Abstract
Since Notch phenotypes in Drosophila melanogaster were identified 100 years, Notch signaling has been extensively characterized as a regulator of cell fate decisions in a variety of organisms and tissues. However, in the past 20 years, accumulating evidence has linked alterations in the Notch pathway to tumorigenesis. In this Perspective, we discuss the pro-tumorigenic and tumor suppressive functions of Notch signaling and dissect the molecular mechanisms that underlie these functions in hematopoietic cancers and solid tumors. Finally, we link these mechanisms and observations to possible therapeutic strategies targeting the Notch pathway in human cancers.
This year will be the centennial of the discovery of a signaling pathway that has fascinated developmental, molecular, and cancer biologists around the world. Mutant Notch phenotypes in the fly wing were characterized by John S. Dexter 100 years ago (Dexter, 1914) and, rapidly after, Thomas Hunt Morgan identified the mutant alleles (Morgan, 1917). Almost seven decades later, after the molecular biology revolution, Spyros Artavanis-Tsakonas and Michael Young cloned the Notch receptor and attributed the wing-notching phenotype to gene haplo-insufficiency (Kidd et al., 1986; Wharton et al., 1985). These studies brought a revolution in a large number of fields including developmental and stem cell biology, neuroscience, and – related to this Perspective – cancer biology (Fortini et al., 1993). Indeed, in the early nineties, gain-of-function mutations of the pathway were identified in cancer (Ellisen et al., 1991; Gallahan and Callahan, 1997; Gallahan et al., 1987; Jhappan et al., 1992). A deluge of reports followed, cementing the role of Notch signaling as oncogenic but also tumor suppressive, depending on the context. In this Perspective, we attempt to provide a detailed characterization of Notch functions in both solid and hematopoietic cancers and discuss the molecular mechanisms explaining such functions as well as approaches to target Notch signaling in human cancers.
A brief description of the Notch signaling pathway
There are four Notch receptors (named Notch1–4) in mammals. Notch1 and Notch2 each have 36 EGF-like repeats, while Notch3 and Notch4 have 34 and 29 repeats, respectively, which affect their affinity for corresponding ligands (Haines and Irvine, 2003; Okajima and Irvine, 2002; Rebay et al., 1991) (Figure 1). Notch receptors are single pass type I transmembrane molecules coded by a single precursor that becomes a non-covalently linked heterodimer consisting of an N-terminal extracellular (NEC) fragment and a C-terminal transmembrane-intracellular subunit (NTM) as a result of cleavage by a furin-like protease in the trans-Golgi network (Blaumueller et al., 1997) (Figures 1 and 2). The Notch pathway is normally activated upon interactions with ligands such as Delta-like and Jagged, which are also transmembrane proteins containing EGF-like repeats. In mammals, there are three Delta-like ligands (Dll1, Dll3, and Dll4) and two Jagged ligands (Jag1 and Jag2). The Notch pathway gets activated in a strictly controlled fashion: ADAM10/17 metalloproteases cause an S2 cleavage in the receptor, followed by a third cleavage (S3 cleavage) mediated by the presenilin–γ-secretase complex, composed of presenilin 1 (PSEN1), PSEN2, nicastrin (NCSTN), presenilin enhancer 2 (PEN2), and anterior pharynx-defective 1 (APH1) (Shah et al., 2005). This series of events releases the intracellular portion of the Notch receptor (termed ICN) that then translocates into the nucleus to mediate target gene activation (De Strooper et al., 1999; Schroeter et al., 1998). Notch-ICN is a transcriptional activator (Bray, 2006) consisting of ankyrin repeats, a RAM (RBP-Jκ associated molecule) domain, a transactivation domain (TAD), a nuclear localization signal (NLS), and a PEST domain regulating protein stability (Figures 1 and 2). Notch ligands are also cleaved by γ-secretase and ADAM metalloprotease complexes, thus providing an additional level of regulation of the pathway (LaVoie and Selkoe, 2003; Six et al., 2003). Despite the overall similarities between the receptors, the differences in the ligand-binding extracellular domains and the transactivation intracellular domains lead to distinct ligand affinities and capacity to activate downstream transcription.
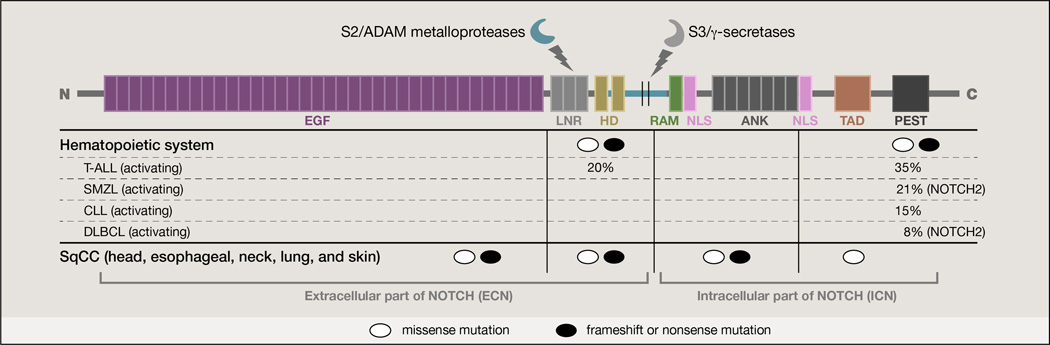
Figure 1. Protein structure and mutations of a typical Notch receptor.
The structure of the NOTCH1 receptor and genetic alterations of the protein in representative types of cancer are depicted. ADAM metalloproteases and the γ-secretase complex cleave the receptor and free the ICN domain. Major mutations are clustered according to their effects on protein activity. Both gain-and loss-of-function mutations are shown. The majority of the T-ALL mutations are clustered in the heterodimerization (HD) and PEST domains controlling processing of the receptors by proteases and the stability of the protein correspondingly. Different characteristic cases of hematopoietic disorders (affecting NOTCH2 as well) are shown. In CLL tumors there is an apparent mutational hotspot at the PEST domain of NOTCH1. In the case of SqCC mutations, they are mainly clustered in the EGF repeat region potentially affecting interaction with the ligands. T-ALL: T-cell acute lymphoblastic leukemia, SMZL: splenic marginal zone lymphoma, CLL: chronic lymphocytic leukemia, DLBCL: diffuse large B cell lymphoma, SqCC: squamous cell carcinoma. Percentages are approximations based on current literature.
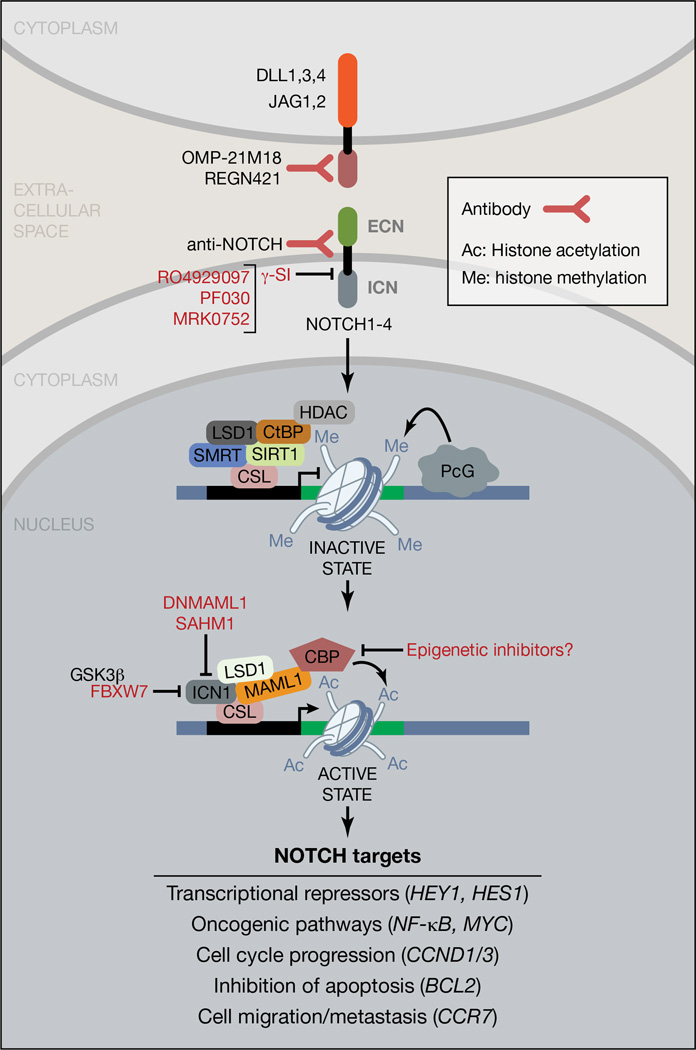
Figure 2. Overview of the Notch signaling pathway.
A visual description of the signaling cascade is shown for the signal-receiving cell (i.e. the cell expressing the Notch receptor). Pathway inhibitors used include antibodies against NOTCH receptors and DLL ligands, γ-secretase complex inhibitors (GSI), and small peptides inhibiting formation of the transcriptional complex. Antibody-based treatments are shown in purple, GSI compounds in pink, peptide-based drugs in red. Potential epigenetic inhibitors (in green) can include BRD inhibitors like JQ1. HDAC: histone deacetylase, ICN1: intracellular part of NOTCH1, LSD1: lysine specific demethylase 1, SMRT: Silencing-Mediator for Retinoid/Thyroid hormone receptors, GSK3β: glycogen synthase kinase 3 beta, DNMAML1: dominant negative MAML1.
In the nucleus, Notch binds to initially inactive CBF1-Su(H)–LAG1 (CSL) (aka RBP-Jκ) complexes and mediates their conversion to a transcriptional activator followed by the recruitment of the co-activator protein mastermind-like 1 (MAML1) (Figure 2) (Nam et al., 2006; Wilson and Kovall, 2006; Wu et al., 2000). The ankyrin repeats seem to play an important role for MAML1 recruitment. The list of target genes regulated by Notch is very much dependent on cell type and can include genes whose products are involved in fundamental aspects of cell biology, such as cell cycle regulation (Joshi et al., 2009; Lewis et al., 2007), cellular differentiation, and metabolism (Palomero et al., 2006). Common targets of the pathway include the HES and HEY (Iso et al., 2001a; Iso et al., 2001b; Jarriault et al., 1995) families of transcription repressors as well as MYC transcription factor (Palomero et al., 2006; Sharma et al., 2006; Weng et al., 2006). The binding and function of Notch on DNA appears to be a rapid and dynamic process controlled by the kinase CDK8 and the ubiquitin ligase Fbxw7 leading to Notch phosphorylation, ubiquitination, and its subsequent proteasomal degradation (Fryer et al., 2004; Mukherjee et al., 2005; O'Neil et al., 2007; Thompson et al., 2007), which shuts off the pathway (Figure 2).
