Abstract
Linoleic acid (18:2n-6) and α-linolenic acid (18:3n-3) are polyunsaturated fatty acids that are essential for mammalian nutrition, because mammals lack the desaturases required for synthesis of Δ12 (n-6) and n-3 fatty acids. Many plants can synthesize these fatty acids and, therefore, to examine the effects of a plant desaturase in mammals, we generated transgenic pigs that carried the fatty acid desaturation 2 gene for a Δ12 fatty acid desaturase from spinach. Levels of linoleic acid (18:2n-6) in adipocytes that had differentiated in vitro from cells derived from the transgenic pigs were ≈10 times higher than those from wild-type pigs. In addition, the white adipose tissue of transgenic pigs contained ≈20% more linoleic acid (18:2n-6) than that of wild-type pigs. These results demonstrate the functional expression of a plant gene for a fatty acid desaturase in mammals, opening up the possibility of modifying the fatty acid composition of products from domestic animals by transgenic technology, using plant genes for fatty acid desaturases.
Linoleic acid (18:2n-6) and α-linolenic acid (18:3n-3) are essential fatty acids that are required for normal growth in mammals. They are also integral components of cell membranes. They appear to play an important role in the structure of eicosanoids that are involved in the regulation of the release of hypothalamic and pituitary hormones (1). Furthermore, highly polyunsaturated fatty acids (PUFAs), namely, arachidonic acid (20:4n-6), docosatetraenoic acid (22:6n-6), and docosahexaenoic acid (22:6n-3), are found in high concentrations in structural lipids of the CNS and are considered to be essential in infant nutrition (2). Mutants of Caenorhabditis elegans lacking genes for fatty acid desaturases and having severe deficiencies in such fatty acids exhibit growth and neurological defects, such as embryonic lethality, abnormal body shape, and behavioral abnormalities (3).
Lower animals, such as American cockroaches, house crickets (4), and nematodes (5), as well as higher plants, can synthesize essential fatty acids/PUFAs. Slugs and snails can also synthesize linoleic acid (18:2n-6; ref. 6) but not docosahexaenoic acid (7). Moreover, the nematode fat-1 and fat-2 genes for n-3 and Δ12 fatty acid desaturases have been cloned and expressed in yeast (5), Arabidopsis (8), and rat cardiac myocytes cultured in vitro (9).
Domestic pigs synthesize and deposit large quantities of fat. This synthesis and deposition mostly occurs in adipose tissue (10). Therefore, if domestic pigs could be induced to synthesize unsaturated fatty acids endogenously, as a result of the introduction of genes for fatty acid desaturases, their meat would be an alternative source of the essential fatty acids and/or PUFAs, which might help to prevent lifestyle-related diseases, such as coronary heart disease and thrombotic disease (11, 12). However, the functional expression in vivo of a plant gene for a fatty acid desaturase has not yet been demonstrated in mammals.
The object of this study was to investigate whether a plant gene for a fatty acid desaturase can be functionally expressed in a mammal and whether the fatty acid composition of lipids in such a mammal can be altered by transgenic technology. We report here the production of transgenic pigs that carry cDNA for the gene for Δ12 fatty acid desaturase (FAD2) from spinach (Spinacia oleracea) under the control of an adipocyte P2 (aP2) promoter, which directs expression of linked marker genes to white and brown adipocytes in transgenic mice (13–15). We detected the functional expression of the FAD2 gene in the transgenic pigs in vivo and also in vitro.
Materials and Methods
Cloning of cDNA for Spinacia FAD2. The coding region of cDNA for a Δ12 desaturase (FAD2) from Arabidopsis thaliana (1,152 base pairs; ref. 16) was cloned into the SmaI site of pBluescript II KS+ (Invitrogen). The plasmid was digested with EcoRI and XbaI, and labeled with 32P-dCTP by using a random primed labeling kit (Takara Bio, Shiga, Japan). The labeled probe was used to screen a Spinacia cDNA library in λZAPII phage (17). Fourteen positive clones were isolated from 48,000 plaques, and the cDNAs were excised from the phage vector with ExAssist helper phage (Stratagene). One plasmid, designated pBS/sfad2-3, contained a DNA fragment of 1,629 base pairs at the EcoRI site of pBluescript SK-. The DNA sequence of this clone was determined with a dye terminator cycle sequencing kit (Applied Biosystems) and a DNA sequencer (model 310A; Applied Biosystems). The extent of the homology between the sequences from Spinacia and Arabidopsis was 68.6%.
Preparation of the Transgene and Production of Transgenic Pigs. We introduced the cDNA for spinach fatty acid desaturase 2 (FAD2, GenBank accession no. AB094415) into the pBluescript II KS plasmid (Stratagene), and we then ligated a 5-kb fragment of the mouse aP2 promoter region (14) and a fragment of the simian virus 40 (SV40) splicing region plus an SV40 poly(A) addition signal (18) were added to the up- and downstream of the cDNA, respectively. A 7.5-kb SacII–XhoI fragment (aP2/FAD2, Fig. 1) of the constructed plasmid was isolated by preparative electrophoresis on an agarose gel, was purified by using GeneClean II (Bio 101), and redissolved in TE buffer (10 mM Tris·HCl/1 mM EDTA/pH 7.4) at a final concentration of 4 μg/ml. Transgenic pigs were produced as described elsewhere (19). Zygotes were collected from superovulated gilts (≈100 kg of live weight), obtained from a cross between Duroc sows and Landrace × Large White boars, by flushing the oviducts through a midventral incision. DNA was injected into the male pronuclei of zygotes after centrifugation of the zygotes at 15,000 × g for 3 min (20). Injected embryos were then transferred into the oviducts of synchronized recipients or the donor pigs. Pregnant recipients were then housed individually and allowed to go to term. At birth, we performed a tail biopsy of each of the piglets. The integrity of the transgene was examined by Southern blotting analysis of genomic DNA extracted from the tail tissues (21). All animal procedures in the present study were approved by the Committee for Experimental Animals of Kinki University.
