Abstract
Due to the difficulties in establishing correspondences between functional regions across individuals and populations, systematic elucidation of functional connectivity alterations in mild cognitive impairment (MCI) in comparison with normal controls (NC) is still a challenging problem. In this paper, we assessed the functional connectivity alterations in MCI via novel, alternative predictive models of resting state networks (RSNs) learned from multimodal resting state fMRI (R-fMRI) and diffusion tensor imaging (DTI) data. First, ICA-clustering was used to construct RSNs from R-fMRI data in NC group. Second, since the RSNs in MCI are already altered and can hardly be constructed directly from R-fMRI data, structural landmarks derived from DTI data were employed as the predictive models of RSNs for MCI. Third, given that the landmarks are structurally consistent and correspondent across NC and MCI, functional connectivities in MCI were assessed based on the predicted RSNs and compared with those in NC. Experimental results demonstrated that the predictive models of RSNs based on multimodal R-fMRI and DTI data systematically and comprehensively revealed widespread functional connectivity alterations in MCI in comparison with NC.
Keywords: mild cognitive impairment, resting state networks, DTI, resting state fMRI, predictive models of networks, functional connectivity
Introduction
Alzheimer’s disease (AD) is a devastating disorder that impairs memory and quality of daily life progressively. The impact of AD on individuals, families and the health care system makes the disease one of the greatest medical, social and economic challenges. The future of healthcare for AD lies in its early diagnosis and treatment, as early intervention helps patients and their families plan for the future (Grundman et al., 2004), and offers the best chance to delay the progression of the disease (Jack et al., 2010). Therefore, this paper focuses on mild cognitive impairment (MCI), which is the precursor of AD and converts to AD at approximately 10% to 15% per year (Grundman et al., 2004). As powerful non-invasive neuroimaging techniques, diffusion tensor imaging (DTI) (e.g., Bozzali et al., 2002; Head et al., 2004; Stahl et al., 2007; Zhang et al., 2007; Salat et al., 2010; Stebbins and Murphy, 2009; Zhu et al., 2013) and resting state fMRI (R-fMRI) (e.g., Greicius et al., 2004; He et al., 2007; Wang et al., 2007; Liu et al., 2008a; Logothetis, 2008; Dickerson and Sperling, 2009; Zhu et al., 2013) are poised to play an increasingly important role in the development of imaging markers for prediction of AD at the early stage of the disease. Earlier studies have shown that MCI/AD pathogenesis involves widespread alterations in structural/functional brain networks revealed by DTI and/or R-fMRI data (Liu et al., 2008a; Dickerson and Sperling, 2009; Stebbins and Murphy, 2009; Salat et al., 2010; Zhang et al., 2011a; Liang et al., 2011; Binnewijzend et al., 2011; Zhu et al., 2013). Therefore, there has been increasingly significant amount of effort in the literature devoted to measuring the hypothesized widespread structural and/or functional connectivity alterations in MCI by DTI and/or R-fMRI. For instance, the NIH-funded ongoing ADNI-2 project (Jack et al., 2010) collects DTI or R-fMRI data for early MCI patients and conducts follow-ups for several years.
Essentially, when mapping brain connectivity, network nodes, or landmarks, provide the structural substrates for mapping connectivities within individual brains and for comparing results across populations. Thus, identification of robust and reproducible landmarks with accurate correspondences across individual brains is critically important for the success of brain connectivity mapping (Liu 2011). Current approaches to identifying network node landmarks in the neuroimaging field can be broadly classified into four categories (Li et al., 2009; Li et al., 2012a). The first category is manual labeling of structural landmark by experts based on their domain knowledge (Dickerson and Sperling, 2009). The second school of methods aims to cluster landmarks from the brain image itself and is typically data-driven (e.g., Zang et al., 2004; Beckmann et al., 2005). The third category usually predefines landmarks in an atlas brain, and then warps them to the individual space using image registration algorithms (Zhang et al., 2011b). The fourth method uses task-based fMRI paradigms, e.g., block-based design, to identify activated brain regions as functional landmarks. This methodology is regarded as a reliable approach for landmark identification (Zhu et al., 2011a; Li et al., 2012a). Despite significant effort from the neuroimaging field, however, it is challenging to identify accurate and reproducible landmarks in diseased brains (e.g. MCI or AD) because the functional connectivities in their brain networks might have already been significantly altered along with the disease progression.
Recently, we developed and validated a novel data-driven discovery approach that identified 358 consistent and corresponding cortical landmarks in over 240 brains (Zhu et al., 2011b; Zhu et al., 2012; Li et al., 2012b), in which each identified landmark was optimized to possess maximal group-wise consistency of DTI-derived fiber shape patterns (Zhu et al., 2011a; Zhu et al., 2011b; Zhu et al., 2012). The neuroscience basis is that each brain’s cyto-architectonic area has a unique set of extrinsic inputs/outputs, named the “connectional fingerprint” (Passingham et al., 2002), which principally determine the functions that each brain area could possibly possess. This close relationship between structural connection pattern and brain function has been confirmed and replicated in a series of our recent works (Li et al., 2010; Zhu et al., 2011a; Zhu et al., 2011b; Zhu et al., 2012; Zhang et al., 2011b; Li et al., 2012a; Li et al., 2012b). These 358 landmarks are named “Dense Individualized and Common Connectivity-based Cortical Landmarks” (DICCCOLs) (Zhu et al., 2011b; Zhu et al., 2012). This set of 358 DICCCOL landmarks has been replicated and reproduced in over 240 brains and its predictions in four different multimodal fMRI/DTI datasets (over 240 brains) have been released online at: http://dicccol.cs.uga.edu.
In this paper, we tackle the challenge of localizing accurate landmarks in MCI brains by DICCCOLs and use the DICCCOL landmarks as predictive models of resting state networks (RSNs) obtained from a machine learning framework to assess possible functional connectivity alterations in MCI. Specifically, first, we identified RSNs using ICA-clustering approach (McKeown et al., 1998; Calhoun et al. 2001; Calhoun et al. 2004; Schmithorst and Holland, 2004) based on R-fMRI data in NC subjects. Second, since some RSNs in MCI are already disrupted and cannot be constructed directly from R-fMRI data (Rombouts et al., 2005; Salvador et al., 2005; Sorg et al., 2007; Bai et al., 2008), the DICCCOL landmarks derived from DTI data were used to encode the RSNs and employed as the predictive models of RSNs. Third, given that the DICCCOL landmarks are structurally consistent and correspondent across NC and MCI, functional connectivities in MCI were assessed based on the predictive models and compared with those in NC.
The major contributions of this paper are summarized in the following two aspects. First, we assessed the widespread functional connectivity alterations in MCI via predictive models of RSNs obtained from a machine learning framework based on multimodal R-fMRI and DTI data. Since some RSNs were disrupted in MCI and can hardly be constructed using data-driven approaches (e.g. ICA-clustering) merely from R-fMRI data, we used robust and consistent structural landmarks from DTI data as predictive models of RSNs. In this way, the correspondences between MCI and NC were established at the fine-scale of network node level, and widespread functional connectivity alterations in MCI could be assessed based on the predictive models. Since the RSNs in MCI are already disrupted, the comparisons of functional connectivities among consistent and corresponding structural DICCCOL landmark-based predictive models of RSNs provide an alternative, fair and meaningful way to assess the widespread RSN dysfunctions in the MCI.
Second, our work offered a general and novel framework to examine large-scale functional connectivity alterations in neurological or psychiatric conditions. In the brain connectivity mapping field, a major barrier is the critical lack of a quantitatively encoded representation of common structural and functional brain landmarks that can be precisely replicated across individuals and populations, especially for diseased populations. This barrier has posed a significant limit to the comparison and cross-validation of brain connectivity mapping results (e.g., Sporns et al., 2005; Lynall et al., 2010; Liu 2011). The predictive models of RSNs learned from multimodal R-fMRI and DTI data in our work potentially offer intrinsically-established structural correspondences across individuals and populations, in particular, for those diseased brains with missing or altered RSNs. These predictive models of RSNs provide a general bridge to compare and cross-validate functional connectivity mapping results across different datasets and labs.
Materials and Methods
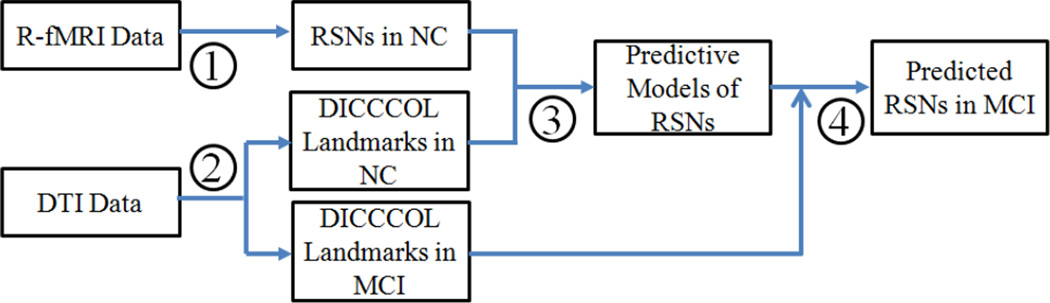
The flowchart of assessing the functional connectivity alterations in MCI via predictive models of RSNs is illustrated in Fig. 1. It consists of four major steps. In step 1, the data-driven ICA-clustering approaches are applied to construct RSNs in normal control (NC) group from R-fMRI data. Step 2 introduces our DICCCOL landmark identification procedure in NC and MCI groups. Step 3 illustrates the machine learning framework that obtains the predictive models of RSNs based on the RSNs in step 1 and DICCCOL landmarks in step 2. In step 4, RSNs are predicted in MCI via the DICCCOL landmark-based predictive models and functional connectivities are assessed based on the predicted RSNs.
Figure 1.
The flowchart of assessing the functional connectivity alterations in MCI via predictive models of RSNs.
