Abstract
Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25), a member of the DEAD-box protein family, is a testis-specific gonadotropin-regulated RNA helicase that is present in Leydig cells and germ cells (meiotic spermatocytes and spermatids). In this study, we observed that GRTH is present in the nucleus, cytoplasm and chromatoid body of germ cells, and is an integral component of messenger ribonuclear protein particles. Male mice with a null mutation in the GRTH gene displayed normal gonadotropin and androgen profiles. However, they were sterile, with azoospermia caused by a complete arrest of spermiogenesis at step 8 of round spermatids and failure to elongate. Round spermatids of the null mice showed marked diminution in the size of chromatoid bodies. The transcription of relevant messages was not altered, but their translation was abrogated in a selective manner. Protein expression of transition proteins 1 and 2 and angiotensin-converting enzyme was completely absent, whereas that of the transcriptional activator cAMP responsive element modulator was intact. These findings indicate that GRTH participates in translational-associated events during germ cell development. Although significant apoptosis was present at the metaphase of meiosis in the GRTH-null mice, spermatogenesis proceeded to step 8 of spermiogenesis when complete arrest occurred. This progression may relate to compensatory gene function(s) and/or the observed up-regulation of DNA repair proteins Rad51 and Dmc1. This study (i) demonstrates that GRTH is essential for completion of spermatogenesis, (ii) provides insights into intrinsic requirements for spermiogenesis, and (iii) establishes a model for studies of male infertility and contraception.
Keywords: messenger ribonuclear protein particle, translation, testis, spermiogenesis, sterility
Spermatogenesis is a complex process that depends on the integrated expression of an array of genes that must operate in a precise temporal sequence to produce normal mature spermatozoa (1, 2). Gene expression in haploid spermatids requires temporal uncoupling of transcription and translation. Two-thirds of the mRNAs in the adult mammalian testes are associated with specific proteins to form messenger ribonuclear protein (mRNP) particles and stored in the cytoplasm of spermatids (3). Translational activation of stored mRNAs at specific times is essential for the completion of spermatogenesis. Modulation of RNA structure by members of the DEAD-box family of RNA helicases is a crucial step in many different fundamental biological processes (4, 5). This class of proteins participates in various aspects of RNA metabolism and translational events. However, little is known about the involvement of DEAD-box proteins in testicular germ cell development.
Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25), a member of the DEAD-box protein family of RNA helicases, was cloned and characterized in our laboratory (6–8). This enzyme, which only shares 30–40% similarity with other helicases, including those predominantly localized in the testis, is the sole family member to be hormonally regulated. GRTH displays ATPase and RNA helicase activities, and increases the in vitro translation of RNA templates. This helicase is a male-specific protein expressed in the rat, mouse, and human testis. It is present in both somatic Leydig cells and germ cells (spermatocytes and round spermatids) and is developmentally regulated. Furthermore, there is cell-specific and hormone-dependent regulation of translation in the rat testis resulting from alternative usage of AUG codons in GRTH mRNA (7). The up-regulation of this enzyme by gonadotropin and androgen and its cell- and stage-specific expression during germ cell development indicate that GRTH could participate in the regulation of androgen-dependent spermatogenesis.
To gain insights into the regulatory actions of GRTH in spermatogenesis, we examined the localization of GRTH and its association with mRNA and subsequently generated GRTH-null mice to determine its functional role in reproduction and testicular function. Our studies demonstrate that GRTH is a mRNA-binding protein that participates in posttranscriptional events concerned with the translation of genes that are essential for the progression of spermatogenesis.
Methods
Generation of GRTH-Null Mice. The 20-kb GRTH gene containing 3.5 kb of the 5′ flanking DNA sequence (accession no. AF326720) and 17 kb of coding region (12 exons and intronic sequences) (accession no. AY380080-380091) was isolated by using a mouse GRTH cDNA probe (6) from a mouse genomic bacterial artificial chromosome library derived from the embryonic stem (ES) 129/SVJ strain (Genome Systems, St. Louis). Five-kilobase BamHI and 14-kb EcoRI genomic clones were used to generate a 5′ and 3′ homologous arm in the targeting pPNT vector (8). The GRTH–pPNT (20 μg) was linearized at the NotI site and electroporated into 129/SVJ (J1) ES cells. Transformants were selected in G418 and ganciclovir. The resistant colonies were isolated and screened by Southern analysis. Homologous recombinant ES clones were identified with BglII digestion. Targeted ES clones were used to generate germ line GRTH+/- and GRTH-/- mice. Genotype screening of the offspring mice for the presence of the GRTH-/- allele were performed by PCR and verified by Southern analysis. PCR primers (P1, P2, and P4) were derived from mouse GRTH cDNA (accession no. AF142630). P1, +315/+337 bp; P2, +424/+ 393 bp; P4, +656/+627 bp; and P3 from Neo gene in pPNT.