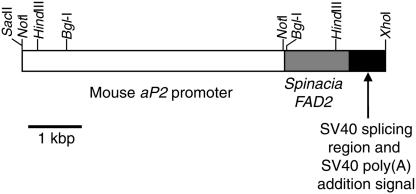
Fig. 1.
Construction of the aP2/FAD2 fusion gene for production of transgenic pigs. A 7.5-kb SacII–XhoI fragment contained a 5-kb mouse aP2 promoter fused to a 1.6-kb cDNA for Spinacia FAD2, a fragment of the SV40 splicing region, and an SV40 poly(A) site.
Analysis of the Founders by RT-PCR and Production of the Next Generation. White adipose tissue was taken from the backfat of the transgenic founder pigs with a cylindrical biopsy device (4 mm in diameter; Fujihira Industry, Tokyo) as described (22). Total RNA was extracted from the tissue with TRIzol (GIBCO) according to the manufacturer's instructions. The RNA was reverse-transcribed by AMV reverse transcriptase (Takara Bio) with random 9-mer primers in a 40-μl reaction. The conditions for amplification of cDNA included 40 cycles of incubation at 94°C for 60 sec, 55°C for 60 sec, and 72°C for 60 sec, with primers specific for the FAD2 gene (5′-CTCTCCAATCTACTCGGAC-3′, and 5′-ATTGGCTTATAGCCTTGGT-3′). The amplified products were subjected to electrophoresis on a 2% agarose gel. Pigs that gave positive results were mated with wild-type pigs for production of the next generation. Transmission of the transgene to piglets was examined by Southern blotting analysis of genomic DNA extracted from tail tissues as described above.
Northern Blotting Analysis. We examined the expression of full-length FAD2 mRNA in the transgenic pigs by using total RNA extracted from white adipose tissue, skeletal muscle, kidney, spleen, liver, lung, heart, brain, and testis or ovary of the transgenic pigs as described above.
Poly(A)+ RNA was purified with Oligotex-dT (Takara Bio) and fractionated on a 1% agarose-formaldehyde gel. Then, bands of RNA were blotted on a nylon membrane (Hybond N+; Amersham Pharmacia) in 20 × SSC. The RNA on the blot was allowed to hybridize with a 32PdCTP-labeled and random-primed cDNA probe for spinach FAD2, as described elsewhere (23).
Preparation of Preadipocytes and Induction of Their Differentiation into Multilocular Adipocytes in Vitro. Mature unilocular adipocytes were isolated from white adipose tissue that had been collected from transgenic pigs as described above (24). Preadipocytes were obtained by the “ceiling culture” method (25). Preadipocytes with a fibroblast-like appearance were cultured for several passages in 25 mM Hepes-buffered DMEM, supplemented with 20% FCS (GIBCO). Lipogenesis by the preadipocytes in vitro was induced by treatment with 5 μg/ml insulin (Wako Pure Chemicals, Osaka), 0.5 mM isobutylmethylxanthine (Wako), and 0.25 μM dexamethasone (Wako) in DMEM supplemented with 20% FCS for 4 days. The medium was then replaced with DMEM supplemented with 20% FCS and cultured for 8 days. After 8 days, most (≈80%) of the cells had differentiated into multilocular adipocytes that contained intracytoplasmic lipid droplets. Cells were then collected and lipids were extracted immediately after collection.
Collection of Adipose Tissue and Blood Samples. Transgenic pigs were fed a normal diet (Spurt G; Nosan, Yokohama, Japan) until they were 5 months old. The pigs were then fed a diet high in oleic acid [diet supplemented with 10% (wt/vol) high-oleic-acid safflower oil, which contained 78% (wt/wt) oleic acid (18:1) (HO diet; Nosan)] for 8 weeks. Wild-type pigs were also fed the HO diet as a control group. The diet and water were available ad libitum. White adipose tissue was collected as described above before and 8 weeks after the start of the HO diet. Blood samples were taken after a 12-h fasting period from pigs that had been fed a normal diet. Blood was collected in heparinized tubes and plasma and blood cells were obtained after centrifugation at room temperature at 1,880 × g for 10 min.
Analysis of Lipids. Lipids were extracted from lipogenic adipocytes generated in vitro, biopsy samples of adipose tissue, plasma, and blood cells with chloroform and methanol as described (26). The fatty acid composition of each sample was then analyzed by gas chromatography (27).
Statistical Analysis. The data obtained from each experiment were subjected to an ANOVA, which was followed by Fisher's least significant difference test with statview 5.0 software (SAS Institute, Cary, NC).