Various tools have been developed to study the transcriptional activity of the pathway, such as ChIP-seq and ChIP-chip to map Notch1 binding on the genome (Castel et al., 2013; Ntziachristos et al., 2012; Palomero et al., 2006; Wang et al., 2011a) and mouse models that allow efficient tracing of receptor cleavage/activity in many different tissues (Hansson et al., 2006; Liu et al., 2011; Mizutani et al., 2007; Souilhol et al., 2006). Recently, the group of Artavanis-Tsakonas (Fre et al., 2011; Sale et al., 2013) and our laboratory (Oh et al., 2013) have traced Notch pathway activity in vivo by using reporter systems for Notch receptors expression and Hes1 activity by coupling them to fluorescent proteins (Figure 3). As there are several unanswered questions regarding Notch ligand expression, even under physiological conditions, an exciting next step could involve development of fluorescent tools to probe ligand expression together with pathway activation in real-time within a living organism.
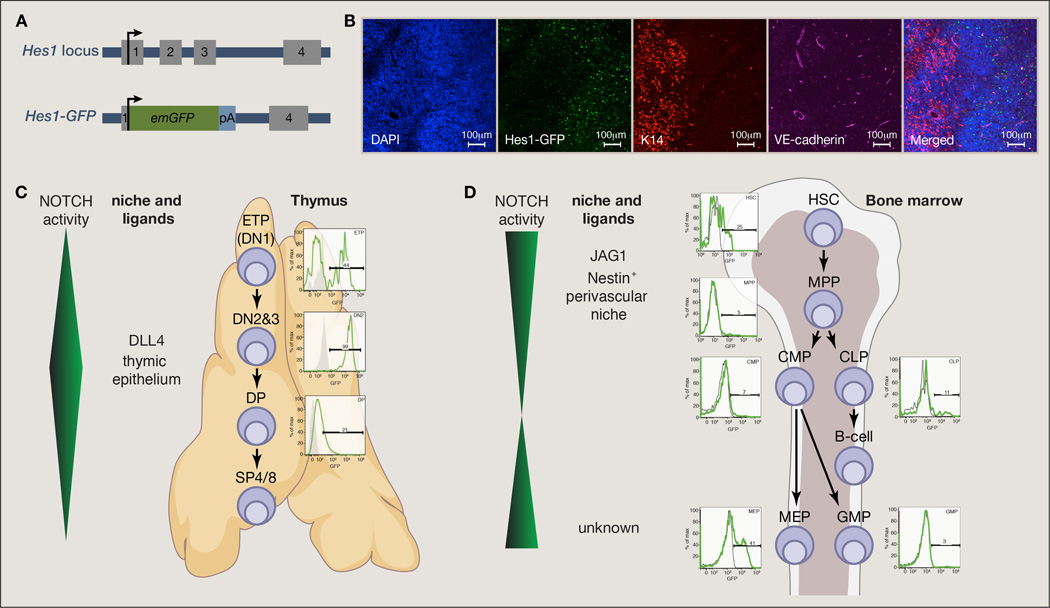
Figure 3. In vivo mapping of Notch pathway activity using a Hes1GFP reporter.
(A) Targeting strategy for the generation of transgenic animals expressing Emerald GFP (emGFP) from the endogenous Hes1 locus. (B) Immuno-fluorescence staining for thymus of the Hes1GFP mice. DAPI stains DNA (nucleus), VE-cadherin is a vascular endothelial marker and K14 is a marker of thymic medullary cells. (C) Increased levels of Notch pathway help differentiation of thymic T cell progenitors through the DN2/3 CD4−8− differentiation stage and the pathway activity is decreased immediately at the DP stage. (D) Activity of the Notch pathway in the mouse bone marrow is detected at the HSC level and is decreased as cells differentiate. Subsequently it is reactivated at the level of a megakaryocytic-erythrocytic progenitor (MEP). HSC: hematopoietic stem cells, MPP: multipotent progenitors, CMP: common myeloid progenitors, CLP: common lymphoid progenitors, MEP: megakaryocyte-erythrocyte progenitor, GMP: granulocyte-macrophage progenitor, DN: double negative (CD4−CD8−), DP: double positive (CD4+CD8+), SP: single positive.
NOTCH signaling pathway in cancer
The Notch pathway is genetically altered in a large number of hematopoietic and solid tumors (Figure 1). Intriguingly, these alterations can lead to either activation or repression of the pathway depending on the context and the activation status of other potentially oncogenic pathways (Table 1 and Figure 4). Interestingly, it appears that there are multiple and distinct modes of aberrant regulation of the pathway and its targets in cancer. They include activating and inactivating mutations, receptor/ligand over-expression, epigenetic regulation, and effects of post-translational modifications, most notably receptor and ligand fucosylation (especially O-fucosylation) (Haines and Irvine, 2003; Lei et al., 2003; Okajima et al., 2003) and ubiquitination (Fryer et al., 2004; Thompson et al., 2007). We initially discuss T cell acute leukemia, a disease in which Notch has a well-characterized oncogenic role. Subsequently we present several other cases of hematopoietic and solid tumors where Notch has tumor suppressive or oncogenic roles, along with its potential mechanisms of action and partners.
Table 1.
Oncogenic and tumor suppressive roles of Notch signaling in human cancers *
| Tumor Type | Oncogene or Tumor Suppressor |
Mutations (%) or noteworthy observations * | References * |
|---|---|---|---|
| T-Cell Acute Lymphoblastic Leukemia (T-ALL) | Oncogene | 50–60% NOTCH1, 30% FBXW7 Role in cancer initiation and maintenance | (Malyukova et al., 2007; Weng et al., 2004) |
| Chronic Lymphocytic Leukemia (CLL) | Oncogene | 5–12% NOTCH1 Role in cancer initiation and survival | (Fabbri et al., 2011; Puente et al., 2011) |
| Melanoma | Oncogenic | ~50% NOTCH1 overexpression in human samples Possible role in metastasis | (Balint et al., 2005; Bedogni et al., 2008) |
| Cholangiocarcinoma (CCC) | Oncogenic | 35% FBXW7 Notch1 promotes tumor initiation and maintenance | (Akhoondi et al., 2007; Zender et al., 2013) |
| Colorectal cancer | Oncogenic | 8–9% FBXW7 Crosstalk with Wnt and Hippo signaling | (Miyaki et al., 2009) (Akhoondi et al., 2007) |
| Lung adenocarcinoma | Oncogenic | 10% NOTCH1 Role in initiation and maintenance (Notch1), and metastasis (Jagged2) Specific role for Notch3 in tumor propagation | (Licciulli et al., 2013; Westhoff et al., 2009; Zheng et al., 2013) |
| Glioblastoma | Oncogenic | Role in tumor propagation and radioresistance | (Chu et al., 2013) (Wang et al., 2010) |
| Renal Cell Carcinoma | Oncogenic | Role in progression and maintenance | (Sjolund et al., 2008) |
| Ovarian cancer | Oncogenic | Role in maintenance and therapy response | (2011; Cancer Genome Atlas Research, 2011; McAuliffe et al., 2012) |
| Prostate | Oncogenic | Activation of the pathway associated with tumor progression, metastasis, and recurrence | (Marignol et al., 2013; Santagata et al., 2004) |
| Breast cancer | Mostly Oncogenic | NOTCH1 and NOTCH4 fusions Potential NOTCH2 dominant-negative truncated mutant Other alterations activating Notch signaling But hyperactive Notch signaling may inhibit cancer growth | (Fu et al., 2010; Imatani and Callahan, 2000; Jhappan et al., 1992) |
| Pancreatic Ductal Adenocarcinoma (PDAC) | Mostly Oncogenic | Notch2 loss inhibits progression and maintenance Overexpression of ligands –Jagged2 (90%), Dll4 (50%) But Notch1 loss may promote tumor initiation | (Hanlon et al., 2010; Mazur et al., 2010; Mullendore et al., 2009) |
| Cervical cancer | Mostly Oncogenic | Pathway activation in human tumors, but dose-dependent effects Possible role in tumor-propagating cells | (Bajaj et al., 2011; Maliekal et al., 2008; Zagouras et al., 1995) |
| Head and neck squamous cell carcinomas (HNSCC) | Mostly oncogenic | Possible bimodal pattern of Notch pathway alterations with a small subset of tumors with inactivating NOTCH1 mutations but a larger group with pathway activation | (Sun et al., 2013) |
| Hepatocellular carcinoma (HCC) | Oncogenic and Tumor Suppressive | Context-dependent effects that may be related to various molecular subtypes | (Qi et al., 2003; Villanueva et al., 2012) |
| Medulloblastoma | Oncogenic and tumor suppressive | Opposite roles for Notch1 and Notch2 | (Fan et al., 2004) |
| B-Cell Acute Lymphoblastic Leukemia (B-ALL) | Tumor Suppressive | No mutations Role in maintenance (activation induces growth arrest and death) | (Zweidler-McKay et al., 2005) |
| Acute Myeloid Leukemia (AML) | Tumor Suppressive | Notch1 and 2 expressed but the pathway is not active Role in cancer initiation and maintenance | (Kannan et al., 2013; Lobry et al., 2013) |
| Small Cell Lung Carcinoma (SCLC) | Tumor Suppressive | No mutations Inhibits tumor maintenance (possible similar role in other neuroendocrine tumor types) | (Sriuranpong et al., 2001) |
| Lung Squamous Cell Carcinoma (SqCC) | Tumor Suppressor | 5–12.5% NOTCH1, NOTCH2 | (2012; Cancer Genome Atlas Research, 2012; Wang et al., 2011b) |
| Cutaneous Squamous Cell Carcinoma (SqCC) | Tumor Suppressor | 60–75% NOTCH1, NOTCH2 | (Wang et al., 2011b) |
| Chronic myelo-monocytic leukemia (CMML) | Tumor Suppressor | 12% various pathway genes (NCSTN, APH1, MAML1, NOTCH2) Role in cancer initiation | (Klinakis et al., 2011) |
Cancers indicated in this table have been selected for historical reasons (first examples of mutations in the Notch pathway), because they affect large populations of cancer patients, or because of the particular insight of some studies to the role of Notch signaling in cancer. Among the selected tumor types, selected observations and references are shown, see text for additional references and details. In particular, data from large cancer genomes efforts indicate that many alterations in the extended Notch pathway exist in human tumors, most of these alterations are awaiting additional analyses and functional validation.
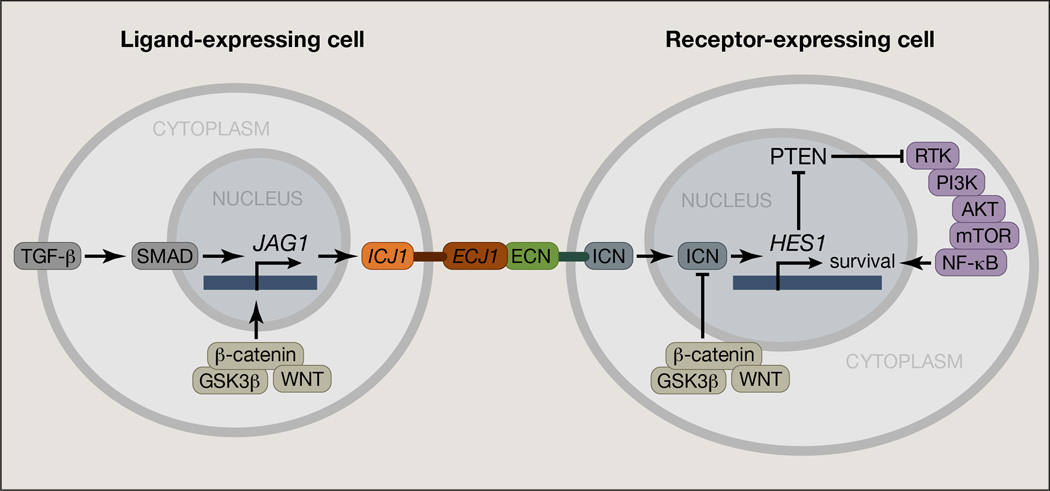
Figure 4. Simplified scheme of Notch interactions with other signaling pathways in cancer.
