Abstract
In humans, microsomal epoxide hydrolase (mEH) contributes important biological functions that underlie both detoxification and bioactivation fates arising from exposures to foreign chemicals. Previously, we discovered that human mEH gene transcription is initiated from alternative promoters. The respective transcripts are programmed with tissue specificity and the upstream E1b promoter contributes predominantly to mEH expression. The results presented demonstrate that exposures to the Nrf2 activators, sulforaphane (SFN) and tert-butylhydroquinone (tBHQ), markedly activate E1b transcription in human lung and liver cells. Genomic analyses identified two major DNase I hypersensitive regions (HS-1 and HS-2) within the ~15 kb intervening sequence separating E1b from the downstream E1 promoter. In BEAS-2B cells, the Nrf2 effectors, SFN and tBHQ, selectively activated the more distal HS-2 through an antioxidant-response element (ARE). An activator protein 1/12-O-tetradecanoylphorbol-13-acetate interaction was further identified within the HS-2 enhancer that functioned to additionally contribute to ARE-mediated induction responsiveness of the E1b promoter. The results demonstrate that ARE modulation, integrated with additional transcriptional complexes, regulates the tissue-specific expression of mEH and that these processes likely coordinate both the protective and bioactivation functions contributed by mEH activities in human tissues.
Keywords: microsomal epoxide hydrolase, human, gene regulation, alternative gene promoters, Nrf2
INTRODUCTION
Chemoprevention is the use of natural or synthetic agents to reverse, prevent or delay carcinogenic progression to invasive cancer [1]. A chemically diverse array of plant-derived phytochemicals or their synthetic derivatives, including the isothiocyanates, have demonstrated effectiveness in the prevention or suppression of carcinogenesis in animal models and human [2]. One key mechanism of their protection against carcinogenesis is to reduce oxidative stress and minimize oxidative DNA damage by inducing Phase II detoxification enzymes and antioxidant proteins [3].
The transcription factor, Nrf2 (NFE2L2), functions as a master regulator of cytoprotective genes that combat cellular oxidative stress. Activation of these genes involves Nrf2 binding to antioxidant response elements (ARE) in their respective promoters. Under normal non-stressed conditions, Nrf2 is constantly subject to proteasomal degradation mediated by KEAP1. KEAP1 functions as a cytoplasmic anchor of Nrf2 and an adaptor of Cullin-3 E3 ubiquitin ligase complex, leading to continuous ubiquitination of Nrf2 and its proteasomal degradation. Under oxidative stress conditions, Nrf2 dissociates from KEAP1 and escapes proteolysis, which in turn leads to rapid accumulation of Nrf2 in the nucleus and the subsequent transcriptional induction of target genes. Constitutive activation of Nrf2-mediated genes has been documented in non-small-cell lung cancer-derived cell lines and tumor samples, as well as other cancers [4-6]. Aberrant activation of Nrf2 was linked to mutations in KEAP1 and Nrf2 that impair the interaction between the two proteins, leading to nuclear accumulation of Nrf2. Consequently, these mutations result in up-regulation of drug metabolizing enzymes and drug efflux pumps and likely confer enhanced resistance of cancer cells to chemotherapeutic agents [4, 5, 7].
Microsomal epoxide hydrolase (mEH, EPHX1) is a critical biotransformation enzyme that catalyzes the hydrolysis of electrophilic epoxides to dihydrodiols [8-10]. Epoxides derived from xenobiotic metabolism by the cytochrome P450 (CYP450) enzymes potentially interact with cellular macromolecules (DNA, RNA and proteins), detrimental processes that include certain mutagenicity. Metabolism of epoxides by mEH often generates less reactive and therefore less toxic dihydrodiol derivatives. Thus, mEH largely contributes a protective role against the deleterious effects of reactive epoxides. Alternatively, mEH may participate in bioactivation functions, for example in the coordinated metabolism of polycyclic aromatic hydrocarbons (PAHs), important components of cigarette smoke and established agents in the etiology of human lung cancers. In these respects, mEH participates in concert with CYP450 enzymes in the formation of stable PAH dihydrodiol epoxide-DNA adducts. The necessity of mEH in the bioactivation of PAHs was established in mEH-null mice, which are highly resistant to PAH-induced carcinogenesis compared with controls [11]. Furthermore, data from epidemiological studies suggest that genetic polymorphisms in the coding regions of mEH contribute to lung cancer risk [12].
In humans, mEH gene transcription is regulated by alternative promoter usage and generates transcripts with unique noncoding first exons, termed E1 and E1b [13]. E1 is initiated from a transcription start site ~3.2kb upstream of coding exon 2, while E1b is driven by a far upstream alternative promoter located ~18.5kb upstream of exon 2. These mRNA isoforms exhibit highly distinctive tissue-specific distribution patterns with the E1 isoform expressed exclusively in liver, while E1b is expressed in all tissues, including liver [13, 14].
Nrf2 has been reported as a regulator of mEH expression in mice, with the knockout of Nrf2 resulting in reduced basal expression of mEH in mouse liver and small intestine [15-19]. Similarly, Nrf2-null mice demonstrated minimal induction of mEH by Nrf2 activators [15-21]. Although a functional ARE is believed to exist in the regulatory region of mouse mEH, as in other phase II detoxifying enzymes [19, 21], its localization has not been discerned. Despite the Nrf2 implication as a regulator of mEH in mice, only minimal data exist regarding human mEH gene regulation. Further, DNA sequence alignment/homology analyses indicate that the 5′ upstream regulatory region comprising the human E1b promoter is poorly conserved between humans and rodents [14], suggesting that the major regulatory mechanisms existing for mEH in human and rodents are quite different.
In the present study, we identified two unique intronic DNA enhancer elements, localized in the intervening region between the human mEH E1 and E1b promoters. These regions demonstrated marked DNase I hypersensitivity and were characterized for their respective functional abilities to modulate mEH expression in human cells. The results generated identify a novel mechanism whereby Nrf2 induces expression of the E1b promoter-derived transcript and provide insight regarding the transcriptional regulation of the human mEH gene and its subsequent activities as a mediator of xenobiotic metabolism.
MATERIALS AND METHODS
Materials
The protease inhibitor mixtures were obtained from Calbiochem, FuGENE 6 Transfection Reagent and the dual luciferase reporter assay system were from Promega (Madison, WI). All cell culture media and supplies and the TRIzol® Reagent were from Invitrogen Life Technologies (Grand Island, NY). Sulforaphane (#574215) was purchased from CalBiochem/EMD Millipore (Billerica, MA); tert-Butylhydroquinone (tBHQ, Cat#B0833) was from TCI America (Philadelphia, PA); TPA (12-O-tetradecanoylphorbol-13-acetate, #445-004-M001) was from Alexis Biochemicals (San Diego, CA). All other chemicals were purchased from Sigma (St. Louis, MO) unless otherwise indicated. Small interfering RNA (siRNA) targeting Nrf2 mRNA (siRNA ID: s9493) or Silencer® Select Negative Control No. 1 siRNA and the Lipofectamine RNAiMAX reagent were purchased from Invitrogen. The mouse monoclonal anti-mEH (sc-135984) and anti-Actin (sc-81178) antibodies and rabbit polyclonal anti-Nrf2 (sc-13032) were from Santa Cruz Biotechnology (Santa Cruz, CA); the rabbit polyclonal anti-GAPDH (G9545) was from Sigma; and normal rabbit IgG was purchased from Cell Signaling Technology (Beverly, MA).
Plasmids
The E1b promoter luciferase reporter constructs containing 300bp, 1kb, 2kb or 2.2kb of the 5′-flanking region upstream of E1b were constructed as described previously [13]. To generate the intronic enhancer/E1b-300 promoter luciferase reporter constructs, the regions containing DNase I hypersensitive sites located between the E1b and E1 gene promoters were amplified by PCR from human genomic DNA using primers as follows: DNase I HS-1(5′-ATGCGCTAGCTCAGGAAAGGAATGTGTAGGAGGG-3′ and 5′-ATGCCTCGAGAAATGCTGGGATTACAGGTGTGCG-3′); and DNase I HS-2 (5′-GATCGGTACCTCCCTTTCCACTGGATGTTCCCTT-3′ and 5′-GATCTCTAGAACCATAAGATGCAGGAAGAGGGCT-3′). The PCR products were inserted to the E1b-300 proximal promoter-containing pGL4 luciferase reporter vector. Site-directed mutagenesis of the putative ARE in DNase I HS-2 was carried out by QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA) with the following primers: mutARE (5′-ATGGCAAGTaaTGAaTtATGGCCGGCTAGCA-3′ and 5′-TGCTAGCCGGCCAtaAtTCAttACTTGCCAT-3′), mutARE-1 (5′-ATGGCAAGTGCTGAaTtATGGCCGGCTAGCA-3′ and 5′-TGCTAGCCGGCCATaAtTCAGCACTTGCCAT-3′), and mutARE-2 (5′-ATGGCAAGTaaTGAGTCATGGCCGGCTAGCA-3′ and 5′-TGCTAGCCGGCCATGACTCAttACTTGCCAT-3′). The mutated nucleotides are in lowercase. Expression plasmids were generated by inserting full length cDNA of Nrf2 (NM_006164.4), KEAP1 (NM_203500.1) and JUN (NM_002228.3) into the p3XFLAG-CMV-10 expression vector (Sigma). DNA primers used for the amplification of these genes were as follows: Nrf2 (5′-GATCGCGGCCGCAATGGACTTGGAGCTGCCGCCG-3′ and 5′-GATCTCTAGACTAGTTTTTCTTAACATCTGG-3′), KEAP1 (5′-ATCGCAGCCAGATCCCAGGCCTAGC-3′ and 5′-GATCTCTAGAATCAACAGGTACAGTTCTGCTGG-3′) and JUN (5′-GATCGAATTCAACTGCAAAGATGGAAACGACC-3′ and 5′-GATCTCTAGAATCAAAATGTTTGCAACTGCTGCG-3′). All constructs were confirmed by DNA sequencing.
Cell culture, transient transfection and luciferase reporter assays
Human lung carcinoma epithelial A549 cells and human bronchial epithelial BEAS-2B cells were purchased from American Type Culture Collection (Manassas, VA). Both cell lines were cultured in 5% CO2 incubator at 37°C in Dulbecco’s modified Eagle medium supplemented with 10% FBS, 2 mM L-glutamine, 0.1 mM Non-Essential Amino Acids, 1.0 mM Sodium Pyruvate, 10 mM HEPES, 0.15% sodium bicarbonate, 100 units/ml penicillin G, and 100 μg/ml streptomycin. Cells were cultured in 24-well or 6-well plates and 100 mm petri dishes and harvested according to the requirements of the experiments. Normal human hepatocytes were obtained and maintained as described previously [22, 23].