Results
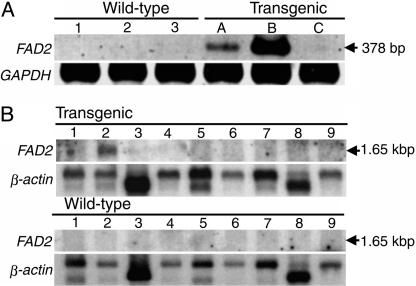
Generation of Transgenic Pigs and Expression of the FAD2 Gene from Spinach. We microinjected a total of 464 pronuclear embryos, collected from superovulated gilts, with the aP2/FAD2 gene. Of 16 recipient animals, 11 (69%) became pregnant and farrowed 70 piglets. Thus, 15% of the DNA-injected embryos developed to term. Southern blotting analysis of the genomic DNA from each piglet indicated that six (9%) of the piglets were transgenic. Of the transgenic pigs, two were stillborn, and one died the day after birth. Two males and one female survived. We examined transgene activity in the three founder pigs when they were 5 months old. We extracted total RNA from biopsies of backfat and analyzed the RNA by RT-PCR with specific primers for FAD2 cDNA. As shown in Fig. 2, we detected FAD2 mRNA in one male and one female founder pig.
Fig. 2.
Expression of Spinacia FAD2 mRNA. (A) Analysis by RT-PCR of biopsies of backfat from transgenic founder pigs. Total RNA was extracted from biopsy samples of backfat adipose tissue taken from transgenic founder pigs (pigs A, B, and C) and wild-type pigs (nontransgenic littermates, pigs 1, 2, and 3). As a control, GAPDH gene was used for analysis by RT-PCR. (B) Northern blotting analysis for detection of full-length FAD2 mRNA in transgenic pigs. Total RNA was extracted from of a transgenic (line A) male pig and a wild-type pig as follows: 1, testis; 2, white adipose tissue; 3, skeletal muscle; 4, kidney; 5, spleen; 6, liver; 7, lung; 8, heart; and 9, brain. As a control, β-actin gene was used for Northern blotting analysis. Three independent experiments were performed in the case of each analysis by RT-PCR and Northern blotting analysis. The respective results were similar in each case.
After puberty, the two transgenic pigs with an active transgene were mated with wild-type pigs. The male (pig A) was mated with two wild-type females that farrowed 32 piglets (21 live and 11 stillborn). Southern blotting analysis indicated that eight (38%) of the live piglets carried the transgene. The female (pig B) was mated with a wild-type male and farrowed 12 piglets (three live and nine stillborn). The three live piglets died within a week because of agalactia. Transmission of the transgene was also confirmed in eight (44%) of 18 dead F1 animals.
Northern blotting analysis revealed a full-length 1.65-kb FAD2 transcript in white adipose tissue only (Fig. 2B). The results indicated that the FAD2 gene was expressed specifically in adipose tissue under the control of the aP2 promoter.
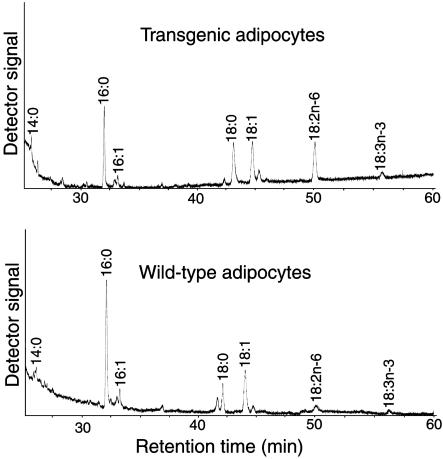
Elevated Level of Linoleic Acid (18:2n-6) in White Adipocytes Differentiated in Vitro. To examine the activity of the FAD2 in white adipocytes of transgenic pigs, we determined the fatty acid composition of lipogenic adipocytes derived from transgenic pigs. Preadipocytes were generated from a ceiling culture (25) of mature adipocytes that had been isolated from the white adipose tissue of transgenic pigs. The preadipocytes were exposed to insulin, isobutylmethylxanthine, and dexamethasone for 4 days to induce differentiation into adipocytes. The adipocytes were then cultured for 8 days to allow them to accumulate lipids. Then, we extracted the lipids from the cells and analyzed their fatty acid composition. As shown in Fig. 3 and Table 1, the level of linoleic acid (18:2n-6) was ≈10 times higher in transgenic adipocytes than that in wild-type cells (20.3% vs. 1.9%; P < 0.01). Because mammalian cells are able to synthesize saturated and Δ9 unsaturated fatty acids, we used the ratio of levels of n-6 unsaturated fatty acids to saturated plus Δ9 unsaturated fatty acids as an index of n-6 desaturation. The index in the transgenic cells (0.27) was much higher than the index in wild-type cells (0.02; P < 0.01). The ratio of n-6 to n-3 PUFAs in the transgenic cells tended to be higher than the ratio in wild-type cells (5.6 vs. 2.5; P < 0.1).
Fig. 3.
Partial gas chromatograms showing fatty acid profiles of total lipid that were extracted from multilocular adipocytes that had differentiated from transgenic (Upper) and wild-type (control; Lower) preadipocytes.
