Abstract
Although many clinicians and researchers work to understand cancer, there has been limited success to effectively combine forces and collaborate over time, distance, data and budget constraints. Here we present a workflow template for multidisciplinary cancer therapy that was developed during the 2nd Annual Workshop on Cancer Systems Biology sponsored by Tufts University, Boston, MA in July 2012. The template was applied to the development of a metronomic therapy backbone for neuroblastoma. Three primary groups were identified: clinicians, biologists, and scientists (mathematicians, computer scientists, physicists and engineers). The workflow described their integrative interactions; parallel or sequential processes; data sources and computational tools at different stages as well as the iterative nature of therapeutic development from clinical observations to in vitro, in vivo, and clinical trials. We found that theoreticians in dialog with experimentalists could develop calibrated and parameterized predictive models that inform and formalize sets of testable hypotheses, thus speeding up discovery and validation while reducing laboratory resources and costs. The developed template outlines an interdisciplinary collaboration workflow designed to systematically investigate the mechanistic underpinnings of a new therapy and validate that therapy to advance development and clinical acceptance.
Introduction
Although numerous dedicated clinicians, biologists, mathematicians, engineers and computational scientists are working towards understanding cancer biology with the goal of improving therapies, there has been no overarching process template to organize how these disciplines may collaborate over time, distance, data and budget constraints. The challenges are many. Cancer biology is considered a complex system and the specialized knowledge needed to understand and treat the disease often resides in non-integrated, isolated research, development and clinical silos.
In July 2012, the Center of Cancer Systems Biology held its 2nd Annual Workshop on Cancer Systems Biology at Tufts University, Boston (1). Researchers with different scientific backgrounds explored the topic of how to advance metronomic drug therapy for cancer from its experimental successes to general clinical acceptance for specific pathologies. Herein we present our approach to the chain of therapeutic research, development, and clinical implementation: a framework for a research process that involves collaborators across disciplines such as biology, medicine, mathematics, engineering, and computer science. This manuscript is a first step towards accelerating the development of optimal metronomic treatment protocols for cancer patients through planned integrative, multi-disciplinary and multi-center research projects that follow a clearly defined iterative workflow from in vitro to in vivo work to clinical trials and back again as new study data and models accumulate.
Background
There is a growing trend for scientists to work together as interdisciplinary teams where each member brings different knowledge and perspectives to address complex challenges. The intent is to deal with the challenges in a new way, and to accelerate implementation of validated solutions. Translational clinical research requires a broad knowledge base from bench to bedside, and, although initially carried out by physician-scientists, it is now moving to collaborative practice (2). There are now tools to support design of translational clinical studies (3) and clinical trial simulation softwares are becoming widely used in drug development (4).
Current interdisciplinary work in cancer therapy development
Interdisciplinary work has been underway in cancer research for some time. For example, cancer control research has evolved during the past 20 years through collaborations between basic science and behavioral researchers (5). The National Cancer Institute has spearheaded the integration of experimentalists and theoreticians through its ‘Integrative Cancer Biology’ and ‘Physical Sciences in Oncology’ programs. The American Association for Cancer Research offers workshops on collaborative translational cancer research as well as an interdisciplinary Team Science Award. Stand Up To Cancer, an initiative of the Entertainment Industry Foundation (EIF) since 2009, funds eight scientific SU2C Dream Teams who must collaborate across specialties, institutions and disciplines to quickly develop innovative therapies.
The need
Although there is a need for interdisciplinary research, there are numerous barriers to overcome including resistance to novelty, communication difficulties across disciplines, and career development outside a single discipline (6). And, interdisciplinary development may raise a number of legal issues from sharing of intellectual property rights to dealing with therapeutic risks (7). Nevertheless, the benefits of early interdisciplinary work have been adapted to collaborations across diagnostic and pharmaceutical industries, where drug research and co-development is now aiming at stratified, or personalized, medicine due to the ever-increasing regulatory demands for drug safety and efficacy (8).
Despite numerous initiatives promoting team science, such as the toolkit offered by the National Cancer Institute, the focus seems to be on multidisciplinary collaborations within each silo of research, development, or clinical practice. There is a need for a systematic interdisciplinary approach that generates not only “educated” research questions, but facilitates commercial development, satisfies regulatory requirements, and speeds up the adoption of new therapies into clinical practice – with feedback into new experimental and preclinical research.