Data Acquisition and Preprocessing
Twenty-eight participants (ten MCI patients and eighteen socio-demographically matched normal controls (NC)) were recruited and scanned in the Duke-UNC Brain Imaging and Analysis Center (BIAC). Informed consent was obtained from all participants, and the experimental protocols were approved by Duke IRB. The criteria for MCI were in accordance with NACC procedures and NINCDS-ADRDA diagnostic guidelines. Detailed inclusion and exclusion criteria have been reported in our previous work (Wee et al., 2011). Confirmation of diagnosis for all subjects was made via expert consensus panels at the Joseph and Kathleen Bryan Alzheimer's Disease Research Center (Bryan ADRC) and the Department of Psychiatry at Duke University Medical Center. Diagnosis was based upon available data from a general neurological examination, neuropsychological assessment evaluation, collateral and subject symptom and functional capacity reports. Confirmation of the diagnosis for all subjects was made by a clinical psychiatrist at Duke Medical Center. Characteristics of the participants in this MCI study are summarized as follows (Table 1).
Table 1.
Demographic and clinical measures of NC and MCI subjects. Data is presented as mean ± SD; NC, normal control; MMSE, Mini-Mental State Exam.
| Parameter | NC | MCI |
|---|---|---|
| n | 18 | 10 |
| Age, years | 72.1 ± 8.2 | 74.2 ± 8.6 |
| Sex, male/female | 8/10 | 5/5 |
| Education | 16.3 ± 2.4 | 17.7 ± 4.2 |
| MMSE | 29.4 ± 0.9 | 28.4 ± 1.5 |
The R-fMRI and DTI datasets were acquired on a 3.0 Tesla scanner (GE Signa EXCITE, GE Healthcare). For R-fMRI imaging, 34 slices were acquired in the same plane (as the low resolution T1-weighted images) using a SENSE inverse-spiral pulse sequence with echo time (TE) = 32 ms, repetition time (TR) = 2000 ms, FOV = 25.6 cm2, matrix = 64 × 64 × 34, 3.8 mm3. For DTI, 25 direction diffusion-weighted whole-brain volumes were acquired axially parallel to the AC-PC line using diffusion weighting values, b = 0 and 1000 s/mm2, flip angle = 90°, TR = 17 s and TE = 78 ms. The imaging matrix was 256 × 256 with a rectangular FOV of 256 × 256 mm2 and 72 slices with a slice thickness of 2.0 mm.
Pre-processing steps of the R-fMRI data included brain skull removal, motion correction, spatial smoothing, slice time correction, global drift removal, and band pass filtering (0.01Hz~0.1Hz) (Li et al., 2010). Pre-processing steps of DTI data included brain skull removal, motion correction, and eddy current correction (Liu et al., 2007). After the pre-processing, fiber tracts, gray matter (GM) and white matter (WM) tissue segmentation (Liu et al., 2007), and the GM/WM cortical surface reconstruction (Liu et al., 2008b) were performed based on the DTI data. Fiber tracking was performed via the MEDINRIA (http://www-sop.inria.fr/asclepios/software/MedINRIA/). The stopping criteria are: FA threshold=0.2 and minimum fiber length=20. Moreover, we adopted other four different fiber tracking software toolkits or parameter settings (Fillard et al., 2011) to examine the identification consistency of our DICCCOL landmarks: DTIStudio (https://www.dtistudio.org/), and MRtrix (Tournier et al., 2012, http://www.nitrc.org/projects/mrtrix/) with three parameter settings (fiber bundle number equals 10,000, 50,000 and 100,000, respectively). Brain tissue segmentation was conducted on the DTI data directly (Liu et al., 2007). Based on the WM tissue map, the cortical surface was reconstructed using the marching cubes algorithm (Liu et al., 2008b). The reconstructed surface has approximately 40,000 vertices and is used as the standard space for predicting DICCCOL landmarks (Zhu et al., 2012) and encoding RSNs.
Construction of RSNs
For the NC subjects, we constructed their RSNs by using the group independent component analysis (gICA) (Schmithorst and Holland, 2004) implemented in the GIFT toolbox (http://mialab.mrn.org/software/gift/index.html). This GIFT toolbox is established for the ICA of group (or single subject) fMRI data (Calhoun et al., 2001; Beckmann et al., 2005; Celone et al., 2006). The basic idea of gICA is that we first concatenated individuals’ data across time, and then computed the subject-specific components and time courses. To be self-contained, the GIFT toolbox performed the gICA via three major steps (Calhoun et al., 2001). In step 1, first, individuals’ R-fMRI data was reduced by principal components analysis (PCA) in order to decrease computing complexity while preserving the useful information. Second, the number of independent components was estimated based on all input fMRI data using the MDL criteria (Li et al., 2007). More details are referred to Li, et al., 2007. Our experiments show that although the estimated total number of ICA components might be slightly different if various sample sizes of the NC group are chosen, all fifteen RSNs are successfully identified among all ICA components. Third, individuals’ R-fMRI data was reduced by using PCA again based on the selected number of components. In step 2, ICA was applied by using the default Infomax algorithm (Linsker, 1997). In step 3, subject-specific components and time courses were back-reconstructed and each mean component was transformed to z-values. For each component, there were two patterns that were correlated or anti-correlated to the time course of this component.
By using the templates and visual inspections similar to those used in Zhu et al., 2012, we determined corresponding group mean independent components that represented RSNs elaborated in previous published works (Damoiseaux et al., 2006; De Luca et al., 2006; Beckmann et al., 2005; van den Heuvel et al., 2008; Salvador et al., 2005; Sorg et al., 2007) and excluded the other components that were artifacts, i.e., in ventricle or cerebrospinal fluid (CSF).
Identification of DICCCOL Landmarks
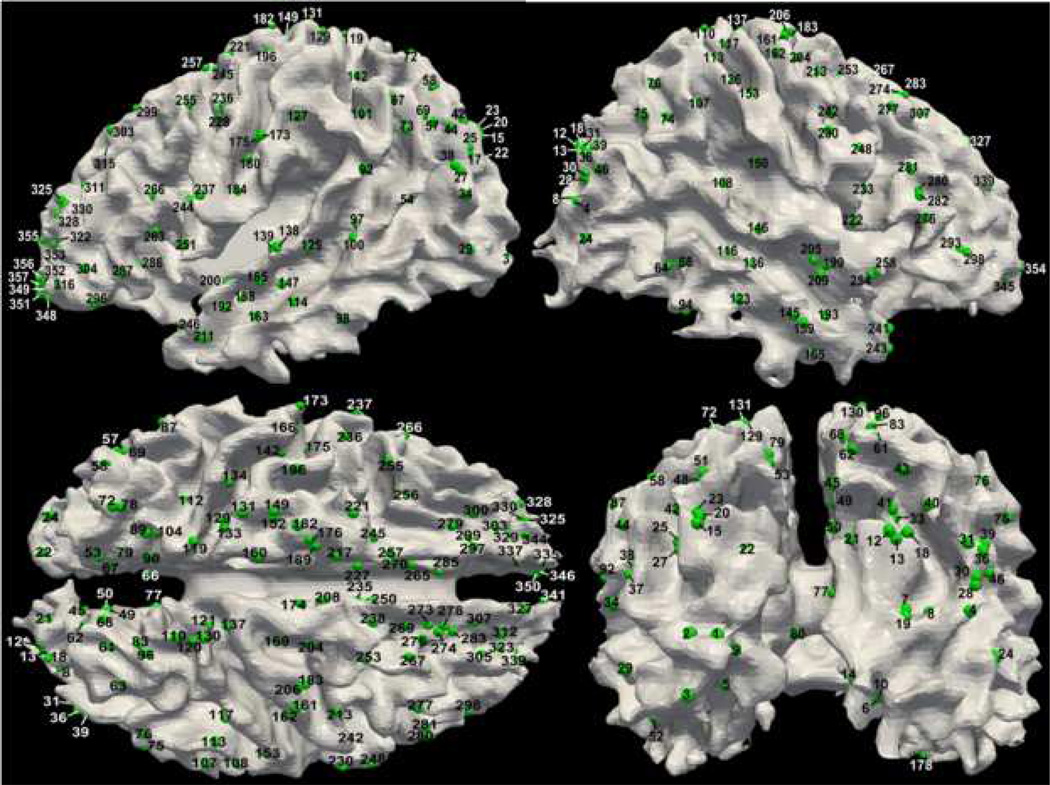
To be self-contained, the DICCCOL landmark discovery and prediction procedures (Zhu et al., 2011b; Zhu et al., 2012) are briefly introduced here. In general, the DICCCOL landmark identification was formulated as an optimization problem. In the first step, we randomly selected one subject as the template brain and generated a dense and regular map of 3D grid points within the boundary box of the reconstructed cortical surface based on DTI data. The intersection locations between the grid map and the cortical surface were used as the initial landmarks. As a result, we generated dense landmarks on the template brain. Then, we registered the landmarks to other subjects using the linear registration via FSL FLIRT to initialize and establish the rough correspondences of the landmarks across different subjects. In the second step, we extracted white matter fiber bundles emanating from small regions around the neighbourhood of each landmark. Each small region served as the candidate for landmark location optimization. Afterwards, we projected the fiber bundles to a standard sphere space, called trace-map (Zhu et al., 2011a; Zhu et al., 2011b; Zhu et al., 2012), and calculated the distance between any pair of trace-maps in different subjects. Finally, we performed a whole-space search to find fiber bundles which gave the best group-wise similarity. By using both quantitative and qualitative methods (Zhu et al., 2011a; Zhu et al., 2011b; Zhu et al., 2012) to evaluate the consistency of converged landmarks in two separate groups of model brains, we determined 358 DICCCOL landmarks. More details can be referred to Zhu et al., 2011a, Zhu et al., 2011b, and Zhu et al., 2012. Fig. 2 shows the anatomical locations and index labelling of these 358 DICCCOL landmarks on one brain.
Figure 2.