The TGFβ, PI3K, NFκB, and WNT pathways are some of the most important pathways interacting with NOTCH. Notably, Jagged 1 is activated by TGFβ pathway and in turn activates NOTCH receptors in neighboring cells. Phosphorylation of NOTCH from the WNT-induced GSK3β leads to ubiquitination through FBXW7 and final degradation. Also, a classical NOTCH target, HES1, represses PTEN, a competitor of another pathway with oncogenic roles, PI3K, which in turn activates NFκB, a pathway important for leukemia progression. Important parameters of the interactions, such as regulation of NFκB pathway by NOTCH through HES1 action, or the interaction of NOTCH with the WNT member DVL (Dishevelled) protein that inhibits both WNT and NOTCH pathways are not shown in this figure. ICJ1: intracellular part of JAG1, ECJ1: extracellular part of JAG1.
T-Cell Acute Lymphoblastic Leukemia (T-ALL)
NOTCH1 is a master transcription factor that controls innate and adaptive immunity and plays an important role in directing hematopoietic development towards T cells (Aifantis et al., 2008; Li and von Boehmer, 2011; Radtke et al., 2013). The very first finding of Notch pathway alterations in cancer comes from the work of Ellisen and colleagues, which was confirmed subsequently by other groups, that revealed a rearrangement between the intracellular part of NOTCH1 (ICN1) and the T cell receptor beta (TRB) locus, leading to high level expression of truncated, constitutively-active NOTCH1 in leukemia (Ellisen et al., 1991). In vitro studies (Capobianco et al., 1997), as well as animal modeling (Girard et al., 1996; Pear et al., 1996) then revealed that ICN1 is a strong oncogenic allele. Most importantly, ten years ago, the Aster and Look laboratories reported the first activating NOTCH1 mutations in human T-ALL, occurring in approximately 50% of all cases (Weng et al., 2004).
The majority of these mutations encompass single amino acid substitutions, insertions and deletions located in exons 26 and 27 of the genetic locus, which encode the N-terminal and C-terminal components of the hetero-dimerization domain respectively. These mutations lead to lower protection of S2 cleavage of Notch, resulting in either ligand independent activation or hypersensitivity of the pathway to ligands. Another rare group of mutations, the Juxtamembrane Expansion Mutants (JME) also augments NOTCH1 activation at the cell membrane (Sulis et al., 2008) (Figure 1). Finally, PEST domain mutants encompass another category of NOTCH1 mutations in 20–25% of T-ALLs. PEST domain alterations lead to truncation or loss of the domain due to frame-shift or nonsense nucleotide substitutions, which impair proteasomal degradation mediated by the ubiquitin ligase FBXW7 and lead to higher ICN1 cellular concentrations (Weng et al., 2004). The importance of ICN1 degradation in physiology becomes more evident by the fact that 15 % of T-ALL cases harbor mutations or deletions in FBXW7 (Asnafi et al., 2009; O'Neil et al., 2007; Thompson et al., 2007). These changes are localized in three arginine residues critical for its interaction with ICN1. Mutations in the PEST domain and FBXW7 do not occur concurrently which implies that they play the same role to increase stability of ICN1 (Asnafi et al., 2009; O'Neil et al., 2007).
The fact that FBXW7 mutations directly affect cells with leukemia-initiating (LIC) properties though the stabilization and overexpression of MYC, another well-characterized substrate of this ubiquitin ligase, further demonstrates that NOTCH and MYC actions are intertwined in cancer cells (King et al., 2013). Interestingly, NOTCH1 mutations in T-ALL were shown to have a favorable prognosis and better outcome post-treatment in a number of studies including the ALL-Berlin-Frankfurt-Munster 2000 study (Breit et al., 2006), a study by the Japan Association of Childhood Leukemia Study that examined NOTCH1 and FBXW7 mutational status in T-ALL and T-cell lymphoblastic lymphoma patients (Park et al., 2009), and the Lymphoblastic Acute Leukemia in Adults (LALA)-94 and the GRAALL-2003 trials (Asnafi et al., 2009). Finally, another report on 134 pediatric patients from the EORTC-CLG 58881 and 58951 protocols concluded that NOTCH1 and FBXW7 mutations associate with improved early chemotherapeutic response and lower minimal residual disease (MRD) levels (Clappier et al., 2010). It remains to be seen whether Notch pathway inhibition will be used successfully to target T-ALL, especially in relapsed disease that is refractory to conventional chemotherapy-based treatments.
Chronic lymphocytic leukemia (CLL)
CLL is the most common leukemia in adults. Recently, it was demonstrated using next generation sequencing-based approaches that 10–12% of CLL cases exhibit activating mutations of NOTCH1, underlining the significance of such mutations as a prognostic marker. The vast majority of these mutations are in the PEST domain, leading to truncated protein variants with a longer half-life (Figure 1). Interestingly, there seems to be a mutational hotspot in this disease, with P2515Rfs being the most prevalent mutation (Fabbri et al., 2011; Puente et al., 2011; Rossi et al., 2012a). Mutations of NOTCH1 are mutually exclusive with TP53 abnormalities and survival outcomes are poor in both cases (Rossi et al., 2012a; Wickremasinghe et al., 2011). NOTCH1 and SF3B1 (a splicing factor) mutations were associated with decreased overall survival, and both retained independent prognostic significance for survival outcomes (Oscier et al., 2013). Mutational activation of NOTCH1 was observed at significantly higher frequency during disease progression towards the high-risk Richter transformation (30%) and chemo-refractory CLL (20%) (Fabbri et al., 2011). This later study and a very recent large-scale clinical analysis of CLL patients (Weissmann et al., 2013) confirmed that NOTCH1 mutations are an adverse prognostic parameter in this disease.
Lymphoma
Non-Hodgkin lymphoma (NHL) is a heterotypic mix of diseases, the most prevalent amongst them consist of Burkitt lymphoma, Follicular lymphoma (FL, the most indolent amongst NHL cases) (Pasqualucci et al., 2014), (Roulland et al., 2011), and diffuse large B cell lymphoma (DLBCL). Burkitt lymphoma, mainly characterized by the upregulation of MYC due to its translocation to the immunoglobulin locus, display recurrent gain-of-function NOTCH1 mutations in 8–9% of patients (Love et al., 2012). FL and DLBCL are malignancies of B cell origin and together comprise 60% of new NHL diagnoses in North America. FL and the germinal center B-cell (GCB) DLBCL subtype are derived from germinal center B cells, whereas the more aggressive activated B-cell (ABC) DLBCL subtype is most likely derived from cells that have exited the germinal center. NOTCH2 is mutated in ~8% of DLBCL cases (Lee et al., 2009). These are mainly gain-of-function mutations affecting the PEST domain (and thus the stability of the protein) as well as copy number alterations (Morin et al., 2011). Interestingly, NOTCH2 is required for B-cell development in the spleen marginal zone environment and has been implicated in splenic marginal zone lymphoma (SMZL) (Kiel et al., 2012; Rossi et al., 2012b), as 20% of SMZL cases exhibit gain-of-function NOTCH2 mutations accompanied by mutations of NOTCH1, SPEN and DTX1 (Rossi et al., 2012b). It was suggested that these genetic changes are associated with adverse prognosis (Kiel et al., 2012). Finally, Jundt and colleagues have characterized an activating role for NOTCH1 in classic Hodgkin lymphoma (Schwarzer et al., 2012; Schwarzer and Jundt, 2011). These authors suggested that NOTCH1 is activated through the upregulation of its ligands within the tumor niche and suppresses genes important for B cell identity, such as E12/E47 and the early B cell factor (EBF) (Jundt et al., 2008). Additional studies are required to better define Notch receptor and ligand expression, targeted signaling pathways in the distinct subtypes of lymphoma.
Acute myeloid leukemia and myelo-monocytic neoplasms
Several years ago, emerging evidence indicated that the Notch pathway could have tumor suppressive roles in various types of tumors, in stark contrast to its oncogenic role in the aforementioned hematopoietic malignancies (Nicolas et al., 2003; Rangarajan et al., 2001). In contrast to the tumorigenic role of NOTCH1 in T-ALL, our laboratory and others have recently characterized a tumor suppressive role of the Notch pathway in myeloid malignancies. We have shown that deletion of nicastrin (Ncstn), an essential component of the γ-secretase complex, leads to the induction of chronic myelomonocytic leukemia (CMML) (Klinakis et al., 2011), a disease characterized by increased extramedullary hematopoiesis, monocytosis, myeloproliferation, and frequent progression to acute myeloid leukemia (AML). This is a Notch-mediated effect, as compound deletion of Notch1/2 in vivo led to similar effects. This was further attested when analysis of the conditional model for the deletion of FX (the homologue of human GDP-L-fucose synthase) or O-fucosyltransferase 1 (Pofut1) showed myeloid hyperplasia (Yao et al., 2011), underlining the importance of Notch receptor fucosylation for ligand binding and pathway activation. Ablation of MAML1 can lead to similar phenotypes (Chen et al., 2008). Mechanistically, the tumor suppressor role of NOTCH in this disease is mediated by direct repression of the PU.1 and CEBPα promoters by HES1. Subsequent screening of primary CMML samples for Notch pathway mutations showed that NCSTN, Mastermind-like 1 (MAML1), APH1A, and NOTCH2 are mutated and genetically inactivated in about 12% of CMML patients. These mutations are unique to CMML and not found in other myeloproliferative disorders such as polycythemia vera (PV) and myelofibrosis (MF). Notch inactivating mutations co-occurred with other described myeloid mutations in genes such as TET2, FLT3, and ASXL1 (Klinakis et al., 2011). Based on these findings we were able to show that combination of Notch pathway and TET2 inactivation leads to acute myeloid leukemia (AML). AML cells specifically express NOTCH2 on their surface but show no signs of pathway activity. Interestingly, re-activation of the Notch pathway in established AML leads to complete disease remission (Kannan et al., 2013; Lobry et al., 2013). This observation provides a rationale for the use of specific NOTCH2 activating antibodies or specific agonists as a viable therapeutic strategy in this type of leukemia. Mechanistically, there might be several ways to suppress Notch pathway activity in AML. Initially, AML cells might reside in microenvironments that lack Notch ligands. Another putative mechanism is epigenetic silencing, achieved by DNA and histone methylation of target gene promoters/transcriptional start sites. In agreement with this possibility, we found that Notch target genes are characterized by H3K27me3 marks (Lobry et al., 2013) and mice carrying the R132H mutation of isocitrate dehydrogenase 1 (IDH1) (Figueroa et al., 2010; Gross et al., 2010; Xu et al., 2011) develop a myeloproliferative disease characterized by marked DNA hyper-methylation of Notch pathway genes such as Lfng, Maml3, and Hes5 (Sasaki et al., 2012).