We propose that this systematic interdisciplinary approach to novel therapeutic discovery can be designed in advance – at least at a conceptual level – and detailed as the work progresses. Communication, sharing of responsibilities, sequence order of deliverables as well as timing for synchronization of process phases are key. Although carried out by different entities, the therapeutic discovery process must be a continuum from lab to patient and back in an ongoing cycle of refinement. Here we present a template for such an interactive/integrative workflow applied to metronomic drug delivery and dosing therapy for high-risk neuroblastoma (NB) as a concrete example of our solution.
Example
Metronomic treatment embraces continuous administration of low-dose chemotherapy vis-à-vis often-practiced maximum tolerable dose (MTD) delivery. The benefits of metronomic chemotherapy have been demonstrated in numerous pre-clinical and clinical studies for breast cancer and pediatric cancers including dynamic treatment of pediatric acute lymphoblastic leukemia (9). In pilot studies of metronomic therapy for pediatric cancers, not only was disease stabilized in some patients but the majority of children experienced less pain and pain medication could be discontinued (10).
High-risk neuroblastoma (NB) is a leading cause of cancer-related deaths in children and patients are treated with a combination of surgery, radiation, induction chemotherapy followed by maintenance therapy and immunotherapy. However, despite very aggressive therapy, only 40% of high-risk NB patients survive, thus prompting the need for more educated, effective therapeutic options.
Why this therapy and this disease?
Despite experimental successes in clinical salvage and maintenance, metronomic therapy is not widely used. There are questions about the best clinical settings for this therapy – induction, maintenance or relapse – and concerns about interactions with other therapeutic agents. How do we validate this therapy for specific uses? How do we optimize the discovery pipeline and “pre-qualify” protocols to improve our chances of success? And how can this be done cost-effectively in laboratory and preclinical research? We suggest that close collaborations among clinicians, biologists and quantitative scientists (including mathematicians, computer scientists, and engineers) throughout the research, development, and implementation process will dramatically increase the chances of success. This process requires an ongoing dialog between theoreticians and experimentalists, and an agreed-upon workflow that outlines responsibilities and deliverables from discovery to clinical trials.
Results: A Template for A Therapeutic Development Pipeline
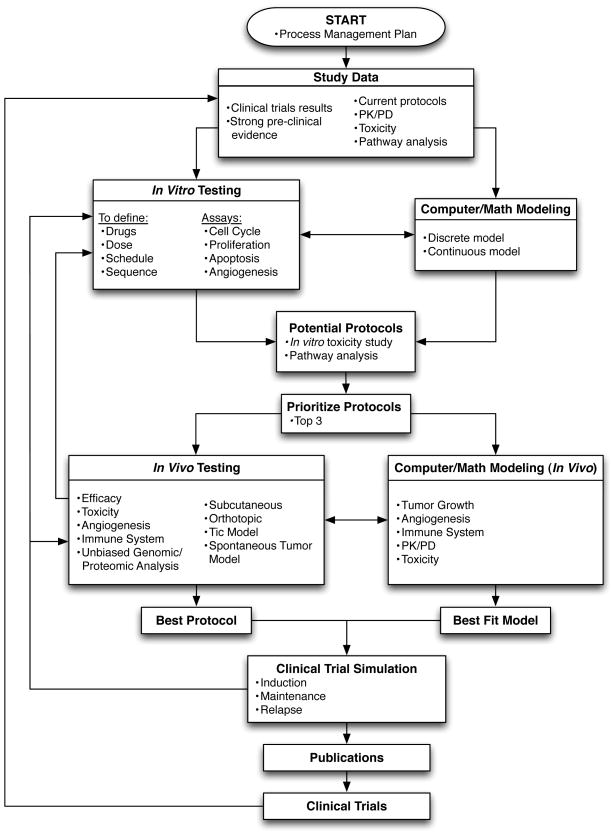
We constructed a flowchart template to formally coordinate the development pipeline and integration of quantitative and life science approaches (Figure 1). Herein we discuss the specific research steps and their respective integration in the overall protocol.
Figure 1.