Table 1. Fatty acid composition of lipids extracted from lipogenic adipocytes generated in vitro.
| Mol % of total fatty acids | Wild-type | Transgenic |
|---|---|---|
| 14:0 | 4.5 ± 1.2 | 3.5 ± 2.0 |
| 14:1 | 0 | 0 |
| 16:0 | 38.2 ± 1.3a | 26.6 ± 0.4b |
| 16:1 | 4.4 ± 0.7 | 6.1 ± 0.5 |
| 18:0 | 21.4 ± 1.8 | 22.0 ± 0.3 |
| 18:1 | 28.0 ± 0.1a | 18.8 ± 1.3b |
| 18:2n—6 | 1.9 ± 0.4a | 20.3 ± 2.1b |
| 18:3n—3 | 1.6 ± 0.6 | 2.7 ± 1.4 |
| 20:0 | 0 | 0 |
| 20:2 | 0 | 0 |
| 20:4n—6 | 0 | 0 |
| 20:5n—3 | 0 | 0 |
| 22:0 | 0 | 0 |
| 22:1 | 0 | 0 |
| 22:5n—3 | 0 | 0 |
| 22:6n—3 | 0 | 0 |
| Sum of saturated plus Δ9 unsaturated | 96.5 ± 0.3a | 77.1 ± 3.5b |
| Sum of n—6 polyunsaturated | 1.9 ± 0.4a | 20.3 ± 2.1b |
| Sum of n—3 polyunsaturated | 1.6 ± 0.6 | 2.7 ± 1.4 |
| n—6/saturated plus Δ9 unsaturated | 0.02 ± 0.04a | 0.27 ± 0.4b |
| n-6/n-3 | 2.5 ± 1.7 | 5.6 ± 0.6 |
Lipids were analyzed in three independent samples from each batch of cells. Results are means ± SEM. The differences between values in the same row with different superscripts are statistically significant (P < 0.01).
Elevated Level of n-6 Fatty Acids in White Adipose Tissue and Blood. To examine the functional expression of the FAD2 gene in pigs, we determined the fatty acid composition of adipose tissue. Pigs were fed an HO diet for 8 weeks to increase the level of oleic acid (18:1) in adipose tissue (28). We examined white adipose tissue, collected from backfat, before (0 weeks) and 8 weeks after feeding pigs the HO diet. As shown in Table 2, levels of linoleic acid (18:2n-6) were ≈1.2-fold higher in transgenic pigs (A line) than those in wild-type pigs (P < 0.05). The level of oleic acid (18:1) increased after feeding the HO diet for 8 weeks in wild-type pigs (P < 0.05), but the level did not increase in transgenic pigs (P > 0.05). The level of saturated and Δ9 unsaturated fatty acids was lower in transgenic pigs than that in wild-type pigs (P < 0.05). The n-6 desaturation index in the transgenic adipose tissue was higher than that in wild-type adipose tissue (P < 0.05; Table 2). The ratio of n-6 to n-3 fatty acids in transgenic adipose tissue at 0 weeks was higher than that in wild-type adipose tissue (P < 0.05; Table 2). We also determined the fatty acid composition of backfat from pig B. This pig contained a higher level of linoleic acid (18:2n-6, 12.3 ± 0.2%, n = 3) than that in wild-type pigs (10.0 ± 0.1%, n = 3; P < 0.01). We examined the fatty acid compositions of blood plasma and blood cells of transgenic pigs because fatty acids are released into the blood by lipolysis of triglycerides stored in adipose tissue. As shown in Table 3, the level of n-6 fatty acids, in particular 20:4n-6, was higher in the plasma of the transgenic pigs than in that of wild-type pigs. The level of saturated plus Δ9 unsaturated fatty acids was lower in transgenic pigs than that in wild-type pigs. The fatty acid composition of the blood cells was the same in transgenic and wild-type pigs (data not shown).
Table 2. Fatty acid composition of lipids extracted from biopsies of white adipose tissue.
| Wild-type
|
Transgenic
|
|||
|---|---|---|---|---|
| Mol % of total fatty acids | Week 0 | Week 8 | Week 0 | Week 8 |
| 14:0 | 2.1 ± 0.1 | 2.2 ± 0.05 | 2.2 ± 0.1 | 2.0 ± 0.1 |
| 14:1 | 0 | 0 | 0 | 0 |
| 16:0 | 31.4 ± 0.9a | 28.6 ± 0.4b | 27.1 ± 1.8b | 27.0 ± 0.2b |
| 16:1 | 3.4 ± 0.4 | 2.9 ± 0.2 | 2.8 ± 0.1 | 2.8 ± 0.2 |
| 18:0 | 11.0 ± 0.8a | 8.8 ± 0.5b | 12.2 ± 0.6c | 9.2 ± 0.2b |
| 18:1 | 41.6 ± 0.8a | 46.0 ± 0.4b | 43.2 ± 2.0ab | 45.5 ± 0.1b |
| 18:2n—6 | 9.3 ± 0.5a | 9.9 ± 0.3a | 11.7 ± 0.6b | 11.6 ± 0.4b |
| 18:3n-3 | 0.8 ± 0.3ab | 1.4 ± 0.1b | 0.6 ± 0.2a | 1.4 ± 0.03b |
| 20:0 | 0.2 ± 0.05 | 0.1 ± 0.04 | 0.1 ± 0.03 | 0.1 ± 0.03 |
| 20:1 | 0.1 ± 0.05 | 0.1 ± 0.04 | 0 | 0.1 ± 0.03 |
| 20:2n—6 | 0 | 0 | 0 | 0 |
| 20:3n-6 | 0 | 0.1 ± 0.04 | 0 | 0.1 ± 0.1 |
| 20:4n-6 | 0.1 ± 0.05 | 0 | 0.1 ± 0.06 | 0.1 ± 0.1 |
| 20:5n-3 | 0 | 0 | 0 | 0 |
| 22:4n-6 | 0 | 0 | 0 | 0 |
| 22:5n-3 | 0 | 0 | 0 | 0 |
| 22:6n-3 | 0 | 0 | 0 | 0 |
| Sum of saturated plus Δ9 unsaturated | 89.5 ± 0.5a | 88.4 ± 0.3ab | 87.8 ± 0.5bc | 86.9 ± 0.5c |
| Sum of n-6 polyunsaturated | 9.4 ± 0.5a | 10.0 ± 0.3a | 11.8 ± 0.5b | 11.8 ± 0.4b |
| Sum of n-3 polyunsaturated | 0.8 ± 0.3ab | 1.4 ± 0.1b | 0.6 ± 0.2a | 1.4 ± 0.03b |
| n-6/saturated plus Δ9 unsaturated | 0.11 ± 0.01a | 0.11 ± 0.003a | 0.14 ± 0.01b | 0.14 ± 0.01b |
| n-6/n-3 | 10.3 ± 2.3a | 7.3 ± 0.4a | 24.2 ± 9.1b | 8.3 ± 0.4a |
Backfat biopsy samples were taken from transgenic (n = 3) and wild-type (n = 5) pigs, 0 and 8 weeks after the start of the HO diet. Values are expressed as means ± SEM. The differences between values in the same row with different superscripts are statistically significant (P < 0.05).
