Abstract
The TZM-bl assay measures antibody-mediated neutralization of HIV-1 as a function of reductions in HIV-1 Tat-regulated firefly luciferase (Luc) reporter gene expression after a single round of infection with Env-pseudotyped viruses. This assay has become the main endpoint neutralization assay used for the assessment of preclinical and clinical trial samples by a growing number of laboratories worldwide. Here we present the results of the formal optimization and validation of the TZM-bl assay, performed in compliance with Good Clinical Laboratory Practice (GCLP) guidelines. The assay was evaluated for specificity, accuracy, precision, limits of detection and quantitation, linearity, range and robustness. The validated manual TZM-bl assay was also adapted, optimized and qualified to an automated 384-well format.
Keywords: Neutralizing Antibody, Assay Validation, HIV, TZM-bl cells, GCLP
1.0 Introduction
The process of assay optimization and validation provides assurance that assay results are as reliable as possible and facilitate compliance with Good Clinical Laboratory Practice (GCLP) (Stiles et al.; Ezzelle et al., 2008; Sarzotti-Kelsoe et al., 2009) and the acceptance of data by regulatory agencies. Here we describe the formal optimization and validation of the TZM-bl assay as it is currently performed in the Laboratory for AIDS Vaccine Research and Development at Duke University Medical Center (Duke Laboratory). This assay is widely used for standardized assessments of vaccine-elicited neutralizing antibodies, for studies of monoclonal antibodies and the neutralizing antibody response in HIV-1 infected people, as well as for studies of SIV and SHIV infected non-human primates. The assay measures neutralization as a function of reductions in HIV-1 Tat-regulated firefly luciferase (Luc) reporter gene expression after a single round of infection with Env-pseudotyped viruses (Montefiori, 2009).
Assay optimization determines how a range of test conditions affect assay parameters and performance. Optimization data, along with scientific judgment, are used to set the acceptance criteria for assay validation. Assay validation provides documented evidence that the method is operating accurately, consistently, is sensitive enough for its intended application and it is suitable for its intended purpose, i.e. the method is “fit for purpose”. Parameters addressed by validation include specificity, precision, linearity, range, accuracy, limit of detection, limit of quantitation, and robustness (Guideline, 2010).
The validated TZM-bl assay was formally transferred to multiple laboratories around the world (Ozaki et al., 2012), preceded by one laboratory, the Vaccine Research Center at the NIH (USA), National Institute of Allergy and Infectious Disease (NIAID) Vaccine Immune T-Cell and Antibody Laboratory (NVITAL), which conducted independent assay validation using the prospectively established acceptance criteria based upon the validation data generated by the Duke Laboratory. The ultimate goal of such independent validation was to demonstrate assay strength and reproducibility, and to perform a further qualification of the assay using an automated 384-well format, designed to provide high throughput results for clinical trial testing. Both laboratories operated in compliance with GCLP guidelines. Combined results of the validation of the manual TZM-bl assay from both Duke Laboratory and NVITAL are presented here, together with the qualification of the assay using an automated 384-well format.
2.0 Materials and Methods
Many of the methods described in this report have been published previously (Montefiori, 2009). Detailed protocols and other supporting materials may be found at http://www.hiv.lanl.gov/content/nab-reference-strains/html/home.htm. The purpose of this report is to describe the key initial elements that went into the formal optimization and validation of the TZM-bl assay and is not meant to be an exhaustive summary of all optimization and validation experiments that have been performed.
2.1 Cell Lines
TZM-bl cells (also called JC53BL-13) were obtained from the NIH AIDS Research and Reference Reagent Program (Cat. no. 8129). The TZM-bl cell line is derived from a HeLa cell clone that was engineered to express CD4, CCR5 and CXCR4 (Platt et al., 1998) and to contain integrated reporter genes for firefly Luc and E. coli β-galactosidase under the control of an HIV-1 long terminal repeat (Wei et al., 2002), permitting sensitive and accurate measurements of infection. The cells are highly permissive to infection by most strains of HIV, SIV and SHIV, including primary or molecularly cloned viral isolates and molecularly cloned Env-pseudotyped viruses. The 293T/17 cell line was obtained from the American Type Culture Collection (catalog no. 11268).
2.2 Culture conditions
TZM-bl and 293T/17 cell lines were maintained in Dulbecco's Modified Eagle's Medium with L-glutamine, sodium pyruvate, glucose, pyridoxine and 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (Gibco BRL Life Technologies) containing 10% heat-inactivated fetal bovine serum (FBS) and 50 μg gentamicin/ml in vented T-75 culture flasks (Corning Costar). Hereafter this complete medium is referred to as growth medium. Cells were incubated at 37°C in a humidified 5% CO2/95% air environment. Unless otherwise specified, all incubations were carried out under these conditions. Cell monolayers were split 1:10 at confluence by treatment with 0.25% trypsin, 1 mM Ethylenediaminetetraacetic acid (EDTA) (Invitrogen) as described (Montefiori, 2009).
2.3 Preparation and Titration of Env-pseudotyped viruses
Env-pseudotyped viruses were prepared by transfecting exponentially dividing 293T/17 cells (5 × 106 cells in 15 ml growth medium in a T-75 culture flask) with 5 μg of rev/env expression plasmid and 10 μg of an env-deficient HIV-1 backbone vector (pSG3Δenv), using PolyFect transfection reagent (QIAGEN) or FuGENE 6 reagent (Promega, USA) in growth medium, as described by the manufacturer. The transfection complexes were allowed to incubate for 30 minutes at room temperature (18°-25°C), after which time they were added to 293T/17 cells and incubated for 48-72 hrs with one change of medium after the first 3-8 hrs. Virus-containing culture fluid was removed from the flasks and filtered through a 0.45μm filter to eliminate cell debris. FBS was added to a final concentration of 20% before storing the virus-containing culture fluid at ≤-70°C in 1.0 ml aliquots in 1.5 ml sterile screw cap polypropylene vials. Env-pseudotyped virus stocks were titrated by performing serial 5-fold dilutions in quadruplicate in growth medium in 96-well culture plates (11 dilution steps total). Freshly trypsinized TZM-bl cells (10,000 cells in 100 ul volume) were added to each well in growth medium containing an optimized concentration of DEAE-dextran (described in the Results section 3.1). The culture plates were incubated for 48 hrs. Virus-induced syncytium formation and cell killing were monitored by microscopic examination. Culture fluid (100 μl) was removed from each well and replaced with a Luc reporter gene assay system reagent (Britelite, PerkinElmer or Brite-Glo, Promega, used as per manufacturer's recommendation). After a 2-min incubation at room temperature to allow cell lysis, 150 μl of cell lysate was transferred to 96-well black solid plates (Corning-Costar) for measurements of luminescence using a Victor 2 luminometer (Perkin-Elmer Life Sciences, Shelton, CT). The recommended virus dilution to use in the TZM-bl assay (expressed as RLU equivalent) was calculated as described in the Results section 3.1 to ensure a standardized virus dose.
2.4 Serologic reagents
All human sera/plasma were obtained with the approval of the Duke University Medical Center Institutional Review Board (IRB) and/or the Intramural IRB of the NIAID, following U.S. Department of Health and Human Services guidelines for conducting clinical research. Blood bank samples were obtained from de-identified donors. Plasma samples were obtained from blood collected using either EDTA or acid-citrate-dextrose (ACD) as anti-coagulants. Serum samples were collected from whole blood that was allowed to coagulate overnight at 4°C. Plasma and serum samples were stored at ≤-70°C. Unless otherwise specified, plasma and serum samples were heat-inactivated at 56°C for 1 h prior to use. Human monoclonal antibod ies (mAbs) 2G12, 2F5, and 4E10 were purchased from Polymun, Inc. (Vienna, Austria). TriMab is a mixture of three mAbs (IgG1b12, 2G12, and 2F5) prepared as a 1mg/ml stock solution containing 333 μg of each mAb in phosphate-buffered saline. Recombinant sCD4 comprising the fulllength extracellular domain of human CD4 produced in Chinese hamster ovary cells was obtained from Progenics Pharmaceuticals, Inc. (Tarrytown, NY). Purified IgG (50 mg/ml in 0.9% saline) from a pool of HIV-1-positive plasma samples (HIVIG) was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID (Catalog no. 3957).
2.5 Sample Pre-Screening
Serum and plasma samples from patients taking antiretroviral drugs often contain residual amounts of the drugs that can inhibit HIV-1 Env-pseudotyped viruses, causing reductions in the RLUs that could be mistaken for neutralizing antibody activity. Specifically, nucleoside and non-nucleoside reverse transcriptase (RT) inhibitors can prevent Tat-induced Luc reporter gene expression in TZM-bl cells. As a control, Env-pseudotyped viruses containing the Env of murine leukemia virus (MLV) but made with the same backbone vector as all HIV-1 Env-pseudotyped viruses (MLV/pSG3Δenv) was used to screen all HIV-1-positive sera and plasma. Since MLV Env-pseudotyped virus contains an irrelevant Env, HIV-1 neutralizing antibodies in sera should not exhibit any reduction in RLU. Activity against the MLV Env-pseudotyped virus is attributed to the presence of drugs blocking the activity of RT expressed by the backbone vector. HIV-1 positive sera and plasma used in these studies were pre-screened by this technique to confirm the absence of MLV Env-pseudotyped virus-specific response.