Start: Process Management Plan
A process management plan must be established prior to initiating the pipeline. This is a formal approved document, under constant revision, that guides management of the projects included in the workflow process, their execution and control. It is agreed upon in advance by the groups involved. This plan is essential to ensure compliance, define responsibilities of the different participants, facilitate communication and troubleshooting, harmonize experimental procedures and ultimately make decisions. Key milestones against which the progress of the project can be measured are defined in the plan, together with actions to take in case of delay in completion of one particular step of the workflow. One aspect of the plan is also risk management: identifying the strengths, weaknesses, opportunities of, and threats to, the project (“SWOT Analysis”). Threats, such as barriers to overcome, may be internal/external, controllable/uncontrollable; if identified in advance, plans may be made to minimize any negative effects on the scientific discovery process. The plan can for instance define upfront the process to follow in case the different groups cannot agree on the best protocol or model to use. In the absence of clear consensus, a vote can be organized among all the participants and, in case of ties, the people responsible for the next step would have the final say (e.g. biologists when moving on to in vivo work, clinicians when moving to clinical trials…). Vote and tie-breaking rules can thus be pre-established as part of the Process Management Plan so as to avoid bottlenecks. Details on project management tools are beyond the scope of this report but more information can be found in several manuscripts specifically for the life sciences (11, 12) as well as the standard Project Management Body of Knowledge (PMBOK Guide) (13).
Study Data
Study Data initiates the pipeline and all research eventually translates back into the clinic with treatment guidelines. At the beginning of novel cancer therapy discovery, there is a wealth of clinical data available in the literature combined with the empirical observations from clinicians and physician-scientists. This growing amount of data has to be mined, integrated and interpreted within the close dialog of clinicians, biologists and computational biologists; working hypotheses and data specifications need to be clearly and ethically communicated (14). Available information includes clinical processes, such as current protocols and guidelines, positive and negative results from completed clinical trials, PK/PD data as well as biological pathway data from molecular analyses of patient and pharmacology data (15). In addition, for rare and/or incurable cancers as well as experimental therapeutics for which clinical data may not be available, strong pre-clinical evidence may also serve as a starting point.
The study data collected from pre-clinical and clinical studies can then be analyzed or modeled using a variety of qualitative and quantitative approaches. In particular, quantitative modeling is a powerful technique to test novel hypotheses, confirm in vitro, in vivo and ex vivo experiments, and simulate the dynamics of complex systems without a priori biases in a relatively fast time without the enormous costs of laboratory experiments and the corresponding biological and technical variation. Quantitative models can be calibrated using experimental or clinical data, and different hypotheses of tumor progression can be evaluated and treatment options thoroughly analyzed before launching costly clinical trials. Techniques for quantitative modeling are plentiful, and an increasing number of theoretical approaches are successfully applied to cancer biology. Molecular data from a patient’s tissues and biofluids can be used to compute the most likely biological network pathways based on existing published molecular interactions and disease associations (16). The evoked pathways can then be compared and contrasted over time, disease, therapy and other stratifications using biomedical analytics methods (17). Such computations can narrow down the set of hypotheses to those most likely to be successfully explored by the biologists. For example, clinical data for NB can include protein concentrations in biofluids and gene expression in tissue biopsies, and can be used to generate a personalized molecular profile of the patient. Brown’s study of glioblastoma multiforme (GBM), based on archived tissues, provided proof of concept that the adaptive hypoxia pathway in GBM was related to Fardin’s outcome-predicting hypoxia gene signature in NB (18), and that the proposed drug therapy for GBM would modulate the pathway network evoked from the tissue data (15).
In Vitro testing and simulation
Interdisciplinary discussions about the disease’s pathophysiology, related clinical information, current approved drugs as well as other investigational drugs, the drugs’ PK/PD profiles, toxicities and possible mechanisms of action can reveal the most promising ways to approach the research question via in vitro experiments and their corresponding simulation. At this point, work reverts to the biologists and quantitative modelers with consultation by clinicians. The various roles modeling plays in cancer research may be integrated with a laboratory research program at all stages. Research on plate cultures may be coupled with models that identify necessary growth and treatment parameters. Hypotheses made by laboratory researchers can be embedded in these models and the results compared with data to provide stronger evidence for the hypothesis or, alternatively to rule it out. A quantitative model of the experimental setup will help to systematically explore the hypothesis and contribution of participating mechanisms as well as alternative mechanisms (19–21).
For our NB application, the in vitro testing would include all the drugs that are currently used in the clinic for the treatment of high-risk NB (i.e. Cyclophosphamide, Doxorubicin, Cisplatin/Carboplatin, Vincristine, Topotecan/Irinotecan, Etoposide, Melphalan, Temozolomide and Retinoic Acid) alongside emerging repositioned drugs such as Cox inhibitors (22), nifurtimox (23), metformin (24), statins (25) and β-blockers (26).