For transient transfection, A549 and BEAS-2B cells were seeded a day before transfection in 24-well plates at a density of 5×104 cells per well. To assess the enhancer properties of intronic regulatory elements on E1b promoter activity, cells were transfected with corresponding reporter plasmids using a FuGENE 6 transfection protocol according to the manufacturer’s instructions. The pRL-CMV plasmid containing Renilla Luciferase cDNA was also co-transfected as an internal control for transfection efficiency. Cells were harvested 24 h post transfection and luciferase activity was measured and analyzed using a Veritas Microplate Luminometer (Turner Biosystems/Promega) using the Dual Luciferase Reporter Assay System (Promega) as described previously [24]. For studying the effect of chemical treatment on E1b promoter activities and the enhancer activities of DNase I HS sites, cells were transfected with E1b promoter reporter or enhancer constructs and pRL-CMV reporter plasmids for 6 h and were incubated for 24 h in culture medium containing the indicated concentration of chemicals or vehicle (0.1% DMSO). For assessing E1b promoter activities in response to ectopic expression of Nrf2 and JUN, cells were co-transfected with reporter plasmids containing E1b proximal promoter and intronic enhancers and the corresponding expression plasmid of the give transcription factor. Luciferase activity was measured in the same manner as described above. All transfections were performed in triplicate and the results were expressed as means ± standard deviations (SD). The experiments were repeated three times with high reproducibility and the most representative results are shown.
RNA isolation, reverse transcription and quantitative real-time PCR
For investigating inducible expression of various genes by chemical treatment, A549 and BEAS-2B cells in 6-well plates were treated with chemicals for 24 h. To assess the effect of overexpression of Nrf2 on E1b expression in BEAS-2B cells, cells were transfected with Nrf2-expressing plasmid for 24 h and 48 h as described above. Total RNA was extracted with TRIzol Reagent according to the manufacturer’s instructions. Total RNA (2 g) was converted to cDNA using the High-Capacity cDNA Archive Kit (Applied Biosystems/Life Technologies). cDNAs were analyzed with CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using PerfeCTa SYBR Green SuperMix (Quanta Biosciences, Gaithersburg, MD). The final concentration of primers in each reaction was 0.2μM. The PCR conditions consisted of an initial denaturation for 3 min at 95°C, followed by 40 cycles of 15s at 95°C and 1 min at 60°C. Each sample was run in duplicate and the results were normalized to the level of β-actin or GAPDH mRNA. The primers used for quantitative real-time PCR were as follows: E1b, 5′-GAGCCTGCGAGCCGAGAC-3′ (forward)/5′-CGTGGATCTCCTCATCTGACGTTT-3′ (reverse); Nrf2, 5′-CAGCGACGGAAAGAGTATGAG-3′ (forward)/5′-GGGCTGGCTGAATTGGGAG-3′ (reverse); HMOX1, 5′-CAGTGCCACCAAGTTCAAGC-3′ (forward)/5′-GTTGAGCAGGAACGCAGTCTT-3′ (reverse); NQO1, 5′-GGCAGAAGAGCACTGATCGTA-3′ (forward)/5′-TGATGGGATTGAAGTTCATGGC-3′ (reverse); β-Actin, 5′-CATGTACGTTGCTATCCAGGC-3′ (forward)/5′-CTCCTTAATGTCACGCACGAT-3′ (reverse); and GAPDH, 5′-CCCATCACCATCTTCCAGGAG-3′ (forward)/5′-GTTGTCATGGATGACCTTGGC-3′ (reverse). The experiments were repeated three times and the most representative results were shown.
Western blotting
To assess the effect of chemical treatments on mEH protein level, A549 and BEAS-2B cells in 100 mm dishes were treated with chemicals at various concentrations for 24 h. To assess the effect of overexpression of Nrf2 on mEH protein level in BEAS-2B cells, cells were transfected with Nrf2-expressing plasmid for 24 h and 48 h as described above. At the time of harvest, cells were washed with PBS, trypsinized and centrifuged at 1000×g for 3 min. For making whole cell lysates, cells were lysed in RIPA buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with 1× protease inhibitor cocktail (#539131, Calbiochem/EMD Millipore). The cell lysates were centrifuged at 16,000×g for 10 min at 4°C and the supernatants were collected as whole-cell lysate. Protein concentrations were determined by Pierce 660 nm Protein Assay (Thermo Scientific. Waltham, MA). The extracted proteins (30 g) were separated on a 10% denaturing polyacrylamide gel (Bio-Rad) and transferred to a PVDF membrane (Bio-Rad). After blocking in 5% skim milk for 30 min, the blots were incubated sequentially with primary antibodies at the dilution of 1:1000 and horseradish peroxidase-conjugated secondary antibodies at the dilution of 1:5000. The membranes were washed three times with 1×TBS/0.1% Tween 20, treated with Pierce ECL Western Blotting Substrate (Thermo Scientific), and exposed to ImageTek-H X-ray films (American X-Ray & Medical Supply, Jackson, CA). The antibodies used for immunoblotting were as follows: anti-mEH (sc-135984, Santa Cruz), anti-Nrf2 (sc-13032, Santa Cruz), anti-β-Actin (sc-81178, Santa Cruz), and anti-GAPDH (G9545, Sigma). The experiments were repeated three times with highly reproducible results and the most representative results are shown.
Nrf2 siRNA knockdown studies
To reduce endogenous Nrf2 and assess its effects on the E1b expression in A549 cells, cells were transfected with either control or Nrf2 siRNA at 25nM using the Lipofectamine RNAiMAX reagent with a Forward Transfection Protocol according to the manufacturer’s instructions. Briefly, cells were seeded a day before transfection in 6-well plates at a density of 3×105 cells per well or in 60 mm petri dishes at a density of 7×105 cells per dish. At the day of transfection, the transfection complexes of the Lipofectamine RNAiMAX reagent and the given siRNA were added to each well containing cells. After 48 h, cells in 6-well plates were harvested for RT-PCR analysis and cells in 60 mm petri dishes were collected for western blotting. To assess the effect of Nrf2 knockdown on tBHQ-mediated induction of E1b expression in BEAS-2B cells, cells in 6-well plates were transfected with 25nM Nrf2 siRNA in the same manner as described above. After 36 h, siRNA-transfected cells were treated with tBHQ at 40 μM for an additional 24 h and then harvested for RT-PCR analysis. The experiments were repeated three times with high reproducibility and the most representative results are shown.
Chromatin immunoprecipitation (ChIP) assay
A549 and BEAS-2B cells were grown to 80-90% confluence in 100 mm dishes. Cells were harvested by trypsinization and fixed in 1% formaldehyde (#252549, Sigma) at 25°C for 10 min with slow agitation. The fixation was stopped by addition of glycine to a concentration of 0.125M. After a 5 min incubation at 25°C, cells were pelleted by centrifugation at 1000×g for 5 min and then washed with ice-cold phosphate-buffered saline twice. Cells were lysed for 10min on ice in SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl, pH 8) with protease inhibitor cocktail (#539131, Calbiochem). Cells were then sonicated with a Bioruptor sonicator (Diagenode, Liège, Belgium) for 5 cycles of 30 sec ON and 30 sec OFF at HIGH setting in a refrigerated water bath. Sheared cross-linked chromatin was centrifuged at 12,000×g for 10 min at 4°C and diluted 10-fold in ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH8, and 167 mM NaCl) with protease inhibitor cocktail. The diluted chromatin was pre-cleaned overnight at 4°C with 35 l protein A/G Plus-agarose beads (sc-2003, Santa Cruz) which were pre-blocked with sonicated salmon sperm DNA (201190, Stratagene) and BSA (#2930, EM Science). Pre-cleaned chromatin was then incubated overnight at 4°C with 4 g of anti-Nrf2 antibodies (sc-13032, Santa Cruz) as well as normal rabbit IgG (#2729s, Cell Signaling Technology). To collect the antibody-chromatin complex, 75 l protein A/G Plus-agarose beads pre-blocked as above were added, incubated for 3 h with rotation at 4°C, and pelleted by centrifugation at 5000×g for 1 min. The pelleted complexes were then washed sequentially with Low Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 150 mM NaCl), High Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8, 500 mM NaCl), and LiCl Immune Complex Wash Buffer (0.25 M LiCl, 1% IGEPAL CA630, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl, pH 8), followed by two washes with TE buffer. Precipitated Protein-DNA complexes were eluted twice with 100 μl elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at 25°C. To reverse crosslinks, the eluates were incubated at 65°C for 4 h in the presence of 8 μl of 5 M NaCl and 1 μl of 10 mg/ml RNase A. Proteins were digested with 2 μl of 10 mg/ml proteinase K for 2h at 45°C in the presence of 4 μl of 0.5 M EDTA, pH8.0, and 8 μl of 1 M Tris-HCl, pH8. DNA was purified with ChIP DNA Clean & Concentrator kit (D5205, Zymo Research). Immunoprecipitated DNA was amplified and the PCR amplicons were analyzed on 1.5% agarose gels. The primer sets used for detection were: E1b proximal promoter (5′-GCGGACCGCCCTTTAAGTAGCC-3′ and 5′-GATCTCTCCGGCTCCCTGGCTC-3′), DNase I HS-1 #1 (5′-GCCACAGGGTAGGGGAGGCAAA-3′ and 5′-GGCTGGATCCTTGGCAACGCTT-3′), DNase I HS-1 #2 (5′-AGCTCACACTGAGCAGCCTCCC-3′ and 5′-ACCCACCAGGCAACAGACGGAA-3′), DNase I HS-2 (5′-CCCAACTGTCGCAGGGCTGG-3′ and 5′-CCGCCTGCCCAAAGACTCCC-3′), E1 proximal promoter (5′-CTGGTAGTGCTGGTGGGGGC-3′ and 5′-CCCACTCCCCACGGCCTTCT-3′), and Exon 3 flanking region (5′-CACCCCACCTTTGGAGGACAGC-3′ and 5′-ACATCCCTCTCTGGCTGGCGTT-3′).
Electrophoretic mobility shift assays (EMSA)
The nuclear extracts from untreated A549 cells or SFN-treated BEAS-2B cells were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer’s instructions. Double-stranded E1b HS-2 ARE probe was end-labeled with [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs, Ipswich, MA). For EMSA the DNA-binding reactions containing 2 μg of nuclear extracts, 20 fmol of labeled probes, 0.01 mg/ml sonicated salmon sperm DNA(201190, Stratagene), 2 μl of 5× binding buffer [20% (v/v) glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol, 250 mM NaCl, 50 mM Tris-HCl (pH 7.5), 0.25 mg/ml poly(dI-dC)] in a final volume of 10μl, were incubated with or without unlabeled competitor for 20 minutes at room temperature. The DNA-protein complexes were resolved by electrophoresis through a nondenaturing 4% polyacrylamide gel in 0.5× TBE buffer. Subsequently, gels were dried and exposed to X-ray film with intensifying screens at −70°C. The probe sequences were as follows: E1b HS-2 ARE: 5′-ATGGCAAGTGCTGAGTCATGGCCGGCTAGCA-3′; mutant E1b HS-2 ARE: 5′-ATGGCAAGTaaTGAaTtATGGCCGGCTAGCA-3′; human NQO1 ARE: 5′-CAGTCACAGTGACTCAGCAGAATCT-3′; and mutant NQO1 ARE: 5′-CAGTCACAtaGttTCAcaAGAATCT-3′. The mutated nucleotides are in lowercase.