Table 3. Fatty acid composition of lipids in blood plasma.
| Mol % of total fatty acids | Wild-type | Transgenic |
|---|---|---|
| 14:0 | 0.4 ± 0.03 | 0.3 (0.3, 0.3) |
| 14:1 | 0 | 0 |
| 16:0 | 14.7 ± 0.9 | 12.8 (13.7, 11.9) |
| 16:1 | 0.8 ± 0.03 | 0.6 (0.6, 0.5) |
| 17:0 | 0.5 ± 0.03 | 0.6 (0.7, 0.4) |
| 17:1 | 0.2 ± 0 | 0.1 (0.1, 0) |
| 18:0 | 15.3 ± 0.9 | 16.4 (15.1, 17.7) |
| 18:1 | 22.0 ± 1.0 | 18.5 (19.0, 18.0) |
| 18:2n-6 | 27.3 ± 1.0 | 30.3 (30.0, 30.6) |
| 18:3n-3 | 0.5 ± 0.03 | 0.4 (0.3, 0.4) |
| 20:1 | 0.2 ± 0.03 | 0.2 (0.2, 0.1) |
| 20:2n-6 | 0.5 ± 0.03 | 0.6 (0.7, 0.4) |
| 20:3n-6 | 0.7 ± 0.03 | 0.5 (0.6, 0.4) |
| 20:4n-6 | 11.6 ± 0.2 | 13.7 (13.5, 13.8) |
| 20:5n-3 | 0.3 ± 0.1 | 0.2 (0.1, 0.3) |
| 22:4n-6 | 0.7 ± 0.1 | 0.5 (0.5, 0.4) |
| 22:5n-3 | 1.5 ± 0.1 | 1.3 (1.3, 1.2) |
| 22:6n-3 | 1.1 ± 0.1 | 0.7 (0.6, 0.8) |
| Unknown | 1.9 ± 0.1 | 2.7 (2.5, 2.8) |
| Sum of saturated plus Δ9 unsaturated | 54.0 ± 1.0 | 49.3 (49.7, 48.9) |
| Sum of n-6 polyunsaturated | 40.7 ± 1.1 | 45.5 (45.3, 45.6) |
| Sum of n-3 polyunsaturated | 3.3 ± 0.2 | 2.5 (2.3, 2.7) |
Blood samples were taken from wild-type (n = 3) and transgenic (n = 2) pigs after they had fasted for 12 h. Values are expressed as means ± SEM and means (raw data) for wild-type and transgenic pigs, respectively.
Discussion
In this study, we demonstrated that a plant FAD2 gene can be functionally expressed and its product can have a significant effect on the fatty acid composition of accumulated lipids in pig adipose cells. There have been several reports on the production of transgenic pigs (29). The efficiency of integration of the transgene (9%) and the rate of expression (two of three) in this study were similar to those in the earlier studies (30). If the product of translation of the transgene is lethal and/or toxic to embryos and/or fetuses, none or few transgenic animals are produced. In this study, the mortality of the F1 piglets was relatively high. However, we have now produced four transgenic pigs of the third generation farrowed by a transgenic female, and all of them grew to a fertile age. Detailed investigations on whether the transgene has an adverse effect on mammalian development or not might be necessary.
Plant FAD2 is an acyl-lipid desaturase, whereas all of the known mammalian fatty acid desaturases are acyl-CoA desaturases. Both FAD2 and acyl-CoA desaturases are localized in the endoplasmic reticulum, and form part of a system composed of cytochrome b5 and NADH-dependent cytochrome b5 reductases (31). An n-3 fatty acid desaturase derived from C. elegans, which is probably an acyl-lipid desaturase, has been shown to convert n-6 fatty acids to n-3 fatty acids efficiently in rat cardiac cells in vitro (9). In addition, a fungal gene for Δ12 fatty acid desaturase functioned in transfected mouse L cells (32). These observations indicate that at least one plant acyl-lipid desaturase can function in mammalian cells.
We demonstrated the functional expression of the aP2/FAD2 transgene in vitro in lipogenic adipocytes from our transgenic pigs: the level of linoleic acid (18:2n-6) increased markedly and levels of saturated and Δ9 unsaturated fatty acids were lower in total lipids from transgenic adipocytes than in those from wild-type cells (Table 1). The FAD2 in adipocytes derived from the transgenic pigs might be expressed during lipogenesis, because the aP2 protein is expressed during the differentiation of adipocytes (33). In lipids of lipogenic 3T3-L1 cells, most triglycerides and phospholipids are formed from newly synthesized fatty acids derived from glucose (34, 35). The major pathway for synthesis of triglycerides involves three sequential transfers of the acyl moiety from acyl-CoA to a glycerol backbone. In addition, triglycerides can be synthesized from fatty acyl-CoAs and diacylglycerols derived from phospholipids after hydrolysis. Therefore, it is very likely that linoleic acid (18:2n-6) synthesized de novo in phospholipids accumulated at high levels in the lipids of the adipocytes that had differentiated in vitro.
