Abstract
Pathological cardiac hypertrophy is characterized by a sustained increase in cardiomyocyte size and re-activation of the fetal cardiac gene program. Previous studies implicated SWI/SNF chromatin remodeling enzymes as regulators of the fetal cardiac gene program in surgical models of cardiac hypertrophy. Although hypertension is a common risk factor for developing cardiac hypertrophy, there has not yet been any investigation into the role of SWI/SNF enzymes in cardiac hypertrophy using genetic models of hypertension. In this study, we tested the hypothesis that components of the SWI/SNF complex are activated and recruited to promoters that regulate the fetal cardiac gene program in hearts that become hypertrophic as a result of salt induced hypertension. Utilizing the Dahl salt-sensitive (S) rat model, we found that the protein levels of several SWI/SNF subunits required for heart development, Brg1, Baf180, and Baf60c, are elevated in hypertrophic hearts from S rats fed a high salt diet compared with normotensive hearts from Dahl salt-resistant (R) rats fed the same diet. Furthermore, we detected significantly higher levels of SWI/SNF subunit enrichment as well as evidence of more accessible chromatin structure on two fetal cardiac gene promoters in hearts from S rats compared with R rats. Our data implicate SWI/SNF chromatin remodeling enzymes as regulators of gene expression in cardiac hypertrophy resulting from salt induced hypertension. Thus we provide novel insights into the epigenetic mechanisms by which salt induced hypertension leads to cardiac hypertrophy.
Keywords: cardiac hypertrophy, SWI/SNF chromatin remodeling enzymes, Dahl model of hypertension, epigenetic, chromatin accessibility, histone covalent modifications
INTRODUCTION
Cardiac hypertrophy occurs in response to pathological as well as physiological conditions that increase cardiac workload (Bernardo et al., 2010). To compensate for the increased workload, there is extensive ventricular remodeling resulting in an increase in heart mass. Pathological cardiac hypertrophy frequently occurs due to hypertension and myocardial infarction (Heineke and Molkentin, 2006). The hypertrophied ventricle is initially able to compensate in the face of an increased workload and maintain cardiac output, but in the later stages, the diastolic and eventually the systolic properties of the left ventricle become impaired causing cardiac dysfunction (Frey and Olson, 2003). Pathological hypertrophy often leads to heart failure, a significant cause of morbidity and mortality worldwide (Franco et al., 2011).
At the cellular level, the increase in heart mass is primarily due to an increase in the size of cardiomyocytes, the contractile cells of the myocardium (Maillet et al., 2013). Cardiomyocytes make up approximately a third of the cells in the myocardium and constitute approximately 70-80% of the heart’s mass. They are terminally differentiated cells that have essentially lost the ability to proliferate. Therefore, they respond to the increased workload by increasing their size. Activation of signaling pathways, changes in gene expression, and increases in protein synthesis lead to cardiomyocyte growth (Bernardo et al., 2010).
Activation of the fetal cardiac gene program is a consistent feature of pathological cardiac hypertrophy (Barry et al., 2008). Genes that are highly expressed in fetal ventricles become silenced at birth, but are then reactivated in the hypertrophic myocardium. Concomitantly, adult isoforms of these genes are silenced. The activated genes include those that encode atrial natiuretic peptide (ANP), brain natriuretic peptide (BNP), as well as genes that encode fetal isoforms of several contractile proteins and cardiac ion channels. The changes in gene expression are critical for promoting the molecular changes that result in cardiac remodeling (Frey and Olson, 2003). Recent studies suggest that chromatin modifications contribute to the changes in gene expression that lead to cardiac remodeling and that inhibition of some of the changes in chromatin structure can reverse cardiac hypertrophy (El-Osta, 2011; McKinsey and Olson, 2004). Thus, a better understanding of the mechanisms by which chromatin is remodeled to promote cardiac hypertrophy may lead to the identification of new therapeutic targets.