Statistical analyses
Data are expressed as means ± standard deviations (SD). The statistical significance of the differences between samples was determined using one-way analysis of variance (ANOVA) in combination with Dunnett’s test or one-tailed Student’s t test, dependent on the design of experiments. Differences were considered significant for samples with p-values <0.05.
RESULTS
Nrf2 activators induce E1b expression in human lung epithelial cell lines
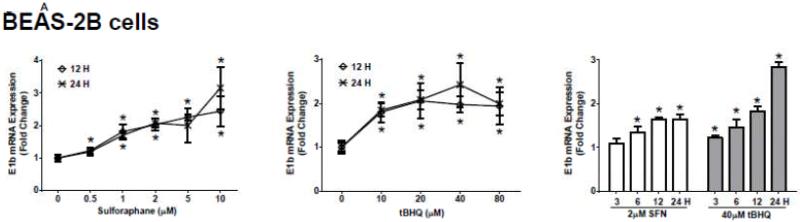
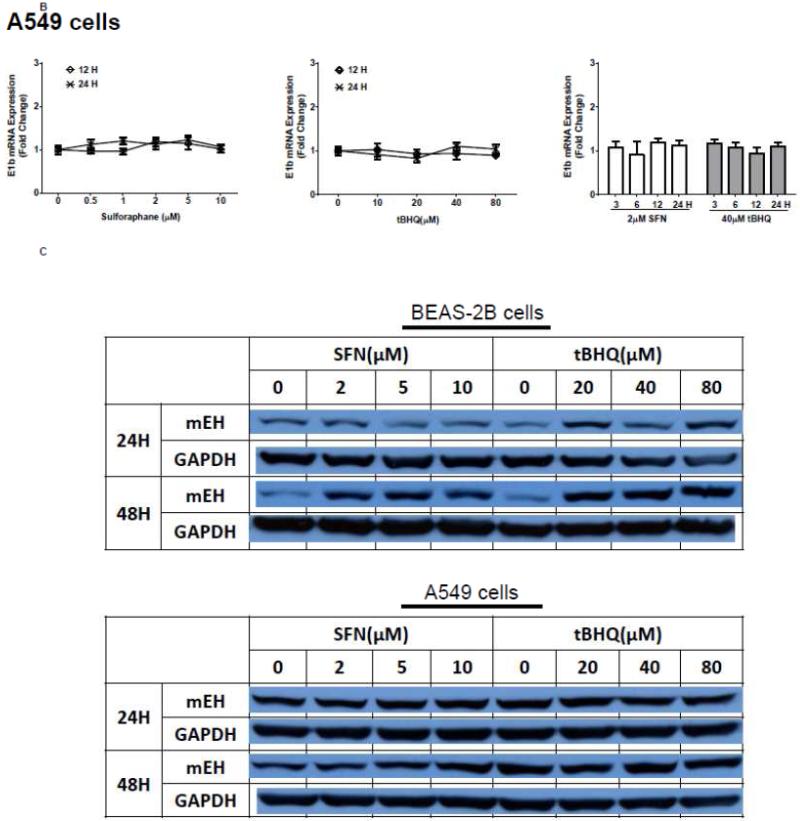
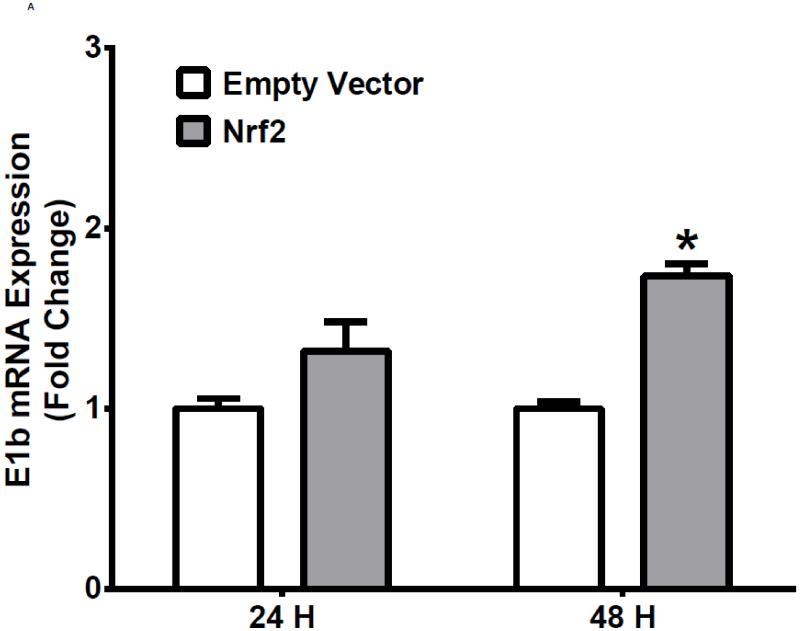
The human bronchial epithelial cell line, BEAS-2B, and the human lung adenocarcinoma epithelial cell line, A549, were selected as experimental models to test the hypothesis that Nrf2 functions as a critical modulator of human mEH gene transcription. A549 cells harbor a point mutation in the KEAP1 gene [6] that creates a loss of function KEAP1, impairing its interaction with Nrf2, resulting in constitutive activation and nuclear accumulation of the Nrf2 protein. In contrast, BEAS-2B cells possess a wild-type KEAP1 and maintain a low level of Nrf2 in cytoplasm due to the tight control imposed by KEAP1. Therefore, A549 cells maintain constitutively activated and high-level Nrf2 while BEAS-2B cells model the typical low level Nrf2 scenario that is responsive to activation with Nrf2 effectors. To investigate the potential role for Nrf2 in human mEH transcriptional regulation, the cells were exposed to various concentrations of Nrf2 inducers for 12 or 24 h. BEAS-2B cells treated with SFN or tBHQ increased E1b transcription in dose- and time-dependent manners (Figure 1A). In A549 cells, SFN and tBHQ did not affect E1b transcription (Figure 1B), an expected result since these cells are constitutively activated by Nrf2 nuclear accumulation. The mEH protein levels in SFN- or tBHQ-treated BEAS-2B and A549 cells correlated well with the observed E1b transcript levels (Figure 1C).
Figure 1. Antioxidants induce E1b expression.
A.) BEAS-2B cells and B.) A549 cells were treated with DMSO, sulforaphane or tBHQ for different time periods and E1b mRNA expression was analyzed by the real time quantitative PCR. GAPDH was used as an internal control. Results are expressed as the mean ± SD. Significant differences from DMSO control are indicated by “*” (p<0.05). C.) The induction of mEH protein in BEAS-2B and A549 cells was evaluated by western blotting. Whole cell lysates were immunoblotted to detect mEH protein. GAPDH was used as a loading control. All experiments were performed three times, and one representative set of data are showed.
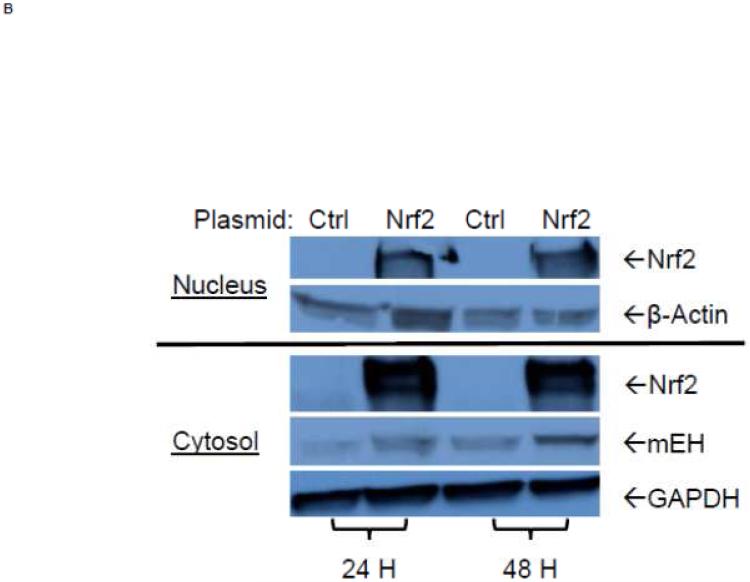
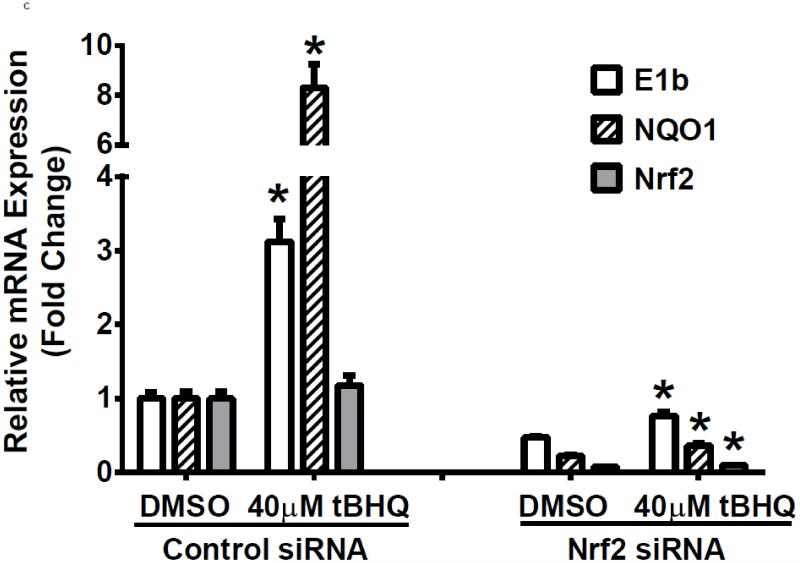
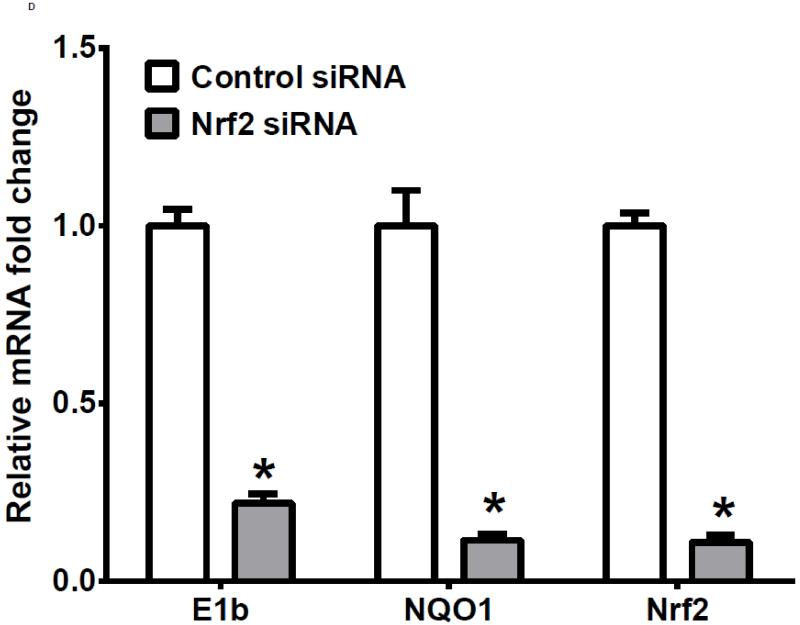
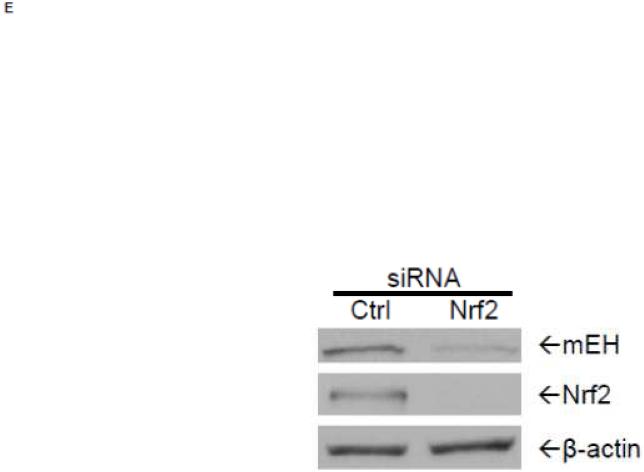
To examine further the mechanistic role for Nrf2 as an E1b expression regulator, BEAS-2B cells were transiently transfected with an Nrf2 expression plasmid. Overexpression of Nrf2 significantly increased E1b, transcription compared with cells transfected with empty vector (Figure 2A). Western blotting analysis demonstrated that transient transfection with the Nrf2-expression plasmid inBEAS-2B cells also resulted in significantly increased levels of Nrf2 cytosolic and nuclear protein (Figure 2B). These data suggested that, although BEAS-2B cells were not treated with any Nrf2 activator, Nrf2 is simultaneously translocated and accumulated in the nucleus apparently, likely because exogenous Nrf2 saturates the normal cytosolic tethering capacity of KEAP1. Accompanying the change in Nrf2 nuclear protein level, protein levels of mEH in cytosol were also significantly increased (Figure 2B). Further, siRNA knockdown of Nrf2 in BEAS-2B cells blocked tBHQ-mediated induction of E1b mRNA, as well as NQO1, a known Nrf2 target gene (Figure 2C). siRNA-mediated knockdown of Nrf2 in A549 cells resulted in a ~5-fold decrease constitutive Nrf2 expression, with similar effects noted for mEH and NQO1 (Figure 2D). mEH and Nrf2 protein expression was also decreased in Nrf2 siRNA-transfected A549 cells (Figure 2E). Together, these results suggested that Nrf2 functions to regulate the tBHQ-mediated E1b induction in BEAS-2B cells and constitutive E1b expression in A549 cells.