2.6 General format for the TZM-bl assay
Neutralizing antibodies were measured as reduction in firefly Luc reporter gene expression after a single round of virus infection in TZM-bl cells as described previously (Montefiori, 2009). This assay is a modified version of the assay used by Wei et al. (Wei et al., 2002). Serologic reagents to be tested for neutralizing activity were serially diluted in 96-well flat-bottom culture plates containing growth medium, followed by the addition of Env-pseudotyped virus that was previously titrated for optimal infectivity. Freshly trypsinized TZM-bl cells in growth medium, containing an optimized concentration of DEAE-dextran, were added to each well. One set of eight control wells received cells plus virus (virus control), and another set of eight wells received cells only (background control). Following a 48-hr incubation, 150 μl of culture medium was removed from each well and replaced with a Luc reporter gene assay system reagent. After a 2-min incubation at room temperature to allow cell lysis, 150 μl of cell lysate was transferred to 96-well black solid plates for measurements of luminescence using a Victor 2 luminometer (Perkin-Elmer). The 50% and 80% inhibitory doses (ID50 and ID80) were defined as the reciprocal of the serologic reagent dilution or sample concentration (IC50 and IC80 in the case of sCD4 and mAbs) that caused either a 50% or 80% reduction in RLU compared to RLU values from virus control wells after subtraction of background RLU (cell control wells).
2.7 Flow Cytometry
Cell surface expression of CD4, CCR5 and CXCR4 was monitored over time by flow cytometry. Cells were stained for CD4, CCR5 and CXCR4 surface expression by using CD4-PE (Cat. No. 555347), CD195-PE (Cat. No. 556042) and CD184-PE (Cat. No. 555974), respectively. Mouse IgG1, κ PE (Cat. No. 556650) was used for background staining in the case of CD4. Rat IgG2a, κ PE (Cat. No. 559317) was used for background staining of CCR5 and CXCR4. All antibodies were purchased from BD/Pharmingen. Approximately 10,000 events were acquired by using a FACSCalibur flow cytometer.
2.8 Automated TZM-bl Assay
Fully automated HIV neutralization assays were performed on an automated workstation consisting of a Beckman Coulter Biomek FX liquid handling system equipped with both 96 and 384 pipette tip heads, Thermo Cytomat Ambient Hotel, Thermo Cytomat 2-8°C Incubator, Thermo Cytomat 37°C Incubator, and Molecular Devices Paradigm Multi-mode detection platform with a luminescence cartridge. All instruments were integrated and operated with Beckman SAMI software. The 384-well Automated Workstation System is designed to perform sample dilution and addition, virus addition, and target cell addition, which are the initial TZM-bl assay procedures. The system is also designed to perform removal of supernatant, addition of substrate, and luminescence measurement, which are the final TZM-bl assay procedures. The standard assay was optimized on this platform in a 384 well plate configuration requiring subsequent modifications of the sample/reagent volumes, as detailed in Table 1.
Table 1. Assay Variations between Manual and Automated Assay.
| Assay Steps/Methods | Manual Assay (96-well plate) | Automated Assay (384-well plate) |
|---|---|---|
| Final volume (μl): | 250 | 80 |
| Sample volume (μl): | 100 | 30 |
| Virus volume (μl): | 50 | 30 |
| Cell density (number / μl): | 10,000 / 100 μl | 3,000 / 20 μl |
| Stock sample volume for 1:10 dilution for 10 viruses (μl): | 440 | 120 |
| Supernatant Removal (μl): | 150 | 50 |
| Substrate Addition (μl): | 100 | 30 |
| No. of sample per 8-plate run | 40 | 160 |
2.9 Statistical Analysis
All inhibitory values were calculated using a formally validated Excel-based macro or web-based neutralizing antibodies tool (Piehler et al., 2011) that utilizes average virus and cell control RLU values as well as replicate test well RLU values to calculate the neutralizing antibody titer as a function of the reduction of luciferase reporter gene expression. All means, standard deviations, and R2 values were calculated using Microsoft Excel formulas. Wilcoxon signed-rank tests were used to assess if neutralization titers against viruses differed between HIV-1-positive plasma pools. For each of the 17 Env-pseudotyped viruses, Wilcoxon rank sum tests were used to assess if neutralization titers differed between vaccine recipients and control subjects. All p-values were adjusted using the Bonferroni method to account for the large number of tests, and adjusted p-values of 0.05 indicated statistical significance. Statistical analysis of the NVITAL results was performed using SAS (version 9.1) software.
3.0 Results
3.1 Optimization of the TZM-bl Assay
3.1.1 Optimization of DEAE-dextran
In order to achieve adequate levels of infection, it is often necessary to supplement the assay medium with the polycation, DEAE-dextran. We first determined the minimum concentration of DEAE-dextran to maximize virus infectivity without causing cell toxicity. To test for cell toxicity, freshly trypsinized TZM-bl cells (10,000/100 μl/well) were added directly to multiple concentrations of DEAE-dextran in growth medium in 96-well plates (triplicate wells for each concentration of DEAE-dextran tested). Cell viability was assessed by neutral red staining (Montefiori et al., 1988) at DEAE-dextran doses of 2.5 - 80 μg/ml. All concentrations were non-toxic after two days of incubation (data not shown).
Next we determined the minimum concentration of DEAE-dextran needed for maximum levels of infectivity in TZM-bl cells. Four different Env-pseudotyped viruses were tested for infectivity in the presence and absence of DEAE-dextran at doses of 5 - 40 μg/ml. Equal amounts of each virus were added to each dose of DEAE-dextran. RLU were measured 48 hrs later. Peak infectivity in all four cases occurred at 20 - 40 μg/ml (data not shown). We chose 30 μg/ml as a standard dose for future assays with this batch of DEAE-dextran. Subsequently we noticed that each new batch of DEAE-dextran needed to be titrated to determine the optimal concentration for infectivity.
We note that addition of a polycation is not always necessary to achieve adequate levels of infection for measurements of neutralization. Use of a polycation permits the use of a larger number of Env-pseudotyped viruses. It also provides a dose-sparing effect for all Env-pseudotyped virus stocks.
3.1.2 Optimal TZM-bl cell number for infectivity
To determine the optimal inoculum density of TZM-bl cells for linear measurements of infection, freshly trypsinized cells were added at various densities to a 96-well plate containing equal amounts of Env-pseudotyped virus QH0692.42. Infection increased linearly over a range of 17 to 37,037 cells/well (Fig. 1). A sharp decrease in infection was observed at higher cell numbers. Background RLU in cell control wells also diminished at high cell densities, suggesting either a loss of cell viability or that density-dependent down-regulation of Luc expression had occurred. Based upon these, and results detailed later in this paper, 10,000 cells/100 μl well were selected as the standard assay density.
Figure 1. Linear range of input TZM-bl cell numbers that support HIV-1 infection.
DEAE-dextran was present at 30 μg/ml. Luminescence was measured in solid black plates after 48 hrs.
3.1.3 Linear relationship between virus inoculum size and RLU
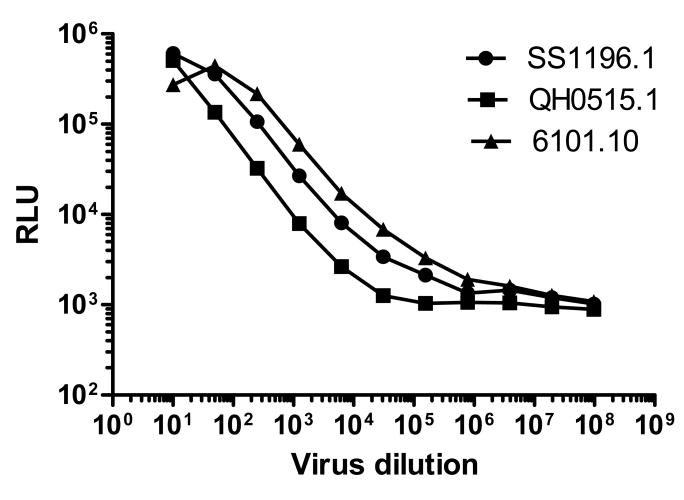
Having defined an optimal number of cells to use for inoculation, we next determined the linear relationship between input virus dose and RLU. Freshly trypsinized TZM-bl cells (10,000/100 μl/well) were added to serially diluted Env-pseudotyped viruses in 96-well plates. Eleven 5-fold dilutions of virus ranging from 1:10 to 1:97,656,250 were made in quadruplicate. An RLU value of ≥2.5 times background of cell control wells was used to score a positive. Curves were constructed by plotting virus dilutions against corresponding RLU on a log-log scale (Fig. 2). RLU increased in a linear fashion over a wide range of infectious input virus doses. This linear range extended from approximately 2,500 - 500,000 RLU.
Figure 2. Linear range of infection in TZM-bl cells.
DEAE-dextran was present at 30 μg/ml after the addition of cells. Luc activity was quantified by luminescence 48 hrs later.
These curves were revealing in two additional ways. First, although the slopes were similar, the magnitude of RLU for virus strain QH0515.1 was lower than the other two strains, indicating that equal doses of different strains may not be expected to yield equivalent absolute RLU values. Second, RLU values for strain 6101.10 peaked and then declined. This decline in RLU was associated with virus-induced syncytium formation and associated cell-killing effects. We have observed even greater cytopathic effects with other strains of virus at high input doses but these cytopathic effects are rare at input RLU values ≤250,000. Thus, a maximum 250,000 RLU inoculum size avoids unwanted cytopathic effects in most cases.