Acute B cell leukemia (B-ALL)
Interestingly, Notch signaling also appears to act as a tumor suppressor in B cell ALL (B-ALL) (Zweidler-McKay et al., 2005). In agreement with the AML findings, Notch pathway re-activation leads to growth inhibition and induces apoptosis in human B-ALL cells. In a recent follow-up publication, it was shown that several Notch pathway targets in B-ALL are suppressed by DNA cytosine hyper-methylation on their promoters followed by histone H3K27 and H3K9 trimethylation (Kuang et al., 2013). The parallel between AML and B-ALL is intriguing and can potentially be explained by a recent Notch activity mapping effort (Oh et al., 2013) that demonstrated activity of the pathway in T cell progenitors and pre-erythrocytes and a lack of pathway activation in the B cell and myelo-monocytic lineages. These findings provide support for a key role for NOTCH as a developmental regulator that can determine the fate of progenitors in the hematopoietic system. In this model, NOTCH action needs the addition of other oncogenic stimuli to transform cells. In agreement with this idea, we found that Notch pathway inactivation can lead to increased frequency of granulocyte-monocyte progenitors (GMP), cells that can initiate diseases like CMML and AML upon further alterations (Klinakis et al., 2011).
Notch signaling in solid tumors
A number of recent reviews have thoroughly summarized our knowledge on Notch signaling in solid tumors (Nowell and Radtke, 2013; Ranganathan et al., 2011; South et al., 2012) (see also Table 1). Recent genomic data, including resources from The Cancer Genome Atlas (TCGA), underscore the prevalence and the complexity of Notch pathway alterations in human cancers, although they do not currently provide detailed functional interpretations for these alterations. Our goal in the section below is not to provide an exhaustive list of the solid tumor types in which Notch signaling is altered and the possible consequences of these alterations. Rather we aim to highlight some key observations in a few prominent tumor types and draw some points of discussion from these studies, including the plethora of partners used by Notch (Figure 4) and the distinct roles of the four Notch receptors.
Breast cancer
Breast cancer is a very prevalent form of cancer in which the Notch pathway may act as a tumor suppressor or an oncogene depending on the subtype. One of the first indications that Notch signaling may play a role in solid tumors actually came from experiments with mouse mammary tumor viruses (MMTV). Integration of the MMTV genome next to the “Int-3” locus resulted in an activating mutation of Notch4, leading to the constitutive activation of the receptor and breast cancer development (Gallahan and Callahan, 1997; Jhappan et al., 1992; Robbins et al., 1992). Since this seminal discovery, a number of studies have confirmed that activation of Notch signaling plays an oncogenic role in breast cancer (Colaluca et al., 2008; Pece et al., 2004; Robinson et al., 2011; Xu et al., 2012). In breast cancer cells, Notch signaling can be activated by functional interactions with other signaling pathways, including the Ras and the Wnt pathways (Ayyanan et al., 2006; Fitzgerald et al., 2000; Izrailit et al., 2013; Meurette et al., 2009; Weijzen et al., 2002). Recent observations indicate that Notch4 may play a more specific role compared to other Notch receptors in breast cancer stem cells (Harrison et al., 2010). In contrast, a recent study indicates that hyperactivation of NOTCH3 may actually be detrimental to breast cancer cells by inducing senescence (Cui et al., 2013). Interestingly, mammary epithelial cells respond differently to different levels of activation of the Notch pathway (Mazzone et al., 2010). Thus, while accumulating evidence indicates that Notch is pro-tumorigenic in breast cancer, in certain contexts, specific (high) levels of activation may be tumor suppressive; alternatively, different Notch receptors may have unique signaling outputs in mammary epithelial cells or in different subtypes of breast cancer. Once more, the notion of a “differentiation switch” could explain the many faces of Notch signaling in this type of tumor.
Lung cancer
Lung adenocarcinoma (LAC) is a major subtype of lung cancer. Initial observations suggested that Notch signaling promotes the expansion of LAC cells in culture (Dang et al., 2003; Eliasz et al., 2010; Haruki et al., 2005). More recent in vivo studies demonstrate that Notch signaling is a key promoter of LAC development and maintenance (Allen et al., 2011; Licciulli et al., 2013; Maraver et al., 2012) and that NOTCH3 plays a unique role in the self-renewal of LAC tumor-propagating cells (Zheng et al., 2013). Expression of JAG2 at the surface of lung adenocarcinoma cells leads to homotypic interactions with Notch receptors and promotes the metastatic potential of these LAC stem cells (Yang et al., 2011). Thus, while mutations and other alterations may not be frequent in LAC (Westhoff et al., 2009), Notch pathway activity correlates significantly with worse survival in lung cancer patients (Hassan et al., 2013; Zheng et al., 2013) and activation of Notch may be important for the sustained growth of LAC. Targeting NOTCH3 and/or JAG2 may benefit a very large number of lung cancer patients worldwide.
Squamous cell lung carcinoma (SqCC) is the second major type of non-small cell lung cancer. In stark contrast to LAC, Notch signaling is thought to be a tumor suppressor of SqCC development, as evidenced by the identification of loss-of-function mutations in human tumors (Wang et al., 2011b). These mutations mainly cluster in the EGF-like repeat region of NOTCH1 and thus have the potential to disrupt ligand binding or to produce truncated receptors (Figure 1). While functional validation for these observations is still missing owing to the current lack of appropriate mouse models, numerous observations indicate that inactivation of Notch signaling promotes the development of squamous cell carcinoma in other tissues, including in cutaneous and head-and-neck tumors (Agrawal et al., 2012; Pickering et al., 2013; Proweller et al., 2006; Rothenberg and Ellisen, 2012; Wang et al., 2011b). These observations suggest that loss of Notch pathway activity may be critical for the growth of tumor cells with squamous differentiation characteristics.
Small cell lung carcinoma (SCLC) is a neuroendocrine subtype of lung cancer, representing a smaller fraction of lung cancer cases (~12–15%) but with the highest mortality rate. Genomic studies have failed to identify recurrent mutations in the Notch pathway in SCLC (Peifer et al., 2012; Rudin et al., 2012). However, early observations indicated that hyperactivation of Notch signaling blocks the cell cycle of SCLC cells (Sriuranpong et al., 2001; Sriuranpong et al., 2002). A tumor suppressive role for Notch in SCLC is supported by evidence that Notch may play a similar role in other neuroendocrine tumors such as medullary thyroid carcinoma (Cook et al., 2010). Thus far, however, no functional evidence has been obtained in vivo that activation of Notch may block SCLC development or maintenance, and it is still possible that subpopulations of cells in SCLC tumors may display some Notch activity and contribute to SCLC growth (Kluk et al., 2013; Salcido et al., 2010).
Thus, three different subtypes of lung cancer display strikingly different roles for Notch signaling in cancer development, from an active oncogenic role with rare genetic alterations in LAC to tumor suppressor with inactivating mutations in SqCC and then possibly tumor suppressive with no sign of mutations in SCLC. It is possible that these differences are related to the role of Notch in cell fate decisions during lung embryonic development.
Liver cancer
Genome sequencing analyses did not reveal recurrent mutations in Notch pathway genes in hepatocellular carcinoma (HCC), a leading cause of cancer-related deaths worldwide (Fujimoto et al., 2012; Guichard et al., 2012). Nevertheless, Notch signaling has been of interest to liver cancer biologists because of the prominent role of Notch signaling in liver development, including mutations in NOTCH2 or JAG1 in patients with Alagille syndrome (syndromic bile duct paucity) (McDaniell et al., 2006; Oda et al., 1997). Haploinsufficient mutations in a specific ligand and a specific receptor in the Notch pathway would suggest that Notch signaling could play very context-dependent and level-dependent roles in liver tumors. Initial observations suggested that low levels of Notch correlates with high activity of the Wnt pathway, a major oncogenic pathway in HCC (Wang et al., 2009). Also, high levels of active Notch1 may inhibit the expansion of HCC cells (Qi et al., 2003), and deletion of Notch1 in the liver of mice results in hyperproliferative hepatocytes, suggesting a tumor suppressive role for Notch in HCC (Croquelois et al., 2005). Similarly, Notch signaling has a tumor suppressive effect in HCC initiated by inactivation of the RB pathway (Viatour et al., 2011). However, other reports have more recently provided evidence that Notch signaling is active and oncogenic in HCC (Dill et al., 2013; Tschaharganeh et al., 2013; Villanueva et al., 2012), and possibly important for the development of tumors following hepatitis B virus infection (Jeliazkova et al., 2013). These observations suggest that the role of Notch signaling in HCC may be different in the distinct molecular subgroups of this cancer type and underscore the need to further explore the molecular contexts associated with tumor suppressive or oncogenic roles of Notch in the liver.
In contrast to the complex roles of Notch signaling in HCC, accumulating evidence supports a pro-tumorigenic role for Notch signaling in cholangiocarcinoma (CCC). Mutations of the Notch repressor FBXW7 are found in a subset of human tumors (Akhoondi et al., 2007). Similar to the disruption of bile ducts in Alagille patients, activation of Notch2 in liver progenitors and adult hepatocytes promotes biliary tubulogenesis (Jeliazkova et al., 2013). Finally, constitutive activation of NOTCH1 is sufficient to initiate CCC development in mice (Zender et al., 2013).
It is likely that the sometimes contradictory consequences of Notch activation in liver cells are due to a combination of the strength of the downstream signal, the timing of the activation, the cell type in which this activation occurs, and the receptor involved (Ortica et al., 2013). There seems to be a consensus that higher Notch levels in liver progenitors favors bile duct differentiation versus hepatocytic differentiation. Possibly activation of Notch (e.g. NOTCH2) in these progenitors promotes CCC while suppressing HCC (Guest et al., 2013). It is also possible that Notch switches from a suppressive role in the early stages of HCC development to a more oncogenic role. Although it was proposed that Notch signaling plays a role in liver cancer invasion and metastasis (Lim et al., 2011; Zhou et al., 2013), more work is required to further support this notion.