Figure 2. Effect of overexpression and siRNA knockdown of NRF2 on E1b expression.
BEAS-2B cells were transfected with either empty expression vector (Ctrl) or Nrf2-expressing plasmid. At 24 and 48 h post-transfection, total RNA and proteins were extracted. A.) Real time qPCR analysis of E1b expression. GAPDH was used as an internal control and results were normalized to empty vector control. *p<0.05 compared to empty vector controls. B.) Western blot analysis of Nrf2 and mEH protein levels with β-actin or GAPDH as a loading control. C.) qPCR analysis of t-BHQ-induced expression of E1b in BEAS-2B cells transfected with Nrf2 siRNA. BEAS-2B cells were transfected with either control or Nrf2 siRNA for 48 h and then treated with tBHQ for another 12 h. The expressions of mEH, NQO1 and Nrf2 genes were measured with Real time quantitative PCR analysis. β-actin was used as an internal control and all values were normalized to DMSO-treated control siRNA values. *p<0.05 compared to DMSO controls. D.) A549 cells were transfected with either control or Nrf2 siRNA. Forty-eight hours after siRNA transfection, cells were harvested for total RNA or total proteins. Significant differences from control siRNA were indicated by “*” (p<0.05). E.) Western blot analysis of mEH protein levels with β-actin as a loading control. All qPCR values are expressed as the mean ± SD.
Identification of an Nrf2-responsive DNA enhancer element
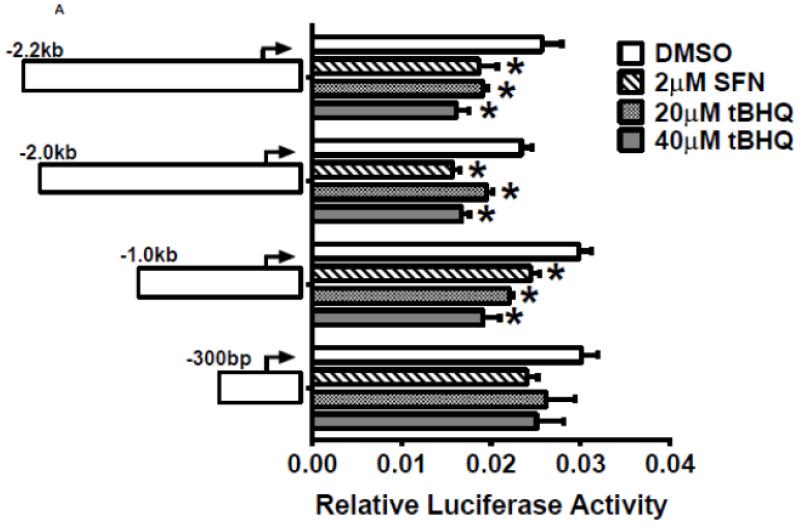
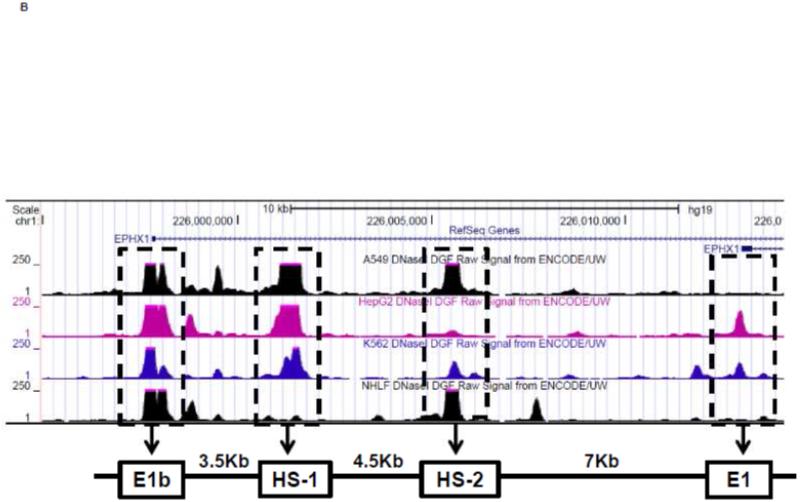
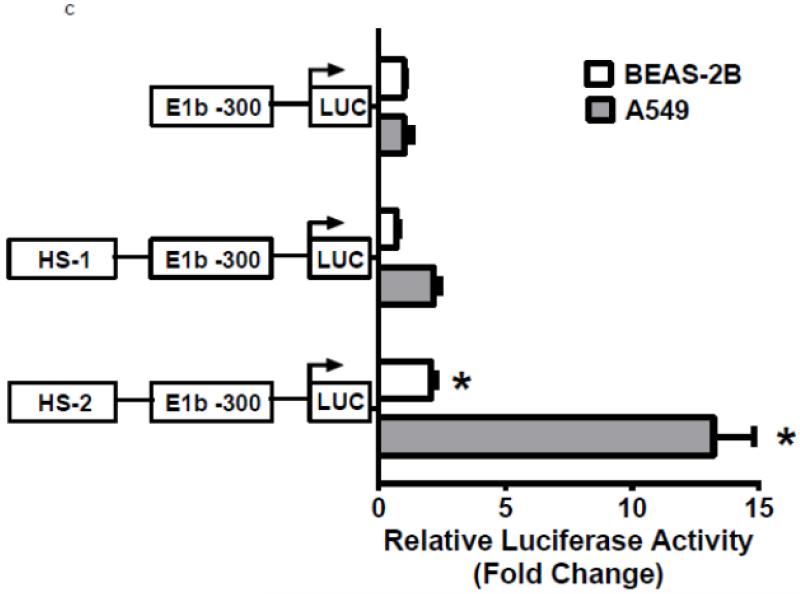
To investigate whether Nrf2 regulatory sites exist in the E1b promoter, we first tested the ability of SFN or tBHQ to activate transcription in a series of truncated E1b promoter-luciferase reporter constructs. Surprisingly, transcriptional activity driven by the E1b promoter was decreased by both SFN and tBHQ in BEAS-2B cells (Figure 3A). The E1b proximal promoter was not able to drive reporter expression in a manner that mimicked the endogenous E1b transcript. Based on these results, we postulated that these E1b promoter regions lack Nrf2 regulatory elements. Thus, we explored the potential for regulatory elements in other regions. Using the UCSC browser (http://genome.ucsc.edu/), genome-wide mapping of DNase I hypersensitive (HS) sites revealed two HS sites (HS-1 and HS-2) located in the 15kb region intervening between the E1b and E1 promoters (Figure 3B). These sites are present in HepG2, normal human lung fibroblasts and other human cell lines. Considering that HS is an indicator of active DNA cis-regulatory elements, we tested if these two elements may restore a more endogenous-like E1b expression pattern to the reporter construct in response to Nrf2 activator. HS-1 and HS-2 were cloned and inserted 5′ of the −300 bp E1b proximal promoter sequence in the pGL4 luciferase reporter plasmid. This promoter was chosen because it yielded the highest basal transcriptional activity in previous studies [13]. In A549 cells, the incorporation HS-1 stimulated transcriptional activity approximately 2-fold, while HS-2 resulted in a 14-fold increase, indicating that both elements were likely functional enhancers (Figure 3C). In BEAS-2B cells, inclusion of HS-1 had no effect, while HS-2 increased transcriptional activity approximately 2-fold. (Figure 3C).
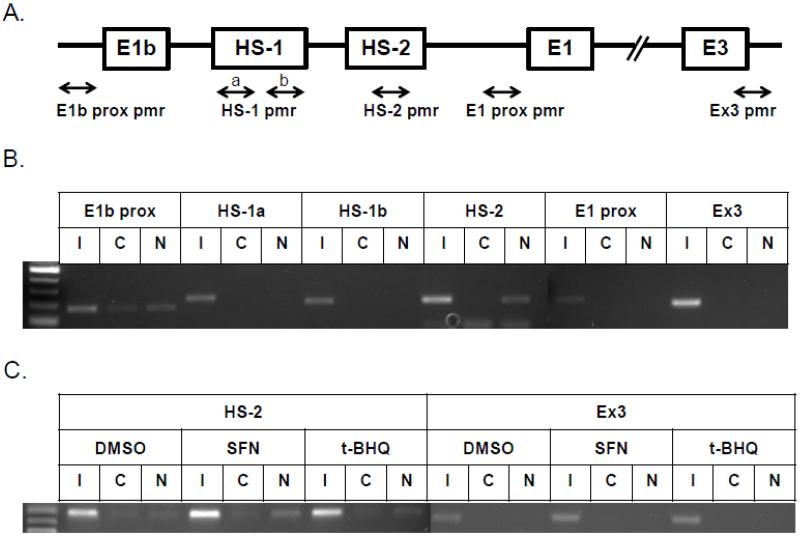
Figure 3. Identification of regulatory elements involved in E1b induction.