In vitro confirmation of drug mechanisms of action and macroscopic cell- and population-level response serves as input for quantitative model design. Calibrated mathematical and computational models are developed. Once they reliably reproduce experimental findings, the models are used to systematically study combinations and scheduling of different drugs at various doses with total cell number, cell proliferation, cell cycle arrest, apoptosis and angiogenesis inhibition as observable endpoints. The potential of the best drug combination(s) is then confirmed by in vitro experiments, which in turns help refine the computational models.
Prioritization of Potential Protocols
The simulation results of different drug combinations, protocols and schedules are prioritized with respect to desired endpoint and pre-defined selection criteria. These include treatment efficacy, anti-angiogenic activity and selectivity towards cancer cells as determined by in vitro experiments, as well as drug availability and safety in pediatric populations. Computational modeling can also help in protocol prioritization. The goal of the numerical modeling aspect is indeed to create a fairly complete and well-tested simulation of in vivo processes that allows the experimenter to extend his reach beyond the experimental animals used in the lab. With modeling, hypotheses generated in the lab may be tested on an arbitrarily large collection of statistically varying virtual animals, quickly and inexpensively. Those that look promising can be followed up in in vivo experiments. The expert opinion of clinicians can also be solicited, and feasibility – as well as resources and budget constraints – determines which protocols proceed to in vivo testing.
In Vivo testing and simulation
In vivo experiments rely on state-of-the-art and clinically relevant models of high-risk NB. These include a combination of cell line- and patient-derived tumor xenografts implanted in a subcutaneous or orthotopic manner (27) (28), implantation of tumor-initiating cells (Tic) (29) and models of spontaneous tumor formation such as the TH-MYCN transgenic mouse model of NB (30). Prioritized prospective treatment protocols will be tested for toxicity and efficacy with focus on the effects on tumor growth, angiogenesis and immune response. Unbiased genomic/proteomic analyses can also be performed on specimens from the tumor, its surrounding tissue and circulating blood to understand changes in the tumor and the microenvironment over time, location, and therapeutic protocol.
Quantitative models of tumor growth that extend the basic cell cycle model to include population interaction with the host and drug response and reflect the higher complexity of in vivo experiments will be developed in parallel. In vivo results will be used to calibrate, validate and predict drug PKs/PDs, toxicity, tumor cell death and cell cycle arrest. The parameterized model of untreated tumor growth coupled with PK/PD profiles enables simulation of population-level treatment effects on the tumor as well as angiogenesis and immune response.
The model parameters calibrated to fit in vivo data can then be adjusted to match human response to simulate treatment in silico (31, 32). The in vitro, in vivo, and in silico results are discussed with clinicians. The best-fit models and protocols can then be used to simulate combination therapies and different treatment modalities. The generated data has to be thoroughly analyzed and meaningful conclusions have to be drawn to advance the most promising protocol(s) for clinical trial. At this point it becomes essential to define the target population including patient-specific parameters of age, gender or ethnicity.
Clinical Trials: Simulated and Real
Clinical Trial Simulations (CTS) are becoming more widely used by the pharmaceutical industry as a tool to help guide clinical trial design (4). Some of the early approaches to the iterative learn and confirm process, used to help in the design and evaluation of clinical trials and CTS, were proposed by L.B. Sheiner (33). Since then CTS have been used: i) to help in study design by identifying design inefficiencies, ii) to help determine the power of proposed clinical trials, and iii) to help determine effective doses and schedules, particularly in pediatric studies (4, 34). CTS have been used in all phases of drug development form early in the process to phase 2, phase 3, and regulatory reviews (35) (4, 34, 36). In addition, CTS are encouraged by regulatory agencies such as the FDA to complement other data to expedite drug approval and label claims (37, 38). CTS are still being evaluated to determine the most effective way to use them and there are various criticisms of these approaches including the need for more detailed model building and validation approaches (39, 40). We propose to use CTS in our workflow to help in study design optimization. Given the data generated by the in vitro and in vivo experimental and modeling studies, along with existing clinical data, the CTS can help determine metronomic dosing levels and schedules that will have a better chance of success. An example of how this can work is shown in a study of topotecan dose and schedule in pediatric neuroblastoma (41). In this study, models of neutropenia, tumor growth, and topotecan pharmacokinetics – based on existing pre-clinical and clinical data – were collaboratively developed by modelers, translational and clinical researchers to simulate how different schedules of topotecan affected both the toxicity and efficacy of treatment. Simulations such as these can then help guide the dose and schedule for the future studies. In this process the simulations can also help suggest which proposed study designs have the highest probability of success. This, as with all the steps in our workflow, is an iterative process---the simulations may suggest additional in vitro or in vivo experiments to clarify very sensitive parameters that are not currently well defined.