Colorectal cancer
The intestinal epithelium possesses an unprecedented self-renewal rate that appears to be linked to a high susceptibility to malignant transformation. Notch signaling has been known for many years now to be involved in both the control of homeostatic self-renewal in stem cell populations and the development of colorectal cancer (CRC) (Fre et al., 2005; Radtke and Clevers, 2005; van Es et al., 2005). While mutations in NOTCH genes are rare, Notch signaling is overexpressed or constitutively activated in CRC in part because of mutations in regulators of Notch signaling, including in FBXW7 (although FBXW7 clearly controls other cellular pathways beyond Notch) (Akhoondi et al., 2007; Babaei-Jadidi et al., 2011; Camps et al., 2013; Miyaki et al., 2009; Sancho et al., 2010; Zhu et al., 2013). In addition, Notch activation has been linked to activation of Wnt signaling and Hippo/YAP signaling in CRC cells, although the various levels of crosstalk between these pathways are still not fully understood (Camargo et al., 2007; Fre et al., 2009; Kim et al., 2012; Kwon et al., 2011; Peignon et al., 2011; Rodilla et al., 2009; Tschaharganeh et al., 2013). In particular, Jagged1, expressed on tumor cells themselves or produced from endothelial cells, is thought to be a key ligand for Notch activation in CRC cells (Lu et al., 2013; Rodilla et al., 2009; Tschaharganeh et al., 2013). Another Notch ligand, DLL4, plays a non-cell autonomous role in CRC development in large part by controlling the development of blood vessels necessary for tumor growth (Fischer et al., 2011; Ridgway et al., 2006). Expression of miR-34a in CRC stem cells may help control Notch output and generate a bimodal Notch response (Bu et al., 2013). Finally, Notch signaling may play a crucial role not only in the early stages of CRC development by controlling the fate of stem cells and cancer stem cells but also at the later stages of tumor invasion and metastasis (Sonoshita et al., 2011).
Pancreatic cancer
The major and most lethal type of pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC). An early study detected evidence of Notch pathway activation in PDAC and showed that Notch lies downstream of TGFβ during ductal metaplasia, an early stage of PDAC development (Miyamoto et al., 2003). Mouse genetics studies have demonstrated that activation of Notch signaling cooperates with oncogenic K-Ras to promote both initiation and dysplastic progression from acinar cells by inducing their rapid reprogramming to a duct-like phenotype (De La et al., 2008). Indeed, pharmacological inhibition of Notch signaling slows the progression of the disease in mutant mice and prevents the expansion of some human PDAC cell lines (Cook et al., 2012; Mizuma et al., 2012; Plentz et al., 2009), possibly in part because of an inhibition of PDAC stem cells (Bailey et al., 2013). Genetic inactivation of Notch2, but not Notch1 (Avila et al., 2012; Mazur et al., 2010), inhibits PDAC development initiated by oncogenic K-Ras. In fact, loss of Notch1 function may even promote PDAC development, although the basis of this observation remains unknown (Hanlon et al., 2010).
Melanoma
The Notch pathway has been found to be active in melanoma (Asnaghi et al., 2012). NOTCH1 appears to promote disease progression (Rangarajan et al., 2001; Zhang et al., 2012) and growth of melanocytes under hypoxic conditions (Bedogni et al., 2008). There are no documented gain-of-function mutations affecting the pathway in this disease (Hodis et al., 2012), suggesting that the pathway might be affected through transcriptional and epigenetic control possibly through contrasting actions of BRN2, a possible activator, and MITF, which acts as a repressor of the Notch pathway (Thurber et al., 2011). Whatever the mechanism of activation, recent pre-clinical studies have reported that γ-secretase inhibitors (GSI) can reduce the tumor initiating potential and suggested that GSI combination with chemotherapy could be a useful new therapeutic approach in melanoma (Huynh et al., 2011).
In conclusion, Notch signaling plays distinct roles in different types of tumors, both solid and liquid (hematopoietic). The presented list is by no means exhaustive but provides us with an extensive overview of Notch signaling in cancer. Most of these studies are relatively recent emphasizing the increased interest in the study of Notch signaling in cancer during the last decade. Mechanistically, more work is required to pinpoint specific molecular pathways and gene targets in each tumor type, but emerging technologies and most notably DNA and RNA next generation sequencing-based approaches will continue to help us further dissect the role of this pathway in tumor initiation and progression.
A perspective on two decades of Notch-centered cancer research: Remaining intriguing questions
This brief overview of some of the most common and/or lethal human cancers, both hematopoietic and solid, highlight several key aspects of Notch signaling in cancer development that hold true in other tumors in which Notch signaling is also altered, including myeloma, prostate, ovarian, skin, and brain cancers. Obviously, there are several outstanding questions that have to be addressed to not only help us better understand pathway function in cancer but also enable more efficient therapeutic targeting (see below). An initial question is whether Notch pathway mutations are tumor-initiating or tumor-propagating. Most likely, both types of mutations can be described, depending on the tumor type. We discussed an intriguing example in myeloid neoplasms where Notch signaling loss of activity seems to expand the frequency of leukemia-initiating cells (LIC) but requires secondary mutational events to lead to full-blown disease (Klinakis et al., 2011; Lobry et al., 2013). A similar scenario might play out in BALL, as it was shown that Notch activity directs lymphocyte progenitors exclusively to the T cell lineage, at the expense of B cell differentiation (Pui et al., 1999; Radtke et al., 1999). On the other hand, it is intriguing to ask whether NOTCH1 activating mutations in T-ALL occur to simply define lineage, by locking cells in a specific differentiation status (T cell in this case), or to truly transform the cells. Further studies that can genetically sequence both leukemia and normal stem cell/progenitor populations, preferably at the single cell level, might address such questions.
One particularly interesting aspect of Notch signaling in cancer progression that has been emerging in the last few years is its potential impact on metastasis, which may be linked to the role of Notch in cancer stem cells (see (Giancotti, 2013) for a recent discussion). Early studies had identified JAG1 expression as a marker of metastatic prostate cancer (Santagata et al., 2004) and found a role for Notch in the control of epithelial-mesenchymal transitions (EMT) (Timmerman et al., 2004). Indeed, JAG1 expression on tumor cells may help promote the spread of breast cancer cells to the bone microenvironment by activating Notch signaling in bone cells (Sethi et al., 2011). Activation of Notch during EMT and metastasis may be under the control of the miR-200 microRNA (Brabletz et al., 2011; Yang et al., 2011). This pro-metastatic function of Notch signaling may be promoted by its crosstalk with the machinery responding to hypoxic environments (Sahlgren et al., 2008; Yeung et al., 2011). Furthermore, an increasing number of studies connect Notch signaling to molecules and pathways involved in tumor invasion and metastatic growth, including Tenascin C (Oskarsson et al., 2011) and regulators of polarity (McCaffrey et al., 2012) in breast cancer.
As discussed above, Notch signaling in tumor cells has been involved in various aspects of angiogenesis in multiple studies, especially via the DLL4 and JAG1 ligands (Benedito et al., 2009; Li and Harris, 2005; Phng and Gerhardt, 2009; Zeng et al., 2005). In particular, the Notch ligand DLL4 is upregulated in the angiogenic vasculature in response to VEGF and blockade of DLL4 was shown to lead to markedly increased non-productive tumor vascularity, which inhibits tumor growth (Noguera-Troise et al., 2006; Ridgway et al., 2006). While these observations seemed promising clinically (Hoey et al., 2009), long-term blockade of DLL4 leads to the development of vascular neoplasms (Yan et al., 2010), potentially limiting the therapeutic potential of DLL4-blocking strategies. Thus, activation of Notch signaling may contribute to tumor spread via multiple mechanisms, including by maintaining the self-renewal of cancer stem cells, by directly contributing to the cellular processes involved in tumor invasion (e.g. EMT and response to hypoxia) (Wang et al., 2011c), by controlling neo-vascularization, as well as by playing a key role in the metastatic niche. Such issues could be more important in solid tumors than leukemia, however, it is intriguing to define Notch-ligand expressing niches in different types of hematopoietic tumors and test whether ligand expression is important for leukemia cell homing to different tissues and response to drug treatments. For example, targeting the expression or function of a specific ligand could affect NOTCH1-expressing T-ALL homing and metastasis. As most cancer patients die from metastatic disease, it will be important in the near future to continue to investigate the molecular and cellular basis of tumor spread in connection with Notch signaling.
Therapeutic targeting of the Notch pathway in tumors
As proteolytic cleavage of NOTCH receptors by the presenilin/γ-secretase complex is a prerequisite for the activation of signaling (in the absence of downstream activating mutations), small molecule GSI efficiently blocks NOTCH1 activity in T-ALL cells and has been proposed as a molecular targeted therapy for the treatment of this disease (Aster and Blacklow, 2012; Palomero and Ferrando, 2009). However, animal studies have shown that systemic inhibition of NOTCH signaling results in “on-target” gastrointestinal toxicity because of the accumulation of secretory goblet cells in the intestine due to alterations in the differentiation of intestinal stem cells following Notch inactivation. Phase 1 clinical trials further confirmed these treatment side effects. As a result, inhibition of the pathway using GSI alone may not be the most viable therapeutic choice in the future. An alternative to the use of single GSI treatment is the combinatorial use of glucocorticoids and GSI, where glucocorticoids ameliorate the GSI-induced gut toxicity by inducing the expression of Cyclin D2, protecting the animals from developing intestinal goblet cell metaplasia (Real et al., 2009).
Notch signaling targeting is, however, not restricted to the usage of GSI. Alpha-secretase inhibitors (ASI) against the ADAM10/17 metalloproteases that mediate receptor S2 cleavage are available (Zhou et al., 2006) and are currently being tested (Purow, 2012). Furthermore, using phage display technology, pharmaceutical companies have generated highly specialized antibodies against NOTCH1 and NOTCH2 that act mainly through stabilization of the negative regulatory region (NRR) of the receptors, and the protection from proteolytic cleavage, thus inhibiting the production of ICN1/2 (Wu et al., 2010). These antibodies lead to lower levels of gastrointestinal toxicity and other side effects emanating from pan-Notch pathway inhibition achieved by GSI. Selective blocking of NOTCH1 inhibits tumor growth in pre-clinical models through at least two mechanisms: inhibition of cancer cell growth and deregulation of angiogenesis. Soluble extracellular fractions of Notch receptors and ligands can also act as decoys and inhibit the pathway in a dominant-negative manner. A Notch1 decoy decreased tumor cell viability in xenograft models (Funahashi et al., 2008). However, under different conditions, a DLL1 decoy can play either an activating or inhibitory role (Hicks et al., 2002). Thus, a better understanding of the dynamics by which decoys work is needed before they can be considered as a viable therapeutic strategy.
Other types of experimental inhibitors entail synthetic peptides that mimic MAML1 but lack its active domains. Despite the use of these peptides to serve basic research purposes, their use for therapeutic purposes is still limited. Moellering et al. generated a synthetic, cell-permeable, alpha-helical peptide (SAHM1) blocking MAML1 recruitment and NOTCH-mediated transcription as it binds with high affinity to the interface on the NOTCH-CSL transactivation complex (Moellering et al., 2009). Treatment of human T-ALL cell lines and a mouse model of NOTCH1-driven T-ALL with SAHM1 resulted in strong, NOTCH-specific inhibition of cell proliferation, and leukemia progression (Moellering et al., 2009).