A.) Reporter analysis on E1b upstream sequence. BEAS-2B cells were transfected with luciferase reporter constructs driven by E1b promoters containing −300bp, −1.0kb, −2.0kb or −2.2kb promoter region. After 24h treatment with Nrf2 inducers, cells were harvested for luciferase reporter assays. Significant differences from DMSO control were indicated by “*” (p<0.05). B.) A map of the intronic region located between E1b and E1 harboring DNase hypersensitive sites in A549, HepG2, K562 and NHLF cells (modified from the UCSC genome browser). C.) Reporter analysis on DNase I hypersensitive sites. A549 and BEAS-2B cells were transfected with the E1b −300 or E1b DNase I HS/E1b-300 plasmids for 24hr and harvested for luciferase assay. Significant differences from the E1b-300 plasmid were indicated by “*” (p<0.05). All luciferase reporter activity values are expressed as the mean ± SD.
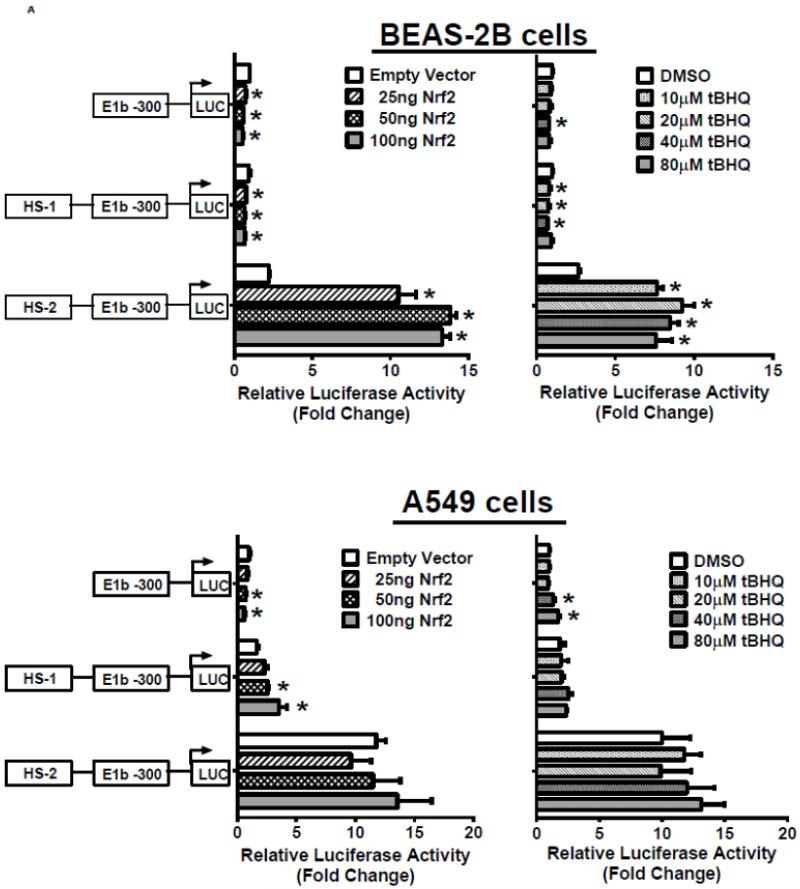
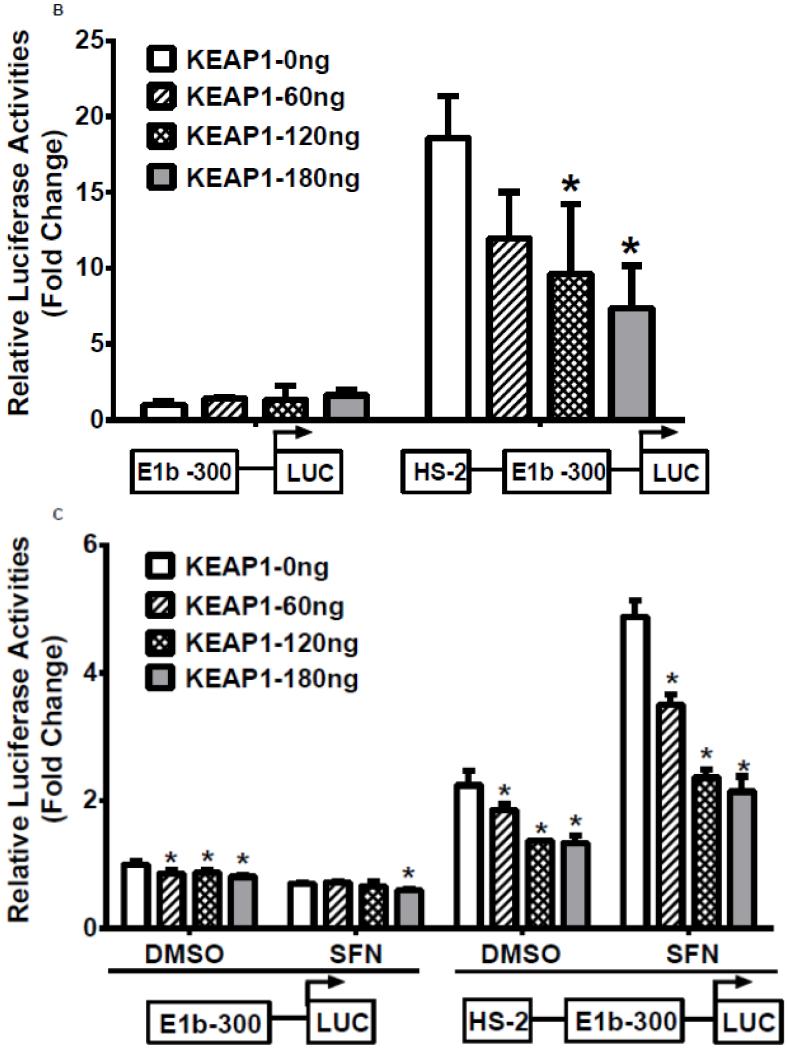
In the next series of experiments, we tested whether the HS-1 and HS-2 DNA elements were responsive to Nrf2 activation. Results of luciferase reporter assays demonstrated that both overexpression of Nrf2 as well as tBHQ treatments produced a significant induction of reporter activity in BEAS-2B cells, and the induction pattern closely resembled expression of endogenous E1b (Figure 4A, upper 2 panels). In contrast, A549 cells did not exhibit any transactivation to Nrf2 overexpression or to tBHQ treatment, which is consistent with the expression profile of E1b transcript in this cell line (Figure 4A, lower panels). Since A549 cells display constitutive Nrf2 activation, we sought to determine whether the high enhancer activity of HS-2 was due to Nrf2 by testing the ability of KEAP1, an Nrf2 inhibitor, to inhibit transactivation. Transient transfection of KEAP1-expression plasmid to A549 cells resulted in significant reduction on the nuclear level of Nrf2 (data not shown). Overexpression of KEAP1 also significantly reduced the enhancer activity in a dose dependent manner, indicating that constitutively activated Nrf2 contributes to increased luciferase activity (Figure 4B). Further, KEAP1 expression abrogated the antioxidant-inducted luciferase activity in BEAS-2B cells (Figure 4C). Based on these results, we projected that the HS-2 enhancer contained a site that confers Nrf2 responsiveness.
Figure 4. Role of DNase HS sites in Nrf2-mediated induction of E1b.
A.) Effect of Nrf2 overexpression or tBHQ treatment on transactivation activity of E1b DNase I HS sites in BEAS-2B and A549 cells. For Nrf2 overexpression, cells were co-transfected with E1b promoter reporter constructs containing HS-1 or HS-2 and varying amounts of Nrf2. Luciferase activity was determined 24 h later. For tBHQ induction experiments, cells were transfected with promoter reporter constructs containing HS-1 or HS-2 and treated with varying amounts of tBHQ 18 h later. Luciferase activity was determined 6 h after treatment. KEAP1 down regulates HS-2 enhancer activity in A549 cells (B) and SFN-treated BEAS-2B cells (C). Cells were co-transfected with the KEAP1 expression plasmid and the reporter vectors containing E1b-300 promoter or E1b DNase I HS-2/E1b-300 promoter for 24 h. A549 cells were harvested for luciferase assay while BEAS-2B cells were treated with 2μM SFN for extra 24 h before luciferase assay. All luciferase values represent mean ± SD. *p<0.05 compared to empty vector controls or DMSO control.
Nrf2 binding to an antioxidant response element (ARE) in HS-2 Enhancer
To investigate Nrf2 binding to HS-2 in vivo, chromatin immunoprecipitation (ChIP) assays were performed. Primers used in ChIP assays were specifically designed to amplify the E1b proximal promoter, the HS regions, the E1 proximal promoter and exon 3 regions (Figure 5A). In A549 cells, an Nrf2 antibody specifically enriched the region containing the HS-2 site in comparison to a control rabbit IgG (Figure 5B). The binding of Nrf2 to the E1b proximal promoter was slightly enhanced over the control IgG. No binding was detected in other regions. In BEAS-2B cells, ChIP assays were performed to determine whether Nrf2 bound to HS-2 in response to Nrf2 activators. As shown in Figure 5C, SFN induced a clear increase in Nrf2 binding to the 2nd enhancer element while tBHQ treatment only produced marginal induction. No binding was detected in the exon 3 region. Taken together, the ChIP analysis indicated an interaction between Nrf2 and HS-2 enhancer element, which further supports a critical role of HS-2 in Nrf2-mediated E1b induction.
Figure 5. Nrf2 binds to HS-2 enhancer element.
A.) Locations of specific primers in mEH gene used for ChIP assay. The binding of Nrf2 to the enhancer regions of human mEH gene was evaluated by ChIP assays in untreated A549 cells (B) and BEAS-2B cells (C) treated with Nrf2 activators for 2 h as described under “Material and Methods”. Cross-linked chromatin was isolated from formaldehyde-fixed A549 and BEAS-2B cells and incubated with anti-Nrf2 or control IgG. Nrf2 binding to E1b and E1 promoters and enhancer regions was analyzed by PCR with specific primers for these regions. I, Input, C, control IgG, N, anti-Nrf2 antibody.
