EHD1–an EH-domain-containing protein with a specific expression pattern. Genomics 59, 66–76. [DOI] [PubMed] [Google Scholar]
- Nagelkerken, B., Van Anken, E., Van Raak, M., Gerez, L., Mohrmann, K., Van Uden, N., Holthuizen, J., Pelkmans, L., and Van Der Sluijs, P. (2000). Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem. J. 346, 593–601. [PMC free article] [PubMed] [Google Scholar]
- Naslavsky, N., Weigert, R., and Donaldson, J.G. (2003). Convergence of non-clathrin and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol. Biol. Cell 14, 417–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, B.J., and Lippincott-Schwartz, J. (2001). Endocytosis without clathrin coats. Trends Cell Biol. 11, 406–412. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., Christoforidis, S., Uttenweiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M., Holflack, B., and Zerial, M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D.N., Pappin, D.J., Creasy, D.M., and Cotrell, J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Picciano, J.A., Ameen, N., Grant, B.D., and Bradbury, N.A. (2003). Rme-1 regulates the recycling of the cystic fibrosis transmembrane conductance regulator. Am. J. Physiol. Cell Physiol. 285, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Pohl, U., Smith, J.S., Tachibana, I., Ueki, K., Lee, H.K., Ramaswamy, S., Wu, Q., Mohrenweiser, H.W., Jenkins, R.B., and Louis, D.N. (2000). EHD2, EHD3, and EHD4 encode novel members of a highly conserved family of EH domain-containing proteins. Genomics 63, 255–262. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman, J., and Scheller, R. H. (2000). A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell 6, 1437–1447. [DOI] [PubMed] [Google Scholar]
- Ren, M., Xu, G., Zeng, J., De Lemos-Chiarandini, C., Adesnik, M., and Sabatini, D.D. (1998). Hydrolysis of GTP on Rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95, 6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem-Yehudar, R., Galperin, E., and Horowitz, M. (2001). Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J. Biol. Chem. 276, 33054–33060. [DOI] [PubMed] [Google Scholar]
- Shao, Y., Akmentin, W., Toledo-Aral, J.J., Rosenbaum, J., Valdez, G., Cabot, J.B., Hilbush, B.S., and Halegoua, S. (2002). Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. J. Cell Biol. 157, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, D.R., Daro, E.A., Hull, M., and Mellman, I. (1999). The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 145, 123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, D., Pelletier, L., O'Connell, C.B., Warren, G., and Mellman, I. (2002). Transferrin receptor recycling in the absence of perinuclear recycling endosomes. J. Cell Biol. 156, 797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., De Renzis, S., Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, O., Reinsch, S., Urbe, S., Zerial, M., and Parton, R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam, E.M., ten Broike, T., Jansen, K., Spijkers, P., and Stoorvogel, W. (2002). Endocytosed transferrin receptors recycle via distinct dynamin and phosphatidylinositol 3-kinase-dependent pathways. J. Biol. Chem. 277, 48876–48883. [DOI] [PubMed] [Google Scholar]
- van der Sluijs, P., Hull, M., Webster, P., Male, P., Goud, B., and Mellman, I. (1992). The small GTP-binding protein Rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729–740. [DOI] [PubMed] [Google Scholar]
- Wallace, D.M., Lindsay, A.J., Hendrick, A.G., and McCaffrey, M.W. (2002a). The novel Rab11-FIP/Rip/RCP family of proteins displays extensive homo- and hetero-interacting abilities. Biochem. Biophys. Res. Commun. 292, 909–915. [DOI] [PubMed] [Google Scholar]
- Wallace, D.M., Lindsay, A.J., Hendrick, A.G., and McCaffrey, M.W. (2002b). Rab11-FIP4 interacts with Rab11 in a GTP-dependent manner and its overexpression condenses the Rab11 positive compartment in HeLa cells. Biochem. Biophys. Res. Commun. 299, 770–779. [DOI] [PubMed] [Google Scholar]
- Yamashiro, D.J., Tycko, B., Fluss, S.R., and Maxfield, F.R. (1984). Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell 37, 789–800. [DOI] [PubMed] [Google Scholar]
- Zeng, J., et al. (1999). Identification of a putative effector protein for rab11 that participates in transferring recycling. Proc. Natl. Acad. Sci. USA 96, 2840–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.