Anatomical locations and index labelling of 358 DICCCOL landmarks on one brain. The DICCCOL landmarks are highlighted by green bubbles. Additional visualizations of these landmarks are available online at: http://dicccol.cs.uga.edu.
The prediction of DICCCOL landmarks on other brains was similar to the optimization procedure. Generally, we warped a new subject to the template brain and performed the optimization procedure to predict its DICCCOL landmarks. The only difference was that since the locations of DICCCOL landmarks in the model brains were already optimized, we kept them unchanged and only optimized the location of the new subject to minimize the trace-map distance among the models and the new subject. This set of 358 DICCCOL landmarks has been predicted and reproduced in four different multimodal fMRI/DTI datasets (over 240 brains) which are released on line at: http://dicccol.cs.uga.edu. In this paper, we predicted 358 DICCCOL landmarks for each brain in both NC and MCI groups.
Predictive Models of RSNs Based on DICCCOL Landmarks
In this section, we used DICCCOL landmarks derived from DTI data to learn the predictive models of RSNs derived from R-fMRI data. As the DICCCOL landmarks were identified in the DTI space, co-registration between DTI and R-fMRI data was performed using the FSL FLIRT, and the resulting global transform matrix was applied to the RSNs to map them into the DTI space. The activity peaks (represented by z-values) of each RSN were then selected in each subject to represent the corresponding RSNs. It was noteworthy that the activity peak location was defined on the DTI-derived cortical surface onto which the RSNs in R-fMRI space were mapped.
For the purpose of encoding the RSNs using DICCCOL landmarks, first, the top 5 closest DICCCOL landmarks (measured by Euclidian distance) in each subject were identified for each corresponding activity peak in a group. Second, the DICCCOL landmark with the most votes (defined as frequencies of being ranked as closest distance to the activity peaks) was determined as the corresponding landmark for that activity peak in a group. Results showed that there was always a DICCCOL landmark which could be found for the corresponding activity peak across subjects (Zhu et al., 2012), suggesting that RSNs were well co-localized with a set of consistent and predictable DICCCOL landmarks across individuals and populations. Therefore, we encoded all RSNs via DICCCOL landmarks, and used these landmarks as predictive models of RSNs. Since the structural brain architectures in MCI brains were relative similar to those in NC group, the nodes of RSNs in MCI were then localized based on the DICCCOL landmark-based predictive models. Afterwards, the functional connectivities among these predicted RSNs were assessed in MCI and compared with those in NC. Since the RSNs in MCI were disrupted or substantially altered, the comparisons of functional connectivities among consistent and corresponding structural DICCCOL landmarks between MCI and NC groups were an alternative, fair and meaningful way to assess the resting state functional connectivity alterations of RSNs in MCI.
Results
Construction of RSNs in NC
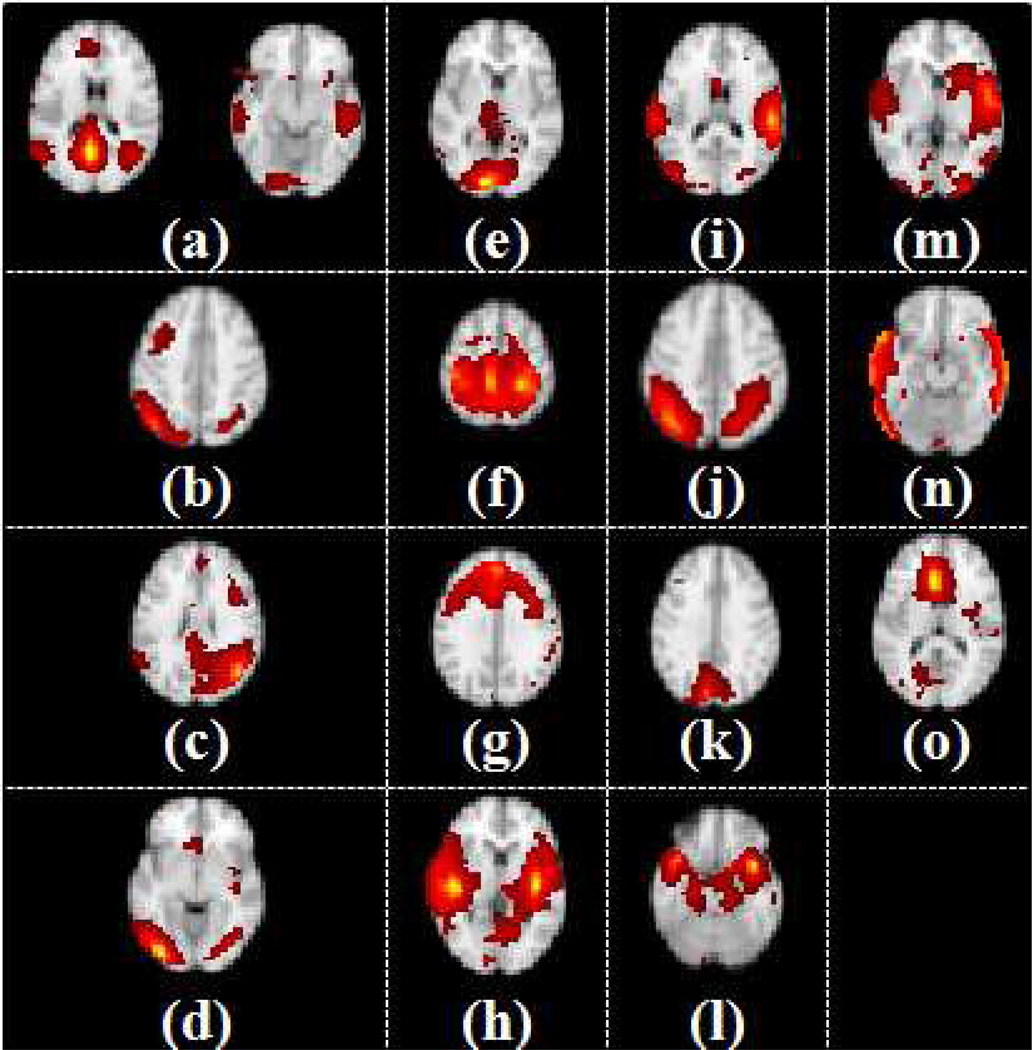
As described above, the group ICA approach was performed to construct RSNs in NC using the GIFT software. Results of the constructed RSNs are depicted in Fig. 3. In total, fifteen independent components representing fifteen separate RSNs were constructed, as shown in Figs. 3a–3o, respectively. Specifically, Fig. 3a includes the medial prefrontal gyrus (BAs 9/10/11), anterior (BAs 12/32) and posterior cingulate cortex (BA 29), bilateral supramarginal gyrus (BA 39), and the inferior temporal gyrus (BA 21). This component corresponds to the default mode network (DMN) (Damoiseaux et al., 2006; Raichle et al., 2001; De Luca et al., 2006; van den Heuvel et al., 2008; Sorg et al., 2007). Figs. 3b–3c show components that have strong lateralization in the right (Fig. 3b) and left (Fig. 3c) hemispheres, including the middle frontal and orbital (BAs 6/9/10) and superior parietal areas (BAs 7/40). These RSNs are known to be involved in memory function (De Luca et al., 2006; Damoiseaux et al., 2006; van den Heuvel et al., 2008; Sorg et al., 2007).
Figure 3.
Fifteen RSNs in the NC subjects. Each group independent components representing RSNs is shown on a T1 structural template brain image. The color scale represents z values ranging from 0 to 23.1.
Two separate components shown in Figs. 3d–3e are in the visual cortex. Fig. 3d consists of the lateral and superior occipital gyrus (BA 19), while Fig. 3e includes the medial visual areas (BAs 17/18) (Damoiseaux et al., 2006; De Luca et al., 2006; van den Heuvel et al., 2008; Salvador et al., 2005; Sorg et al., 2007). Fig. 3f shows the sensory-motor systems including pre- and post-central gyrus from the sylvian fissure to the medial wall of the inter-hemispheric fissure (BAs 1/2/3/4), and the Supplementary Motor Area (SMA) (BA 6). This RSN can be seen in the bimanual motor tasks (Damoiseaux et al., 2006; De Luca et al., 2006; van den Heuvel et al., 2008; Salvador et al., 2005). Fig. 3g includes the prefrontal cortex (BA 11) and dorsal anterior cingulate (BA 32) (Damoiseaux et al., 2006). This RSN has been found in previous research to be involved in the execution and working memory functions (Beckmann et al., 2005; Miller and Cohen, 2001; Salvador et al., 2005).
Fig. 3h shows the auditory system encompassing the Heschl’s gyrus, lateral superior temporal gyrus and posterior insular cortex (Rivier and Clarke, 1997; Rademacher et al., 2001; Damoiseaux et al., 2006; van den Heuvel et al., 2008; Salvador et al., 2005). Fig. 3i shows the superior temporal gyrus (BA 22) as the main part of this RSN. This RSN shows the occipito-temporal pathway (ventral stream) (Damoiseaux et al., 2006; De Luca et al., 2006; Sorg et al., 2007). Fig. 3j involves the superior parietal cortex (BA 7), occipito-temporal (BA 37) and precentral gyrus (BA 4) (Damoiseaux et al., 2006; Salvador et al., 2005; Sorg et al., 2007). Fig. 3k consists of a posterior part of superior parietal cortex (BA 7) (van den Heuvel et al., 2008; Salvador et al., 2005). Fig. 3l shows temporal polar cortex of the medial temporal system (Salvador et al., 2005). Fig. 3m shows medial temporal regions of the medial temporal system (Salvador et al., 2005). Fig. 3n shows the temporal association cortex of the lateral temporal/auditory-verbal system (Salvador et al., 2005). Fig. 3o represents the anterior part of the cingulate cortex (van den Heuvel et al., 2008).