3.1.4 Optimal viral dose for maximum assay sensitivity
A neutralization assay was designed (Fig. 3) in which different input virus doses of Env-pseudotyped virus SS1196.01 were incubated with serial dilutions of either an HIV-1-positive serum pool, broadly neutralizing antibodies (IgG1b12 and 2G12) or sCD4 in a total volume of 150 μl of growth medium. SS1196.01 was chosen for this experiment because little or no cytopathic effects were observed at relatively high doses of this virus. Each dilution of antibody or sCD4 was tested in triplicate wells of a 96-well plate. Freshly trypsinized TZM-bl cells in 100 μl growth medium containing DEAE-dextran (75 μg/ml; 30 μg/ml final concentration) were added to each well at a density of 10,000 cells/well. One set of 8 control wells received cells only (cell control). Another set of 8 wells received cells and virus but no antibody (virus control). The final volume of control wells was adjusted to 250 μl with growth medium. Luc activity was quantified by luminescence 48 hrs later. Percent reduction in RLU (% neutralization) was determined by calculating the difference in average RLU between test wells (cells + sample + virus) and cell control wells (cells only), dividing this result by the difference in average RLU between virus control (cell + virus) and cell control wells, subtracting from 1 and multiplying by 100.
Figure 3. Neutralization curves in TZM-bl cells at different input virus doses.
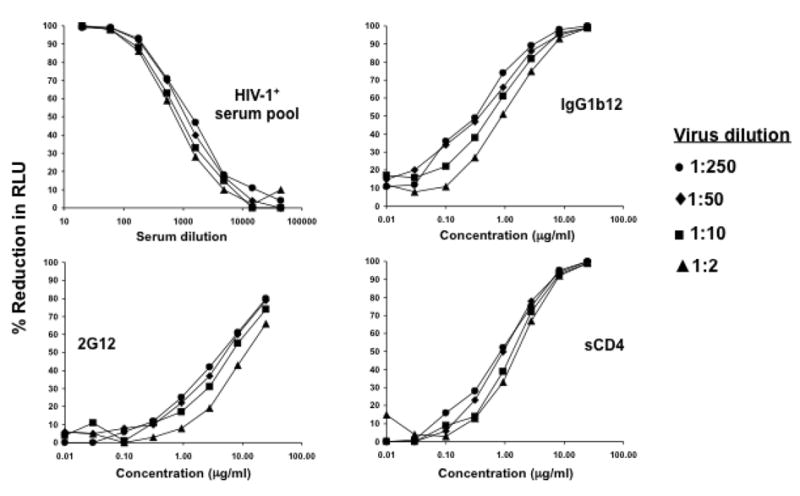
Average RLU in cell control and virus control wells, respectively: 1:250 dilution (926 vs. 11,813; range = 10,887); 1:50 dilution (997 vs. 55,285; range = 54,287); 1:10 dilution (1,059 vs. 242,013; range = 240,954); 1:2 dilution (1,194 vs. 551,651; range = 550,457).
Positive neutralization was detected with all four reagents regardless of input virus dose (Fig. 3). Dose-response curves were linear between approximately 20% and 80% reductions in RLU in all cases. A value of 50% reduction in RLU was chosen to determine inhibitory doses (ID50) because this value was midway in the linear portion of the neutralization curve. The relative potency of each test reagent increased with decreasing input virus, indicating that assay sensitivity is dependent on virus dose, where greater sensitivity is achieved with lower virus doses. However, this effect on assay sensitivity was not dramatic, since the ID50 values of each serologic reagent differed by <3-fold over a 100-fold range of virus doses. Thus, minor differences in the dose of virus used in this neutralization assay are unlikely to alter the results significantly, as long as the virus dose does not cause unwanted cytopathic effects.
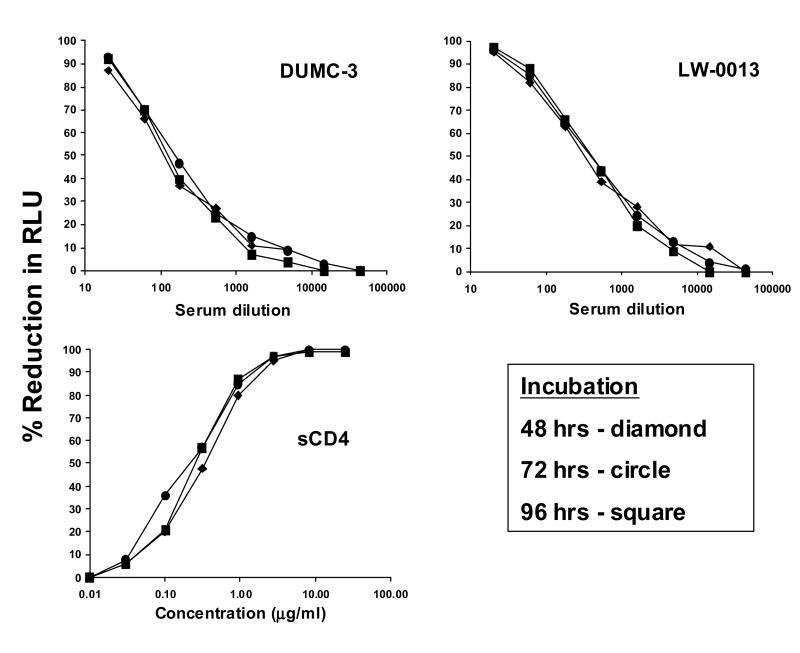
3.1.5 Optimal TZM-bl cell number for assay sensitivity
We had previously determined that linear measurements of virus infectivity in a 48 hr assay were only possible when <40,000 cells were added per well in 96-well plates (Fig. 1). Using these results as a guide, we aimed to determine the effect of input cell number on the sensitivity of the assay for detecting neutralization. In this experiment, Env-pseudotyped virus SS1196.01 was assayed with two HIV-1-positive serum samples (DUMC-3 and LW-0013) and sCD4 as described for the experiment in Fig. 3 with the exception that three different densities of TZM-bl cells were used (5,000, 10,000, and 20,000 cells/well) (data not shown). Separate assay plates were set-up for each cell density. Neutralization curves and ID50 values were nearly identical for all three serologic reagents when 5,000 and 10,000 cells/well were used in the assay. A loss in assay sensitivity of 25-50% in neutralization titers was seen when 20,000 cells were used. We therefore chose 10,000 cells/well as a standardized, optimal input cell number for the assay.
3.1.6 Impact of minor species of replication competent virus (RCV) in Env-pseudotyped virus preparations
The backbone vector most commonly used for Env-pseudotyped virus production, pSG3Δenv, is a full-length HIV-1 subtype B genome in plasmid pTZ19U containing a four nucleotide insertion that inactivates the env gene. Because most of env is preserved, potential exists for homologous recombination between this backbone plasmid and a functional env plasmid during either transfection or infection that could produce RCV. The presence of RCV could compromise the neutralization assay by representing an unintended viral target for neutralization. In addition, RCV could allow the virus to multiply and kill cells. Our results showed that 16/60 (27%) of subtype B Envs made with the SG3Δenv backbone plasmid tested positive for RCV (data not shown). Notably, subtype A, C, BC and AG viruses made with the subtype B backbone (SG3Δenv) were rarely RCV positive (1/48 cases). These results indicate that the potential for RCV is greater when using a backbone plasmid that is clade-matched to the Env clone. However, when we next tested whether RCV impacted the outcome of TZM-bl assays, equivalent neutralization results were obtained with a wide variety of antibodies regardless of RCV status. These results indicate that RCV has no measurable effect on the TZM-bl assay. Nonetheless, the occasional presence of RCV raises the level of biosafety compliance when working with Env-pseudotyped viruses, to conform with the requirements for working with fully replication-competent HIV (Rosa Borges, A., Wieczorek, L., Bilska, M., Li, M., Sanders-Buell, E., Wesberry, M., Brown, B.K., Michael, N.L., McCutchan, F.E., Montefiori, D.C., and Polonis, V.R. Detection of low levels of replication-competent virus (RCV) in HIV-1 Env-pseudotyped virus stocks prepared by co-transfection of HIV-1 env and backbone DNA. Manuscript In Preparation)
3.2 Validation of the TZM-bl Assay
The assay conditions and acceptance criteria defined in the optimization studies and described in Standard Operating Procedures (SOPs) were utilized by the Duke Laboratory and NVITAL in the assay validation testing experiments described in the next sections. The following pass/fail criteria for the TZM-bl assay were also included in the SOP: 1) the average RLU of virus control wells is >10 times the average RLU of cell control wells; 2) the % CV of RLU in the virus control wells is ≤30%; 3) the % CV for replicate wells is ≤30% for sample dilutions that yield at least 40% neutralization; 4) the neutralization curves are smooth and linear around the 50% neutralization cut-off; 5) the value of the positive control is within 3-fold of the average of the Levey-Jennings values for that particular control-virus combination.
3.2.1 Specificity
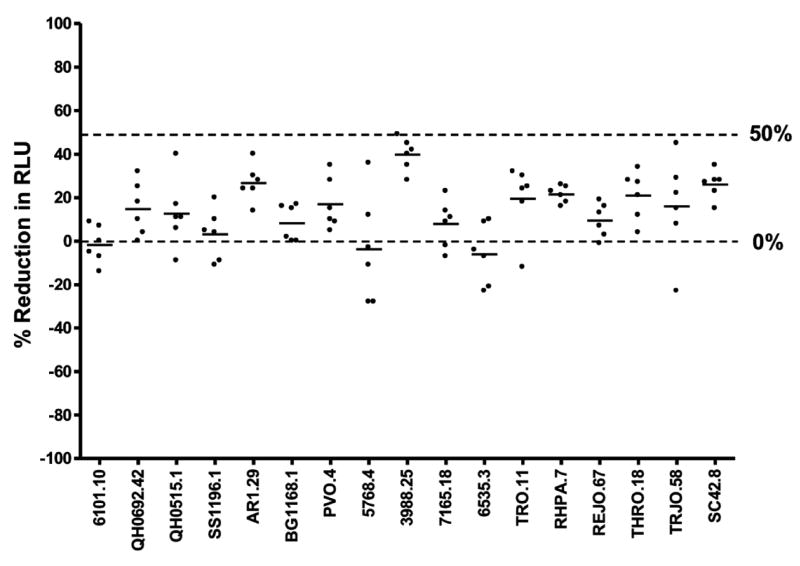
An assay is specific when it can unambiguously detect the analyte in the presence of other components. The specificity of the TZM-bl assay may be affected by cellular toxicity and/or non-specific antiviral activity of normal serum components that produce false-positive artifacts. To determine the ability of the assay to discriminate between true neutralizing antibody activity and possible artifacts, the nonspecific background activity of normal human serum samples was assessed with multiple strains of virus. Serum samples from six healthy HIV-1-negative subjects were assayed against 17 strains of Env-pseudotyped HIV-1 (Fig. 4). Serum samples were tested at 3-fold dilutions of 1:20-1:43,740. Only 1 of 102 (0.9%) serum/virus combinations tested positive using 50% reduction in RLU as cut-off (ID50). The ID50 titer of this one positive test result (ID50=21) was just above the lowest serum dilution tested (1:20). Based on this analysis, a 0.9% false positive response rate for an ID50=20 was seen for normal human serum samples.