Results from the simulated clinical trials will be published for peer review and scientific discussion together with the in vitro and in vivo data prior to moving to clinical trials. The clinical trial finishes the interdisciplinary development pipeline, and trial results initiate subsequent iterations thereof until an optimal metronomic backbone for the treatment of NB is identified and validated.
Discussion
A cross-disciplinary research pipeline promises to optimize and advance the clinical adoption of metronomic therapy for specific diseases and conditions (cancer type, drug, treatment schedule, dose, patient demographics, and maintenance). In fact, the use of mathematical modeling has been identified as essential to progress in cancer (42).
Mathematical modeling has been widely applied in different areas of cancer research including cell cycle, drug resistance, PK/PD, angiogenesis and anti-angiogenic therapy, single cell level models, circadian rhythms, and modeling cancer in three dimensions (43–45). Unfortunately, many of these models are purely theoretical and thus difficult to translate to or penetrate experimental research. However, successful collaboration and integration of quantitative approaches is possible, if not necessary. We developed a template flowchart of the integrated dialog, highlighting what has to be done, when, and by whom, to facilitate collaboration across disciplines, sharing of resources, and the use of cost-effective computational research methods where appropriate, with the ultimate goal to improve cancer therapies. Ideally, the process template is discussed in advance, to jointly schedule prospective research rather than rely on retrospective studies that may have design deficiencies for their particular usage.
As is well known in the development of computer systems, interfaces must be clearly defined for a successful project. Here, the interfaces are not between components but between processes carried out by researchers from different disciplines who use different tools, different scientific vocabulary and with different goals. Valuable insights gained during research may be lost because they are not visible to other scientists with a very different and possibly useful perspective. Opportunities to collect auxiliary data may be missed because the researcher does not recognize its importance, whereas a different pair of eyes might see its value. An interdisciplinary team is therefore likely to get more information out of the same experiment than a specialist.
We believe that our integrative, multi-disciplinary approach to cancer treatment development would positively impact on clinical practice on two levels: 1) by speeding up the development of optimal treatment protocols and 2) by decreasing the rate of clinical trial failure. Promising results were recently reported for a metronomic chemotherapy protocol in the treatment of recurrent embryonal brain tumors (46). It took more than ten years to empirically and incrementally develop the sophisticated 8-drug combination regimen based on the results of previous pre-clinical and clinical studies (47–49). By involving clinicians, biologists and modelers throughout the entire process, our workflow has the potential to significantly reduce the time required to develop optimal metronomic protocols. Furthermore, it is increasingly recognized that a major factor responsible for the high rate of clinical trial failure is the lack of robustness of pre-clinical studies. Lowenstein and Castro thus recently proposed that pre-clinical experimentations should combine the most advanced mathematical and biological models to account for the heterogeneity of patient populations and the complexity of tumors (50). Our integrative workflow represents a significant effort in this direction.
Conclusions
A defined research/development/clinical process framework facilitates interdisciplinary collaboration by clarifying up front what has to be done, when and by whom. An initial framework, such as that developed during a workshop meeting, can be as simple as a flowchart graph that can later be extended into an overall action plan. As this paper demonstrates, an orderly progression of experiments and simulations, developed together with shared insights and proceeding from simple to complex, has the potential to streamline the therapeutic pipeline. A defined framework can reduce the number of options that must be tested by expensive in vivo methods and accelerate the impact on clinical practice and outcome.
Acknowledgments
The authors thank the organizers of the 2nd Annual Workshop on Cancer System Biology: Tumor Metronomics held at Tufts University, Boston, MA in July 2012 for giving us the opportunity to meet and brainstorm about how we might better collaborate to advance cancer research and therapy. The workshop was sponsored by the Integrative Cancer Biology program of the National Cancer Institute, the NIH Center of Cancer Systems Biology, and Steward/St. Elizabeth’s Medical Center at the Tufts University School of Medicine.
Grant Support
This research was supported by the Children’s Cancer Institute Australia for Medical Research, which is affiliated with the University of New South Wales and Sydney Children’s Hospital and grants from the Balnaves Foundation to E. Pasquier. M. F. McGuire was supported by a UT Health Innovation for Cancer Prevention Research Post-doctoral Fellowship - CPRIT grant #RP101503 and the Harvey S Rosenberg, Endowed Chair in Pathology for the Morphoproteomics Initiative, University of Texas Medical School at Houston.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts were disclosed.