All activity peaks (represented by z-values) of RSNs in NC were then selected and summarized in Table 2. Overall, there were 38 peaks in all of the fifteen RSNs. All results were reported in the MNI_152 template space and Brodmann Area (BA). The atlases used to label the activity peaks were the Harvard-Oxford cortical and subcortical structural atlases and Talairach Daemon Labels provided by FSL.
Table 2.
Activity peaks of RSNs. RSNs #1–#15 are corresponding to Figs. 3a-3o, respectively. X, Y and Z (units: mm), peak coordinates in MNI_152 template space; BA, Brodmann Area.
| RSN | X | Y | Z | Atlas Label | BA | X | Y | Z | Atlas Label | BA |
|---|---|---|---|---|---|---|---|---|---|---|
| #1 | −2 | 50 | 26 | Medial Frontal Gyrus | 9 | 7 | 43 | 26 | Medial Frontal Gyrus | 9 |
| −9 | −51 | 8 | Posterior Cingulate | 29 | 6 | −55 | 11 | Posterior Cingulate | 29 | |
| −58 | −18 | −12 | Middle Temporal Gyrus | 21 | 48 | −12 | −12 | Middle Temporal Gyrus | 21 | |
| −41 | −62 | 39 | Angular Gyrus | 39 | 43 | −61 | 39 | Angular Gyrus | 39 | |
| #2 | 48 | −44 | 39 | Supramarginal Gyrus | 40 | 41 | 17 | 34 | Middle Frontal Gyrus | 9 |
| #3 | −42 | −45 | 39 | Supramarginal Gyrus | 40 | −49 | 21 | 33 | Middle Frontal Gyrus | 9 |
| #4 | −27 | −90 | −5 | Inferior Occipital Gyrus | 18 | 27 | −92 | −6 | Inferior Occipital Gyrus | 18 |
| #5 | −8 | −98 | 4 | Lingual Gyrus | 17 | 21 | −95 | 2 | Lingual Gyrus | 17 |
| #6 | −37 | −20 | 56 | Precentral Gyrus | 4 | 37 | −20 | 53 | Precentral Gyrus | 4 |
| −8 | −19 | 55 | Medial Frontal Gyrus | 6 | 13 | −18 | 52 | Medial Frontal Gyrus | 6 | |
| #7 | −17 | 54 | 27 | Superior Frontal Gyrus | 9 | 9 | 51 | 25 | Superior Frontal Gyrus | 9 |
| #8 | −37 | 5 | 7 | Insular Cortex | 13 | 45 | 3 | 7 | Insular Cortex | 13 |
| #9 | −52 | −6 | 0 | Superior Temporal Gyrus | 22 | 53 | −8 | −2 | Superior Temporal Gyrus | 22 |
| #10 | −36 | −41 | 57 | Inferior Parietal Lobule | 40 | 31 | −48 | 50 | Superior Parietal Lobule | 7 |
| #11 | −10 | −67 | 48 | Precuneus | 7 | 10 | −64 | 41 | Precuneus | 7 |
| #12 | −33 | 5 | −20 | Temporal Pole | 38 | 30 | 13 | −27 | Temporal Pole | 47 |
| #13 | −46 | 5 | −29 | Middle Temporal Gyrus | 21 | 51 | 5 | −25 | Superior Temporal Gyrus | 38 |
| #14 | −55 | −10 | −20 | Middle Temporal Gyrus | 21 | 60 | −13 | −21 | Middle Temporal Gyrus | 21 |
| #15 | −3 | 35 | 1 | Cingulate Gyrus | 24 | 4 | 37 | 8 | Cingulate Gyrus | 24 |
DICCCOL Landmark Prediction in NC and MCI
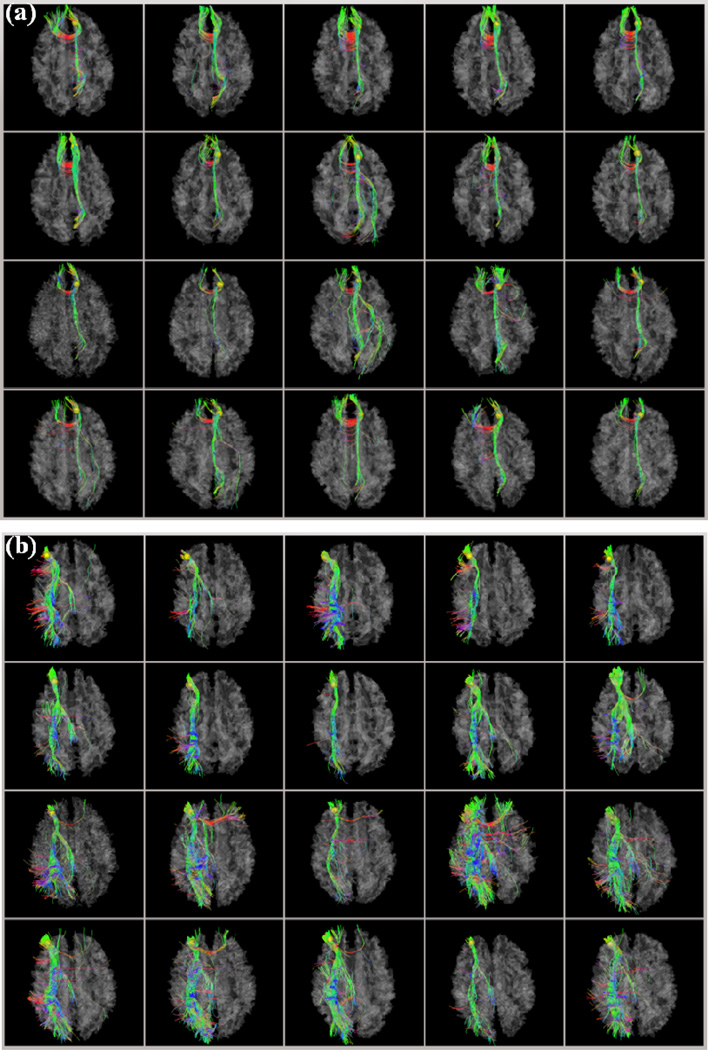
In this section, we analyzed the DICCCOL landmark prediction results in NC and MCI qualitatively and quantitatively. Figs. 4a–4b show two examples of DICCCOL landmark prediction results in NC and MCI, respectively. For Figs. 4a–4b, the first two rows are ten model brains (Zhu et al., 2012), the third and fourth rows are the predicted results in five randomly selected NC and MCI brains, respectively. The example DICCCOL landmarks are #311 (Fig. 4a) and #315 (Fig. 4b) as shown in Fig. 2. From visual inspection, we can see that the fiber bundle patterns of corresponding DICCCOL landmarks in both predicted NC and MCI brains are quite similar with those in the model brains. More results are referred to http://dicccol.cs.uga.edu.
Figure 4.
Two examples of DICCCOL landmark prediction results in NC and MCI. The DICCCOL landmarks are represented by the yellow bubbles and their indices are (a) #311 and (b) #315 as shown in Fig. 2. For (a) and (b), the first two rows are 10 model brains, the third and fourth rows are the predicted results of five randomly selected brains in NC and MCI, respectively. More results are referred to http://dicccol.cs.uga.edu.
To quantitatively evaluate the similarity of fiber bundle patterns of 358 DICCCOL landmarks between NC and MCI, we first calculated the trace-maps of 358 DICCCOL landmarks in the model, NC, and MCI groups, respectively. Then, for each of the three groups (model, NC and MCI), we calculated the mean trace-maps of 358 DICCCOL landmarks. At last, distances of the mean trace-maps of 358 DICCCOL landmarks between any two groups were calculated via the methods in Zhu et al., 2012.
Table 3 shows the distances of the mean trace-maps of sixteen randomly selected DICCCOL landmarks between any pair of the three groups. As shown in Table 3, the distances of the mean trace-map of these sixteen DICCCOL landmarks among all three groups are all very small: 22.29×10−5 between model and NC, 20.87×10−5 between model and MCI, and 6.29×10−5 between NC and MCI. To further demonstrate that the above distances are indeed small, we randomly selected ten vertices on the entire cortical surface of one model brain, extracted their fiber bundles and calculated the trace-maps. Then, the distances between each of the ten trace-maps and the trace-map of DICCCOL #327 were calculated as an example in Table 4. We can see that the mean distance is 337.06×10−5 which is obviously much larger than the above three mean distances. For all of the rest DICCCOL landmarks, we have the same finding.
Table 3.
Distances (×10−5) of the mean trace-maps of sixteen randomly selected DICCCOL landmarks between any two pairs of model, NC and MCI groups.
| DICCCOL ID | #327 | #77 | #145 | #46 | #303 | #80 | #156 | #73 |
| Model vs. NC | 35.29 | 14.11 | 20.62 | 33.41 | 38.88 | 15.25 | 14.85 | 4.90 |
| Model vs. MCI | 24.19 | 15.66 | 18.91 | 22.97 | 34.05 | 12.26 | 17.68 | 8.76 |
| NC vs. MCI | 8.66 | 5.75 | 1.96 | 4.29 | 4.52 | 2.66 | 1.91 | 4.23 |
| DICCCOL ID | #87 | #255 | #205 | #139 | #193 | #211 | #159 | #114 |
| Model vs. NC | 9.82 | 34.23 | 30.17 | 29.52 | 20.43 | 7.77 | 28.30 | 19.12 |
| Model vs. MCI | 15.27 | 15.81 | 22.67 | 24.72 | 5.29 | 13.80 | 13.81 | 26.54 |
| NC vs. MCI | 7.80 | 11.98 | 4.83 | 10.25 | 10.03 | 2.84 | 11.53 | 7.47 |
Table 4.