Northern and Western Analyses. Poly(A)+ RNA samples were extracted from adult (16-week-old) mice testes (GRTH+/+, GRTH+/-, and GRTH-/-) were resolved on 1% agarose gel and hybridized to a full-length GRTH cDNA probe (6, 7). Protein samples (20 μg) extracted from adult mice testes were subjected to Western analysis with GRTH antibody (7) or other antibodies indicated (Santa Cruz Biotechnology). Polyclonal antisera for rat transition proteins 1 (Tp1) and 2 (Tp2) were kindly provided by S. Kistler (University of South Carolina, Columbia) (9), and a 17α-hydroxylase antiserum was generated in our laboratory (10).
Northwestern Analysis. A 3′ UTR downstream of the termination codon of mouse transition protein 2 (nucleotide of 386–555 bp, accession no. NM_013694) and mouse protamine 1 (nucleotide of 234–385 bp, accession no. K02926) were cloned into pGEM3Zf(-) vector (Promega) and used for generating 32P-labeled riboprobes. Purified GRTH–GST fusion protein or GST (0.5 μg) separated by SDS/PAGE were hybridized to RNA probes.
Coimmunoprecipitation and RT-PCR Analysis. Coimmunoprecipitation of the GRTH–RNP complex was performed by incubating 100 mg of testicular extracts with 2 μg of affinity-purified anti-GRTH antiserum (7) and 50 μl of protein A-agarose beads for 16 h at 4°C. GRTH–RNP complexes were recovered by centrifugation followed by washing. The RNA from the complexes was extracted by phenol and subjected to RT-PCR analysis. First-strand cDNA reverse transcribed by using a oligo(dT) was further amplified by PCR with specific primer sets for the genes of interest.
Isolation of mRNP Particles. Poly(A)+ RNP particles were isolated from homogenates of testis by affinity chromatography by using oligo(dT)-cellulose (Pharmacia) as described (11). mRNP particles were eluted in 5 mM Tris·HCl, pH 7.5, and evaluated by Western blot analysis with specific GRTH antibody.
Histological, Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL), and Electron Microscopic (EM) Analysis. Testes from GRTH (+/+, +/-, and -/-) males from 16-week-old mice were fixed in 4% paraformaldehyde and embedded in paraffin (6). Five-micron sections were stained with hematoxylin/eosin or periodic acid/Schiff reagent or processed for TUNEL analysis. TUNEL-positive cells were counted in at least 10 tubule sections per testis. Tissues for EM analyses were fixed in 2.5% glutaraldehyde, postfixed in 1% osmium tetroxide, en bloc-stained with 2% uranyl acetate, dehydrated, and embedded in Spurr's epoxy. Thin sections were poststained with lead citrate. For immuno-gold EM, ultrathin sections (≈80 nm) were incubated with rabbit anti-GRTH antibody or IgG (control) (1:50 dilution) followed by anti-rabbit biotinylated Ig antibody (Vector Laboratories; 1:100) and in streptavidin-gold (10 nm, 1:20; EM Sciences). The sections were stained with uranyl acetate and lead citrate and examined under transmission EM.
Confocal Microscopy Analysis. The full length of GRTH cDNA coding region was subcloned into the pEGFP-N2 vector (7) and transfected into COS-1 cells by using Lipofectamine (Invitrogen) for 24 h. The cells were fixed and examined in a Bio-Rad laser confocal microscope system (MRC-1024).
Hormone Measurements. Blood samples were collected from tail veins of individual animals. Serum levels of testosterone, luteinizing hormone, and follicle-stimulating hormone were measured by ELISA kits from ALPCO Diagnostics (Windham, NH).