Another intriguing idea for the treatment of tumors that are induced by NOTCH and depend on pathway activity is to not target the Notch pathway itself but focus on its signaling targets. Several such efforts are currently underway. Briefly, we have recently demonstrated in vivo T-ALL remission when we target: a) the NOTCH1-induced IKK kinase complex with a pivotal role in controlling the NF-kB pathway which-in turn-is strongly related to NOTCH in leukemia (Figure 4) (Dan et al., 2008; Espinosa et al., 2010; Vilimas et al., 2007), b) the CyclinD:CDK4/6 kinase complex, hyperactivated in this type of acute leukemia (Sawai et al., 2012), and c) the bromodomain-containing protein BRD4 (King et al., 2013). BRD proteins can be transcriptional co-activators and share common binding patterns with T-ALL oncogenes NOTCH1 and MYC in promoters and enhancers of key genes for the induction and progression of the disease. Bradner and colleagues recently modified a thienodiazepine molecule so that it inhibits binding of BRD to the acetylated residues of histone H4 (Filippakopoulos et al., 2010). We were able to show that such drugs can target both NOTCH1- and MYC-regulated transcription in T-ALL, leading to complete disease remission in vivo. Such, “epigenetically”-targeted therapies might be particularly attractive considering the ability of Notch to alter locus accessibility and initiate transcription. We have recently connected NOTCH1 binding to loss of H3K27me3 on target promoters and demonstrated an antagonism between NOTCH1 binding and polycomb complex 2 (PRC2) recruitment and activity (Ntziachristos et al., 2012). Based on these findings, H3K27me3 demethylation inhibitors might be an attractive therapy option in NOTCH1-induced T-ALL (or CLL). Finally, recent evidence suggests that it may be possible to inhibit Notch signaling by interfering with its trafficking in cancer cell secretory pathways (Ilagan and Kopan, 2013; Kramer et al., 2013).
While a number of “anti-Notch” strategies are emerging, it may be as important to specifically activate Notch in tumors where activation of the Notch pathway is tumor suppressive. As discussed above for AML, in the case where tumor cells express a Notch receptor (NOTCH2) but do not show signs of pathway activation, providing a ligand for these receptors, or treating with activating antibodies may be sufficient to activate the pathway in certain contexts and inhibit tumor growth. Furthermore, in cases where Notch receptors are not expressed (e.g. they are transcriptionally silenced), or if some of their key target genes are silenced, approaches to de-repress the expression of these genes may be useful to slow cancer growth (Stockhausen et al., 2005).
Future directions in the understanding and treatment of Notch-induced tumors
The NOTCH pathway has been the intense focus of cancer researchers for the last two decades. Unfortunately, there are still no FDA-approved, Notch-targeted therapies. Retrospectively, this is not surprising, as we now know that the pathway plays key roles in several tissues, including adult differentiating and regenerating tissues, explaining the potential side-effects of general inhibitors of the Notch pathway such as GSI. The critical question is whether one can successfully target the Notch pathway to significantly inhibit cancer growth. Another major conundrum comes from possible distinct roles for Notch at several stages of the tumorigenic process, an idea that was not been thoroughly examined. Furthermore, it is likely that inhibition of Notch signaling in tumors initiated by Notch activating mutations will have a therapeutic effect, as tumors are often addicted to early initiating events. However, in tumors where alterations in Notch pathway members occur late during tumor evolution, tumors may rapidly invent ways around the targeting of Notch.
We would suggest that specificity should be the key for future attempts to target Notch activity in cancer cells: one should have a complete map of both Notch receptor and ligand expression in different cancers and their microenvironments to be able to use antibodies or other small molecules that specifically inhibit only the relevant molecules. Targeted (Notch-focused) sequencing of tumors is also important to provide a clear idea of the type of mutation and its potential impact on pathway activity. Importantly, a large number of tumors containing Notch activating mutations, like the ICN1 translocation, cannot be treated with GSI. In contrast, receptor-specific antibody agonists could be of significance clinical value for tumors in which Notch signaling has a tumor suppressive function. Myeloid neoplasms are a cancer subtype that could benefit from targeted pathway activation as we have shown that in such tumors, the pathway is inactive but the NOTCH2 receptor is expressed on the surface of the cells and can be activated by ligand binding, leading to cell death. Strategies to activate Notch in some cancer are worth testing, first in pre-clinical models and hopefully in the near future in patients. Moreover, both Notch agonists and antagonists could also be used in combination with current treatments, including chemotherapy and more recent targeted therapies. A recent intriguing example of such treatments is the combination of anti-DLL4 antibodies with either chemotherapy or Avastin or VEGF traps to target tumor angiogenesis (Lobov et al., 2011; Noguera-Troise et al., 2006). Another one is the combination of Notch receptor inhibition (using GSI or antibodies) and glucocorticoids for the treatment of T-ALL (Real et al., 2009).
As with other signaling pathways involved in cancer, identification and targeting of Notch-interacting partners and targets could be pivotal for the development of anti-tumor therapy protocols. Some attempts have been made to identify such genes/proteins using whole proteome (mass spectrometry) (Yatim et al., 2012) and genome/transcriptome (RNA-seq, gene array, ChIP-seq for NOTCH1 and HES1) (Ntziachristos et al., 2012; Wang et al., 2011a) approaches. These studies suggest that, apart from a small fraction of “universal” targets, including members of the HES and HEY families, Notch pathway activity controls the expression of a large number of tissue and cell-type specific gene targets. Indeed, we have previously shown that NOTCH2-HES1 signaling can regulate the expression of CEBPA and PU1, two key regulators of myeloid differentiation, but these genes are not affected by Notch pathway regulation in T-ALL. Thus, potential targeting of the function of tissue-specific Notch pathway targets could offer more targeted therapies with fewer side effects. In a similar fashion, it will be intriguing to define the biochemical composition of the nuclear Notch complex in different tissues to see whether there is specificity that can guide small molecule inhibition efforts.
Overall, it is fair to say that it took the scientific community almost a century to reach to the point that the basic molecular tenets of Notch signaling are well understood. Similarly, although we know for the last two decades that Notch signaling is involved in cancer, only recently we developed the means (small molecules, antibodies) to effectively target pathway activation in this disease. There is still a significant need for further research efforts that will better define the pathway and propose drugs or drug combinations that can affect Notch signaling specifically in cancer, avoiding harmful side-effects and improving both survival and quality of life for patients with Notch-induced tumors.
ACKNOWLEDGEMENTS
We would like to thank the members of the Aifantis laboratory for helpful discussions and especially Anastasia Tikhonova and Philmo Oh for sharing Hes1GFP data. The Aifantis Lab is supported by the National Institutes of Health (1R01CA169784, 1R01CA133379, 1R01CA105129, 1R01CA149655, 5R01CA173636), the William Lawrence and Blanche Hughes Foundation, The Leukemia & Lymphoma Society (TRP#6340-11, LLS#6373-13), The Chemotherapy Foundation, The V Foundation for Cancer Research, the Alex’s Lemonade Stand Foundation for Childhood Cancer and the St. Baldrick’s Cancer Research Foundation. I.A. is grateful for the continuous support of the Feinberg Foundation. P.N. has been supported by the Lady Tata Memorial Trust and the American Society for Hematology. I.A. is a Howard Hughes Medical Institute Early Career Scientist. Work on Notch signaling in the Sage lab is supported by the National Institute of Health (1R01CA172560), the American Cancer Society (118898-RSG-10-071-01-TBG), and the Leukemia & Lymphoma Society (LLS#1029-10). JS is the Harriet and Mary Zelencik Scientist in Children’s Cancer and Blood Diseases. JSL is funded by the A*STAR in Singapore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer discovery. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nature reviews Immunology. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer research. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer research. 2011;71:6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, Reman O, Witz F, Fagot T, Tavernier E, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113:3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- Asnaghi L, Ebrahimi KB, Schreck KC, Bar EE, Coonfield ML, Bell WR, Handa J, Merbs SL, Harbour JW, Eberhart CG. Notch signaling promotes growth and invasion in uveal melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:654–665. doi: 10.1158/1078-0432.CCR-11-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Blacklow SC. Targeting the Notch pathway: twists and turns on the road to rational therapeutics. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:2418–2420. doi: 10.1200/JCO.2012.42.0992. [DOI] [PubMed] [Google Scholar]
- Avila JL, Troutman S, Durham A, Kissil JL. Notch1 is not required for acinar-to-ductal metaplasia in a model of Kras-induced pancreatic ductal adenocarcinoma. PloS one. 2012;7:e52133. doi: 10.1371/journal.pone.0052133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, DEK for degradation. The Journal of experimental medicine. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, et al. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells With Stem Cell Properties in Preinvasive Pancreatic Cancer. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj J, Maliekal TT, Vivien E, Pattabiraman C, Srivastava S, Krishnamurthy H, Giri V, Subramanyam D, Krishna S. Notch signaling in CD66+ cells drives the progression of human cervical cancers. Cancer research. 2011;71:4888–4897. doi: 10.1158/0008-5472.CAN-11-0543. [DOI] [PubMed] [Google Scholar]
- Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. The Journal of clinical investigation. 2005;115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. Notch1 is an effector of Akt and hypoxia in melanoma development. The Journal of clinical investigation. 2008;118:3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, Brabletz T. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. The EMBO journal. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nature reviews Molecular cell biology. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, Happich M, Muckenthaler MU, Kulozik AE. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell stem cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Current biology : CB. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, Beissbarth T, et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer research. 2013;73:2003–2013. doi: 10.1158/0008-5472.CAN-12-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Molecular and cellular biology. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PM, Yen CC, Wang WS, Lin YJ, Chu CJ, Chiou TJ, Liu JH, Yang MH. Down-regulation of Notch-1 expression decreases PU.1-mediated myeloid differentiation signaling in acute myeloid leukemia. International journal of oncology. 2008;32:1335–1341. doi: 10.3892/ijo_32_6_1335. [DOI] [PubMed] [Google Scholar]
- Chu Q, Orr BA, Semenkow S, Bar EE, Eberhart CG. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:3224–3233. doi: 10.1158/1078-0432.CCR-12-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G, Kaltenbach S, Yakouben K, Mazingue F, Robert A, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24:2023–2031. doi: 10.1038/leu.2010.205. [DOI] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Cook M, Yu XM, Chen H. Notch in the development of thyroid C-cells and the treatment of medullary thyroid cancer. American journal of translational research. 2010;2:119–125. [PMC free article] [PubMed] [Google Scholar]
- Cook N, Frese KK, Bapiro TE, Jacobetz MA, Gopinathan A, Miller JL, Rao SS, Demuth T, Howat WJ, Jodrell DI, Tuveson DA. Gamma secretase inhibition promotes hypoxic necrosis in mouse pancreatic ductal adenocarcinoma. The Journal of experimental medicine. 2012;209:437–444. doi: 10.1084/jem.20111923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croquelois A, Blindenbacher A, Terracciano L, Wang X, Langer I, Radtke F, Heim MH. Inducible inactivation of Notch1 causes nodular regenerative hyperplasia in mice. Hepatology. 2005;41:487–496. doi: 10.1002/hep.20571. [DOI] [PubMed] [Google Scholar]
- Cui H, Kong Y, Xu M, Zhang H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer research. 2013;73:3451–3459. doi: 10.1158/0008-5472.CAN-12-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes & development. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TP, Eichenberger S, Gonzalez A, Olson S, Carbone DP. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Dexter JS. The analysis of a case of continuous variation in Drosophila by a study of its linkage relationships. The American Naturalist. 1914:712–758. [Google Scholar]