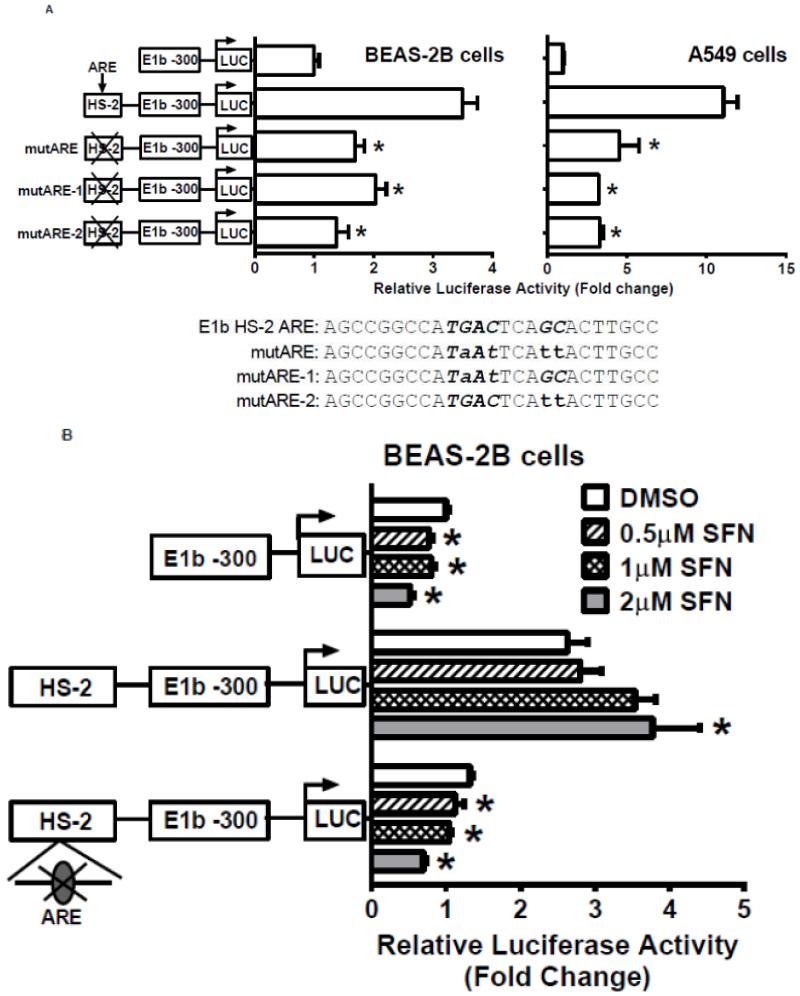
We next used the Genomatix Software Suite (http://www.genomatix.de/solutions/genomatix-software-suite.html) to identify a putative ARE in the HS-2 enhancer element. DNA sequence alignment with known AREs from several human genes confirmed the presence of a perfect ARE (Table 3-1). Site-directed mutagenesis of the ARE site resulted in significantly decreased luciferase reporter activity in the A549 and BEAS-2B cells (Fig. 6A). Interestingly, mutation of either the nucleotides “TGA” at the 5′-end or “GC” at the 3′ end in the core ARE motif abolished the luciferase activity (Figure 6A). The results indicated that both motifs are essential for ARE function. In addition, mutation of the ARE abolished the responsiveness of BEAS-2B cells to SFN treatments (Figure 6B). These data indicate that the ARE within the HS-2 enhancer is essential for not only the enhancer activity, but also for Nrf2-mediated induction of the E1b transcript.
Table 1. Alignment of HS-2 ARE with known AREs.
| ARE | Sequence |
|---|---|
|
| |
| E1b DNaseI HS-2 | AGCCGGCCATGACTCAGCACTTGCC |
| human NQO1 | CAGTCACAGTGACTCAGCAGAATCT |
| human GCLC | CCTCCCCGTGACTCAGCGCTTTG |
| human GCLM | GAAGACAATGACTAAGCAGAAAA |
| ARE core motif | GTGACnnnGC |
| ARE consensus | TMAnnRTGAYnnnGCRwwww |
| AP-1 (TRE) | TGACTCA |
| MAF-recognition element | TGCTGACTCAGCA |
Figure 6. Identification of Antioxidant response element (ARE) within HS-2.
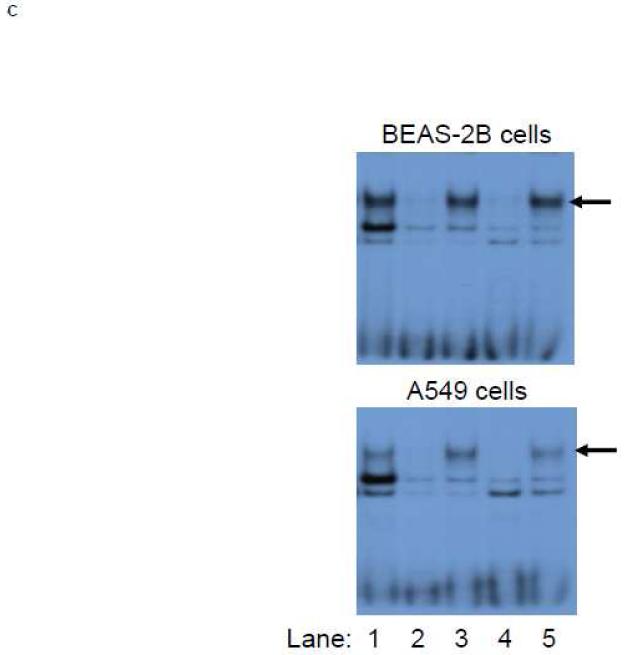
A.) BEAS-2B and A549 cells were transfected with E1b promoter constructs containing HS-2 (wild-type) or HS-2 with mutations in the core ARE motif. Luciferase values were determined 24 h later. *p<0.05 compared to wild-type HS-2 construct. B.) Mutation of the putative Nrf2-binding site influences SFN responsiveness of BEAS-2B cells. Cells were transfected with the reporter vectors containing E1b-300 promoter, E1b DNase I HS-2/E1b-300 promoter, or E1b DNase I mutARE/E1b-300 promoter for 18hr and treated with DMSO or SFN for 6hr before harvested for luciferase assay. All luciferase values represent mean ± SD. *p<0.05 compared to DMSO controls. C.) EMSA analysis of Nrf2 binding to the putative ARE in E1b DNase I HS-2 in SFN-treated BEAS-2B and A549 cells. E1b DNase I HS-2 ARE was end-labeled with [γ-32P]ATP and T4 kinase and incubated with nuclear extracts from untreated A549 cells or BEAS-2B cells treated with 2 M SFN for 2 h. The binding of Nrf2 to E1b DNase I HS-2 ARE was competed with 50× molar excess of cold E1b DNase I HS-2 ARE (Lane 2), mutated E1b DNase I HS-2 ARE (Lane 3), ARE from human NQO1 gene (Lane 4), or mutated NQO1 ARE (Lane 5). The arrows show specific binding.
To verify the specific interaction of Nrf2 with this ARE in the 2nd enhancer, EMSA analysis was performed. Incubation of nuclear extracts prepared from untreated A549 cells and SFN-treated BEAS-2B cells with double stranded oligonucleotides containing this ARE, resulted in three protein-DNA complexes (Figure 6C, lane 1). Unlabeled probe containing the ARE from the 2nd enhancer competed out the first and third complex completely and weakened the intensity of the second complex substantially (Figure 6C, lane 2), whereas a mutant ARE probe was not able to compete with the wild-type probe in the formation of the first complex, but weakened the intensity of the second band (Figure 6C, lane 3). These results suggested that the first complex was specific for Nrf2 binding whereas the two other binding sites were nonspecific. Further, an oligonucleotide containing human NQO1 ARE prevented the formation of the first and second complexes, but not the third (Figure 6C, lane 4), while the mutated NQO1 ARE oligonucleotide had no effect on the first complex formation (Figure 6C, lane 5). In summary, these data strongly suggest a specific Nrf2 interaction with the ARE in the HS-2 element.
Characterization of a TPA-response element overlapping with the ARE within HS-2
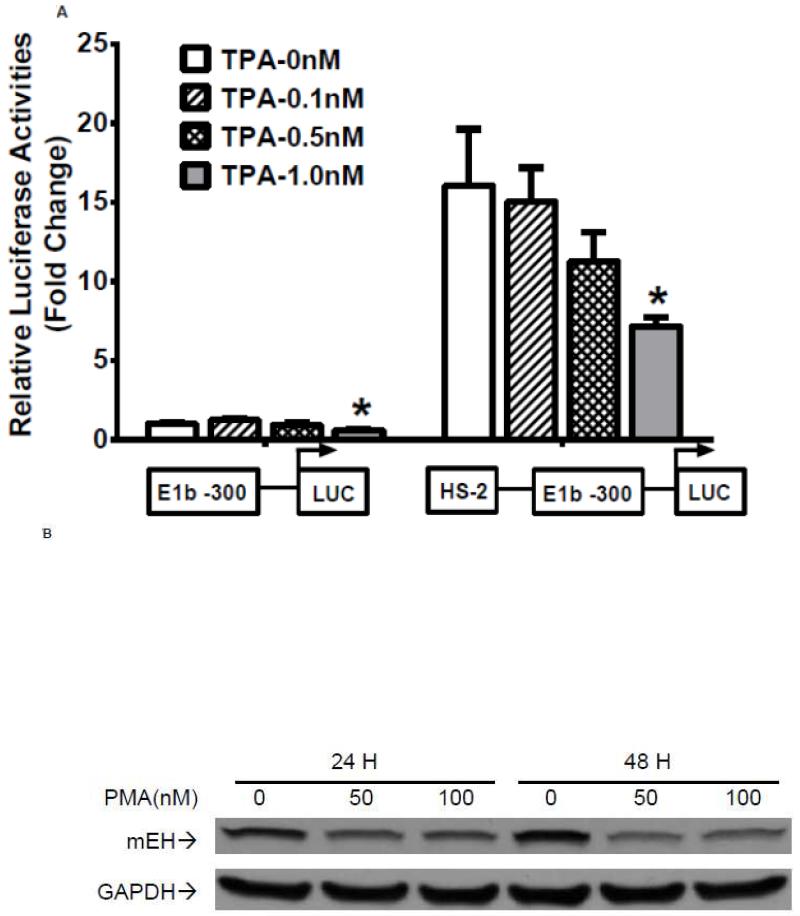
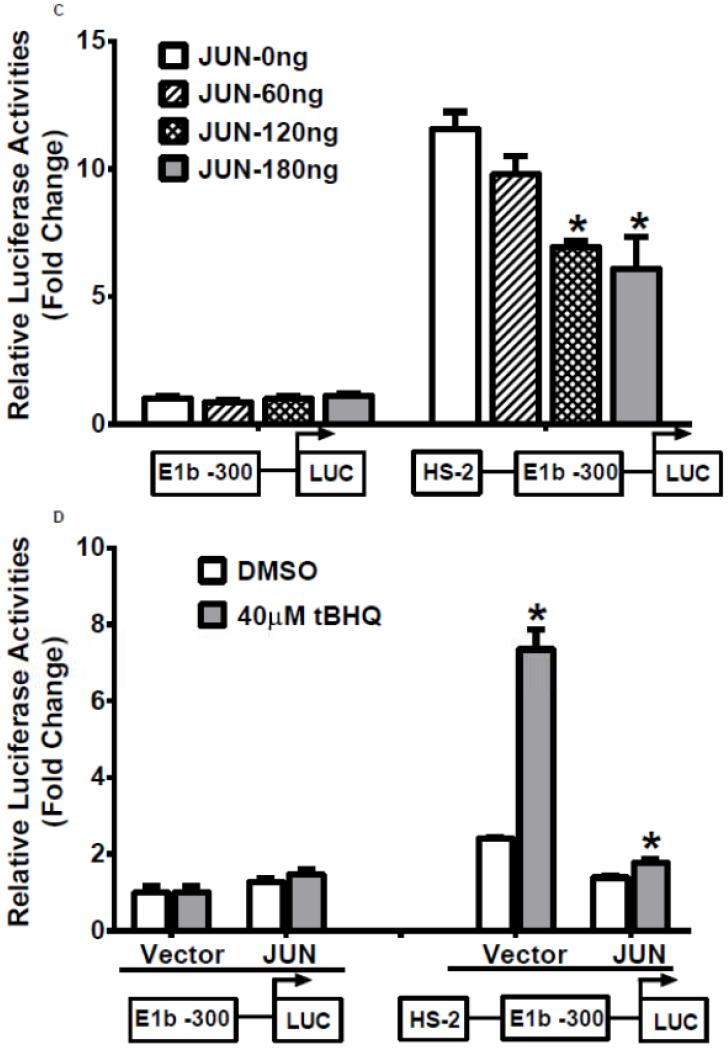
Our analysis of the HS-2 element also revealed a consensus TPA (12-O-tetradecanoylphorbol-13-acetate)-response element (TRE) that overlaps with the ARE site. Thus, we tested if this TRE affects Nrf2-mediated reporter activity in A549 cells. TPA treatments significantly decreased the transactivation of the HS-2 enhancer in a dose-dependent manner (Figure 7A). Furthermore, A549 cells treated with TPA exhibited a decreased level of mEH (Figure 7B). Overexpression of JUN (AP-1), a transcription factor that binds TRE, repressed the transcriptional activity of the HS-2/E1b-300 luciferase reporter plasmid in A549 cells (Figure 7C). In addition, JUN blocked the tBHQ-induced transactivation of the HS-2/E1b-300 reporter construct in BEAS-2B cells (Figure 7D). These data suggested that the binding of JUN to the TRE embedded in the ARE interfered with the binding activity of Nrf2 to its target motif in HS-2 and prevented the up-regulation of E1b promoter activity and E1b expression.