Results
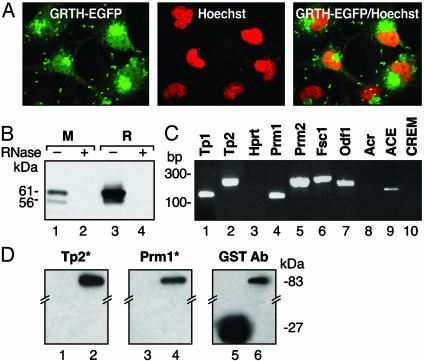
GRTH Localization and Association with mRNA. Our recent studies revealed a cell-specific utilization of GRTH translation initiation codons in the rat testis that resulted in the expression of two main protein forms (7). To gain insights into the function of this protein, we have initially investigated the cellular localization of the predominantly expressed GRTH long form and its association with mRNA particles in the testis. GRTH was localized in both cytoplasm and nuclear sites by confocal analysis of over-expressed GRTH–GFP fusion protein in COS-1 cells (Fig. 1A) and endogenously by Western analysis in rodent testis (data not shown), suggesting that GRTH could function in both cellular compartments. To identify whether GRTH is a RNA-binding protein, Western analysis of isolated poly(A)+ RNA–protein complexes from germ cell homogenates of both mouse and rat revealed the association of the 61/56-kDa GRTH protein species with mRNAs (Fig. 1B, lanes 1 and 3). GRTH-binding activity was absent in the negative control with RNase treatment (Fig. 1B, lanes 2 and 4). RT-PCR analysis of GRTH complexes from testes of adult mice, obtained by immunoprecipation of homogenates with GRTH antiserum, demonstrated that GRTH bound to selective mRNA messages whose proteins are expressed at different steps of spermiogenesis. These proteins include Tp1, Tp2, protamines 1 and 2 (Prm1 and Prm2), angiotensin converting enzyme (ACE), fibrous sheath component 1 and outer dense fiber. However, messages for acrosin, cAMP responsive element modulator (CREM), and house-keeping gene hypoxanthine guanine phosphoribosyl transferase were not detected (Fig. 1C). In addition, among subsets of mRNAs from a testicular 3′ UTR library, germ cell-specific Tp2 and Prm1 mRNAs were isolated by GRTH protein screening (data not shown). These two messages of relevant basic proteins involved in chromatin remodeling were used as representatives to reveal the potential function of the GRTH as a binding protein in spermatogenesis. Northwestern analysis showed that GRTH–GST protein bound to 32P-labeled riboprobes derived from the 3′ UTR of Tp2 and Prm1 mRNAs (Fig. 1D, lanes 2 and 4). No binding was present with GST only (lanes 1 and 3). Western analysis, used as reference, displayed the 83-kDa fusion protein (GRTH–GST, lane 6) and 27-kDa GST protein (lane 5) used in the study. These results clearly demonstrated that GRTH is an integral component of mRNA protein particles in the testis.
Fig. 1.
Cellular localization of GRTH protein and its association with mRNP. (A) Confocal analysis of overexpressed GRTH–GFP fusion protein in COS-1 cells (Left), nuclear staining by Hoechst 33342 (Middle), and merge image (Right). (B) Western analysis of oligo(dT)-purified mRNA–protein complex from mouse (M) and rat (R) testis. (C) RT-PCR analysis of immunoprecipitated testicular GRTH complexes with germ cell-specific mRNAs and hypoxanthine guanine phosphoribosyl transferase (Hprt; ubiquitously expressed). Fsc1, fibrous sheath component-1. Odf1, outer dense fiber. Acr, acrosin. (D) Northwestern analysis (Left) of GRTH–GST fusion protein binding to in vitro transcribed 3′UTR 32P-labeled RNA probe (*) of Tp2 (lane 2) and Prm1 (lane 4). GST, negative control (lanes 1 and 3). Western blot (Right) with a GST antibody shows the presence of GRTH–GST (83 kDa) and GST (27 kDa).
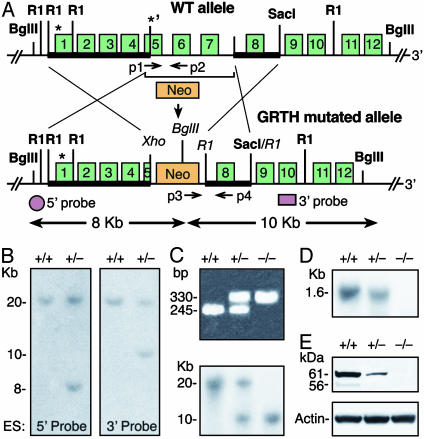
Insights from GRTH-Null Mice. To understand the functional role of GRTH in the male reproduction, the GRTH/Ddx25 gene was disrupted by mutation of the second ATG codon in exon 5, partial deletion of exon 5, and complete deletion of exons 6–7 containing the conserved ATPase motif, G-GKT, and the PTRELA motif of the RNA helicase family (Fig. 2A). Genotyping of the offspring was verified by using Southern and PCR analyses with expected DNA fragment size (Fig. 2 B and C). The expression of GRTH mRNA and its protein products was significantly reduced in the testes of GRTH+/- mice, and no expression was detectable in GRTH-/- mice (Fig. 2 D and E). Interbreeding of heterozygous mice produced offspring with Mendelian segregation ratios. Newborn GRTH-/- mice of both sexes were phenotypically normal. GRTH-/- females and GRTH+/- males were fertile. In contrast, the GRTH-/- male mice exhibited normal sexual behavior but were sterile. The weight of testes from adult GRTH-/- mice was 25% smaller than those of GRTH+/+ and GRTH+/- (P < 0.05). No change in weight was observed in accessory sex organs and prostate gland. The basal serum levels of luteinizing hormone, follicle-stimulating hormone and testosterone were similar in all groups. The normal serum levels of testosterone observed in null mice excluded abnormal steroidogenesis responsible for the arrest of spermiogenesis revealed by light and EM (see below).