Figure 4. Nonspecific activity of normal human serum samples.
Six serum samples assayed at a 1:20 dilution against 17 Env-pseudotyped viruses.
When NVITAL tested 27 sera from HIV-negative donors, starting at a 1:10 dilution, against eight strains of Env-pseudotyped viruses, only 1.4% (3 of 216) positive responses were observed. All three positive responses were from the same sample (data not shown). Overall, the results are within acceptance criteria and indicate that ID50 is a specific measurement of neutralization at serum dilutions ≥1:10, with a false positive rate of ≤ 2%. Because of occasional variation between serum samples and virus strains, specificity should be re-examined in each study by including corresponding pre-immune samples or samples from placebo recipients as controls.
Heat-inactivation is needed to destroy complement, which might otherwise enhance virus infection in cells that express appropriate complement receptors (Montefiori et al., 1994). The infection-enhancing effect of complement is facilitated by the presence of Env-specific antibodies and may mask the activity of neutralizing antibodies. TZM-bl cells are not known to express complement receptors, and therefore might not be susceptible to these complement effects. Nonetheless, if heat-inactivation is used, it is necessary to establish the effect it has on neutralizing activity of serum samples as measured in the TZM-bl assay. As shown in Table 2, when we assessed the neutralizing activity of serum samples from HIV-1-infected individuals before and after heat-inactivation at 56°C for 1 hour, a minor decrease in neutralizing activity was detected after heat-inactivation in each case examined (1.2-1.4-fold). This change could be due to either a minor enhancing effect of complement under non-heat-inactivated conditions or a small loss in antibody stability under conditions of heat-inactivation. All ID50 values were within the pre-established acceptable limits (3-fold variation with a 20% error rate).
Table 2.
Effect of heat-inactivation on serum neutralizing antibody activity.
| Serum Sample | ID50 against SS1196.1 | Fold variation | |
|---|---|---|---|
| Heat-inactivated | Non-heat-inactivated | ||
| DUMC-2 | 1,267 | 1,062 | 1.2 |
| DUMC-3 | 163 | 127 | 1.3 |
| LW.0013 | 599 | 431 | 1.4 |
| TH.10.03 | 353 | 268 | 1.3 |
The possibility that DEAE-dextran might alter the activity of neutralizing antibodies was examined by performing neutralization assays with two Env-pseudotyped viruses that exhibit adequate infectivity in the absence of the polycation. HIV-1-specific monoclonal antibodies (sCD4, IgG1b12, 2G12, 2F5, 4E10, TriMab), as well as serum samples from five HIV-1 seropositive individuals, tested against two Env-pseudotyped viruses in the presence and absence of DEAE-dextran showed that the ID50 values were within the pre-established acceptable limits (3-fold variation with a 20% error rate) (Table 3). The largest difference (>3-fold increase) in sensitivity was observed in the presence of DEAE-dextran when HIV-1-positive serum DUMC-3 was tested against Env-pseudotyped virus QH0692.42 (1/22, or 4.5% serum/virus combination, within 20% acceptable error rate). We conclude that DEAE-dextran has negligible effects on neutralizing antibody titers.
Table 3.
Effect of DEAE-dextran on serum neutralizing antibody activity.
| Serum Sample | ID50 against BG1168.1 | ID50 against QH0692.42 | ||
|---|---|---|---|---|
| +DEAE-dextran | -DEAE-dextran | +DEAE-dextran | -DEAE-dextran | |
| DUMC-1 | 305 | 127 | 462 | 603 |
| DUMC-2 | <20 | <20 | 40 | <20 |
| DUMC-3 | 21 | <20 | 68 | <20 |
| LW.0013 | <20 | <20 | 54 | 57 |
| TH.10.03 | <20 | <20 | 53 | 33 |
| Abs |
IC50 against BG1168.1 (μg/ml) |
IC50 against QH0692.42 (μg/ml) |
||
| sCD4 | 10 | 22.7 | 0.5 | 1.0 |
| IgG1b12 | >50 | >50 | 0.3 | 0.3 |
| 2G12 | >50 | >50 | 3.2 | 2.7 |
| 2F5 | 0.8 | 1.7 | 1.5 | 1.2 |
| 4E10 | 1.8 | 1.5 | 2.1 | 1.8 |
| TriMab | 2.4 | 5.7 | 0.3 | 0.6 |
In order to discern the ability of the assay to discriminate between neutralizing activity of HIV-1 and viral antibodies present due to other disease states, a panel of heat-inactivated sera containing detectable antibody concentrations to various viruses was evaluated at NVITAL. To determine if the presence of other viral antibodies interferes with or enhances the neutralizing activity of HIV-1, heat-inactivated serum from an HIV-1 positive donor was spiked (1:10) with an aliquot of each member of the antisera panel described below. This HIV-1-positive serum sample was also spiked with pooled normal human sera (NHS) and assayed in parallel. The “contaminant” antisera panel included HTLV-I, HTLV-II, HAV, HBV, HCV, HSV, and CMV (Table 4). Samples were assayed against two strains of Env-pseudotyped virus. Only one sample of the seven tested (200-138-01-S-HI + HTLV II), had a 3.3 fold increase over the sample spiked with NHS, thus addressing the acceptance criteria that no more than 20% of the samples can exceed a 3-fold increase for a given virus. Therefore, there was no effect of the “contaminant” on the neutralization of the virus in the assay.
Table 4. Effect of contaminating antiviral antibodies in the anti-HIV sera.
| Specimen ID | SF162.LS | HXB2 | ||||||
|---|---|---|---|---|---|---|---|---|
| ID50 | Fold | ID80 | Fold | ID50 | Fold | ID80 | Fold | |
| 200-138-01-S-HI + Anti HBs Abs | 2280.0 | 1.7 | 349.7 | 1.3 | 653.0 | 1.2 | 169.5 | 1.0 |
| 200-138-01-S-HI + Anti CMV Abs | 2539.0 | 1.9 | 534.1 | 2.1 | 608.5 | 1.2 | 196.0 | 1.1 |
| 200-138-01-S-HI + Anti HAV Abs | 1504.4 | 1.1 | 329.7 | 1.3 | 468.4 | 0.9 | 143.6 | 0.8 |
| 200-138-01-S-HI + Anti HCV Abs | 2744.0 | 2.1 | 351.6 | 1.4 | 647.3 | 1.2 | 186.5 | 1.1 |
| 200-138-01-S-HI + Anti HSV1 Abs | 3396.6 | 2.5 | 407.9 | 1.6 | 560.7 | 1.1 | 176.9 | 1.0 |
| 200-138-01-S-HI + Anti HTLV I Abs | 3870.8 | 2.9 | 482.3 | 1.9 | 586.6 | 1.1 | 176.2 | 1.0 |
| 200-138-01-S-HI + Anti HTLV II Abs | 4405.3 | 3.3 | 541.5 | 2.1 | 625.5 | 1.2 | 205.8 | 1.2 |
| 200-138-01-S-HI + NHS | 1336.7 | - | 260.4 | - | 526.6 | - | 175.3 | - |
3.2.2 Precision
Precision expresses the degree of scatter between a series of measurements obtained from multiple testing of the same homogeneous sample under the prescribed conditions. Precision can be studied at three levels: repeatability, intermediate precision, and reproducibility.
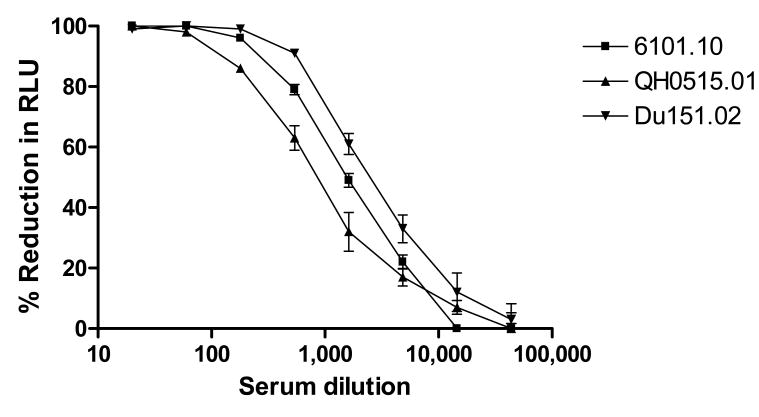
As a representation of intra-assay variation (repeatability), standard deviations of triplicate values for each dilution of an HIV-1-positive serum sample (DUMC-1) tested against three different Env-pseudotyped viruses are shown in Fig. 5. The range of standard deviations was 0 - 11% of the experimental values. Examination of large data sets has revealed that standard deviation increases as percent reductions in RLU decrease (i.e., standard deviations are greatest at low neutralization potencies). Standard deviations rarely exceeded 8% of experimental values in the linear portion of the neutralization curve. In addition, when the same operator tested a serum sample 8 times on different dates, concordance was achieved in each case, with a mean %CV for IC50/ID50 of ≤45%. Intra-plate variation (well-to-well variation) in virus control wells (8 wells/plate) and in cell control wells (8 wells/plate) had %CV values of ≤12% (data not shown).