Figure 7. A TRE binding site influences ARE-driven enhancer activity.
A.) TPA treatments down-regulated the enhancer activity in A549 cells. Cells were transfected with the E1b −300 or E1b DNase I HS-2/E1b-300 plasmids for 18hr, treated with TPA for 6hr and harvested for luciferase assay. *p<0.05 compared to DMSO controls. B.) Western blot analysis of mEH expression in A549 cells after treated with TPA for 24 and 48 h. GAPDH was used as a loading control. C.) Regulation of the E1b DNase HS-2 enhancer activity by JUN overexpression in A549 cells. Cells were co-transfected with the JUN expressing plasmid and the reporter vectors containing E1b-300 promoter or E1b DNase I HS-2/E1b-300 promoter. After 24 h, luciferase activity was measured. *p<0.05 compared to empty vector controls. D.) JUN blocks tBHQ induced activation of E1b DNase I HS-2 enhancer activity in BEAS-2B cells. Cells were co-transfected with the JUN expressing plasmid and the reporter vectors containing E1b-300 promoter or E1b DNase I HS-2/E1b-300 promoter for 18 h and treated with DMSO or tBHQ for 6 h before harvested for luciferase assay. *p<0.05 compared to DMSO controls. All luciferase values represent mean ± SD.
Sulforaphane induces mEH expression in human normal primary hepatocytes
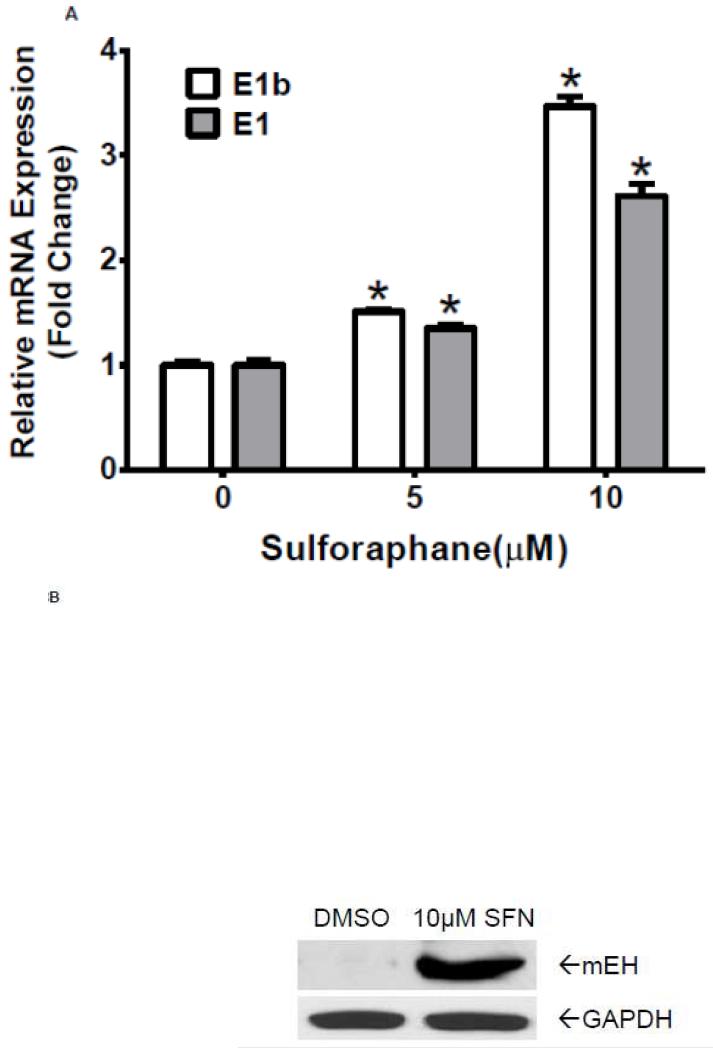
Having demonstrated that antioxidants induce E1b expression in human lung cancer cell lines, we asked if this regulation is seen in normal cells. Normal human primary hepatocytes were treated with sulforaphane at various concentrations for 24 h. Results of quantitative RT-PCR revealed a strong upregulation of E1b and E1 mRNA levels in the primary hepatocytes after sulforaphane treatments (Figure 8A). We also observed that the total mEH protein expression was elevated in these cells following treatment with 10 μM sulforaphane (Figure 8B). These findings are consistent with the results in BEAS-2B cells and suggest that the role of Nrf2 on E1b induction in lung cancer cells holds true for normal primary cells.
Figure 8. Sulforaphane induces mEH expression in human normal primary hepatocytes.
A.) Quantitative RT-PCR analysis of E1b and E1 expression in normal human primary hepatic cells treated with DMSO or Sulforaphane for 24 h. GAPDH was used as an internal control. *p<0.05 compared to DMSO controls. B.) Western blot analysis of mEH expression in normal human primary hepatic cells after treated with DMSO or sulforaphane for 24 h. GAPDH was used as a loading control.
DISCUSSION
Our previous studies characterized the expression of the human mEH transcripts, E1 and E1b, which result from the use of alternative gene promoters that are separated by >18kb. These transcripts are expressed in a tissue-specific manner, yet little is known about their regulation. The results presented in the current study demonstrate a role for the transcription factor Nrf2 in the regulation of the human mEH E1b transcript, which is the predominant transcript detected in all tissues and derived from the far upstream mEH gene promoter. The regulatory elements involved in Nrf2-mediated regulation of E1b transcript are unique in that they are located in the intronic region between the two alternative first exons, and localized downstream of the E1b promoter. A specific HS site identified in the intronic region, HS-2, present in several human cell lines as well as in normal human liver cells, conferred responsiveness to Nrf2 inducers and contained an Nrf2 ARE binding element.
AREs are cis-acting regulatory elements present in the promoter region of cytoprotective genes targeted by Nrf2. Functional AREs contain a core sequence of RTGACnnnGC, where ‘n’ represents any nucleotide [25]. Further characterization of the nucleotides flanking the core ARE sequences demonstrate that the flanking sequences are required for both basal and inducible activity of AREs. Thus, the consensus sequence of AREs was extended to TMAnnRTGAYnnnGCRwwww [26]. Other minor revisions of this consensus sequence have also been suggested [27, 28]. It is notable that some AREs contain a JUN binding site (also known as TPA-response element or TRE, 5′-TGAGTCA-3′). Based on the length of ARE sequences and the presence of a JUN binding site, AREs have been divided into four categories [3]. Class 1 AREs have an extended ARE consensus sequence, as well as an embedded JUN-binding site, while Class 2 AREs have an extended ARE sequence without a JUN site. Class 3 and 4 AREs have a minimal core ARE sequence. The difference between Class 3 and 4 is that a JUN site is present in Class 3 AREs, but not in Class 4 AREs. The intronic ARE of mEH identified in this study is a Class 3 ARE which contains only minimal core sequence and an embedded JUN site.
AREs in different classes display differential responses to chemopreventive agents. Class 1 and 2 AREs are likely more responsive than those in Class 3 and 4 [3]. These classifications are consistent with the effects of the antioxidant treatments noted here, inducing a moderate ~2-4 fold change on mEH gene transcription, characteristic of a Class 3 ARE. The embedded JUN binding sites in Class 1 and 3 AREs may also respond to other factors that activate JUN, such as UV radiation and TPA. Indeed, we found that JUN overexpression in A549 cells attenuated the transactivation activity of the HS-2 enhancer in luciferase reporter assays. Therefore, further study of mEH regulation by bZIP transcriptional factors and AP-1 activators through this enhancer element is warranted. Although site-directed mutagenesis of the ARE in HS-2 completely abolished its responsiveness to Nrf2 activators, the mutated ARE still enhanced the promoter activity of E1b proximal promoter. These results indicate that other transcription factors likely bind to the HS-2 region and confer transcriptional enhancement of the E1b proximal promoter. Further studies are needed to explore this possibility.
Thus far, identified AREs have been localized to upstream regions of transcriptional start sites [3, 29]. Interestingly, the HS-2 ARE identified in this study is located in the intronic region between the E1b and E1 promoters, and downstream of the E1b promoter that it appears to regulate. The large distance (~7kb) separating the HS-2 from the E1b transcription start site suggests that an intrachromosomal interaction resulting from chromatin looping is involved in promoting the transcriptional activation of E1b mediated by Nrf2. It is well established that chromatin looping functions to bring transcription factors that bind distal regulatory elements into close proximity to the general transcription machinery at transcription start sites [30]. This process enables the distal response element and its associated transcription factors to direct the transcription activity of proximal promoter. Many studies have demonstrated interactions between promoters and intronic regulatory elements [31]. In these latter studies, the Chromatin Conformation Capture (3C) assay was an essential tool used to dissect this mechanism [32], and laboratory efforts are currently focused on pursuing this technology to define the exact nature of these interactions within the context of mEH gene regulation.