Fig. 2.
Targeted disruption of the GRTH gene. (A) Targeted gene construction. (Upper) Wild-type mouse GRTH gene allele. *, ATGs. Bold line indicates the homologous 5′ arm (exons 1–4 and part of exon 5) and the 3′ arm (2.9-kb fragment with exon 8, 5′ and 3′ intron sequences) were subcloned at NotI/Xho and R1 site, respectively, in the targeting vector pPNT with (Neo) and (TK) selection markers. (Lower) The GRTH mutated homologus recombinant allele. Shown are the 5′ and 3′ probes used for Southern analysis and the PCR primers used for genotype screening (p1-p4). (B) Screening ES clones by Southern analysis. Positive homologous recombinant clones were identified with BglII digestion (predicted size: wild-type, 20 kb; recombinant allele with 5′ probe, 8 kb, and 3′ probe, 10 kb). (C) Genotyping of offspring mice by PCR (Upper). Predicted size: GRTH+/+, 245 bp (p1/p2); GRTH+/-, 245 bp (p1/p2) and 330 bp (p3/p4); and GRTH-/- mice, 330 bp (p3/p4). Positive clones were confirmed by Southern analysis by using a 3′ probe (Lower). (D and E) Northern (D) and Western (E) analyses of testis from adult GRTH+/+, GRTH+/-, and GRTH-/- mice.
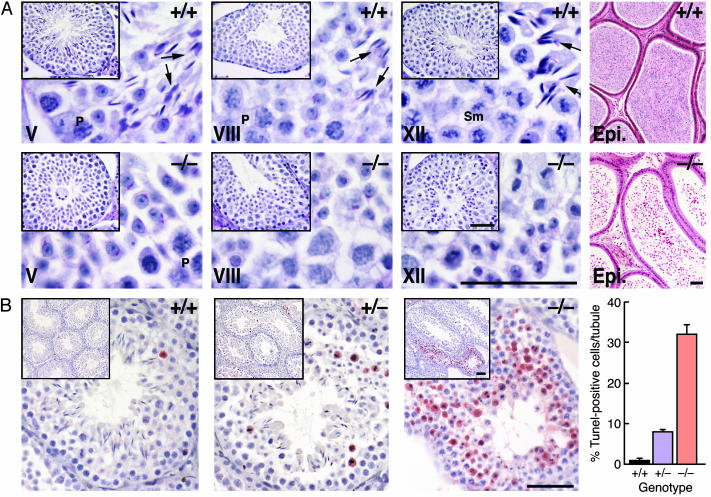
No morphological changes in germ cell development were observed from the time of birth to 21 days after birth in GRTH-/- mice. Subsequently, round spermatids were arrested at step 8, and no progression to elongating germ cells was observed (Fig. 3A, Lower versus Upper). Only degenerating spermatids were present in the epididymis. Germ cell development in the testis of GRTH+/- was normal. Interstitial Leydig cells in the GRTH-/- animals appeared to be normal by light microscopy. TUNEL analysis showed a significant increase in apoptotic germ cells in GRTH+/- compared to GRTH+/+ mice (Fig. 3B). In GRTH -/- mice, apoptosis was striking, most abundantly (>30% per tubule) at stage XII of the spermatogenic cycle, but limited to spermatocytes entering the metaphase of meiosis. This result suggested that germ cell development in the GRTH-/- mice was already impaired before the appearance of round spermatids and that spermatid development up to step 8 could arise from the remaining viable cells.
Fig. 3.
Complete arrest of spermatogenesis in GRTH-/- mice. (A) Periodic acid/Schiff reagent-stained sections from 16-week-old GRTH+/+ and GRTH -/- mice testis. (Scale bar, 0.5 μm.) Elongated spermatids and mature sperm were completely absent in the testis of GRTH-/- animals, and no mature sperm was observed in the epididymis. Arrows indicate elongated spermatids. P, pachytene spermatocytes. Sm, spermatocytes in the metaphase of meiosis. (B) TUNEL assay. Error bars indicate mean ± SEM positive cells per tubule. (Scale bar, 0.5 μm.)