SWI/SNF chromatin remodeling enzymes have emerged as key regulators of gene expression during cardiac development and have also recently been implicated in regulating gene expression profiles in hypertrophic ventricles (Chang et al., 2011; Hang et al., 2010; Huang et al., 2008; Lei et al., 2012; Lickert et al., 2004; Wang et al., 2004). SWI/SNF enzymes are multi-subunit complexes that use the energy derived from ATP hydrolysis to alter chromatin structure (de la Serna et al., 2006). Mammalian SWI/SNF complexes contain either Brg1 or Brm as the catalytic subunit and nine to twelve associated factors (Bafs). Brg1, Baf180, and Baf60c have been shown to have critical roles in cardiac development (Lickert et al., 2004; Takeuchi et al., 2011; Wang et al., 2004). Interestingly, Brg1 is turned off in terminally differentiated cardiomyocytes but is reactivated by cardiac stresses and forms a complex with its embryonic partners to induce a pathological shift in gene expression (Hang et al., 2010). Furthermore, Brg1, Brm, and Baf57 occupancy on the regulatory regions of several hypertrophy related genes has been demonstrated in a transverse aortic constriction model of cardiac hypertrophy (TAC) (Chang et al., 2011; Hang et al., 2010). Thus, emerging evidence strongly indicates that SWI/SNF enzymes activate the fetal cardiac gene expression profile that underlies cardiac hypertrophy. However, there have not yet been any studies investigating SWI/SNF enzymes in genetic models of hypertension. These models are particularly informative because they result in a gradual onset of cardiac hypertrophy that closely recapitulates the human disease process.
In this study, we tested the hypothesis that components of the SWI/SNF complex are activated and recruited to promoters that regulate the fetal cardiac gene program in hearts that become hypertrophic as a result of salt induced hypertension. For these studies, we utilized the Dahl salt-sensitive (S) and salt-resistant (R) rat models of hypertension (Dahl et al., 1962a; Dahl et al., 1962b). The Dahl model we used was developed from inbred Spraque Dawley rats that were fed a high salt diet and then selected for sensitivity or resistance to developing hypertension (Rapp, 2000). They have been utilized extensively for identifying genetic determinants of hypertension (Joe and Shapiro, 2012) and in a limited number of epigenetic studies (Kaneda et al., 2009; Mu et al., 2011). In this well established model of salt induced hypertension, S rats develop hypertension and cardiac hypertrophy when placed on a high salt diet while R rats fed the same diet do not (Dahl et al., 1962a; Dahl et al., 1962b; Patten and Hall-Porter, 2009). Under these conditions, we found that the protein levels of several SWI/SNF subunits required for heart development, Brg1, Baf180, and Baf60c, are elevated in hypertrophic hearts from hypertensive S rats fed a high salt diet compared with hearts from normotensive R rats fed the same diet. Furthermore, we detected significantly higher levels of SWI/SNF subunit enrichment as well as evidence of more accessible chromatin structure on the ANP and BNP promoters in hearts from S rats compared with R rats. Our data implicate SWI/SNF chromatin remodeling enzymes as regulators of gene expression in cardiac hypertrophy resulting from salt induced hypertension. Thus we provide novel insights into the epigenetic mechanisms by which salt induced hypertension leads to cardiac hypertrophy.
MATERIAL AND METHODS
Animals
Male, Dahl salt-sensitive (S) and salt-resistant (R) rats were from the colony maintained in our institution. Rats were housed in pathogen-free conditions and maintained with 12 h dark/light cycle and had ad libitum access to food and water. Age-matched, 6 week-old male rats for both R and S group were given a 2% NaCl containing diet for 6 weeks, then weighed and euthanized with carbon dioxide. The hearts were excised, weighed and also the tibia length of each animal was measured. All animal experimentation was conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals using protocols approved by the University of Toledo Institutional Animal Use and Care Committee.
RNA isolation and quantitative real-time PCR
Total RNA was isolated from 10 to 15 μg of heart tissue using a total RNA purification kit (Qiagen, Valencia, CA). Heart tissue was minced under liquid nitrogen and homogenized using a sterile pestle in Qiagen RLT lysis buffer. In the next step, 0.01% v/v proteinase K was added to the homogenate, followed by incubation at 55°C for 10 minutes. RNA was then isolated according to the manufacturer’s instructions. Quantitative real-time PCR was performed in SYBR Green Master Mix (Qiagen, Germantown, Maryland) with an Applied Biosystems Prism 7500 PCR system and analyzed with the SDS software as described (Keenen et al., 2010). Primer sequences for ANP and BNP were as follows: ANP 5′-ACCTGGAGGAGAAGATGCCG-3′; 5′-TGTTGCAGCCTAGTCCGCTC-3′ and BNP 5′-GTGCTGCCCCAGATGATT-3′ 5′-GGTCTATCTTCTGCCCAAAG-3′(Gaspar-Pereira et al., 2012). Primer sequences for the control 18S rRNA were 5′-AGTCCCTGCCCTTTGTACACA-3′ and 5′-GATCCGAGGGCCTCACTAAAC-3′.