Figure 5. Neutralization curves with s.d. bars.
HIV-1-positive serum DUMC-1 was assayed against molecularly cloned Env-pseudotyped viruses 6101.10, QH0515.01 and Du151.02 in triplicate wells for each dilution of serum sample.
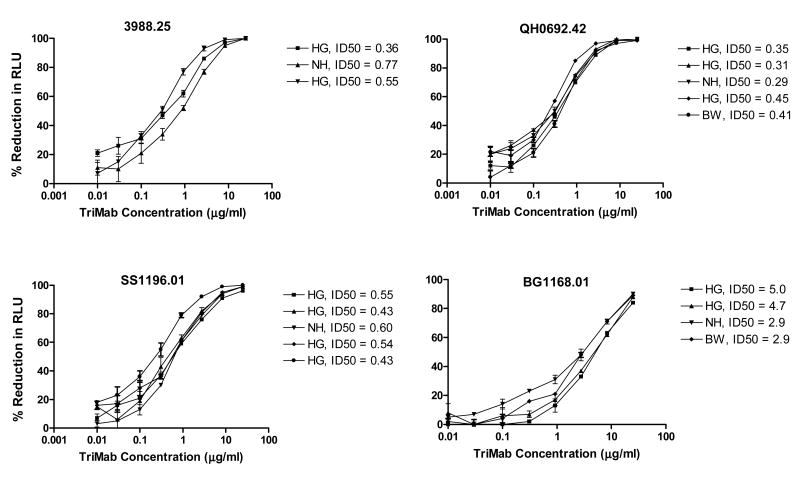
Optimization experiments performed prior to assay validation indicated that, for an acceptable range of precision, the experimental values should be within 3-fold variation from the mean, with an error rate of 20% of the repeat assays. Inter-assay variation (intermediate precision) was assessed by testing a positive control reagent (TriMab) multiple times by different operators against different strains of Env-pseudotyped viruses. Each assay was performed on a different date in independent experiments. Results are shown in Fig. 6. IC50 values of replicate assays agreed within 3-fold of the mean in each case. As an additional example of intermediate precision, Env-pseudotyped virus Du152.02 was assayed with sera from HIV-1-infected individuals, sCD4, four mAbs and TriMab at different times by two different operators. A high level of concordance was achieved between operators. Only 1/15 tests (7%) was discordant (8-fold difference for mAb 4E10). All other repeat assay results agreed within 3-fold of the mean value (data not shown).
Figure 6. Inter-assay and inter-operator variability.
TriMab was assayed multiple times against Env-pseudotyped viruses 3988.25, QH0692.42, SS1196.01 and BG1168.01. ID50 values of each curve are shown next to the operator who performed the assay.
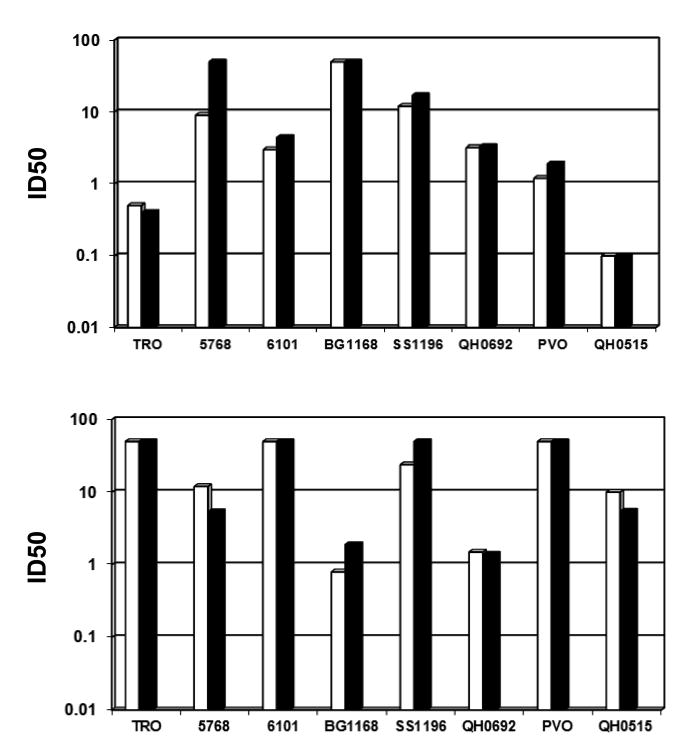
The ultimate and most difficult measure of assay precision is inter-laboratory precision (reproducibility). Results of assay reproducibility were obtained by the analysis of data obtained by 15 domestic and international laboratories participating in the TZM-bl Standardized Proficiency Testing Program over four years since its inception (Todd et al., 2012). Fig. 7 provides the distribution of neutralization titers obtained from repeat assays performed by the participating laboratories on the same two representative serologic reagents (provided in a blinded format, as reagent 1 and 2) against six Env-pseudotyped viruses. Panels A and C show the reproducibility data obtained for reagent 1 and 2 respectively, by only the Duke Laboratory and NVITAL, while the bottom panel shows data for these same neutralizing reagents/Env-pseudotyped virus combinations from all labs analyzed. The horizontal lines indicate the acceptance criteria ranges established by the Program (Todd et al., 2012). Overall, 100% (A), 98.4% (B), 92.8% (C) and 83.3% (D) of the total number of serologic reagent/virus combinations were within the pre-set acceptance criteria, indicating strong reproducibility of the assay between the Duke Laboratory and NVITAL (> 98.4%) and between all participating laboratories (> 83.3%).
Figure 7. Reproducibility of the TZM-bl assay in domestic and international laboratories.
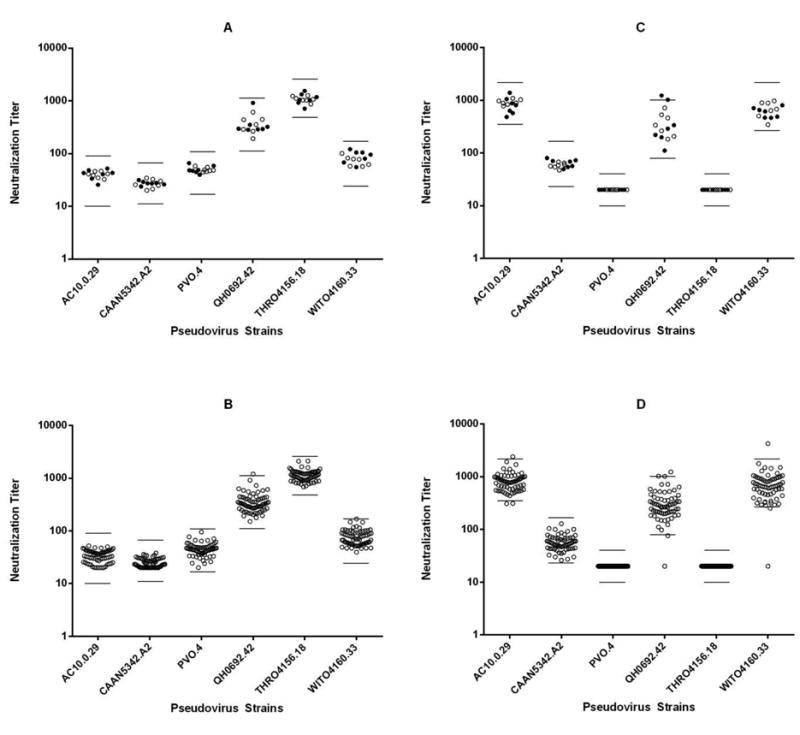
Proficiency testing results (ID50 titers) from participating laboratories testing two representative serologic reagents (A, B: reagent 1) (C, D: reagent 2), against six Env-pseudotyped viruses, over a four-year period. Panels A (n=14) and C (n=14) show the reproducibility data obtained from repeat testing by the Duke Laboratory and NVITAL, panels B (n=66) and D (n=66) show data from repeat testing by all participating labs analyzed. Bars indicate the upper and lower limits of the acceptance ranges.
3.2.3 Accuracy
Accuracy is the closeness of agreement between the value, which is accepted as a conventional true value, or an accepted reference value, and the value found. Efforts to assess the accuracy of HIV-1 neutralizing antibody assays are hindered by the absence of gold standard reference reagents. In the absence of these reagents, we have compared the performance of the TZM-bl assay to the performance of a similar assay conducted in other laboratories.
Two monoclonal antibodies, 2G12 and 2F5, were assayed against eight different strains of clade B HIV-1 in TZM-bl cells at the Duke Laboratory, and in U87.CD4.CCR5 cells at Monogram Bioscience, Inc. The U87.CD4.CCR5 assay (Richman et al., 2003) utilized a quasispecies of Env-pseudotyped viruses prepared from peripheral blood mononuclear cell (PBMC)-grown stocks of each virus, whereas corresponding molecularly cloned Env-pseudotyped viruses were used in the TZM-bl assay. As shown in Fig. 8, relative potencies were similar in the two assays. In particular, the TZM-bl and U87.CD4.CCR5 assays exhibited striking equivalency in 14 of 16 cases, where the titers in each assay were within 3-fold agreement.