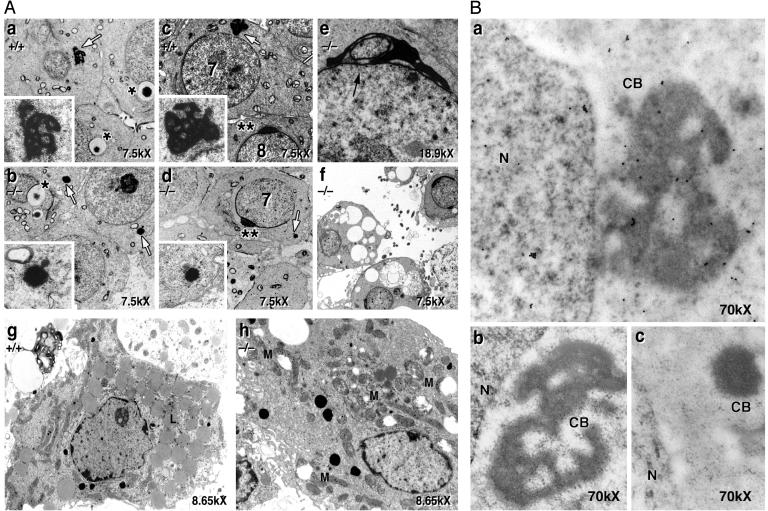
The nucleus, Golgi apparatus, mitochondria, and acrosome were morphological normal by EM analysis up to step 7 of spermiogenesis (Fig. 4 Aa and Ac versus Ab and Ad). However, chromatoid bodies (CB) of GRTH-/- mice were unusually condensed, greatly reduced in size, and lacking the typical amorphous “nuage” texture throughout all steps of spermiogenesis (Fig. 4 Aa and Ac versus Ab and Ad). After step 8, the acrosome and nucleus oriented toward the basement membrane of the tubule, but the nucleus maintained a spherical shape. There was an abnormal indentation of the nucleus in the region covered by the acrosome, part of the nucleus was separated by the acrosome (Fig. 4Ae), spermiogenesis ceased, and spermatids degenerated. Degenerating cells with abundant vacuoles, loss of nuclear chromatin, and abnormal acrosome formation were present at stage XII (Fig. 4Af). Leydig cells from null mice revealed reduced lipid droplets, swollen mitochondria without normal central cristae (Fig. 4Ah) when compared to cells from normal (Fig. 4Ag), and heterozygous littermates (data not shown). Despite these morphological changes, Leydig cells were able to sustain normal basal circulating levels of testosterone. This result was also reflected by the findings of normal luteinizing hormone plasma levels and weight of secondary accessory organs and prostate. However, it is conceivable that changes in steroid hormones production in plasma and or Leydig cells from null mice could be revealed upon acute gonadotropin stimulation in vivo and or in vitro. Also, age progression could provide further information about the steroidogenic function of Leydig cells from null mice.
Fig. 4.
EM analysis of GRTH+/+ and GRTH-/- mouse testis. (A) Spermatids of GRTH+/+ and GRTH-/- at step 2/3 (a and b) and at step 7/8 (c and d) of spermiogenesis. White arrows indicate round spermatid CB. (Inset) CB at higher magnification (×40,000). *, Acrosomal vesicles noted in step 2/3. **, Typical acrosomal structure noted in step 7/8. (e) Black arrow indicates abnormal acrosome in GRTH -/- mice after arrest. (f) Degenerating round spermatids at stage XII. (g and h) Leydig cells of GRTH+/+ (g) and GRTH-/- mice (h). L, lipid. M, swollen mitochondria. (B) Immuno-gold labeling analysis. GRTH associates with the nuclear (N), cytoplasmic, and CB of round spermatids from adult wild-type mice (a) and is absent in the GRTH-/- (c). Gold particles are shown as small intense dark spots. (b) IgG as control in the GRTH+/+ testis. (Magnification, ×70,000.)
Further immuno-EM studies of the adult mice testis confirmed that GRTH protein is expressed in both nuclear and cytoplasm of testicular cells, consistent with our findings from confocal microscopy and Western analysis. In addition, GRTH is present in CBs of wild-type mice round spermatids (Fig. 4Ba). No immunolabeling is present either in the wild-type spermatids devoid of primary GRTH antibody (Fig. 4, Bb) or the abnormal CB of knockout mice testis (negative control) (Fig. 4Bc).