Antibodies
Antisera to Brg1 (de La Serna et al., 2000) was previously described. Baf180 antibody was from Bethyl laboratories (Montgomery, TX). The Baf60c antibody was from Abcam (Cambridge, MA). The tri-methylated histone H3 at lysine 4 (H3K4me3), tri-methylated histone H3 at lysine 9 (H3K9me3) and tetra-acetylated histone H4 (AcH4) antibodies were from Active Motif (Carlsbad, CA). Control IgG and antibodies to tri-methylated histone H3 at lysine 27 (H3K27me3) were from Millipore. The α/βTubulin antibody was from Sigma (St. Louis, Missouri).
Western Blotting
For protein isolation, left ventricles were minced under liquid nitrogen and immediately resuspended in lysis buffer and processed as described (de La Serna et al., 2000). Total proteins were run on SDS polyacrylamide gels and Western blotted. Signal detection was performed with an enhanced chemiluminescence super signal kit (Pierce, Rockford, IL). The signal density was determined using Image J, image-processing program developed at the National Institutes of Health (Bethesda, MD).
Chromatin Immunoprecipitations (ChIPs)
Left ventricles were minced and homogenized using a sterile pestle before fixing with 1% formaldehyde. After crosslinking, nuclei were isolated by disruption using a dounce homogenizer in a buffer containing 50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% TritonX, and protease inhibitors. Chromatin immunoprecipitations (ChIPs) were performed and subjected to quantitative (q) PCR analysis as described (Keenen et al., 2010). PCR primers used to amplify regions of the BNP and ANP promoter were: BNP site 1 Forward 5′-CTATACAAGGCCTGCGGTTT- 3′ and reverse 5′-TGCCTCTGCTTTATCCTG- 3′; ANP forward 5′-GAGAGGAGCTGGACCATGAG-3′ and reverse 5′-CCCAGCATCCACATAAAAGC-3′ as previously described in (Gaspar-Pereira et al., 2012). The myelin protein zero intron was detected with 5′-TAGTTATGAGCCCCCAGCAC-3′ and 5′-GTGGTCCAATCATCCTCACC-3′.
Chromatin accessibility
Left ventricles were minced and homogenized using a sterile pestle. Nuclei were isolated and digested with 3U/ml MNase I (Worthington Biochemical Corp., Lakewood, NJ) as described (Saladi et al., 2013). Purified genomic DNA was subjected to qPCR with the same primers as used in ChIPs and analyzed as described in (Cruickshank et al., 2008; Rao et al., 2001).
Statistical analysis
P values of <0.05 obtained with a two-tailed Student t-test were considered significant. Values are expressed as means ±SEM.
RESULTS
Genetically hypertensive S rats fed a high salt diet develop cardiac hypertrophy
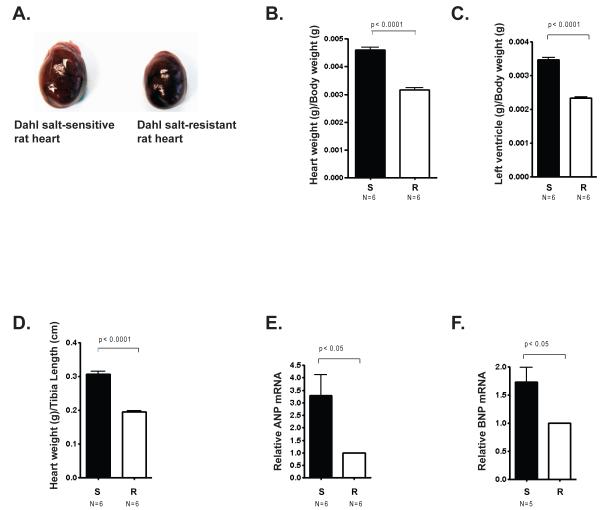
In this study, age-matched, 6 week-old Dahl salt-sensitive (S) rats and salt-resistant (R) rats were fed a high salt diet for six weeks and then evaluated for several parameters of cardiac hypertrophy. Representative pictures of hearts from S rats and R rats reveal that hearts from S rats were visibly bigger than hearts from R rats (Fig. 1A). S rats had a significantly greater heart weight to body weight ratio than R rats (Fig. 1B) as well as a greater left ventricle to body weight ratio (Fig. 1C) and a greater left ventricle to tibia length ratio (Fig. 1D). Thus, as expected, several features indicative of cardiac hypertrophy were markedly more pronounced in the genetically hypertensive S rats that were fed a high salt diet compared with the genetically normotensive R rats fed the same diet. Furthermore, we found that expression of two genes in the fetal cardiac program, ANP and BNP, was elevated in S rats compared to R rats (Figs. 1E and 1F). These results demonstrate differential development of cardiac hypertrophy in S rats fed a high salt diet compared to R rats fed a high salt diet with characteristic differences in gene expression.