Distances (×10−5) between each of the trace-maps of ten randomly selected vertices and trace-map of DICCCOL #327.
| Vertex | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 | #9 | #10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Distance | 197.97 | 424.49 | 251.96 | 241.75 | 155.14 | 485.82 | 361.04 | 514.73 | 482.05 | 255.66 |
Moreover, we adopted other four different fiber tracking software toolkits or parameter settings (Fillard et al., 2011) to examine the identification consistency of our DICCCOL landmarks: DTIStudio (https://www.dtistudio.org/), and MRtrix (Tournier et al., 2012, http://www.nitrc.org/projects/mrtrix/) with three parameter settings (fiber bundle number equals 10,000, 50,000 and 100,000, respectively). The basic idea is that in the DICCCOL landmark identification part, we predicted all 358 DICCCOL landmarks in all 28 subjects based on the extracted fibers in DTI image via five different fiber tracking software toolkits or parameter settings, respectively. Then, for each of the predicted corresponding DICCCOL landmark across subjects in the NC/MCI group, we calculated the mean distance of any pair of the predicted locations based on the extracted fibers via five different fiber tracking software toolkits or parameter settings. The overall mean distance for all 358 DICCCOL landmarks is 4.25 mm ± 0.89 mm in NC group, and 4.87 mm ± 1.02 mm in MCI group, which is relatively small. The results show that our DICCCOL landmark locations are consistent across different subjects using different fiber tracking software toolkits and thus are not affected by different tracking software toolkits or parameters.
From above qualitative and quantitative results, we can conclude that: 1) given the DICCCOL landmarks in the model brains, we can effectively and accurately predict their corresponding counterparts in a new brain in NC and MCI groups with DTI data; 2) the patterns of fiber bundles of corresponding DICCCOL landmarks in both NC and MCI brains are quite consistent with model brains. We have examined all of the 358 predicted DICCCOL landmarks in NC and MCI subjects, and found the similar conclusion (http://dicccol.cs.uga.edu), suggesting that the DICCCOL landmarks indeed reveal the common structural connectivity patterns of the human brains including MCI subjects, which is also the foundation that we can use these structural DICCCOL landmarks to encode RSNs and use them as predictive models to elucidate the functional connectivity alterations in MCI.
Predictive Models of RSNs Based on DICCCOL Landmarks
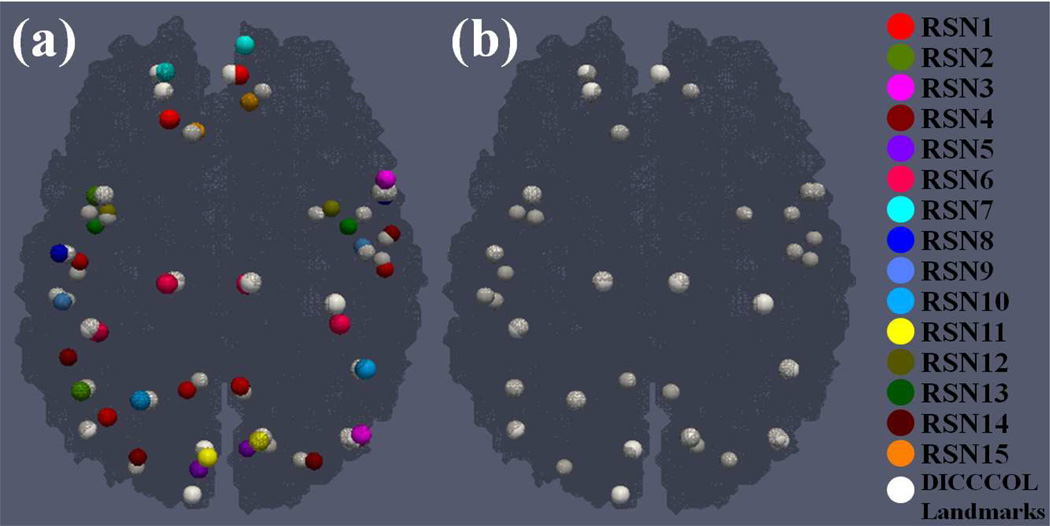
Once RSNs were identified based on R-fMRI data and DICCCOL landmarks were predicted based on DTI data, we mapped all RSNs to the DTI space and encoded them via DICCCOL landmarks by the approaches described in above sections. The learned set of DICCCOL landmarks was employed as the predictive models of RSNs for MCI. Fig. 5a visualizes all 38 activity peaks of 15 RSNs in NC and their corresponding predictive DICCCOL landmarks on one NC subject. Fig. 5b shows the DICCCOL landmark-based predicted RSNs on one MCI subject.
Figure 5.
Predictive models of RSNs in NC and MCI. (a) All 38 activity peaks of 15 RSNs and corresponding predictive DICCCOL landmarks in NC; (b) DICCCOL landmark-based predicted RSNs in MCI. Activity peaks in different RSNs are represented by the bubbles with different colors as shown in the right panel. Corresponding predictive DICCCOL landmarks are represented by the white bubbles.
To quantitatively evaluate the RSN encoding accuracy by DICCCOL landmarks, we measured the Euclidean distance between the centers of activity peaks within each RSN and corresponding predictive DICCCOL landmarks in the NC group, respectively. All results are reported in Table 5. The average distances for the fifteen RSNs in the NC group are 5.54 mm, 5.99 mm, 4.86 mm, 7.31 mm, 8.79 mm, 4.11 mm, 5.39 mm, 6.02 mm, 4.92 mm, 5.99 mm, 5.91 mm, 5.54 mm, 5.60 mm, 5.51 mm and 5.73 mm, respectively. On average, the distance is 5.81 mm. The results in Table 5 demonstrate that DICCCOL landmarks are consistently co-localized with RSNs, and can encode RSNs effectively and accurately.
Table 5.
Euclidean distance between the center of activity peak within each RSN and corresponding predictive DICCCOL landmark. Distance is measured in mm; Stdev, standard deviation.
| RSN | #1 | |||||||
| Activity Peak | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 |
| DICCCOL ID | #327 | #77 | #145 | #46 | #303 | #80 | #156 | #73 |
| Distance: Mean | 5.87 | 7.63 | 3.97 | 6.05 | 5.88 | 6.35 | 3.99 | 4.54 |
| Distance: Stdev | 3.02 | 3.94 | 2.14 | 2.61 | 2.84 | 3.22 | 1.24 | 2.51 |
| RSN | #2 | #3 | #4 | #5 | ||||
| Activity Peak | #1 | #2 | #1 | #2 | #1 | #2 | #1 | #2 |
| DICCCOL ID | #87 | #255 | #76 | #276 | #35 | #3 | #14 | #15 |
| Distance: Mean | 4.60 | 7.37 | 4.77 | 4.94 | 5.93 | 8.68 | 8.99 | 8.58 |
| Distance: Stdev | 3.14 | 2.84 | 2.34 | 2.92 | 3.85 | 2.53 | 4.81 | 3.85 |
| RSN | #6 | #7 | #8 | |||||
| Activity Peak | #1 | #2 | #3 | #4 | #1 | #2 | #1 | #2 |
| DICCCOL ID | #161 | #197 | #196 | #189 | #341 | #329 | #233 | #190 |
| Distance: Mean | 4.06 | 3.48 | 3.74 | 5.17 | 6.09 | 4.68 | 6.48 | 5.56 |
| Distance: Stdev | 1.98 | 1.27 | 1.83 | 3.90 | 2.56 | 2.61 | 3.75 | 3.74 |
| RSN | #9 | #10 | #11 | #12 | ||||
| Activity Peak | #1 | #2 | #1 | #2 | #1 | #2 | #1 | #2 |
| DICCCOL ID | #205 | #139 | #117 | #78 | #68 | #47 | #185 | #168 |
| Distance: Mean | 5.62 | 4.21 | 5.79 | 6.18 | 4.81 | 7.01 | 6.04 | 5.04 |
| Distance: Stdev | 2.73 | 2.85 | 3.17 | 2.92 | 2.02 | 3.47 | 4.30 | 3.20 |
| RSN | 13 | 14 | 15 | |||||
| Activity Peak | #1 | #2 | #1 | #2 | #1 | #2 | ||
| DICCCOL ID | #193 | #211 | #159 | #114 | #314 | #285 | ||
| Distance: Mean | 5.34 | 5.86 | 4.65 | 6.36 | 6.84 | 4.62 | ||
| Distance: Stdev | 2.55 | 3.92 | 2.53 | 2.30 | 2.78 | 1.79 | ||
Furthermore, we adopted a 10-fold cross-validation to validate the learned functional network on DICCCOL landmarks. Specifically, we divided 18 subjects in the NC group into ten folds (eight folds have two subjects and the other two folds have one subject respectively). Each time one fold was used as the testing set, and the rest of the folds were used as the training set to perform RSN encoding using DICCCOL landmarks. For each corresponding activity peak, the learned DICCCOL landmark was examined in the testing set to see if it is closest to the activity peak. Then the accuracy rate was calculated across ten times for each activity peak. The mean accuracy rate of all activity peaks is 80.26%, indicating that the learned RSNs based on DICCCOL landmarks are reliable.