Figure 8. Comparison of the TZM-bl assay to the U87.CD4.CCR5 assay (Monogram) using 2G12 and 2F5 as standards.
Values on the y-axis are μg/ml. TZM-bl assay (white); U87.CD4.CCR5 assay (black).
In the absence of reference reagents or gold standard assays, accuracy was also inferred at NVITAL by linearity, precision and range, using HIVIG as a model. An estimate of accuracy of this assay was obtained by evaluating the response (ID50 and ID80 titers) and by ensuring that all observed values fell within the 95% confidence intervals (Table 5). The percent difference for the ID50 values for both viruses assayed ranged from -9.4 to 12.5%, while those for the ID80 values for both viruses ranged from -8.6 to 2.0%. Based on these results, a value ascertained from the TZM-bl assay will be within 12.5% of the true value for a sample.
Table 5. Accuracy analysis based 95% Confidence Interval.
| Virus | ID50 or ID80 |
Dilution | Target | 95% CI | Difference from Theoretical | Percent Difference | |||
|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Lower | Upper | ||||
| HBX2 | 50 | 1 | 1 | 0.993 | 1.125 | -0.007 | 0.125 | -0.7% | 12.5% |
| 2 | 0.5 | 0.496 | 0.562 | -0.004 | 0.062 | -0.7% | 12.5% | ||
| 4 | 0.25 | 0.248 | 0.281 | -0.002 | 0.031 | -0.7% | 12.5% | ||
| 8 | 0.125 | 0.124 | 0.141 | -0.001 | 0.016 | -0.7% | 12.5% | ||
| 16 | 0.0625 | 0.062 | 0.07 | 0.000 | 0.008 | -0.8% | 12.5% | ||
| 32 | 0.03125 | 0.031 | 0.035 | 0.000 | 0.004 | -0.7% | 12.4% | ||
| 64 | 0.015625 | 0.016 | 0.018 | 0.000 | 0.002 | -0.7% | 12.4% | ||
| 128 | 0.0078125 | 0.008 | 0.009 | 0.000 | 0.001 | -0.8% | 12.5% | ||
| HBX2 | 80 | 1 | 1 | 0.974 | 1.02 | -0.026 | 0.020 | -2.6% | 2.0% |
| 2 | 0.5 | 0.487 | 0.51 | -0.013 | 0.010 | -2.6% | 2.0% | ||
| 4 | 0.25 | 0.244 | 0.255 | -0.007 | 0.005 | -2.6% | 2.0% | ||
| 8 | 0.125 | 0.122 | 0.127 | -0.003 | 0.002 | -2.6% | 2.0% | ||
| 16 | 0.0625 | 0.061 | 0.064 | -0.002 | 0.001 | -2.6% | 2.0% | ||
| 32 | 0.03125 | 0.03 | 0.032 | -0.001 | 0.001 | -2.6% | 2.0% | ||
| 64 | 0.015625 | 0.015 | 0.016 | 0.000 | 0.000 | -2.6% | 2.0% | ||
| 128 | 0.0078125 | 0.008 | 0.008 | 0.000 | 0.000 | -2.6% | 2.0% | ||
| SF162.LS | 50 | 1 | 1 | 0.906 | 0.993 | -0.094 | -0.007 | -9.4% | -0.7% |
| 2 | 0.5 | 0.453 | 0.496 | -0.047 | -0.004 | -9.4% | -0.7% | ||
| 4 | 0.25 | 0.226 | 0.248 | -0.024 | -0.002 | -9.4% | -0.7% | ||
| 8 | 0.125 | 0.113 | 0.124 | -0.012 | -0.001 | -9.4% | -0.7% | ||
| 16 | 0.0625 | 0.057 | 0.062 | -0.006 | 0.000 | -9.4% | -0.7% | ||
| 32 | 0.03125 | 0.028 | 0.031 | -0.003 | 0.000 | -9.4% | -0.7% | ||
| 64 | 0.015625 | 0.014 | 0.016 | -0.001 | 0.000 | -9.4% | -0.7% | ||
| 128 | 0.0078125 | 0.007 | 0.008 | -0.001 | 0.000 | -9.4% | -0.7% | ||
| SF162.LS | 80 | 1 | 1 | 0.914 | 0.998 | -0.086 | -0.002 | -8.6% | -0.2% |
| 2 | 0.5 | 0.457 | 0.499 | -0.043 | -0.001 | -8.6% | -0.2% | ||
| 4 | 0.25 | 0.229 | 0.25 | -0.021 | 0.000 | -8.6% | -0.2% | ||
| 8 | 0.125 | 0.114 | 0.125 | -0.011 | 0.000 | -8.6% | -0.2% | ||
| 16 | 0.0625 | 0.057 | 0.062 | -0.005 | 0.000 | -8.6% | -0.2% | ||
| 32 | 0.03125 | 0.029 | 0.031 | -0.003 | 0.000 | -8.6% | -0.2% | ||
| 64 | 0.015625 | 0.014 | 0.016 | -0.001 | 0.000 | -8.5% | -0.2% | ||
| 128 | 0.0078125 | 0.007 | 0.008 | -0.001 | 0.000 | -8.6% | -0.2% | ||
3.2.4 Limits of Detection and Quantitation
The limit of detection of an analytical procedure is the lowest amount of analyte in a sample that can be detected, but not necessarily quantified, as an exact value. The lowest amount of analyte that can be accurately quantified defines the limit of quantitation. The lower limit of quantitation of neutralizing antibodies is the highest dilution of serum or lowest concentration of mAb that reduces RLU by 50% relative to either the virus control wells or an appropriate negative test sample. This cut-off was chosen because it lies midway in the linear portion of neutralization curves (20-80% reductions in RLU) and is above the level of background of the assay. The upper limit (lowest dilution) of quantitation of serum neutralizing antibodies is determined in large part by the nonspecific virus inhibitory activity and cell toxicity of normal serum samples. Referring to the data in Fig. 4, and to additional assays with 1:10-diluted normal human serum against different strains of HIV-1 (data not shown), 50% reductions in RLU are rare for normal human serum diluted >1:10. Since background activity may vary between serum samples and virus strains, it is strongly recommended that the upper limit of quantitation be confirmed with pre-immune serum samples in every study for each virus tested.
The detection limit was also determined by NVITAL using the ICH Q2 (R1) guidance method of ± 3.3 s.d. of the background (cell control wells) (Guideline, 2010). The detection limit is the maximum percent inhibition that can be detected by the assay using the following formula:
Using this method with cell control and virus control values from a randomly selected neutralization plate it was determined that this assay can detect a sample with neutralizing antibodies that would have 14-100% viral inhibition (data not shown). This is an acceptable limit of detection for both IC50/ID50 and IC80/ID80 specific values.
The limit of quantitation was similarly determined by NVITAL using the ICH Q2 (R1) guidance method of ± 10 s.d. of the background (Guideline, 2010). Using the method described above for the limit of detection, it was determined that the TZM-bl assay can quantify a sample with antibodies that would have 41 - 99% viral inhibition, an acceptable limit of quantitation for both IC50/ID50 and IC80/ID80 specific values (data not shown).
3.2.5 Linearity and Range
Linearity is the ability of an analytical procedure to obtain test results, within a given range, that are directly proportional to the concentration of analyte in the sample. The range of an assay is the interval between the upper and lower concentration of an analyte in the sample for which it has been demonstrated that the analytical procedure has a suitable level of precision, accuracy, and linearity. Neutralization curves generated with positive serum samples and mAbs show a consistent pattern of linearity over an approximate range of 20-80% reductions in RLU (Figs. 5, 6). Values in this range are directly proportional to the concentration of neutralizing antibodies in the sample.
Linearity was also determined by NVITAL using dilution series of constructed samples. Eight constructed samples were prepared using the control reagent HIVIG pre-diluted 1:2, 4, 8, 16, 32, 64, and 128 in pooled NHS. Each constructed sample was assayed starting at a 1:8 dilution, and serially diluted 3-fold against two Env-pseudotyped viruses. Three runs were performed by multiple analysts (Table 6). A least squares regression model with no intercept was fit to the data using a simple linear model with ID50 and/or ID80 of the undiluted sample as the dependent variable (y-axis) and the dilution factor as the independent variable (x-axis). The acceptance criteria for the ID50 and ID80 graphs required that the F-test for lack of fit would have a p-value >0.01, standardized residuals would be within ± 3.5, and R2 would exceed 0.85 for the model with an intercept. Table 6 demonstrates that the lack of fit test had p-values ranging from 0.11 to 0.99. All standardized residuals were within ±3.5, ranging from 3.31 to 3.48. The R2 value for each IC50 and IC80 graph for both viruses assayed exceeded 0.85 with values ranging from 0.957 to 0.975. Since the relationship between ID50/ID80 and dilution is proportional, the assay was shown to be linear.
Table 6. Lack of Fit, R2 and Standardized Residuals for Linearity Analysis.
| Virus | ID50/ID80 | p-value (Lack of Fit) | R2 | Maximum Standardized Residual |
|---|---|---|---|---|
| HXB2 | ID50 | 0.1110 | 0.957 | 3.31 |
| ID80 | 0.9987 | 0.960 | 3.48 | |
| SF162.LS | ID50 | 0.1829 | 0.975 | 3.34 |
| ID80 | 0.3917 | 0.967 | 3.36 |
3.2.6 Robustness
The focus of robustness is to determine the consistency of the assay under real-life changes that can occur in standard laboratory conditions, such as assay incubation time, cell surface marker expression by TZM-bl cells, reagent stability, type of luminescence reader, backbone vectors used to generate Env-pseudotyped viruses, and others.