In the adipose tissues examined in this study, levels of linoleic acid (18:2n-6) in the tissues from transgenic pigs were 1.2-fold higher than those from wild-type animals (Table 2). The elevated level of linoleic acid (18:2n-6) in the adipose tissues of the transgenic pigs indicated the synthesis of linoleic acid (18:2n-6) de novo by FAD2 expressed from the transgene. Fatty acid desaturases introduce an unsaturated bond at a specific position of fatty acids, and the order in which desaturases operate is very strictly determined. A Δ12 desaturase introduces an unsaturated bond into fatty acids that have an unsaturated bond at the Δ9 position (31). Therefore, the FAD2 in the transgenic pigs might covert only oleic acid (18:1) to linoleic acid (18:2n-6). In mammals, oleic acid (18:1) is converted from stearic acid (18:0) by their Δ9 desaturase activity, and stearic acid (18:0) is elongated from palmitic acid (16:0) by their acyl chain elongation system (36). These sequential reactions might be induced by the synthesis of linoleic acid (18:2n-6) de novo by the FAD2, resulting in a decrease in the level of the saturated plus Δ9 unsaturated fatty acids in the transgenic pigs. Levels of oleic acid in samples of adipose tissue from pigs fed the HO diet for 8 weeks rose, as reported earlier (28, 37). However, the elevated level of oleic acid (18:1) did not enhance the accumulation of linoleic acid (18:2n-6) in the white adipose tissue of the transgenic pigs. Moreover, the increases were not as great as those observed in our study in vitro (Table 1). The primary function of adipocytes is to store triglycerides as energy substrates within the body. The fat stored in adipocytes is not static but changes dynamically under the complementary forces of lipogenesis and lipolysis (38). The triglyceride pool in adipocytes is mostly derived from the uptake of free fatty acids from the blood lipoproteins, which are synthesized in hepatocytes and are derived from dietary fats (38). In the present study, linoleic acid (18:2n-6) was synthesized only within the adipocytes of the transgenic pigs because FAD2 mRNA was expressed only in the adipose tissue (Fig. 2B). We infer that linoleic acid (18:2n-6) synthesized de novo in adipocytes is stored in the adipose tissue and that, after lipolysis, the fatty acid is released into the blood-stream. The levels of n-6 fatty acids were elevated in the blood plasma of the transgenic pigs (Table 3). An elevated level of n-6 fatty acids, in particular, 20:4n-6, indicates that linoleic acid (18:2n-6), synthesized in the adipocytes, was released into the blood subsequent to and/or before its elongation, because the level of a longer n-6 fatty acid, 20:4n-6, rose in the plasma of pigs fed a diet rich in linoleic acid (18:2n-6; ref. 39). The level of n-6 fatty acids did not rise in the blood cells in the present study, as was the case in the cells of pigs fed a diet rich in linoleic acid (18:2n-6; ref. 39). The complex steps involved in the derivation and circulation of fatty acids in vivo might decrease the concentration of linoleic acid (18:2n-6) in the adipose tissue of the transgenic pigs.
The substrates for FAD2 from spinach in mammals remain to be identified, but FAD2 might desaturate glycerolipids, rather than fatty acyl-CoAs, as it does in spinach. In plant seeds, triglycerides are formed mainly from pools of phosphatidylcholine in the endoplasmic reticulum (40). By contrast, in mature adipocytes that are the main cellular component of mammalian white adipose tissue (41), the majority of triglycerides are formed from fatty acyl-CoAs, rather than from membrane phospholipids (42). The limited enhancement of the level of linoleic acid (18:2n-6) in the adipose tissue of the transgenic pigs might have been due to differences among substrates recruited for the synthesis of triglycerides between plants and mammals. On the other hand, we detected much higher level of linoleic acid (18:2n-6) in transgenic adipocytes than in the adipose tissue of the transgenic pigs. The difference in the level of linoleic acid (18:2n-6) of our results in vitro and in vivo might be raised from the different characters of adipocytes in culture in vitro and in adipose tissue in vivo. The adipocytes differentiated in vitro, such as 3T3-L1 cells, accumulate lipids mainly derived from fatty acids de novo-synthesized from glucose in a culture medium (34, 35), whereas mature adipocytes in adipose tissue accumulate and release lipids from and into blood circulation simultaneously (38). In addition, we speculated that more amounts of phospholipids containing linoleic acid (18:2n-6), which had been synthesized by the FAD2, might contribute to the accumulation of lipids in the adipocytes differentiated in vitro than in the adipose tissue of the transgenic pigs. Further investigations of the involvement of FAD2 in membrane fluidity might provide novel clues to an understanding of the underlying molecular mechanisms that control the normal functions of membranes.