Measurement of Functional Connectivity in NC and MCI
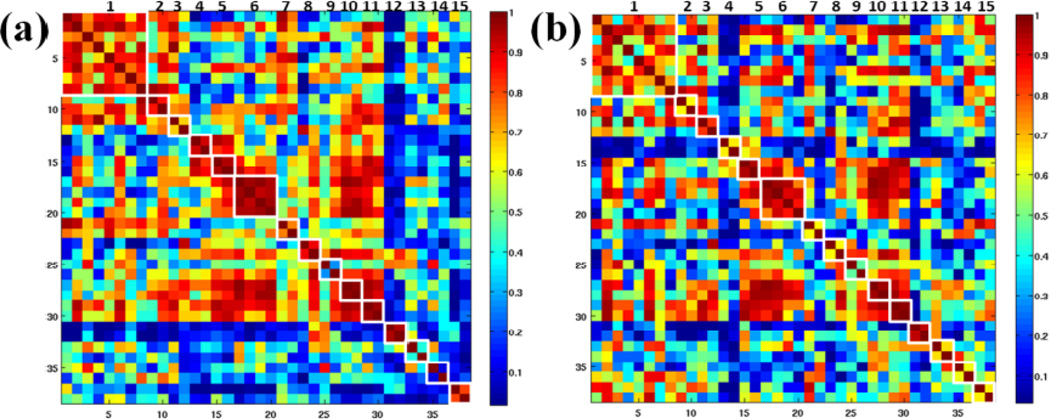
Our previous studies in Zhu et al., 2011b and Zhu et al., 2012 and the results in above sections have demonstrated that the DICCCOL landmarks are consistently co-localized with RSNs (e.g., default mode network) across subjects. That is, the functional connectivities based on (1) activity peaks of RSNs and (2) DICCCOL landmark-based predictive models of RSNs should be similar. Typically, the functional connectivity between two peaks/landmarks was defined as the Pearson correlation coefficient between their fMRI time series (Li et al., 2010; Li et al., 2012a; Zhu et al., 2013). Figs. 6a–6b show the average functional connectivities based on activity peaks of R-fMRI derived RSNs (Fig. 6a) in the NC group and DICCCOL landmark-based predictive models of RSNs (Fig. 6b), respectively. They are similar as highlighted by the white frames in the principal diagonal direction. Furthermore, for some RSNs (#9, 13 and 14), the average functional connectivity based on the predictive models of RSNs is even higher than that based on activity peaks of RSNs, suggesting that the DICCCOL landmarks generated by DTI data might help localize individual RSNs more accurately than using the group ICA approach merely based on R-fMRI data. This is in agreement with our previous results (Zhu et al., 2011a) that have shown that the maximization of group-wise consistency of structural connection patterns across functional landmarks could benefit functional connectivity profile as well.
Figure 6.
Average functional connectivities based on (a) activity peaks of R-fMRI derived RSNs and (b) DICCCOL landmark-based predictive models of RSNs, respectively. RSNs are highlighted by the white frames in the principal diagonal direction and indexed in the top panel.
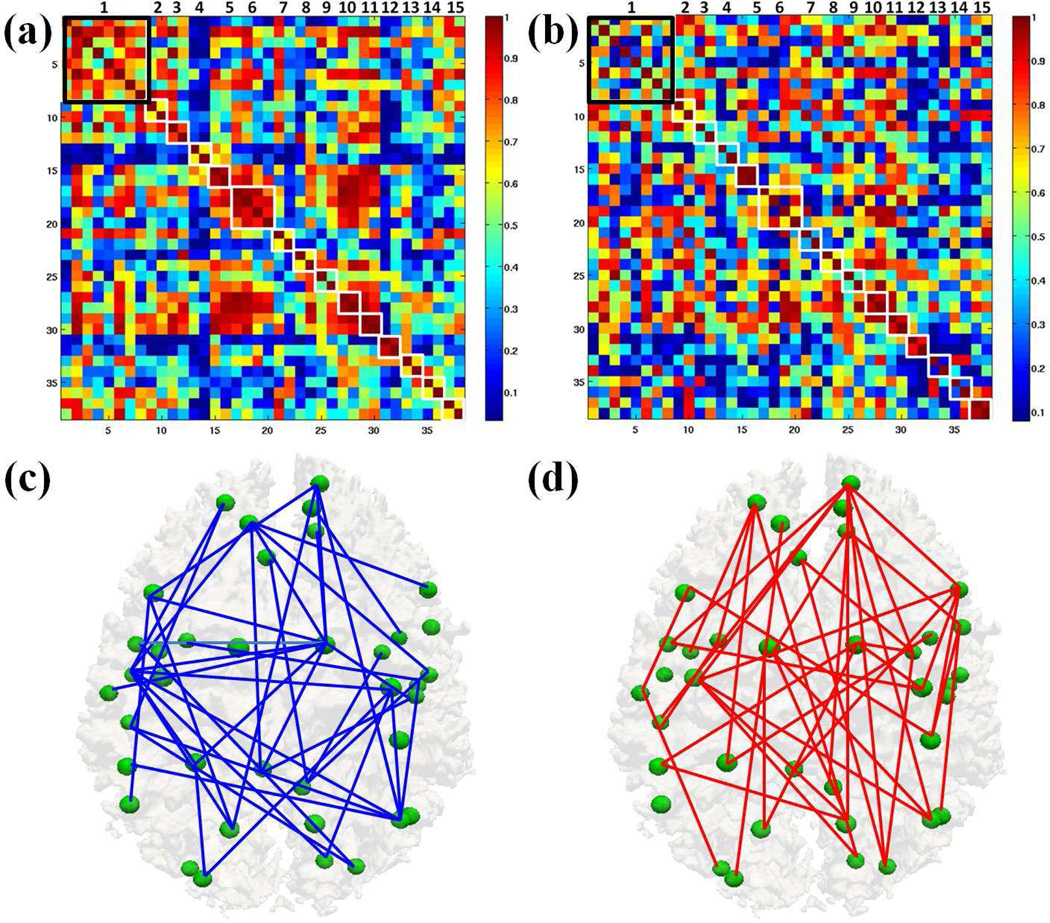
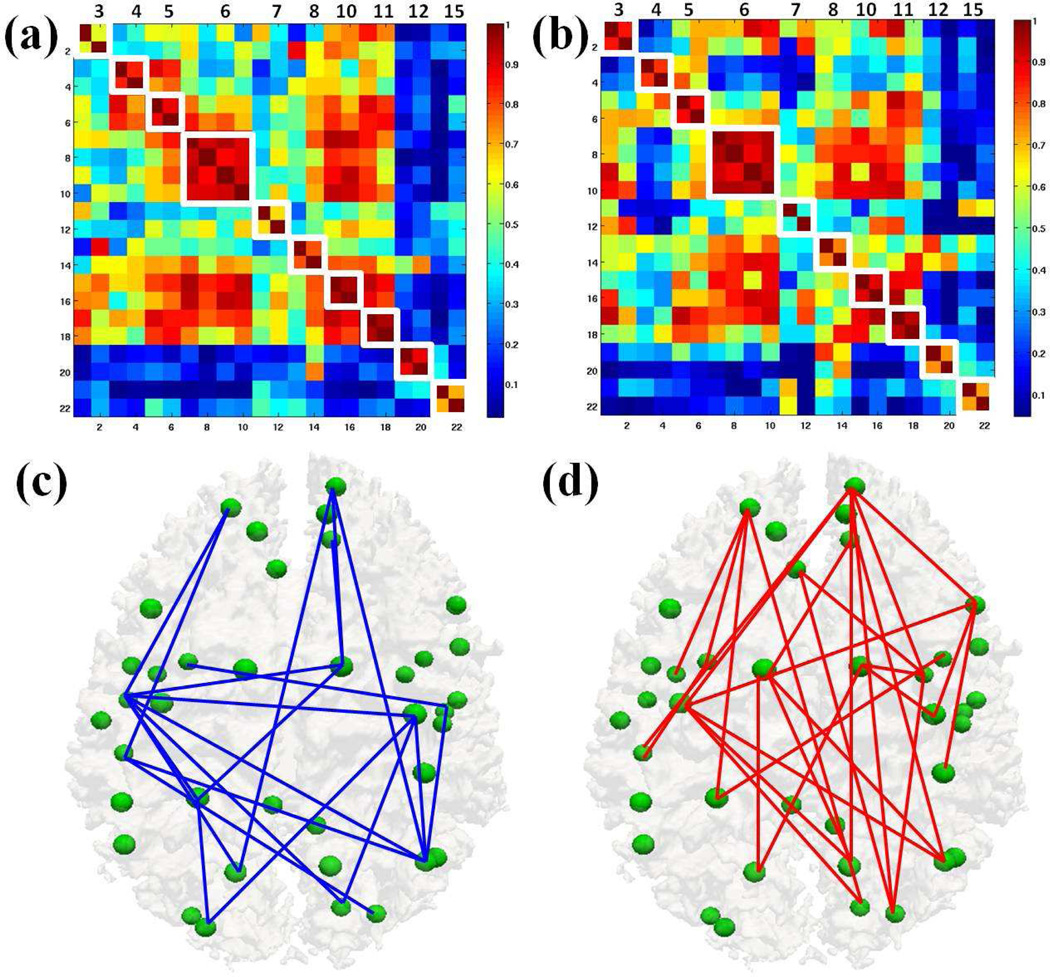
Then, we used the DICCCOL landmark-based predictive models to assess the functional connectivity alterations in MCI compared with those in NC. Figs. 7a– 7b show the average functional connectivities based on the predictive models of RSNs in NC (Fig. 7a) and MCI (Fig. 7b), respectively. We can see that the functional connectivities between NC and MCI are substantially different, suggesting the widespread alterations of functional connectivity in MCI. To statistically examine significant between-group difference, we performed a two-sample t-test of functional connectivities in Figs. 7a–7b. Significantly decreased (in blue) and increased (red) (p-value<0.05, false discovery rate (FDR) corrected for multiple comparisons) functional connectivities in MCI in comparison with NC are shown in Figs. 7c–7d, respectively. These results show that the DICCCOL landmark-based predictive models of RSNs can reveal widespread functional connectivity alterations in MCI. First, there are functional connectivity alterations within a specific RSN. For instance, there are functional connectivity alterations between specific node pairs within the default mode network (RSN #1) in MCI, as highlighted by the black boxes in Figs. 7a–7b. There are also functional connectivity alterations within RSN #3, #4, #6, #7, #13 and #14 in MCI, as highlighted white boxes respectively along the diagonal in Figs. 7a–7b. Second, if we examine the upper/lower triangular matrices in Figs. 7a–7b, we can see that there are wide spread functional connectivity alterations between node pairs which belong to two different RSNs. In conclusion, Fig. 7 demonstrates that there are widespread functional connectivity alterations not only within specific RSNs, but also among different RSNs.
Figure 7.
Widespread functional connectivity alterations in MCI. (a)–(b) represent average functional connectivities based on the DICCCOL landmark-based predictive models of RSNs in NC and MCI, respectively; (c)–(d) show significantly decreased (in blue) and increased (red) functional connectivities in MCI in comparison with NC (p-value < 0.05, FDR corrected for multiple comparisons).