Authors’ Contributions
Conception and design: M. F. McGuire, H. Enderling, D. I. Wallace, J. Batra, M. Jordan, S. Kumar, J. C. Panetta, E. Pasquier
Development of Methodology: M. F. McGuire, H. Enderling, D. I. Wallace, J. Batra, M. Jordan, S. Kumar, J. C. Panetta, E. Pasquier
Writing, review, and/or revision of the manuscript: M. F. McGuire, H. Enderling, D. I. Wallace, J. Batra, M. Jordan, S. Kumar, J. C. Panetta, E. Pasquier
Study supervision: E. Pasquier
Disclaimers
The content is solely the responsibility of the authors and does not necessarily represent the official views of their employers or supporters.
References
- 1.Hahnfeldt P, Hlatky L, Klement GL. Center of cancer systems biology second annual workshop--tumor metronomics: timing and dose level dynamics. Cancer research. 2013;73:2949–54. doi: 10.1158/0008-5472.CAN-12-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathan DG. The several Cs of translational clinical research. J Clin Invest. 2005;115:795–7. doi: 10.1172/JCI24753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne PR, Borlawsky TB, Kwok A, Greaves AW. Supporting the design of translational clinical studies through the generation and verification of conceptual knowledge-anchored hypotheses. AMIA Annu Symp Proc. 2008:566–70. [PMC free article] [PubMed] [Google Scholar]
- 4.Holford N, Ma SC, Ploeger BA. Clinical trial simulation: a review. Clinical pharmacology and therapeutics. 2010;88:166–82. doi: 10.1038/clpt.2010.114. [DOI] [PubMed] [Google Scholar]
- 5.Best A, Hiatt RA, Cameron R, Rimer BK, Abrams DB. The evolution of cancer control research: an international perspective from Canada and the United States. Cancer Epidemiol Biomarkers Prev. 2003;12:705–12. [PubMed] [Google Scholar]
- 6.IOM. Barriers to Interdisciplinary Research and Training. In: Pellmar TC, Eisenberg L, editors. Bridging Disciplines in the Brain, Behavioral, and Clinical Sciences. Washington (DC): The National Academies; 2000. pp. 41–57. [PubMed] [Google Scholar]
- 7.IOM. Facilitating Collaborations to Develop Combination Investigational Cancer Therapies: Workshop Summary. The National Academies Press; 2012. [PubMed] [Google Scholar]
- 8.Mittra J, Tait J. Analysing stratified medicine business models and value systems: innovation-regulation interactions. New biotechnology. 2012;29:709–19. doi: 10.1016/j.nbt.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Pasquier E, Kavallaris M, Andre N. Metronomic chemotherapy: new rationale for new directions. Nature reviews Clinical oncology. 2010;7:455–65. doi: 10.1038/nrclinonc.2010.82. [DOI] [PubMed] [Google Scholar]
- 10.Andre N, Abed S, Orbach D, Alla CA, Padovani L, Pasquier E, et al. Pilot study of a pediatric metronomic 4-drug regimen. Oncotarget. 2011;2:960–5. doi: 10.18632/oncotarget.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beukers MW. Project management of life-science research projects: project characteristics, challenges and training needs. Drug discovery today. 2011;16:93–8. doi: 10.1016/j.drudis.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Milton JG, Radunskaya AE, Lee AH, de Pillis LG, Bartlett DF. Team research at the biology-mathematics interface: project management perspectives. CBE life sciences education. 2010;9:316–22. doi: 10.1187/cbe.10-03-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.PMI. A Guide To The Project Management Body Of Knowledge (PMBOK Guide) Newtown Square, Pa: Project Management Institute, Inc; 2008. [Google Scholar]
- 14.Sodeke S, Turner T, Tarver W. The ethics of good communication in a complex research partnership. Journal of health care for the poor and underserved. 2010;21:35–45. doi: 10.1353/hpu.0.0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown RE, McGuire MF. Oncogenesis Recapitulates Embryogenesis via the Hypoxia Pathway: Morphoproteomics and Biomedical Analytics Provide Proof of Concept and Therapeutic Options. Annals of clinical and laboratory science. 2012;42:243–57. [PubMed] [Google Scholar]