In conclusion, we have demonstrated the synthesis of linoleic acid (18:2n-6) de novo in domestic pigs after introduction of a plant FAD2 gene. At present, addition to an animal's diet of oils rich in PUFAs is widely used to increase levels of PUFAs in the meat (43). However, such a diet contains fairly expensive purified plant and/or fish oils (44), and the resultant meat occasionally has a fishy flavor (43). In the present study, we did not detect any evidence of an undesirable oxidation status of the transgenic adipose tissue, such as a rancid odor and/or “yellow fat.” However, if the levels of PUFAs are raised in meat, methods for prevention of autooxidation by the meat should be considered. Vitamin E, as a dietary supplement, has been reported to be effective in delaying the oxidative deterioration of the meat (43).
In this study, the higher n-6 desaturation index in adipocytes of transgenic pigs, both in vitro and in vivo, indicated significant enhancement of the synthesis of linoleic acid (18:2n-6) de novo, and the enhancement also resulted in an increase in the ratio of n-6 to n-3 fatty acids (Tables 1 and 2). The n-3 fatty acids are associated with a lower risk of morbidity and mortality from atherosclerosis and coronary heart disease, according to epidemiological investigations (11, 12, 45). In the Western diet, the ratio of n-6 to n-3 fatty acids is very high (20 to 30), and appears to increase the risk of coronary heart disease (46). Therefore, further studies are needed to introduce the gene for an n-3 fatty acid desaturase, such as FAD3, into domestic animals. Furthermore, chimeric genes should now be designed that encode desaturases that recognize fatty acyl-CoAs and catalyze the introduction of unsaturated bonds beyond the Δ9 position to increase the levels of PUFAs in mammalian adipose tissue.
Acknowledgments
We thank Dr. Leslie P. Kozak (The Jackson Laboratory) for generously providing the aP2 promoter; Dr. Shin Abe (College of Bioresource Sciences, Nihon University) for the analysis of lipids in lipogenicadipocytes; and Ms. Hiroko Fujimoto, Ms. Toshiko Otsui, and Ms. Chikako Okamoto for technical assistance. This research was supported by the Research for the Future Program of the Japan Society for the Promotion of Science Grant 97L00903 and by a Grant-in-Aid for the 21st Century Center of Excellence Program of the Japan Ministry of Education, Culture, Sports, Science, and Technology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FAD2, fatty acid desaturation 2; aP2, adipocyte P2; HO diet, diet supplemented with 10% (wt/vol) high-oleic-acid safflower oil, which contained 78% (wt/wt) oleic acid (18:1); PUFA, polyunsaturated fatty acid; SV40, simian virus 40.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB094415).
References
- 1.Pond, W., Church, D. & Pond, K. (1995) in Basic Animal Nutrition And Feeding (Wiley, New York), pp. 95-117.
- 2.Innis, S. M. (1991) Prog. Lipid Res. 30, 39-103. [DOI] [PubMed] [Google Scholar]
- 3.Watts, J. L. & Browse, J. (2002) Proc. Natl. Acad. Sci. USA 99, 5854-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borgeson, C. E., Kurtti, T. J., Munderloh, U. G. & Blomquist, G. J. (1991) Experientia 47, 238-241. [DOI] [PubMed] [Google Scholar]
- 5.Peyou-Ndi, M. M., Watts, J. L. & Browse, J. (2000) Arch. Biochem. Biophys. 376, 399-408. [DOI] [PubMed] [Google Scholar]
- 6.Weinert, J., Blomquist, G. J. & Borgeson, C. E. (1993) Experientia 49, 919-921. [Google Scholar]
- 7.Zhu, N., Dai, X., Lin, D. S. & Connor, W. E. (1994) Lipids 29, 869-875. [DOI] [PubMed] [Google Scholar]
- 8.Spychalla, J. P., Kinney, A. J. & Browse, J. (1997) Proc. Natl. Acad. Sci. USA 94, 1142-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang, Z. B., Ge, Y., Chen, Z., Cluette-Brown, J., Laposata, M., Leaf, A. & Kang, J. X. (2001) Proc. Natl. Acad. Sci. USA 98, 4050-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Hea, E. & Leveille, G. (1969) J. Nutr. 99, 338-344. [DOI] [PubMed] [Google Scholar]
- 11.Dyerberg, J. & Bang, H. (1979) Lancet 2, 433-435. [DOI] [PubMed] [Google Scholar]
- 12.Kinsella, J. E., Lokesh, B. & Stone, R. A. (1990) Am. J. Clin. Nutr. 52, 1-28. [DOI] [PubMed] [Google Scholar]
- 13.Graves, R. A., Tontonoz, P., Ross, S. R. & Spiegelman, B. M. (1991) Genes Dev. 5, 428-437. [DOI] [PubMed] [Google Scholar]
- 14.Ross, S. R., Graves, R. A., Greenstein, A., Platt, K. A., Shyu, H. L., Mellovitz, B. & Spiegelman, B. M. (1990) Proc. Natl. Acad. Sci. USA 87, 9590-9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross, S. R., Graves, R. A. & Spiegelman, B. M. (1993) Genes Dev. 7, 1318-1324. [DOI] [PubMed] [Google Scholar]
- 16.Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E. & Browse, J. (1994) Plant Cell 6, 147-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishida, I., Beppu, T., Matsuo, T. & Murata, N. (1992) Plant Mol. Biol. 19, 711-713. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto, K., Anzai, M., Nakagata, N., Takahashi, A., Takahashi, Y. & Miyata, K. (1994) Mol. Reprod. Dev. 39, 136-140. [DOI] [PubMed] [Google Scholar]
- 19.Hammer, R. E., Pursel, V. G., Rexroad, C. E., Jr., Wall, R. J., Bolt, D. J., Ebert, K. M., Palmiter, R. D. & Brinster, R. L. (1985) Nature 315, 680-683. [DOI] [PubMed] [Google Scholar]
- 20.Wall, R. J., Pursel, V. G., Hammer, R. E. & Brinster, R. L. (1985) Biol. Reprod. 32, 645-651. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto, K., Kakidani, H., Takahashi, A., Nakagata, N., Anzai, M., Matsuzaki, Y., Takahashi, Y., Miyata, K., Utsumi, K. & Iritani, A. (1993) Mol. Reprod. Dev. 36, 53-58. [DOI] [PubMed] [Google Scholar]
- 22.Warnants, N., Van Oeckel, M. J. & Boucque, C. V. (1999) J. Anim. Sci. 77, 2478-2490. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, K., Nakayama, T., Sakai, H., Tanemura, K., Osuga, H., Sato, E. & Ikeda, J. E. (1999) Mol. Reprod. Dev. 54, 103-111. [DOI] [PubMed] [Google Scholar]
- 24.Rodbell, M. (1964) J. Biol. Chem. 239, 375-380. [PubMed] [Google Scholar]
- 25.Sugihara, H., Yonemitsu, N., Miyabara, S. & Yun, K. (1986) Differentiation (Berlin) 31, 42-49. [DOI] [PubMed] [Google Scholar]
- 26.Kates, M. (1986) in Techniques of Lipidology: Isolation, Analysis, and Identification of Lipids (Elsevier, Amsterdam).