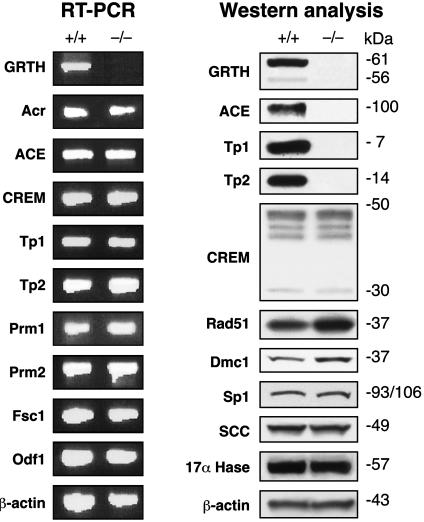
RT-PCR analysis revealed no changes in mRNA from stage-relevant germ cell genes in GRTH-/- mice (Fig. 5). Western analysis with available antibodies revealed that the ACE testicular isoform (normally expressed from steps 4–12 of spermiogenesis), and Tp1 and Tp2 chromatin remodeling proteins (present after step 9) were completely absent in the GRTH-/- mice testis (Fig. 5). No change in any of the CREM isoforms was observed, including the germ cell transcriptional activator CREM τ isoforms, which are expressed in haploid spermatids at steps before the spermatogenic arrest. No difference in basal protein levels was found for the transcriptional factor Sp1, or proteins expressed in other than germ cells, including steroidogenic enzymes, P450scc and P450c17 (Leydig cells). DNA repair proteins Rad51 and Dmc1, which are both expressed at the meiotic prophase preceding the apoptotic events observed in the GRTH-/- were significantly increased in the GRTH-/- mice testis. These findings indicate that the function of GRTH in germ cell development is predominantly posttranscriptional and that selectively exhibits its regulation.
Fig. 5.
Analysis of gene expression by RT-PCR and Western in GRTH+/+ and GRTH-/- mouse testis. mRNA or protein was prepared from adult mouse testis. The indicated genes were analyzed by RT-PCR reaction and Western blot. Fsc1, fibrous sheath component-1. Odf1, outer dense fiber. Acr, acrosin.
Discussion
We have demonstrated that the testis-specific helicase GRTH/Ddx25 is required for progression of spermiogenesis beyond round spermatids step 7/8. Consequently, GRTH-null mice are sterile because of spermatogenic arrest with failure of spermatids to elongate. Also, significant structural abnormalities of CBs were observed in the round spermatids at steps before arrest. Furthermore, considerable apoptosis was observed in germ cells entering the metaphase of meiosis. However, this process was sufficiently compensated to permit germ cells to further develop to haploid cells and to the arrest point. We have also shown that in addition to its helicase unwinding and ATPase activities demonstrated in our previous study, GRTH is an mRNA-binding protein and an integral component of RNP particles that may play a role in the regulation of translational processes during germ cell development. Moreover, the nuclear and cytoplasmic colocalization of GRTH demonstrated by confocal, Western, and immuno-gold EM analyses support the notion that GRTH may function as a shuttle mRNA-binding protein between cellular compartments and participate in the assembly of stored mRNP particles. Preservation of the expression of relevant mRNAs with failure of protein expression in the GRTH-null mice indicate that this protein is involved in posttranscriptional events. Given that GRTH has been shown to selectively bind to sets of germ cell-specific RNAs that could be required for posttranscriptional regulation of protein expression, loss of this function may contribute to the spermatogenic arrest in GRTH-null mice.
Cells lacking genes that are critical for spermatogenesis usually degenerate after the arresting step. The first checkpoint of GRTH function appears to be localized at the transition of prophase to metaphase during meiosis, when marked apoptosis was observed. This apoptosis could have resulted from mismatched chromosome pairing and/or recombination events during meiotic prophase (12, 13). Despite the significant proportion of metaphase-arrested spermatocytes in GRTH-/- mice (≈30%), the remaining cells proceeded through early steps of spermiogenesis. This indicated that some degree of compensation was operative at meiosis. The smaller increase in apoptotic cells observed in GRTH+/- mice is related to the reduced GRTH protein expressed in the testis. In these animals, the subsequent germ cell development was normal throughout spermiogenesis. It is conceivable that GRTH either directly or indirectly influences the apoptosis pathway and the dynamic changes of chromosome events through the regulation of protein expression of relevant genes. It is interesting to note that DNA repair proteins that are necessary for the maintenance of chromosome integrity, such as Rad51 and Dmc1 (14, 15), were increased in GRTH-/- testis. Rad51 is involved in mitotic and meiotic double-strand DNA break repair and Dmc1 is a germline meiotic-specific gene essential for chromosome synapsis in the first meiotic prophase. The increase in both proteins in the GRTH-/- testis, may reflect the activation of survival mechanism(s) in response to increased apoptosis. This mechanism could facilitate recombination events for progression of meiotic to haploid germ cells in GRTH-null mice.