- Dill MT, Tornillo L, Fritzius T, Terracciano L, Semela D, Bettler B, Heim MH, Tchorz JS. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57:1607–1619. doi: 10.1002/hep.26165. [DOI] [PubMed] [Google Scholar]
- Eliasz S, Liang S, Chen Y, De Marco MA, Machek O, Skucha S, Miele L, Bocchetta M. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene. 2010;29:2488–2498. doi: 10.1038/onc.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Espinosa L, Cathelin S, D'Altri T, Trimarchi T, Statnikov A, Guiu J, Rodilla V, Ingles-Esteve J, Nomdedeu J, Bellosillo B, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer cell. 2010;18:268–281. doi: 10.1016/j.ccr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. The Journal of experimental medicine. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer research. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Yen WC, Kapoun AM, Wang M, O'Young G, Lewicki J, Gurney A, Hoey T. Anti-DLL4 inhibits growth and reduces tumor-initiating cell frequency in colorectal tumors with oncogenic KRAS mutations. Cancer research. 2011;71:1520–1525. doi: 10.1158/0008-5472.CAN-10-2817. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, Kissel H, Louvi A, Greve J, Louvard D, Artavanis-Tsakonas S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, Artavanis-Tsakonas S, Louvard D. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Molecular cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fu YP, Edvardsen H, Kaushiva A, Arhancet JP, Howe TM, Kohaar I, Porter-Gill P, Shah A, Landmark-Hoyvik H, Fossa SD, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Molecular cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Funahashi Y, Hernandez SL, Das I, Ahn A, Huang J, Vorontchikhina M, Sharma A, Kanamaru E, Borisenko V, Desilva DM, et al. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer research. 2008;68:4727–4735. doi: 10.1158/0008-5472.CAN-07-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4) Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Kozak C, Callahan R. A new common integration region (int-3) for mouse mammary tumor virus on mouse chromosome 17. Journal of virology. 1987;61:218–220. doi: 10.1128/jvi.61.1.218-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes & development. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. The Journal of experimental medicine. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest RV, Boulter L, Kendall TJ, Minnis-Lyons SE, Walker R, Wigmore SJ, Sansom OJ, Forbes SJ. Cell lineage tracing reveals a biliary origin of intrahepatic cholangiocarcinoma. Cancer research. 2013 doi: 10.1158/0008-5472.CAN-13-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature genetics. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nature reviews Molecular cell biology. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Hanlon L, Avila JL, Demarest RM, Troutman S, Allen M, Ratti F, Rustgi AK, Stanger BZ, Radtke F, Adsay V, et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer research. 2010;70:4280–4286. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson EM, Teixeira AI, Gustafsson MV, Dohda T, Chapman G, Meletis K, Muhr J, Lendahl U. Recording Notch signaling in real time. Developmental neuroscience. 2006;28:118–127. doi: 10.1159/000090758. [DOI] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer research. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, Carbone DP, Dang TP. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer research. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Wang L, Korkaya H, Chen G, Maillard I, Beer DG, Kalemkerian GP, Wicha MS. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:1972–1980. doi: 10.1158/1078-0432.CCR-12-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C, Ladi E, Lindsell C, Hsieh JJ, Hayward SD, Collazo A, Weinmaster G. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J Neurosci Res. 2002;68:655–667. doi: 10.1002/jnr.10263. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Yen WC, Axelrod F, Basi J, Donigian L, Dylla S, Fitch-Bruhns M, Lazetic S, Park IK, Sato A, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell stem cell. 2009;5:168–177. doi: 10.1016/j.stem.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Huynh C, Poliseno L, Segura MF, Medicherla R, Haimovic A, Menendez S, Shang S, Pavlick A, Shao Y, Darvishian F, et al. The novel gamma secretase inhibitor RO4929097 reduces the tumor initiating potential of melanoma. PloS one. 2011;6:e25264. doi: 10.1371/journal.pone.0025264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MX, Kopan R. Selective blockade of transport via SERCA inhibition: the answer for oncogenic forms of Notch? Cancer cell. 2013;23:267–269. doi: 10.1016/j.ccr.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Imatani A, Callahan R. Identification of a novel NOTCH-4/INT-3 RNA species encoding an activated gene product in certain human tumor cell lines. Oncogene. 2000;19:223–231. doi: 10.1038/sj.onc.1203295. [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Molecular and cellular biology. 2001a;21:6071–6079. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Poizat C, Iezzi S, Wu HY, Chung G, Kedes L, Hamamori Y. HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Molecular and cellular biology. 2001b;21:6080–6089. doi: 10.1128/MCB.21.17.6080-6089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izrailit J, Berman HK, Datti A, Wrana JL, Reedijk M. High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFbeta pathways as fundamental Notch regulators in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1714–1719. doi: 10.1073/pnas.1214014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jeliazkova P, Jors S, Lee M, Zimber-Strobl U, Ferrer J, Schmid RM, Siveke JT, Geisler F. Canonical Notch2 signaling determines biliary cell fates of embryonic hepatoblasts and adult hepatocytes independent of Hes1. Hepatology. 2013;57:2469–2479. doi: 10.1002/hep.26254. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes & development. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, Osborne BA. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113:1689–1698. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jundt F, Acikgoz O, Kwon SH, Schwarzer R, Anagnostopoulos I, Wiesner B, Mathas S, Hummel M, Stein H, Reichardt HM, Dorken B. Aberrant expression of Notch1 interferes with the B-lymphoid phenotype of neoplastic B cells in classical Hodgkin lymphoma. Leukemia. 2008;22:1587–1594. doi: 10.1038/leu.2008.101. [DOI] [PubMed] [Google Scholar]
- Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, Akers LJ, Hammitt RA, McMurray JS, Kornblau SM, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. The Journal of experimental medicine. 2013;210:321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Molecular and cellular biology. 1986;6:3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Velusamy T, Betz BL, Zhao L, Weigelin HG, Chiang MY, Huebner-Chan DR, Bailey NG, Yang DT, Bhagat G, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. The Journal of experimental medicine. 2012;209:1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Koo BK, Cho JH, Kim YY, Seong J, Chang HJ, Oh YM, Stange DE, Park JG, Hwang D, Kong YY. Notch1 counteracts WNT/beta-catenin signaling through chromatin modification in colorectal cancer. The Journal of clinical investigation. 2012;122:3248–3259. doi: 10.1172/JCI61216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluk MJ, Ashworth T, Wang H, Knoechel B, Mason EF, Morgan EA, Dorfman D, Pinkus G, Weigert O, Hornick JL, et al. Gauging NOTCH1 Activation in Cancer Using Immunohistochemistry. PloS one. 2013;8:e67306. doi: 10.1371/journal.pone.0067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Mentrup T, Kleizen B, Rivera-Milla E, Reichenbach D, Enzensperger C, Nohl R, Tauscher E, Gorls H, Ploubidou A, et al. Small molecules intercept Notch signaling and the early secretory pathway. Nature chemical biology. 2013;9:731–738. doi: 10.1038/nchembio.1356. [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Fang Z, Zweidler-McKay PA, Yang H, Wei Y, Gonzalez-Cervantes EA, Boumber Y, Garcia-Manero G. Epigenetic inactivation of Notch-Hes pathway in human B-cell acute lymphoblastic leukemia. PloS one. 2013;8:e61807. doi: 10.1371/journal.pone.0061807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D. Notch post-translationally regulates beta-catenin protein in stem and progenitor cells. Nature cell biology. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. The Journal of biological chemistry. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, Yamamoto G, Nannya Y, Suzuki R, Ota S, Ota Y, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer science. 2009;100:920–926. doi: 10.1111/j.1349-7006.2009.01130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits Notch activation. Development. 2003;130:6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- Lewis HD, Leveridge M, Strack PR, Haldon CD, O'Neil J, Kim H, Madin A, Hannam JC, Look AT, Kohl N, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chemistry & biology. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer cell. 2005;8:1–3. doi: 10.1016/j.ccr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Li X, von Boehmer H. Notch Signaling in T-Cell Development and T-ALL. ISRN hematology. 2011;2011:921706. doi: 10.5402/2011/921706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licciulli S, Avila JL, Hanlon L, Troutman S, Cesaroni M, Kota S, Keith B, Simon MC, Pure E, Radtke F, et al. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer research. 2013;73:5974–5984. doi: 10.1158/0008-5472.CAN-13-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SO, Kim HS, Quan X, Ahn SM, Kim H, Hsieh D, Seong JK, Jung G. Notch1 binds and induces degradation of Snail in hepatocellular carcinoma. BMC biology. 2011;9:83. doi: 10.1186/1741-7007-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Turkoz A, Jackson EN, Corbo JC, Engelbach JA, Garbow JR, Piwnica-Worms DR, Kopan R. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. The Journal of clinical investigation. 2011;121:800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobov IB, Cheung E, Wudali R, Cao J, Halasz G, Wei Y, Economides A, Lin HC, Papadopoulos N, Yancopoulos GD, Wiegand SJ. The Dll4/Notch pathway controls postangiogenic blood vessel remodeling and regression by modulating vasoconstriction and blood flow. Blood. 2011;117:6728–6737. doi: 10.1182/blood-2010-08-302067. [DOI] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. The Journal of experimental medicine. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliekal TT, Bajaj J, Giri V, Subramanyam D, Krishna S. The role of Notch signaling in human cervical cancer: implications for solid tumors. Oncogene. 2008;27:5110–5114. doi: 10.1038/onc.2008.224. [DOI] [PubMed] [Google Scholar]
- Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer research. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- Maraver A, Fernandez-Marcos PJ, Herranz D, Canamero M, Munoz-Martin M, Gomez-Lopez G, Mulero F, Megias D, Sanchez-Carbayo M, Shen J, et al. Therapeutic effect of gamma-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer cell. 2012;22:222–234. doi: 10.1016/j.ccr.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marignol L, Rivera-Figueroa K, Lynch T, Hollywood D. Hypoxia, notch signalling, and prostate cancer. Nature reviews Urology. 2013;10:405–413. doi: 10.1038/nrurol.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur PK, Einwachter H, Lee M, Sipos B, Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Kloppel G, et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, et al. Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5012–5017. doi: 10.1073/pnas.1000896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, Chin KT, Partridge JC, Poole BB, Cheng KH, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2939–E2948. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Montalbano J, Mihai C, Macara IG. Loss of the Par3 polarity protein promotes breast tumorigenesis and metastasis. Cancer cell. 2012;22:601–614. doi: 10.1016/j.ccr.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. American journal of human genetics. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurette O, Stylianou S, Rock R, Collu GM, Gilmore AP, Brennan K. Notch activation induces Akt signaling via an autocrine loop to prevent apoptosis in breast epithelial cells. Cancer research. 2009;69:5015–5022. doi: 10.1158/0008-5472.CAN-08-3478. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Yamaguchi T, Iijima T, Takahashi K, Matsumoto H, Mori T. Somatic mutations of the CDC4 (FBXW7) gene in hereditary colorectal tumors. Oncology. 2009;76:430–434. doi: 10.1159/000217811. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- Mizuma M, Rasheed ZA, Yabuuchi S, Omura N, Campbell NR, de Wilde RF, De Oliveira E, Zhang Q, Puig O, Matsui W, et al. The gamma secretase inhibitor MRK-003 attenuates pancreatic cancer growth in preclinical models. Molecular cancer therapeutics. 2012;11:1999–2009. doi: 10.1158/1535-7163.MCT-12-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. The theory of the gene. The American Naturalist LI. 1917:513–544. [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nature cell biology. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Mullendore ME, Koorstra JB, Li YM, Offerhaus GJ, Fan X, Henderson CM, Matsui W, Eberhart CG, Maitra A, Feldmann G. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nature genetics. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Nowell C, Radtke F. Cutaneous notch signaling in health and disease. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. The Journal of experimental medicine. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nature genetics. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Oh P, Lobry C, Gao J, Tikhonova A, Loizou E, Manent J, van Handel B, Ibrahim S, Greve J, Mikkola H, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell stem cell. 2013;13:190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein Ofucosyltransferase 1 and fringe. The Journal of biological chemistry. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Ortica S, Tarantino N, Aulner N, Israel A, Gupta-Rossi N. The 4 Notch receptors play distinct and antagonistic roles in the proliferation and hepatocytic differentiation of liver progenitors. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013 doi: 10.1096/fj.13-235903. [DOI] [PubMed] [Google Scholar]