- 16.McGuire MF, Iyengar MS, Mercer DW. Computational Approaches for Translational Clinical Research in Disease Progression. Journal of investigative medicine: the official publication of the American Federation for Clinical Research. 2011;59:893–903. doi: 10.231/JIM.0b013e318224d8cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire MF, Iyengar MS, Mercer DW. Data driven linear algebraic methods for analysis of molecular pathways: application to disease progression in shock/trauma. J Biomed Inform. 2012;45:372–87. doi: 10.1016/j.jbi.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fardin P, Barla A, Mosci S, Rosasco L, Verri A, Versteeg R, et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol Cancer. 2010;9:185. doi: 10.1186/1476-4598-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherar MD, Noss MB, Foster FS. Ultrasound backscatter microscopy images the internal structure of living tumour spheroids. Nature. 1987;330:493–5. doi: 10.1038/330493a0. [DOI] [PubMed] [Google Scholar]
- 20.Folkman J. What is the evidence that tumors are angiogenesis dependent? Journal of the National Cancer Institute. 1990;82:4–6. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 21.Freyer JP. Role of necrosis in regulating the growth saturation of multicellular spheroids. Cancer research. 1988;48:2432–9. [PubMed] [Google Scholar]
- 22.Johnsen JI, Lindskog M, Ponthan F, Pettersen I, Elfman L, Orrego A, et al. NSAIDs in neuroblastoma therapy. Cancer letters. 2005;228:195–201. doi: 10.1016/j.canlet.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 23.Saulnier Sholler GL, Bergendahl GM, Brard L, Singh AP, Heath BW, Bingham PM, et al. A phase 1 study of nifurtimox in patients with relapsed/refractory neuroblastoma. Journal of pediatric hematology/oncology. 2011;33:25–30. doi: 10.1097/MPH.0b013e3181f47061. [DOI] [PubMed] [Google Scholar]
- 24.Dowling RJ, Niraula S, Stambolic V, Goodwin PJ. Metformin in cancer: translational challenges. Journal of molecular endocrinology. 2012;48:R31–43. doi: 10.1530/JME-12-0007. [DOI] [PubMed] [Google Scholar]
- 25.Gonyeau MJ, Yuen DW. A clinical review of statins and cancer: helpful or harmful? Pharmacotherapy. 2010;30:177–94. doi: 10.1592/phco.30.2.177. [DOI] [PubMed] [Google Scholar]
- 26.Pasquier E, Street J, Pouchy C, Carre M, Gifford AJ, Murray J, et al. beta-blockers increase response to chemotherapy via direct antitumour and anti-angiogenic mechanisms in neuroblastoma. British journal of cancer. 2013;108:2485–94. doi: 10.1038/bjc.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francia G, Cruz-Munoz W, Man S, Xu P, Kerbel RS. Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nature reviews Cancer. 2011;11:135–41. doi: 10.1038/nrc3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–50. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nature reviews Cancer. 2012;12:133–43. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 30.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. The EMBO journal. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Nguyen MH, Cheng S, Schmidt S, Zhong L, Derendorf H, et al. A pharmacokinetic/pharmacodynamic mathematical model accurately describes the activity of voriconazole against Candida spp. in vitro. International journal of antimicrobial agents. 2008;31:369–74. doi: 10.1016/j.ijantimicag.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pery AR, Brochot C, Zeman FA, Mombelli E, Desmots S, Pavan M, et al. Prediction of dose-hepatotoxic response in humans based on toxicokinetic/toxicodynamic modeling with or without in vivo data: A case study with acetaminophen. Toxicology letters. 2013;220:26–34. doi: 10.1016/j.toxlet.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 33.Sheiner LB. Learning versus confirming in clinical drug development. Clin Pharmacol Ther. 1997;61:275–91. doi: 10.1016/S0009-9236(97)90160-0. [DOI] [PubMed] [Google Scholar]
- 34.Manolis E, Osman TE, Herold R, Koenig F, Tomasi P, Vamvakas S, et al. Role of modeling and simulation in pediatric investigation plans. Paediatric anaesthesia. 2011;21:214–21. doi: 10.1111/j.1460-9592.2011.03523.x. [DOI] [PubMed] [Google Scholar]