Spermatid development progresses through steps 1–7 with no apparent overall ultra-structural change, however a striking reduction in the size of CB was observed following the early stage of round spermatids to the arrest point. CBs, electron-dense cytoplasmic structures that are located in the vicinity of the nucleus, may serve as storage organelles of long-lived spermatid mRNAs associated as mRNA ribonucleoprotein particles (mRNP) awaiting translation during spermiogenesis (16, 17). The evidence in our studies of GRTH immunoactivity present in the CB of wild-type mice, and its absence in the spermatids of null mice suggests its direct association with this organelle. Tp2 mRNA has been reported to localize in the CB (16). The marked structural changes of the CB in GRTH-null mice and the biochemical evidence of GRTH protein–Tp2 mRNA interaction in wild-type mice suggest that the participation of GRTH as a translational regulator may take place at least in part in the CB. The mouse Vasa homolog (18), which is a specific RNA helicase of germ cells, was also reported to be associated with CBs in normal mice. However, because the mouse Vasa homolog gene deletion resulted in premeiotic arrest at the zygotene stage (19), information about the CB in this case could not be obtained. Our study provides the first evidence of a CB abnormality associated with failure of germ cells development. The striking reduced size of CBs in spermatids of GRTH-/- mice was not accompanied by premature translation of messages (i.e., Tp1 and Tp2) or affect steady-state mRNA levels. This pointed to a role of CB-associated GRTH on translation rather than mRNA stabilization. In GRTH-/- mice, abnormal indentation of the nucleus covered by acrosome was also observed. This finding may have resulted from the absence of some structural elements involved in nuclear shaping, such as the manchette, a cytoskeletal complex formed around the nucleus by a sleeve of microtubules (20). It is possible that the lack of this structure in the cytoplasm of spermatids when the straightening forces emanating from the acrosomal complex are present induced the nuclear deformation.
In an attempt to identify the possible target genes regulated by GRTH, we examined sets of genes known to play a role in spermatogenesis. The disruption of the GRTH gene caused no significant changes on the expression of testicular mRNAs. In contrast, the selective absence of proteins, Tp1, Tp2, and ACE during spermiogenesis clearly indicated that the function of GRTH is posttranscriptional. Because the testicular ACE protein is first detected during steps 4–7 (21), its lack of expression well before spermiogenesis arrest indicates that ACE is one of the candidate genes regulated by GRTH. The association of GRTH with ACE mRNA message demonstrated in our coimmunoprecipitation analysis also suggests that the translation process of ACE is governed by GRTH. It is not possible to determine whether Tp1 and Tp2 are GRTH target genes at the translation level because both proteins are normally expressed after the spermatid arrest step observed in GRTH-/- mice. We cannot exclude the possibility that in normal testis, GRTH association with Tp1 and Tp2 mRNA in CB may serve as a mechanism to regulate the translation process of these proteins during spermiogenesis. On the other hand, the absence of Tp1 and Tp2 proteins in the null mice excludes the possibility that the spermiogenesis arrest was induced by premature translation. Male mice lacking individual Tp1, Tp2, or ACE showed either no effect or reduced fertility with normal sperm parameters (22–24). However, the fact that Tp1 and Tp2 double-knockout mice were infertile (25) further points to GRTH as a important regulator of spermiogenesis. Thus, GRTH may serve as a master translational regulator of a selective panel or cascade of genes that are crucial for spermiogenesis. The germ cell essential transcriptional regulator, CREM (26), is not among the GRTH-regulated genes because its mRNA or protein levels were not changed in the GRTH-/- mice. This finding is further supported by the evidence that the mRNA level of CREM-targeted genes, including Tp1, Tp2, Prm1, and Prm2, were not altered in the GRTH-/- mice testis.
Here we propose that GRTH protein may function as a component of mRNP and/or may be required for the formation of CB. As a binding protein and a component of mRNP, GRTH could be important in the translation of crucial genes at specific times during spermatogenesis. GRTH could also affect transport of poly(A)+ mRNA to the cytoplasm for storage in CB of spermatids, to be released for translation in a time-specific manner during spermiogenesis. Furthermore, GRTH could be associated with polyribosomes and influence the translation of genes. During spermiogenesis, all genes must be transcribed before the condensation of the spermatid nucleus, and some transcripts are stored in the cytoplasm for later translation (1). The expression of sets of genes determining the progression of spermiogenesis may be governed by GRTH. Mice deficient in this protein could serve as a model system for the study of male reproduction and fertility control.
Acknowledgments
We thank Dr. Heiner Westphal (Laboratory of Mammalian Genes and Development, National Institute of Child Health and Human Development) for the support of his laboratory in the development of the null mice.
Abbreviations: CB, chromatoid body; Tp1 and Tp2, transition proteins 1 and 2; Prm1 and Prm2, protamines 1 and 2; CREM, cAMP responsive element modulator; mRNP, messenger ribonuclear protein; GRTH, gonadotropin-regulated testicular RNA helicase; ES, embryonic stem; EM, electron microscopy; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; ACE, angiotensin converting enzyme.