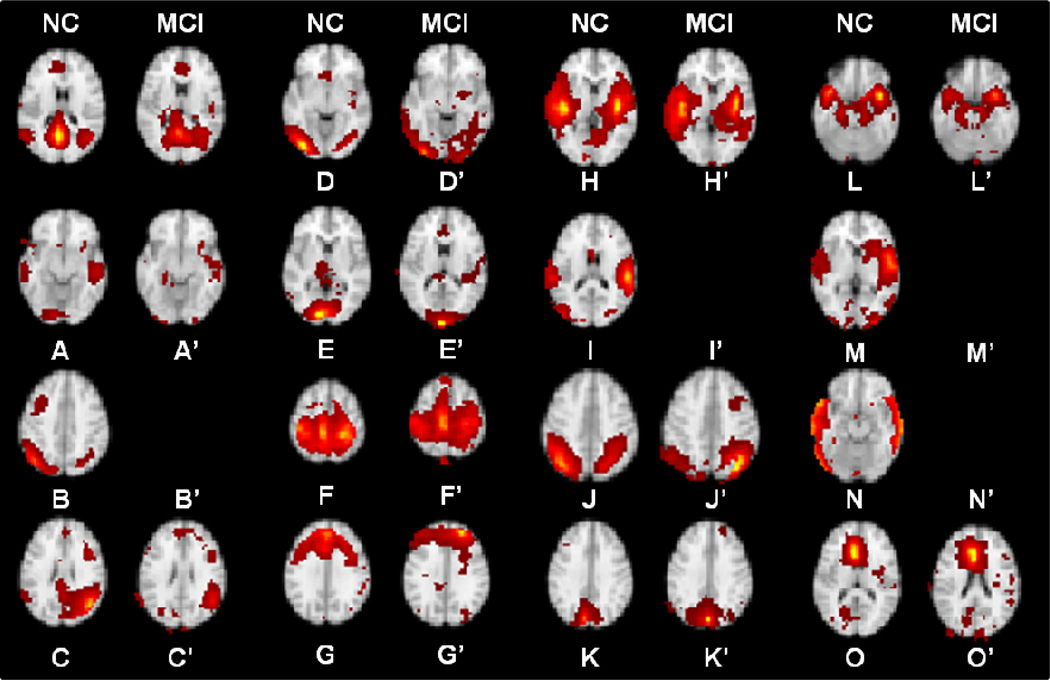
Finally, to demonstrate that our predictive models can improve the detection power of identifying functional connectivity alterations in MCI, we calculated the functional connectivity alterations in MCI based on the activity peaks merely derived from R-fMRI data and compared the results with those in Fig. 7. First, we adopted the same gICA approach to identify the RSNs in MCI group. As shown in Fig. 8, we can see that 10 corresponding RSNs were identified in MCI group, while the other five RSNs (RSN #1, #2, #9, #13 and #14 in Fig. 5) in NC were not found or altered in MCI. This shows the advantage of our proposed predictive models to locate those altered RSNs in MCI which cannot be identified using gICA. Second, we calculated the functional connectivity alterations in MCI based on the activity peaks of the ten RSNs derived merely from R-fMRI data. Figs. 9a–9b show the average functional connectivities based on the activity peaks of the ten RSNs merely derived from R-fMRI data. Similar as in Fig. 7, we performed a two-sample t-test of functional connectivities in Figs. 9a–9b and showed the significantly decreased (in blue) and increased (in red) (p-value<0.05, FDR corrected for multiple comparison) functional connectivities in MCI in comparison with NC in Figs. 9c–9d, respectively. Figs. 9c–9d demonstrate that we can also identify some functional connectivity alterations in MCI merely based on R-fMRI data. However, many decreased/increased connectivity alterations were not identified compared with Figs. 7c–7d. In conclusion, our DICCCOL landmark-based predictive models of RSNs not only can locate those RSNs which are already altered and thus cannot be identified via gICA, but also have more detection power of identifying widespread functional connectivity alterations in MCI.
Figure 8.
Ten identified RSNs in MCI group. Each pair of group-independent components is shown on a T1 structural template brain image. The identified fifteen RSNs in NC group (Fig. 3) are also shown here for comparison. Four RSNs in NC group (B, I, M and N) are not identified in MCI. One RSN (DMN) in MCI as shown in A’ partially alters. The color scale represents z values ranging from 0 to 23.1.
Figure 9.
Functional connectivity alterations in MCI merely based on R-fMRI data. (a)–(b) represent average functional connectivities merely based on the R-fMRI data in NC and MCI, respectively. It should be noted that RSN #1, #2, #9, #13 and #14 are altered and not identified via gICA approach in MCI group. (c)–(d) show significantly decreased (in blue) and increased (red) functional connectivities in MCI in comparison with NC (p-value < 0.05, FDR corrected for multiple comparison).
Discussion and Conclusion
Based on the predictive models of RSNs learned from multimodal R-fMRI and DTI data of MCI and NC groups, our results revealed widespread functional connectivity alterations across the whole brain in MCI. Since RSNs in MCI are disrupted and can hardly be constructed merely from R-fMRI data, structurally consistent DICCCOL landmark-based predictive models of RSNs learned from NC were used to assess the functional connectivity alterations in MCI, which is one of the major methodological contributions of this paper. In a broader sense, our work demonstrated the importance of using multimodal DTI and R-fMRI data to localize and assess the resting state connectivities in brain conditions.
In this work, fifteen independent RSNs were identified by group-wise ICA method in the NC group. It should be noted that these fifteen RSNs are by no means representing all possible RSNs that could be inferred from R-fMRI data. Instead, these fifteen RSNs are just among the most reproducible and frequently reported ones in the literature, and there could be other RSNs that are potentially missed in our study. In the future, other alternative RSNs identification approaches, such as cortical surface based RSN identification methods (Li et al., 2010) or identification of RSNs directly from the DICCCOL systems (Zhu et al., 2012; Zhu et al., 2013), could be applied on our dataset to examine the reproducibility of the RSNs reported in this paper and to include potentially other additional RSNs. In this case, even larger scale assessment of altered functional connectivities in MCI could be possibly performed.
The results in this study were based on ten MCI patients and eighteen normal controls. Although this number of subjects is reasonably sufficient for report valid results (Wee et al., 2011), it does not allow us to perform reproducibility study in separate groups. Therefore, in the future, we plan to apply similar approaches in larger scale multimodal DTI/R-fMRI datasets of MCI and control subjects to further replicate the experimental results reported in this paper. In the current stage, the pair-wise functional connectivity reported in this work was not statistically corrected yet. In the future, it is possible to apply the network-based statistics (NBS) methods (Zalesky et al., 2010) for the statistical correction of the connectivity measurements. In addition, we can use different levels of t thresholds (Zalesky et al., 2010) to assess the connectivity difference between MCI brains and controls at the network-levels.
In the future, we plan to correlate the altered resting state functional connectivity in MCI with clinical parameters. For instance, longitudinal studies of the functional connectivities in the RSNs in MCI brains are very much desired, given the availability of such longitudinal R-fMRI datasets e.g., the ADNI-2 R-fMRI datasets (Jack et al., 2010) being released to the community. Then, the measured functional connectivity changes along the temporal domain could be associated with psychological and clinical parameters of MCI such as the MCI to AD conversion. We premise that resting state functional connectivity alterations in MCI brains could possibly be a useful biomarker for differential and early diagnosis of AD in the future.
Finally, we envision that the proposed methodology of predictive models of RSNs based on multimodal R-fMRI/DTI data could be possibly used to assess functional connectivities in other neurological or psychiatric conditions such as Alzheimer’s disease, Schizophrenia and Autism, in which resting state functional brain networks might have already been significantly disrupted during the disease progression. The experimental results in this paper demonstrated that although the functional connectivities between DICCCOL-based predictive models of RSNs in MCI were substantially altered, their structural connection patterns were relatively intact. Therefore, the proposed DICCCOL landmarks with intrinsic structural correspondences across subjects and populations (Zhu et al., 2012; Li et al., 2012b) and their prediction framework (Zhang et al., 2011b; Zhu et al., 2012) offer a fair, effective and meaningful way to assess the potentially widespread RSNs dysfunctions in many brain diseases/conditions in the future.
Acknowledgements
T Liu was supported by NIH Career Award (NIH EB-006878), NSF CAREER Award (IIS-1149260), NIH R01 DA-033393, NIH R01 AG-042599, and NSF BME-1302089. L Guo was supported by the NWPU Foundation for Fundamental Research. K Li and T Zhang were supported by the China Government Scholarship. L Wang was supported by the Paul B. Beeson Career Developmental Awards (K23-AG028982) and a National Alliance for Research in Schizophrenia and Depression Young Investigator Award. The authors would like to thank the anonymous reviewers for their constructive comments.