- Oscier DG, Rose-Zerilli MJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, Parker H, Parker A, Gardiner A, Collins A, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121:468–475. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nature medicine. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clinical lymphoma & myeloma. 2009;9(Suppl 3):S205–S210. doi: 10.3816/CLM.2009.s.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Taki T, Oda M, Watanabe T, Yumura-Yagi K, Kobayashi R, Suzuki N, Hara J, Horibe K, Hayashi Y. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. British journal of haematology. 2009;145:198–206. doi: 10.1111/j.1365-2141.2009.07607.x. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, Ouillette P, Trifonov V, Rossi D, Tabbo F, et al. Genetics of Follicular Lymphoma Transformation. Cell reports. 2014 doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. The Journal of experimental medicine. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. The Journal of cell biology. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peignon G, Durand A, Cacheux W, Ayrault O, Terris B, Laurent-Puig P, Shroyer NF, Van Seuningen I, Honjo T, Perret C, Romagnolo B. Complex interplay between beta-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Developmental cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond J, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer discovery. 2013;3:770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plentz R, Park JS, Rhim AD, Abravanel D, Hezel AF, Sharma SV, Gurumurthy S, Deshpande V, Kenific C, Settleman J, et al. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2009;136:1741–1749e1746. e1746. doi: 10.1053/j.gastro.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, Millar SE, Pear WS, Parmacek MS. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer research. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Purow B. Notch inhibition as a promising new approach to cancer therapy. Adv Exp Med Biol. 2012;727:305–319. doi: 10.1007/978-1-4614-0899-4_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, Guo Z, Cheng T, Cao X. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer research. 2003;63:8323–8329. [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nature reviews Immunology. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature reviews Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. The EMBO journal. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nature medicine. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- Robbins J, Blondel BJ, Gallahan D, Callahan R. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. Journal of virology. 1992;66:2594–2599. doi: 10.1128/jvi.66.4.2594-2599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F, Marasca R, Laurenti L, Bruscaggin A, Cerri M, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012a;119:521–529. doi: 10.1182/blood-2011-09-379966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, Spina V, Monti S, Vaisitti T, Arruga F, Fama R, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. The Journal of experimental medicine. 2012b;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg SM, Ellisen LW. The molecular pathogenesis of head and neck squamous cell carcinoma. The Journal of clinical investigation. 2012;122:1951–1957. doi: 10.1172/JCI59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulland S, Faroudi M, Mamessier E, Sungalee S, Salles G, Nadel B. Early steps of follicular lymphoma pathogenesis. Advances in immunology. 2011;111:1–46. doi: 10.1016/B978-0-12-385991-4.00001-5. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcido CD, Larochelle A, Taylor BJ, Dunbar CE, Varticovski L. Molecular characterisation of side population cells with cancer stem cell-like characteristics in small-cell lung cancer. British journal of cancer. 2010;102:1636–1644. doi: 10.1038/sj.bjc.6605668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale S, Lafkas D, Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nature cell biology. 2013;15:451–460. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Jandke A, Davis H, Diefenbacher ME, Tomlinson I, Behrens A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139:929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer research. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, Genesca L, Trimarchi T, Kelliher MA, Clark M, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer cell. 2012;22:452–465. doi: 10.1016/j.ccr.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Schwarzer R, Dorken B, Jundt F. Notch is an essential upstream regulator of NF-kappaB and is relevant for survival of Hodgkin and Reed-Sternberg cells. Leukemia. 2012;26:806–813. doi: 10.1038/leu.2011.265. [DOI] [PubMed] [Google Scholar]
- Schwarzer R, Jundt F. Notch and NF-kappaB signaling pathways in the biology of classical Hodgkin lymphoma. Current molecular medicine. 2011;11:236–245. doi: 10.2174/156652411795243423. [DOI] [PubMed] [Google Scholar]
- Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, 3rd, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, Krishnamoorthy V, Bhasin M, Capobianco AJ, Kelliher MA. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Molecular and cellular biology. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund J, Johansson M, Manna S, Norin C, Pietras A, Beckman S, Nilsson E, Ljungberg B, Axelson H. Suppression of renal cell carcinoma growth by inhibition of Notch signaling in vitro and in vivo. The Journal of clinical investigation. 2008;118:217–228. doi: 10.1172/JCI32086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, Hashida H, Takabayashi A, Sasaki M, Robine S, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer cell. 2011;19:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Souilhol C, Cormier S, Monet M, Vandormael-Pournin S, Joutel A, Babinet C, Cohen-Tannoudji M. Nas transgenic mouse line allows visualization of Notch pathway activity in vivo. Genesis. 2006;44:277–286. doi: 10.1002/dvg.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South AP, Cho RJ, Aster JC. The double-edged sword of Notch signaling in cancer. Seminars in cell & developmental biology. 2012;23:458–464. doi: 10.1016/j.semcdb.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, Ball DW. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer research. 2001;61:3200–3205. [PubMed] [Google Scholar]
- Sriuranpong V, Borges MW, Strock CL, Nakakura EK, Watkins DN, Blaumueller CM, Nelkin BD, Ball DW. Notch signaling induces rapid degradation of achaete-scute homolog 1. Molecular and cellular biology. 2002;22:3129–3139. doi: 10.1128/MCB.22.9.3129-3139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhausen MT, Sjolund J, Manetopoulos C, Axelson H. Effects of the histone deacetylase inhibitor valproic acid on Notch signalling in human neuroblastoma cells. British journal of cancer. 2005;92:751–759. doi: 10.1038/sj.bjc.6602309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, Barnes K, Zuurbier L, Meijerink JP, Ferrando AA. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer research. 2013 doi: 10.1158/0008-5472.CAN-13-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. The Journal of experimental medicine. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber AE, Douglas G, Sturm EC, Zabierowski SE, Smit DJ, Ramakrishnan SN, Hacker E, Leonard JH, Herlyn M, Sturm RA. Inverse expression states of the BRN2 and MITF transcription factors in melanoma spheres and tumour xenografts regulate the NOTCH pathway. Oncogene. 2011;30:3036–3048. doi: 10.1038/onc.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelialmesenchymal transition during cardiac development and oncogenic transformation. Genes & development. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542. e1512. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Viatour P, Ehmer U, Saddic LA, Dorrell C, Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A, et al. Notch signaling inhibits hepatocellular carcinoma following inactivation of the RB pathway. The Journal of experimental medicine. 2011;208:1963–1976. doi: 10.1084/jem.20110198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S, Alegre ML, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nature medicine. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- Villanueva A, Alsinet C, Yanger K, Hoshida Y, Zong Y, Toffanin S, Rodriguez-Carunchio L, Sole M, Thung S, Stanger BZ, Llovet JM. Notch signaling is activated in human hepatocellular carcinoma and induces tumor formation in mice. Gastroenterology. 2012;143:1660–1669. e1667. doi: 10.1053/j.gastro.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Xue L, Cao Q, Lin Y, Ding Y, Yang P, Che L. Expression of Notch1, Jagged1 and beta-catenin and their clinicopathological significance in hepatocellular carcinoma. Neoplasma. 2009;56:533–541. doi: 10.4149/neo_2009_06_533. [DOI] [PubMed] [Google Scholar]
- Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, Leigh IM, Collisson EA, Gordon PB, Jakkula L, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu Y, Malek SN, Zheng P, Liu Y. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell stem cell. 2011c;8:399–411. doi: 10.1016/j.stem.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature medicine. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Weissmann S, Roller A, Jeromin S, Hernandez M, Abaigar M, Hernandez-Rivas JM, Grossmann V, Haferlach C, Kern W, Haferlach T, et al. Prognostic impact and landscape of NOTCH1 mutations in chronic lymphocytic leukemia (CLL): a study on 852 patients. Leukemia. 2013;27:2393–2396. doi: 10.1038/leu.2013.218. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & development. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, et al. Alterations of the Notch pathway in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Wickremasinghe RG, Prentice AG, Steele AJ. p53 and Notch signaling in chronic lymphocytic leukemia: clues to identifying novel therapeutic strategies. Leukemia. 2011;25:1400–1407. doi: 10.1038/leu.2011.103. [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nature genetics. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, et al. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basallike breast cancer. Cancer cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, Niessen K, Plowman GD. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010;463:E6–E7. doi: 10.1038/nature08751. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. The Journal of clinical investigation. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB, Zhou L. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Molecular cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung TM, Gandhi SC, Bodmer WF. Hypoxia and lineage specification of cell linederived colorectal cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4382–4387. doi: 10.1073/pnas.1014519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender S, Nickeleit I, Wuestefeld T, Sorensen I, Dauch D, Bozko P, El-Khatib M, Geffers R, Bektas H, Manns MP, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer cell. 2013;23:784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Li S, Chepeha DB, Giordano TJ, Li J, Zhang H, Polverini PJ, Nor J, Kitajewski J, Wang CY. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer cell. 2005;8:13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong P, Zhang L, Jacobs B, Borden EC, Aster JC, Bedogni B. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene. 2012;31:4609–4618. doi: 10.1038/onc.2011.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, de la Cruz CC, Sayles LC, Alleyne-Chin C, Vaka D, Knaak TD, Bigos M, Xu Y, Hoang CD, Shrager JB, et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer cell. 2013;24:59–74. doi: 10.1016/j.ccr.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, Lo Y, Baribaud F, Mikami I, Reguart N, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang N, Song W, You N, Li Q, Sun W, Zhang Y, Wang D, Dou K. The significance of Notch1 compared with Notch3 in high metastasis and poor overall survival in hepatocellular carcinoma. PloS one. 2013;8:e57382. doi: 10.1371/journal.pone.0057382. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu J, Wang J, Shi Z, Franklin JL, Deane NG, Coffey RJ, Beauchamp RD, Zhang B. Deciphering Genomic Alterations in Colorectal Cancer through Transcriptional Subtype-Based Network Analysis. PloS one. 2013;8:e79282. doi: 10.1371/journal.pone.0079282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, Aster JC, Allman D, Pear WS. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]