- 35.Simulation for Designing Clinical Trials: A Pharmacokinetic-Pharmacodynamic Modeling Perspective. New York, NY: Marcel Dekker; 2003. [Google Scholar]
- 36.Miller R, Ewy W, Corrigan BW, Ouellet D, Hermann D, Kowalski KG, et al. How modeling and simulation have enhanced decision making in new drug development. Journal of pharmacokinetics and pharmacodynamics. 2005;32:185–97. doi: 10.1007/s10928-005-0074-7. [DOI] [PubMed] [Google Scholar]
- 37.Gobburu JV, Marroum PJ. Utilisation of pharmacokinetic-pharmacodynamic modelling and simulation in regulatory decision-making. Clinical pharmacokinetics. 2001;40:883–92. doi: 10.2165/00003088-200140120-00001. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Bhattaram AV, Jadhav PR, Lesko LJ, Madabushi R, Powell JR, et al. Leveraging prior quantitative knowledge to guide drug development decisions and regulatory science recommendations: impact of FDA pharmacometrics during 2004–2006. Journal of clinical pharmacology. 2008;48:146–56. doi: 10.1177/0091270007311111. [DOI] [PubMed] [Google Scholar]
- 39.Boessen R, Knol MJ, Groenwold RH, Roes KC. Validation and predictive performance assessment of clinical trial simulation models. Clin Pharmacol Ther. 2011;89:487–8. doi: 10.1038/clpt.2010.277. author reply 8. [DOI] [PubMed] [Google Scholar]
- 40.Holford N, Ma SC, Ploeger BA. Response to Validation and Assessment of Predictive Performance in Simulation Models of Clinical Trials. Clin Pharmacol Ther. 2011;89:488. doi: 10.1038/clpt.2010.277. [DOI] [PubMed] [Google Scholar]
- 41.Panetta JC, Schaiquevich P, Santana VM, Stewart CF. Using pharmacokinetic and pharmacodynamic modeling and simulation to evaluate importance of schedule in topotecan therapy for pediatric neuroblastoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:318–25. doi: 10.1158/1078-0432.CCR-07-1243. [DOI] [PubMed] [Google Scholar]
- 42.Cantley LC, Dalton WS, DuBois RN, Finn OJ, Futreal PA, Golub TR, et al. AACR Cancer Progress Report 2012. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:S1–100. doi: 10.1158/1078-0432.CCR-12-2891. [DOI] [PubMed] [Google Scholar]
- 43.Swierniak A, Kimmel M, Smieja J. Mathematical modeling as a tool for planning anticancer therapy. European journal of pharmacology. 2009;625:108–21. doi: 10.1016/j.ejphar.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byrne HM. Dissecting cancer through mathematics: from the cell to the animal model. Nature reviews Cancer. 2010;10:221–30. doi: 10.1038/nrc2808. [DOI] [PubMed] [Google Scholar]
- 45.Wallace DI, Winsor P. Sensitive Dependence on the Threshold for TAF Signaling in Solid Tumors. In: Mondaini R, editor. BIOMAT 2011. World Scientific Publishing; 2011. pp. 264–78. [Google Scholar]
- 46.Peyrl A, Chocholous M, Kieran MW, Azizi AA, Prucker C, Czech T, et al. Antiangiogenic metronomic therapy for children with recurrent embryonal brain tumors. Pediatr Blood Cancer. 2012;59:511–7. doi: 10.1002/pbc.24006. [DOI] [PubMed] [Google Scholar]
- 47.Kieran MW, Turner CD, Rubin JB, Chi SN, Zimmerman MA, Chordas C, et al. A feasibility trial of antiangiogenic (metronomic) chemotherapy in pediatric patients with recurrent or progressive cancer. J Pediatr Hematol Oncol. 2005;27:573–81. doi: 10.1097/01.mph.0000183863.10792.d4. [DOI] [PubMed] [Google Scholar]
- 48.Panigrahy D, Kaipainen A, Butterfield CE, Chaponis DM, Laforme AM, Folkman J, Kieran MW. Inhibition of tumor angiogenesis by oral etoposide. Exp Ther Med. 2010;1:739–46. doi: 10.3892/etm.2010.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robison N, Campigotta F, Chi S, Manley P, Turner C, Zimmerman MA, et al. A phase II trial of a multi-agent oral antiangiogenic (metronomic) regimen in children with recurrent or progressive cancer. Pediatr Blood Cancer. 2011;57:815. doi: 10.1002/pbc.24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowenstein PR, Castro MG. Uncertainty in the translation of preclinical experiments to clinical trials. Why do most phase III clinical trials fail? Curr Gene Ther. 2009;9:368–74. doi: 10.2174/156652309789753392. [DOI] [PMC free article] [PubMed] [Google Scholar]