Our optimization experiments determined that RLU increased dramatically between 24 and 48 hrs of infection, where 48 hrs was adequate to measure as much as a 2-3-log reduction in virus infectivity. This led to the use of a 48 hrs infection period for assay performance. Unexpected delays due to scheduling conflicts may sometimes require assays to be processed after a prolonged period of infection. We therefore compared assay results after 48, 72 and 96 hrs of infection. In this experiment, Env-pseudotyped virus SS1196.01 was assayed and RLU were measured after 48, 72 and 96 hrs. Separate assay plates were set-up for each time point. Identical results were obtained at all three time points for each reagent (Fig. 9). This result is not unexpected given that Env-pseudotyped viruses are only capable of a single round of infection. In the case of replication competent viruses, the assay incubation should not exceed 48 hrs, to minimize replication that might modify neutralization results.
Figure 9. Effect of altering the length of time of infection prior to measuring luminescence.
RLU in cell control (background) and SS1196.01 Env-pseudotyped virus control wells were, respectively: 48 hrs = 1,068 vs 65,158; 72 hrs = 1,224 vs 70,756; 96 hrs = 1,604 vs 60,621.
Variation in the fraction of cells expressing HIV-1 fusion receptors (CD4 and either CCR5 or CXCR4) may affect the performance of the assay by altering the magnitude and kinetics of infection. The expression of CD4, CCR5 and CXCR4 by cultured TZM-bl cells was monitored over time by flow cytometry. As shown in Table 6, all three fusion receptors were stably expressed on TZM-bl cells for at least 24 weeks in culture (77 passages). A precipitous drop in CD4 and CCR5 expression, and more gradual decrease in CXCR4 expression, occurred soon afterward. These results indicate that TZM-bl cells must be discarded after either 60 passages or a period of five months, whichever comes first.
Env-pseudotyped virus preparations are routinely stored in aliquots at ≤-70°C and thawed immediately before assay. In cases where a frozen stock of Env-pseudotyped virus has a high concentration of infectious particles, as a matter of convenience thawed aliquots may be refrozen for later use. We examined the infectivity of several different strains of Env-pseudotyped viruses after a second thaw cycle. The infectivity of most Env-pseudotyped viruses was reduced after the second thaw cycle (data not shown). The amount of reduction varied between strains, where no reduction was seen in one case and a 10-fold reduction was seen in the most dramatic case. In 8 of 9 cases the infectivity remained at an adequate level to be used in neutralization assays. However, it is recommended that viruses be titrated after a second thaw cycle, to ensure the appropriate selection of input virus dose in the TZM-bl assay.
Other variables tested included the use of multiple luminometers of the same model (Victor 2 luminometer, Perkin-Elmer Life Sciences, Shelton, CT), the use of two different manufacturers of Luc reaction kits (Britelite, PerkinElmer or Brite-Glo, Promega) and the use of different backbone vectors to generate Env-pseudotyped viruses (pSG3Δenv or Q23-17Δenv, kindly proviδed by Dr. J. Overbaugh, Fred Hutchinson Cancer Research Center, Seattle, WA). All results were within acceptance criteria (3-fold, 20% error rate) (data not shown).
Either white or black 96-well plates may be used when measuring luminescence. Overall luminescence values will be approximately 10-fold lower with black plates, but still provide adequate RLU values for the assay. Black plates are preferred, because in cases of very high luminescence, some bleeding of light to adjacent wells may occur and cause artificially high readings. This artifact is avoided by using back plates.
The TZM-bl assay was originally optimized to test each dilution of test sample in triplicate wells. A duplicate well assay format would increase throughput, conserve sample and reduce costs. Before adopting the duplicate format, it was necessary to determine how the neutralization curves, standard deviation and IC50 values compared to the triplicate format. In this experiment, three operators assayed five positive serologic reagents (sCD4, IgG1b12, 2G12, 2F5 and TriMab) against two strains of Env-pseudotyped viruses (SF162.LS and QH0692.42) in TZM-bl cells, using duplicate and triplicate formats. All IC50 values agreed within 3-fold per operator and no difference was seen in the range of standard deviation among duplicate and triplicate wells (range was 0 - 8% at ID50 values ≥50%) (data not shown).
3.3 Automated TZM-bl Neutralization Assay
The automated 384-well format TZM-bl assay is designed to perform high throughput automated virus neutralization assays. The optimized assay volumes and dilution methods are different from those of the manual TZM-bl assay (Table 1). Following optimization and validation of the manual TZM-bl assay, the automated assay was qualified using the pre-acceptance criteria developed for the manual assay. The results are summarized in Table 7. The automated assay successfully met the acceptance criteria for specificity, precision and equivalency to the manual assay. Although the linearity evaluation only passed 5 out of 6 p-value criteria (Table 7), the graphical analysis of the HXB2 values (data not shown) showed acceptable linearity. Using a dilution model with HIVIG to define linearity, the limit of detection for the automated assay was determined to be 21-100% virus inhibition. The limit of quantitation was determined to range from 63% to 99% virus inhibition, which is an acceptable limit of quantitation for ID80 values. In this linearity model the IC50 values fall below the limit of quantitation, but within the limit of detection.
Table 7.
Flow cytometric analysis of CD4, CCR5 and CXCR4 cell surface expression on TZM-bl cells.
| Weeks in culture | Passage No. | % CD4 positive | % CCR5 positive | % CXCR4 positive |
|---|---|---|---|---|
| 2 | 17 | 99 | 100 | 99 |
| 4 | 21 | 100 | 98 | 95 |
| 6 | 26 | 98 | 98 | 89 |
| 8 | 30 | 99 | 98 | 89 |
| 10 | 34 | 99 | 100 | 90 |
| 12 | 39 | 100 | 100 | 98 |
| 14 | 44 | 99 | 99 | 98 |
| 18 | 54 | 100 | 99 | 90 |
| 22 | 64 | 99 | 99 | 83 |
| 24 | 77 | 97 | 97 | 72 |
| 28 | 89 | 35 | 37 | 61 |
| 33 | 99 | 1 | 22 | 65 |
4.0 Discussion
Here we describe key experiments that were performed to optimize and validate the TZM-bl assay for standardized assessments of HIV-1-specific neutralizing antibodies. The procedures associated with this validated assay have been developed into robust, centrally controlled SOPs, which have been utilized by GCLP-compliant laboratories around the world for HIV-1 vaccine development efforts (Ozaki et al., 2012). A formal proficiency testing program was established in 2009 (Todd et al., 2012) to facilitate compliance with GCLP when performing this assay.
The validated, manual TZM-bl Nab assay was also adapted, optimized and qualified to an automated 384-well format for use as a high throughput assay in testing sera, plasma, and monoclonal antibody samples. The automated assay successfully met the criteria for specificity, precision, and equivalency to the manual assay.
Several antibodies have been shown to exhibit much greater neutralization potency in the PBMC assay than in the TZM-bl assay (Binley et al., 2004) (Brown et al., 2007) (Choudhry et al., 2007) (Polonis et al., 2008), raising concern that the TZM-bl assay falsely underestimates the neutralizing activity of some reagents and creating uncertainty as to which assay provides a more meaningful assessment of antibody-mediated virus neutralization. Much of this discrepancy was demonstrated to be an artifact of endotoxin contamination affecting the PBMC assay (Geonnotti et al., 2010). Endotoxin, which is bacterial lipopolysaccharide stimulates susceptible PBMCs to produce a complex array of soluble HIV-1 inhibitors, including beta-chemokines and IFN-gamma, and can cause false-positive results in neutralizing antibody assays done in PBMCs, while assays performed in TZM-bl cells are unaffected (Geonnotti et al., 2010).
Furthermore, the presence of MLV was detected in the TZM-bl cell line (Takeuchi et al., 2008) raising questions of MLV's potential interference with HIV-1 neutralization in the assay. However, when such interference was directly addressed, no evidence was found that ecotropic MLV contamination had a measurable effect on HIV-1 neutralization (Platt et al., 2009).
This assay has been compared to other neutralizing antibody assays and shown to be one of the most sensitive of the assays evaluated (Fenyo et al., 2009). However, as emphasized in that report, multiple assays often gave different results, making it difficult to decide a single assay that gives the most reliable results, and where it may be necessary to use more than one assay, especially when evaluating vaccine elicited responses. In this regard, TZM-bl cells express a much higher density of CD4 and CCR5 than is expressed on mitogen-stimulated human PBMCs. In particular, the sensitivity for detecting antibody-mediated HIV-1 neutralization is increased in cells that express lower levels of CCR5 (Choudhry et al., 2006). This has led to the development of an alternate HIV-neutralization assay in A3R5 cells, which express much lower levels of CD4 and CCR5 than are found on TZM-bl cells, and where the detection of neutralization of Tier 2 viruses is dramatically improved compared to the TZM-bl assay (Montefiori et al., 2012) (McLinden, 2013) (see Sarzotti-Kelsoe et al. in this issue). The TZM-bl assay has great utility for studies of mAbs and the neutralizing antibody responses in HIV infected individuals, where the increased levels of sensitivity are not required (Seaman et al., 2010). It also remains a valuable assay for strong vaccine-elicited neutralizing antibody responses (Montefiori et al., 2012). Because of its lower cost, it remains the assay of choice for measurements of vaccine-elicited neutralizing antibodies to Tier 1 viruses (Mascola et al., 2005).
5.0 Conclusions
In conclusion, the TZM-bl assay was optimized and validated for use as an easily transferrable, high throughput assay for the evaluation of HIV-1 neutralizing antibody activity. The assay was tested for specificity, accuracy, precision, limits of detection and quantitation, linearity, range and robustness. The validated, manual TZM-bl assay was also adapted, optimized and qualified to an automated 384-well format.