The intronic enhancers in human mEH gene identified here are likely important drivers of the transcriptional activity exhibited by the E1b proximal promoter. In the proximal and core promoters of E1b, we characterized putative binding sites for Sp1/Sp3 [33]. In other reports, Sp1 has been shown to regulate chromatin looping between intronic enhancers and upstream promoter regions of several genes [31, 34]. Thus, we speculate that Sp1 participates in the formation of a chromatin loop between the intronic enhancers and the E1b proximal promoter by interacting with Nrf2 or other factors binding to these enhancers. However, other E1b promoter-binding factors may also mediate the DNA looping interaction. Therefore, although the factors contributing to the E1b chromatin looping remain to be elucidated, they represent unique targets as E1b expression modulators that may in turn be regulated through the action of xenobiotics at this distal enhancer.
Nrf2 is a therapeutic target for cancer chemoprevention [35]. Nrf2 induces the expression of cytoprotective genes, including NAD(P)H:quinone oxidoreductase-1, several UDP-glucuronosyl and glutathione transferases, heme oxygenase-1, multidrug resistance-associated proteins and others [36]. Nrf2 is essential in protecting cells from the toxic and carcinogenic effects of electrophiles, oxidants and inflammatory agents, as it has been demonstrated that Nrf2-deficient mice are more sensitive to oxidative stress and reactive electrophiles than their wild type counterparts [37]. The protective role of Nrf2 has also been established in many type of diseases, including cancers, Alzheimer’s and Parkinson’s diseases, chronic obstructive pulmonary disease, asthma, diabetes, and inflammatory diseases [38]. However, recent clinical studies using lung tumor tissues and cell lines demonstrate that Nrf2 is excessively expressed due to mutations in KEAP1 or Nrf2 [4-6]. Further, enhanced Nrf2 activity promotes cancer cell survival and contributes to chemo- and radio-resistance [38]. Therefore, the consequences of Nrf2 activation in carcinogenesis are biological context-dependent. In these respects, Nrf2 appears as s clinical target to direct the inhibition of tumor growth and for enhancing the efficacy of anti-cancer drugs.
Similarly, mEH functions in a dual role in the metabolism of xenobiotics. On one hand, mEH activity is pivotal in the protection against toxicities elicited by reactive epoxide intermediates. On the other, for example in the metabolism of polycyclic aromatic hydrocarbons, mEH activity facilitates the generation of DNA adducts and the subsequent processes of mutagenesis and cancer development. In these respects, mEH enzymatic activities contribute toward a delicate balance in xenobiotic metabolism, determining both cellular defenses against xenobiotic exposures as well as chemical bioactivation. Selective activation, or alternatively, inhibition, of mEH activities, perhaps through Nrf2 targeting, may provide a strategy for the prevention or treatment of toxicities resulting from certain xenobiotic exposures. Further detailed mechanistic study of human mEH gene regulation may enable the needed insight for fine tuning mEH expression to favor its protective functions.
To fully understand the consequence of mEH up-regulation by Nrf2 activation in human cancers, it will be necessary to establish a model that closely mimics the in vivo human situation. In this regard, the current rodent models available are not suitable for human predictive study in that the respective rodent mEH regulatory regions do not conserve the human alternative gene promoter regulatory features or DNA structures [14]. Specifically with respect to the current investigation, the presence of the two functional enhancers identified are apparently not maintained in rodents and thus contribute selectively to human-specific responses to transcriptional modulators, such as exposures to antioxidant chemicals as described here. For example, previous reports have indicated that rodents often exhibit high mEH inducibility following exposures to prototypical enzymatic inducers such as phenobarbital and polyaromatic hydrocarbons [39], whereas human primary hepatocytes from multiple donors exhibit only modest response to the same agents [40]. Thus, there is a strong rationale for developing an appropriate humanized mEH rodent model for use in better characterizing the contributions of mEH as a determinant of biological, toxicological and carcinogenic responses arising from xenobiotic exposures.
In summary, the results of the studies presented demonstrate that Nrf2 contributes a critical role in the antioxidant mediated transcriptional regulation of human mEH, through the gene’s distal enhancer region. The data obtained from these investigations provide new insights regarding the nature of human mEH gene regulation, its modulation by xenobiotic agents and the identification of Nrf2 in the context of human mEH as a potential therapeutic target for cancer chemoprevention.
Highlights.
Human microsomal epoxide hydrolase is transcribed from alternative gene promoters
Nrf2 contributes as a critical transcriptional regulator of the E1b transcript
A novel intronic enhancer element for Nrf2 is identified
Nrf2 regulation drives mEH transcription in both lung cells and primary hepatocytes
ACKNOWLEDGEMENTS
This work was supported by a USPHS grant from the National Institute of Environmental Health Sciences, ES016358. The authors are appreciative of Dr. Elizabeth Laurenzana for providing critical review and discussion
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Hong WK, Sporn MB. Recent advances in chemoprevention of cancer. Science. 1997;278:1073–1077. doi: 10.1126/science.278.5340.1073. [DOI] [PubMed] [Google Scholar]
- [2].Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- [4].Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 Function Activates Nrf2 and Provides Advantages for Lung Cancer Cell Growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- [5].Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang X-J, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fretland AJ, Omiecinski CJ. Epoxide hydrolases: biochemistry and molecular biology. Chem. Biol. Interact. 2000;129:41–59. doi: 10.1016/s0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- [9].Decker M, Arand M, Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch. Toxicol. 2009;83:297–318. doi: 10.1007/s00204-009-0416-0. [DOI] [PubMed] [Google Scholar]
- [10].Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu. Rev. Pharmacol. Toxicol. 2005;45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920. [DOI] [PubMed] [Google Scholar]
- [11].Miyata M, Kudo G, Lee YH, Yang TJ, Gelboin HV, Fernandez-Salguero P, Kimura S, Gonzalez FJ. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J. Biol. Chem. 1999;274:23963–23968. doi: 10.1074/jbc.274.34.23963. [DOI] [PubMed] [Google Scholar]
- [12].Kiyohara C, Yoshimasu K, Takayama K, Nakanishi Y. EPHX1 Polymorphisms and the Risk of Lung Cancer: A HuGE Review. Epidemiology. 2006;17:89–99. doi: 10.1097/01.ede.0000187627.70026.23. [DOI] [PubMed] [Google Scholar]
- [13].Liang SH, Hassett C, Omiecinski CJ. Alternative promoters determine tissue-specific expression profiles of the human microsomal epoxide hydrolase gene (EPHX1) Mol. Pharmacol. 2005;67:220–230. doi: 10.1124/mol.104.005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang X, Liang SH, Weyant DM, Lazarus P, Gallagher CJ, Omiecinski CJ. The expression of human microsomal epoxide hydrolase is predominantly driven by a genetically polymorphic far upstream promoter. J. Pharmacol. Exp. Ther. 2009;330:23–30. doi: 10.1124/jpet.109.150870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu X, Roberts JR, Apopa PL, Kan YW, Ma Q. Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol. Cell. Biol. 2006;26:940–954. doi: 10.1128/MCB.26.3.940-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol. Med. 2001;7:135–145. [PMC free article] [PubMed] [Google Scholar]
- [17].Ma Q, Battelli L, Hubbs AF. Multiorgan Autoimmune Inflammation, Enhanced Lymphoproliferation, and Impaired Homeostasis of Reactive Oxygen Species in Mice Lacking the Antioxidant-Activated Transcription Factor Nrf2. Am. J. Pathol. 2006;168:1960–1974. doi: 10.2353/ajpath.2006.051113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reisman SA, Yeager RL, Yamamoto M, Klaassen CD. Increased Nrf2 Activation in Livers from Keap1-Knockdown Mice Increases Expression of Cytoprotective Genes that Detoxify Electrophiles more than those that Detoxify Reactive Oxygen Species. Toxicol. Sci. 2009;108:35–47. doi: 10.1093/toxsci/kfn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated Genes Induced by the Chemopreventive Agent Sulforaphane by Oligonucleotide Microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- [20].Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong A-NT. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- [21].Lee J-M, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related Factor-2-dependent Genes Conferring Protection against Oxidative Stress in Primary Cortical Astrocytes Using Oligonucleotide Microarray Analysis. J. Biol. Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- [22].Goyak KM, Johnson MC, Strom SC, Omiecinski CJ. Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol. Appl. Pharmacol. 2008;231:216–224. doi: 10.1016/j.taap.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laurenzana EM, Chen T, Kannuswamy M, Sell BE, Strom SC, Li Y, Omiecinski CJ. The Orphan Nuclear Receptor DAX-1 Functions as a Potent Corepressor of the Constitutive Androstane Receptor (NR1I3) Mol. Pharmacol. 2012;82:918–928. doi: 10.1124/mol.112.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Auerbach SS, Stoner MA, Su S, Omiecinski CJ. Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3) Mol. Pharmacol. 2005;68:1239–1253. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- [26].Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Erickson AM, Nevarea Z, Gipp JJ, Mulcahy RT. Identification of a Variant Antioxidant Response Element in the Promoter of the Human Glutamate-Cysteine Ligase Modifier Subunit Gene. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- [28].Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochemical Journal. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim. Biophys. Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Deshane J, Kim J, Bolisetty S, Hock TD, Hill-Kapturczak N, Agarwal A. Sp1 regulates chromatin looping between an intronic enhancer and distal promoter of the human heme oxygenase-1 gene in renal cells. J. Biol. Chem. 2010;285:16476–16486. doi: 10.1074/jbc.M109.058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miele A, Dekker J. Mapping Cis - and Trans - Chromatin Interaction Networks Using Chromosome Conformation Capture (3C) 2009. pp. 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Su S, Omiecinski CJ. Sp1 and Sp3 transcription factors regulate the basal expression of human microsomal epoxide hydrolase (EPHX1) through interaction with the E1b far upstream promoter. Gene. 2014;536:135–144. doi: 10.1016/j.gene.2013.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. USA. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kwak M-K, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharmacol. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Klaassen CD, Reisman SA. Nrf2 the rescue: Effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010;244:57–65. doi: 10.1016/j.taap.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Slocum S, Kensler T. Nrf2: control of sensitivity to carcinogens. Arch. Toxicol. 2011;85:273–284. doi: 10.1007/s00204-011-0675-4. [DOI] [PubMed] [Google Scholar]
- [38].Zhang DD. The Nrf2-Keap1-ARE signaling pathway: The regulation and dual function of Nrf2 in cancer. Antioxid. Redox Signal. 2010;13:1623–1626. doi: 10.1089/ars.2010.3301. [DOI] [PubMed] [Google Scholar]
- [39].Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog. Lipid Res. 2005;44:1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- [40].Hassett C, Laurenzana EM, Sidhu JS, Omiecinski CJ. Effects of Chemical Inducers on Human Microsomal Epoxide Hydrolase in Primary Hepatocyte Cultures. Biochem. Pharmacol. 1998;55:1059–1069. doi: 10.1016/s0006-2952(97)00679-5. [DOI] [PubMed] [Google Scholar]