- 27.Sato, N. & Murata, N. (1988) Methods Enzymol. 167, 251-259. [Google Scholar]
- 28.Myer, R., Johnson, D., Knauft, D., Gorbet, D., Brendemuhl, J. & Walker, W. (1992) J. Anim. Sci. 70, 3734-3741. [DOI] [PubMed] [Google Scholar]
- 29.Pursel, V. (1998) in Animal Breeding: Technology for the 21st Century, ed. Clark, A. (Harwood, Amsterdam), pp. 183-200.
- 30.Pursel, V. G., Pinkert, C. A., Miller, K. F., Bolt, D. J., Campbell, R. G., Palmiter, R. D., Brinster, R. L. & Hammer, R. E. (1989) Science 244, 1281-1288. [DOI] [PubMed] [Google Scholar]
- 31.Los, D. & Murata, N. (1998) Biochim. Biophys. Acta 1394, 3-15. [DOI] [PubMed] [Google Scholar]
- 32.Kelder, B., Mukeji, P., Kirchner, S., Hovanec, G., Leonard, A. E., Chuang, L. T., Kopchick, J. J. & Huang, Y. S. (2001) Mol. Cell. Biochem. 219, 7-11. [DOI] [PubMed] [Google Scholar]
- 33.Spiegelman, B. M. & Green, H. (1980) J. Biol. Chem. 255, 8811-8818. [PubMed] [Google Scholar]
- 34.Green, H. & Kehinde, O. (1975) Cell 5, 19-27. [DOI] [PubMed] [Google Scholar]
- 35.Mackall, J. C., Student, A. K., Polakis, S. E. & Lane, M. D. (1976) J. Biol. Chem. 251, 6462-6464. [PubMed] [Google Scholar]
- 36.Cook, H. W. (1985) in Biochemistry of Lipids and Membranes, eds. Vance, D. E. & Vance, J. E. (Benjamin/Cummings Publishing Co. Inc., Menlo Park, CA), pp. 181-212.
- 37.Klingenberg, I., Knabe, D. & Smith, S. (1995) Comp. Biochem. Physiol. 110, 183-192. [DOI] [PubMed] [Google Scholar]
- 38.Ramsay, T. G. (1996) Endocrinol. Metab. Clin. North Am. 25, 847-870. [DOI] [PubMed] [Google Scholar]
- 39.Seiquer, I., Martinez-Victoria, E., Manas, M., Huertas, J. R., Ballesta, M. C. & Mataix, F. J. (1996) Arch. Physiol. Biochem. 104, 20-29. [DOI] [PubMed] [Google Scholar]
- 40.Thelen, J. J. & Ohlrogge, J. B. (2002) Metab. Eng. 4, 12-21. [DOI] [PubMed] [Google Scholar]
- 41.Gregoire, F. M., Smas, C. M. & Sul, H. S. (1998) Physiol. Rev. 78, 783-809. [DOI] [PubMed] [Google Scholar]
- 42.Brindlay, D. N. (1985) in Biochemistry of Lipids and Membranes, ed. Vance, D. E. & Vance, J. E. (Benjamin/Cummings Publishing Co. Inc., Menlo Park, CA), pp. 213-298.
- 43.Wood, J. & Enser, M. (1997) Br. J. Nutr. 78, Suppl. 1, 49-60. [DOI] [PubMed] [Google Scholar]
- 44.Morgan, C., Noble, R., Cocchi, M. & McCartney, R. (1992) J. Sci. Food Agri. 58, 357-368. [Google Scholar]
- 45.de Lorgeril, M., Renaud, S., Mamelle, N., Salen, P., Martin, J. L., Monjaud, I., Guidollet, J., Touboul, P. & Delaye, J. (1994) Lancet 343, 1454-1459. [DOI] [PubMed] [Google Scholar]
- 46.Bemelmans, W. J., Muskiet, F. A., Feskens, E. J., de Vries, J. H., Broer, J., May, J. F. & Jong, B. M. (2000) Eur. J. Clin. Nutr. 54, 865-871. [DOI] [PubMed] [Google Scholar]