Table 8. Validation of the Automated TZM-bl Assay.
| Validation Parameter | Experimental Design | Acceptance Criteria | Observed Results |
|---|---|---|---|
| Specificity | 19 HIV- Sera Samples, tested against eight strains of Env-pseudotyped virus. | <2.0% positive responses (≤3 of 152 responses) was allowed from the sera sample panel | 0 positive responses were observed. |
| Precision: Intra-plate effect | HIVIG used in all 20 sample positions of an assay plate, tested against two strains of Env-pseudotyped virus, SF162.LS and SS1196.1 |
|
|
| Precision: Inter-assay and Interplate | Five control reagents (4E10, 2F5, 2G12, sCD4, HIVIG), tested against three Env-pseudotyped viruses (SF162.LS, SS1196.1, HXB2). Each control reagent run in triplicate within each assay and each assay run in triplicate | % CV of the overall mean ID50 values must be ≤45% with a 20% error rate (12 of 15) of the determinations. | -15 of 15 of the calculated %CVs for ID50 were ≤45% (range from 0 to 30.04 %CV). |
| Precision: Manual vs Automated -Assay equivalency | 19 HIV+ Sera and Plasma Samples and HIVIG, each tested against six strains of Env-pseudotyped virus. |
|
|
| Linearity | Eight constructed samples prepared from HIVIG prediluted 1:1, 2, 4, 8, 16, 32, 64, 128 in NHS. Each constructed sample tested against three Env-pseudotyped viruses (SF162.LS, SS1196.1, HXB2). Each virus assayed in three runs. |
|
|
| Range | Linearity experiments were analyzed to determine the range of this analytical procedure. | Range was determined by the range of concentrations for which:
|
|
| Accuracy | In absence of standard reference reagents, accuracy was inferred based upon precision, linearity and the determined range. | Using the linearity results and 95% confidence intervals, an estimate of the accuracy was obtained by evaluating the response (ID50 and ID80) to ensure the 95% confidence intervals for each dilution do not overlap (the maximum bias of 5.7% for ID50 and 9.3% for ID80). |
|
| Detection Limit | Linearity experiments were analyzed to determine the detection limit of this analytical procedure. | The detection limit is determined by ±3.3 standard deviations of the background. | This assay can detect a sample with antibodies that would have 21-100% inhibition. This is an acceptable limit as ID/IC50 and ID/IC80 values are reported. |
| Quantitation Limit | Linearity experiments were analyzed to determine the Quantitation limit of this analytical procedure. | The quantitation limit is determined by ±10 standard deviations of the background. | This assay can quantify a sample with antibodies that would have 63-99% inhibition. This is an acceptable limit for reporting IC/ID80 values. |
Acknowledgments
This study was supported in part by the Bill & Melinda Gates Foundation's Collaboration for AIDS Vaccine Discovery / Comprehensive Antibody Vaccine Immune Monitoring Consortium (Grant IDs: 38619 & 1032144), the HIV Vaccine Trials Network (AI46705 & AI068618), NIH (Contract IDs: AI30034 & HHSN272200110016C), and Duke University Center for AIDS Research (CFAR), a NIH funded program (Program ID: 5P30 AI064518), Flow Cytometry Core. The authors thank Monogram Bioscience, Inc. for performing the U87.CD4.CCR5 assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. Journal of virology. 2004;78:13232–52. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BK, Karasavvas N, Beck Z, Matyas GR, Birx DL, Polonis VR, Alving CR. Monoclonal antibodies to phosphatidylinositol phosphate neutralize human immunodeficiency virus type 1: role of phosphate-binding subsites. Journal of virology. 2007;81:2087–91. doi: 10.1128/JVI.02011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry V, Zhang MY, Harris I, Sidorov IA, Vu B, Dimitrov AS, Fouts T, Dimitrov DS. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochemical and biophysical research communications. 2006;348:1107–15. doi: 10.1016/j.bbrc.2006.07.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry V, Zhang MY, Sidorov IA, Louis JM, Harris I, Dimitrov AS, Bouma P, Cham F, Choudhary A, Rybak SM, Fouts T, Montefiori DC, Broder CC, Quinnan GV, Jr, Dimitrov DS. Cross-reactive HIV-1 neutralizing monoclonal antibodies selected by screening of an immune human phage library against an envelope glycoprotein (gp140) isolated from a patient (R2) with broadly HIV-1 neutralizing antibodies. Virology. 2007;363:79–90. doi: 10.1016/j.virol.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzelle J, Rodriguez-Chavez I, Darden J, Stirewalt M, Kunwar N, Hitchcock R, Walter T, D'souza M. Guidelines on good clinical laboratory practice: bridging operations between research and clinical research laboratories. Journal of pharmaceutical and biomedical analysis. 2008;46:18–29. doi: 10.1016/j.jpba.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyo EM, Heath A, Dispinseri S, Holmes H, Lusso P, Zolla-Pazner S, Donners H, Heyndrickx L, Alcami J, Bongertz V, Jassoy C, Malnati M, Montefiori D, Moog C, Morris L, Osmanov S, Polonis V, Sattentau Q, Schuitemaker H, Sutthent R, Wrin T, Scarlatti G. International network for comparison of HIV neutralization assays: the NeutNet report. PloS one. 2009;4:e4505. doi: 10.1371/journal.pone.0004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geonnotti AR, Bilska M, Yuan X, Ochsenbauer C, Edmonds TG, Kappes JC, Liao HX, Haynes BF, Montefiori DC. Differential inhibition of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and TZM-bl cells by endotoxin-mediated chemokine and gamma interferon production. AIDS research and human retroviruses. 2010;26:279–91. doi: 10.1089/aid.2009.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guideline IHT. Validation of Analytical Procedures: Text and Methodology Q2 (R1) (2005) 2010 Websites: http://www.ich.org/cache/compo/363-272-1.html.
- Mascola JR, D'Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. Journal of virology. 2005;79:10103–7. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLinden RJ, L CC, Chenine AL, Polonis VR, Eller MA, Wieczorek L, Ochsenbauer C, Kappes JC, Perfetto S, Montefiori DC, Michael NL, Kim JH. Detection of HIV-1 neutralizing antibodies in a human CD4+/CXCR4+/CCR5+ T-lymphoblastoid cell assay system. PloS one. 2013 doi: 10.1371/journal.pone.0077756. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. In: Vinayaka GVK, Prasad R, editors. Methods Mol Virol, HIV Protocols. Second. Vol. 485. Humana Press; 2009. pp. 395–405. [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Cornell RJ, Zhou JY, Zhou JT, Hirsch VM, Johnson PR. Complement control proteins, CD46, CD55, and CD59, as common surface constituents of human and simian immunodeficiency viruses and possible targets for vaccine protection. Virology. 1994;205:82–92. doi: 10.1006/viro.1994.1622. [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Karnasuta C, Huang Y, Ahmed H, Gilbert P, de Souza MS, McLinden R, Tovanabutra S, Laurence-Chenine A, Sanders-Buell E, Moody MA, Bonsignori M, Ochsenbauer C, Kappes J, Tang H, Greene K, Gao H, LaBranche CC, Andrews C, Polonis VR, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Self SG, Berman PW, Francis D, Sinangil F, Lee C, Tartaglia J, Robb ML, Haynes BF, Michael NL, Kim JH. Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. The Journal of infectious diseases. 2012;206:431–41. doi: 10.1093/infdis/jis367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Robinson WE, Jr, Schuffman SS, Mitchell WM. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. Journal of clinical microbiology. 1988;26:231–5. doi: 10.1128/jcm.26.2.231-235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki DA, Gao H, Todd CA, Greene KM, Montefiori DC, Sarzotti-Kelsoe M. International technology transfer of a GCLP-compliant HIV-1 neutralizing antibody assay for human clinical trials. PloS one. 2012;7:e30963. doi: 10.1371/journal.pone.0030963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler B, Nelson EK, Eckels J, Ramsay S, Lum K, Wood B, Greene KM, Gao HM, Seaman MS, Montefiori DC, Igra M. LabKey Server NAb: A tool for analyzing, visualizing and sharing results from neutralizing antibody assays. Bmc Immunol. 2011;12 doi: 10.1186/1471-2172-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. Journal of virology. 2009;83:8289–92. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonis VR, Brown BK, Rosa Borges A, Zolla-Pazner S, Dimitrov DS, Zhang MY, Barnett SW, Ruprecht RM, Scarlatti G, Fenyo EM, Montefiori DC, McCutchan FE, Michael NL. Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology. 2008;375:315–20. doi: 10.1016/j.virol.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzotti-Kelsoe M, Cox J, Cleland N, Denny T, Hural J, Needham L, Ozaki D, Rodriguez-Chavez IR, Stevens G, Stiles T. Evaluation and recommendations on good clinical laboratory practice guidelines for phase I–III clinical trials. PLoS medicine. 2009;6:e1000067. doi: 10.1371/journal.pmed.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of virology. 2010;84:1439–52. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles TG, Mawby V, BARQA N. Good Clinical Laboratory Practice (GCLP): A Quality System for Laboratories that undertake the Analyses of Samples from Clinical Trials. 2003 [Google Scholar]
- Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. Journal of virology. 2008;82:12585–8. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd CA, Greene KM, Yu X, Ozaki DA, Gao H, Huang Y, Wang M, Li G, Brown R, Wood B, D'Souza MP, Gilbert P, Montefiori DC, Sarzotti-Kelsoe M. Development and implementation of an international proficiency testing program for a neutralizing antibody assay for HIV-1 in TZM-bl cells. Journal of immunological methods. 2012;375:57–67. doi: 10.1016/j.jim.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrobial agents and chemotherapy. 2002;46:1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]