References
- Bai F, Zhang Z, Yu H, Shi Y, Yuan Y, Zhu W, et al. Default-mode network activity distinguishes amnestic type mild cognitive impairment from healthy aging: a combined structural and resting-state functional MRI study. Neurosci Lett. 2008;438(1):111–115. doi: 10.1016/j.neulet.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, De Luca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewijzend MA, Schoonheim MM, Sanz-Arigita E, Wink AM, van der Flier WM, Tolboom N, et al. Resting-state fMRI changes in Alzheimer's disease and mild cognitive impairment. Neurobiol Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Bozzali M, Falini A, Franceschi M, Cercignani M, Zuffi M, Scotti G, et al. White matter damage in Alzheimer’s disease assessed in vivo using diffusion tensor magnetic resonance imaging. J Neurol Neurosurg Psychiatry. 2002;72(6):742–746. doi: 10.1136/jnnp.72.6.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Pekar JJ, Pearlson GD. Alcohol intoxication effects on simulated driving: Exploring alcohol-dose effects on brain activation using functional MRI. Neuropsychopharmacology. 2004;29:2097–3017. doi: 10.1038/sj.npp.1300543. [DOI] [PubMed] [Google Scholar]
- Celone KA, Calhoun VD, Dickerson BC, Atri A, Chua EF, Miller SL, et al. Alterations in memory networks in mild cognitive impairment and Alzheimer's disease: an independent component analysis. J Neurosci. 2006;26(40):10222–10231. doi: 10.1523/JNEUROSCI.2250-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, et al. Consistent resting-state networks across healthy subjects. PNAS. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29(4):1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Sperling RA. Large-scale functional brain network abnormalities in Alzheimer’s disease: Insights from functional neuroimaging. Behav Neurol. 2009;21(1):63–75. doi: 10.3233/BEN-2009-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillard P, Descoteaux M, Goh A, Gouttard S, Jeurissen B, Malcolm J, Ramirez-Manzanares A, Reisert M, Sakaie K, Tensaouti F, Yo T, Mangin JF, Poupon C. Quantitative evaluation of 10 tractography algorithms on a realistic diffusion MR phantom. Neuroimage. 2011;56(1):220–234. doi: 10.1016/j.neuroimage.2011.01.032. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101(13):4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, et al. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Arch Neurol. 2004;61(1):59–66. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]
- He Y, Wang L, Zang Y, Tian L, Zhang X, Li K, et al. Regional coherence changes in the early stages of Alzheimer’s disease: A combined structural and resting-state functional MRI study. Neuroimage. 2007;35(2):488–500. doi: 10.1016/j.neuroimage.2006.11.042. [DOI] [PubMed] [Google Scholar]
- Head D, Buckner RL, Shimony JS, Williams LE, Akbudak E, Conturo TE, et al. Differential vulnerability of anterior white matter in nondemented aging with minimal acceleration in dementia of the Alzheimer type: evidence from diffusion tensor imaging. Cereb Cortex. 2004;14(4):410–423. doi: 10.1093/cercor/bhh003. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, et al. Predicting human resting-state functional connectivity from structural connectivity. PNAS. 2009;106(6):2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM, et al. Update on the magnetic resonance imaging core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement. 2010;6(3):212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp. 2007;28(11):1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Guo L, Nie J, Li G, Liu T. Review of Methods for Functional Brain Connectivity Detection Using fMRI. Comput Med Imaging Graph. 2009;33(2):131–139. doi: 10.1016/j.compmedimag.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Guo L, Faraco CC, Zhu D, Deng F, Zhang T, et al. Individualized ROI Optimization via Maximization of Group-wise Consistency of Structural and Functional Profiles. Neural Information Processing Systems (NIPS) 2010 [Google Scholar]
- Li Kaiming, Guo Lei, Zhu Dajiang, Hu Xintao, Han Junwei, Liu Tianming. Individual Functional ROI Optimization via Maximization of Group-wise Consistency of Structural and Functional Profiles. Neuroinformatics. 2012 doi: 10.1007/s12021-012-9142-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Kaiming, Zhu Dajiang, Guo Lei, Li Zhihao, Lynch Mary Ellen, Coles Claire, Hu Xiaoping, Liu Tianming. Connectomics Signatures of Prenatal Cocaine Exposure Affected Adolescent Brains. Human Brain Mapping. 2012b doi: 10.1002/hbm.22082. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Wang Z, Yang Y, Jia X, Li K. Functional Disconnection and Compensation in Mild Cognitive Impairment: Evidence from DLPFC Connectivity Using Resting-State fMRI. PLoS One. 2011;6(7):e22153. doi: 10.1371/journal.pone.0022153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsker R. A local learning rule that enables information maximization for arbitrary input distributions. Neural Computation. 1997;9(8):1661–1665. [Google Scholar]
- Liu T, Li H, Wong K, Tarokh A, Guo L, Wong ST. Brain Tissue Segmentation Based on DTI Data. NeuroImage. 2007;38(1):114–123. doi: 10.1016/j.neuroimage.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang K, Yu C, He Y, Zhou Y, Liang M, et al. Regional homogeneity, functional connectivity and imaging markers of Alzheimer’s disease: A review of resting-state fMRI studies. Neuropsychologia. 2008;46(6):1648–1656. doi: 10.1016/j.neuropsychologia.2008.01.027. [DOI] [PubMed] [Google Scholar]
- Liu T, Nie J, Tarokh A, Guo L, Wong ST. Reconstruction of Central Cortical Surface from MRI Brain Images: Method and Application. NeuroImage. 2008;40(3):991–1002. doi: 10.1016/j.neuroimage.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. A few thoughts on Brain ROIs. Brain Imaging Behav. 2011;5(3):189–202. doi: 10.1007/s11682-011-9123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453(7197):869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, et al. Functional connectivity and brain networks in schizophrenia. J Neurosci. 2010;30(28):9477–9487. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, et al. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6(3):160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3(8):606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Rademacher J, Morosan P, Schormann T, Schleicher A, Werner C, Freund HJ, et al. Probabilistic mapping and volume measurement of human primary auditory cortex. Neuroimage. 2001;13(4):669–683. doi: 10.1006/nimg.2000.0714. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. PNAS. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier F, Clarke S. Cytochrome oxidase, acetylcholinesterase, and NADPH-diaphorase staining in human supratemporal and insular cortex: evidence for multiple auditory areas. NeuroImage. 1997;6(4):288–304. doi: 10.1006/nimg.1997.0304. [DOI] [PubMed] [Google Scholar]
- Rombouts SA, Barkhof F, Goekoop R, Stam CJ, Scheltens P. Altered resting state networks in mild cognitive impairment and mild Alzheimer's disease: an fMRI study. Hum Brain Mapp. 2005;26(4):231–239. doi: 10.1002/hbm.20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat DH, Tuch DS, van der Kouwe AJ, Greve DN, Pappu V, Lee SY, et al. White matter pathology isolates the hippocampal formation in Alzheimer’s disease. Neurobiol Aging. 2010;31(2):244–256. doi: 10.1016/j.neurobiolaging.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005;15(9):1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Schmithorst VJ, Holland SK. Comparison of three methods for generating group statistical inferences from independent component analysis of functional magnetic resonance imaging data. J Magn Reson Imaging. 2004;19(3):365–368. doi: 10.1002/jmri.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Mühlau M, Calhoun VD, Eichele T, Läer L, et al. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. PNAS. 2007;104(47):18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1(4):e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl R, Dietrich O, Teipel SJ, Hampel H, Reiser MF, Schoenberg SO. White matter damage in Alzheimer’s disease and mild cognitive impairment: assessment with diffusion tensor MR imaging and parallel imaging techniques. Radiology. 2007;243(2):482–492. doi: 10.1148/radiol.2432051714. [DOI] [PubMed] [Google Scholar]
- Stebbins GT, Murphy CM. Diffusion tensor imaging in Alzheimer’s disease and mild cognitive impairment. Behavioural Neurology. 2009;21(1):39–49. doi: 10.3233/BEN-2009-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol. 2012;22(1):53–66. [Google Scholar]
- van den Heuvel M, Mandl R, Hulshoff Pol H. Normalized cut group clustering of resting-state FMRI data. PLoS One. 2008;3(4):e2001. doi: 10.1371/journal.pone.0002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Liang M, Wang L, Tian L, Zhang X, et al. Altered functional connectivity in early Alzheimer's disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28(10):967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee Chong-Yaw, Yap Pew-Thian, Li Wenbin, Denny Kevin, Browndyke Jeffrey N, Potter Guy G, Welsh-Bohmer Kathleen A, Wang Lihong. Enriched White Matter Connectivity Networks for Accurate Identification of MCI Patients. NeuroImage. 2011 Feb;54(3):1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET. Network-based statistic: Identifying differences in brain networks. NeuroImage. 2010;53(4):1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. NeuroImage. 2004;22(1):394–400. doi: 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, Schad L, et al. Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer’s disease. Neurology. 2007;68(1):13–19. doi: 10.1212/01.wnl.0000250326.77323.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal Classification of Alzheimer’s Disease and Mild Cognitive Impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Tuo, Guo Lei, Li Kaiming, Jing Changfeng, Yin Yan, Zhu Dajing, Cui Guangbin, Li Lingjiang, Liu Tianming. Predicting functional cortical ROIs based on fiber shape models. Cerebral Cortex. 2011;22(4):854–864. doi: 10.1093/cercor/bhr152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Li K, Faraco CC, Deng F, Zhang D, Guo L, et al. Optimization of functional brain ROIs via maximization of consistency of structural connectivity profiles. NeuroImage. 2011;59(2):1382–1393. doi: 10.1016/j.neuroimage.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Zhang D, Faraco C, Li K, Deng F, Chen H, et al. Discovering Dense and Consistent Landmarks in the Brain. IPMI. 2011;22:97–110. doi: 10.1007/978-3-642-22092-0_9. [DOI] [PubMed] [Google Scholar]
- Zhu Dajiang, Li Kaiming, Guo Lei, Jiang Xi, Zhang Tuo, Zhang Degang, Chen Hanbo, Deng Fan, Faraco Carlos, Jin Changfeng, Wee Chong-Yaw, Yuan Yixuan, Lv Peili, Yin Yan, Hu Xiaolei, Duan Lian, Hu Xintao, Han Junwei, Wang Lihong, Shen Dinggang, Miller L Stephen, Li Lingjiang, Liu Tianming. DICCCOL: Dense Individualized and Common Connectivity-based Cortical Landmarks. Cerebral Cortex. 2012 doi: 10.1093/cercor/bhs072. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Dajiang, Li Kaiming, Terry Douglas P, Puente A Nicholas, Wang Lihong, Shen Dinggang, Miller L Stephen, Liu Tianming. Connectome-scale Assessments of Structural and Functional Connectivity in MCI. Human Brain Mapping. 2013 doi: 10.1002/hbm.22373. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]