References
- 1.Steger, K. (2001) Anat. Embryol. (Berlin) 203, 323-334. [DOI] [PubMed] [Google Scholar]
- 2.Eddy, E. M. (2002) Recent Prog. Horm. Res. 57, 103-128. [DOI] [PubMed] [Google Scholar]
- 3.Schmid, S. R. & Linder, P. (1992) Mol. Microbiol. 6, 283-291. [DOI] [PubMed] [Google Scholar]
- 4.de la Cruz, J., Kressler, D. & Linder, P. (1999) Trends Biochem. Sci. 24, 192-198. [DOI] [PubMed] [Google Scholar]
- 5.Silverman, E., Edwalds-Gilbert, G. & Lin, R. J. (2003) Gene 312, 1-16. [DOI] [PubMed] [Google Scholar]
- 6.Tang, P. Z., Tsai-Morris, C. H. & Dufau, M. L. (1999) J. Biol. Chem. 274, 37932-37940. [DOI] [PubMed] [Google Scholar]
- 7.Sheng, Y., Tsai-Morris, C. H. & Dufau, M. L. (2003) J. Biol. Chem. 278, 27796-27803. [DOI] [PubMed] [Google Scholar]
- 8.Tsai-Morris, C. H., Lei, S., Jiang, Q., Sheng, Y. & Dufau, M. L. (2004) Gene 331, 83-94. [DOI] [PubMed] [Google Scholar]
- 9.Kistler, W. S., Henriksen, K., Mali, P. & Parvinen, M. (1996) Exp. Cell Res. 225, 374-381. [DOI] [PubMed] [Google Scholar]
- 10.Koh, Y., Buczko, E. & Dufau, M. L. (1993) J. Biol. Chem. 268, 18267-18271. [PubMed] [Google Scholar]
- 11.Marello, K., LaRovere, J. & Sommerville, J. (1992) Nucleic Acids Res. 20, 5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipkin, S. M., Moens, P. B., Wang, V., Lenzi, M., Shanmugarajah, D., Gilgeous, A., Thomas, J., Cheng, J., Touchman, J. W., Green, E. D., Schwartzberg, P., et al. (2002) Nat. Genet. 31, 385-390. [DOI] [PubMed] [Google Scholar]
- 13.Baarends, W. M., Wassenaar, E., Hoogerbrugge, J. W., van Cappellen, G., Roest, H. P., Vreeburg, J., Ooms, M., Hoeijmakers, J. H. & Grootegoed, J. A. (2003) Mol. Cell. Biol. 23, 1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita, T., Yoshimura, Y., Yamamoto, A., Murata, K., Mori, M., Yamamoto, H. & Matsushiro, A. (1993) Proc. Natl. Acad. Sci. USA 90, 6577-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida, K., Kondoh, G., Matsuda, Y., Habu, T., Nishimune, Y. & Morita, T. (1998) Mol. Cell 1, 707-718. [DOI] [PubMed] [Google Scholar]
- 16.Saunders, P. T., Millar, M. R., Maguire, S. M. & Sharpe, R. M. (1992) Mol. Reprod. Dev. 33, 385-391. [DOI] [PubMed] [Google Scholar]
- 17.Figueroa, J. & Burzio, L. O. (1998) Cell Tissue Res. 291, 575-579. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., Yokoyama, M. & Noce, T. (2000) Genes Dev. 14, 841-853. [PMC free article] [PubMed] [Google Scholar]
- 19.Noce, T., Okamoto-Ito, S. & Tsunekawa, N. (2001) Cell Struct. Funct. 26, 131-136. [DOI] [PubMed] [Google Scholar]
- 20.Meistrich, M. L. (1993) Nuclear Morphogenesis During Spermiogenesis (Academic, San Diego), pp. 67-97.
- 21.Sibony, M., Segretain, D. & Gasc, J. M. (1994) Biol. Reprod. 50, 1015-1026. [DOI] [PubMed] [Google Scholar]
- 22.Yu, Y. E., Zhang, Y., Unni, E., Shirley, C. R., Deng, J. M., Russell, L. D., Weil, M. M., Behringer, R. R. & Meistrich, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adham, I. M., Nayernia, K., Burkhardt-Gottges, E., Topaloglu, O., Dixkens, C., Holstein, A. F. & Engel, W. (2001) Mol. Hum. Reprod. 7, 513-520. [DOI] [PubMed] [Google Scholar]
- 24.Esther, C. R., Marino, E. M., Howard, T. E., Machaud, A., Corvol, P., Capecchi, M. R. & Bernstein, K. E. (1997) J. Clin. Invest. 99, 2375-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullas, K. S. & Rao, M. R. (2003) J. Biol. Chem. 278, 52673-52680. [DOI] [PubMed] [Google Scholar]
- 26.Beissbarth, T., Borisevich, I., Horlein, A., Kenzelmann, M., Hergenhahn, M., Klewe-Nebenius, A., Klaren, R., Korn, B., Schmid, W., Vingron, M. & Schutz, G. (2003) Mol. Cell. Endocrinol. 212, 29-39. [DOI] [PubMed] [Google Scholar]