Figure 1. Cardiac hypertrophic parameters in Dahl salt-sensitive rats and salt-resistant rats.
Male Dahl salt-sensitive (S) and salt-resistant (R) rats were given a 2% NaCl diet at 6 weeks of age for another 6 weeks. A. Image of hearts from representative S and R rats after 6 weeks on a high salt diet. B. Whole heart weight/body weight ratio (g/g) was measured in both S and R animals. C. Left ventricle (LV)/ body weight ratio (g/g) was measured in both S and R animals. (D) Left ventricle (LV)/tibia length ratio (g/cm) was measured in both S and R animals. (E) ANP and (F) BNP mRNA levels in the left ventricles of S rats and R rats measured by quantitative (q) PCR. Bars represent the average obtained from independent experiments performed with the indicated number of rat hearts (N). Error bars represent SEM.
Elevated expression of SWI/SNF components is associated with cardiac hypertrophy in genetically hypertensive S rats
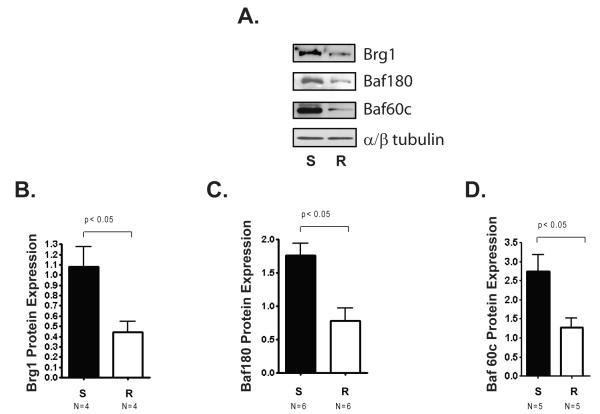
In order to determine if the expression of Brg1 and other SWI/SNF subunits is correlated with cardiac hypertrophy induced by a high salt diet in S rats, we assessed the protein levels of Brg1, Baf180, and Baf60c (Fig. 2A). We found that Brg1, Baf180, and Baf60c levels were significantly higher in hearts from S rats compared with those in hearts from R rats (Figure 2 B, C, and D). Thus, elevated expression of Brg1 and two other SWI/SNF subunits was associated with cardiac hypertrophy in this salt induced model of hypertension.
Figure 2. Differential expression of SWI/SNF components in left ventricles from Dahl salt-sensitive rats and Dahl salt-resistant rats.
Male Dahl salt-sensitive (S) and salt-resistant (R) rats were given a 2% NaCl diet at 6 weeks of age for another 6 weeks. A. Representative Western blot showing protein levels of Brg1, Baf180, and Baf 60c in left ventricles from S and R rats. α/β Tubulin is a loading control. B. Quantification of Brg1 protein levels. C. Quantification of Baf180 protein levels. D. Quantification of Baf60c protein levels. SWI/SNF subunit expression was normalized to expression of α/β Tubulin. Bars represent the average obtained from independent experiments performed with the indicated number of rat hearts (N). Error bars represent SEM.
SWI/SNF recruitment to fetal cardiac gene promoters is elevated in hearts from genetically hypertensive S rats
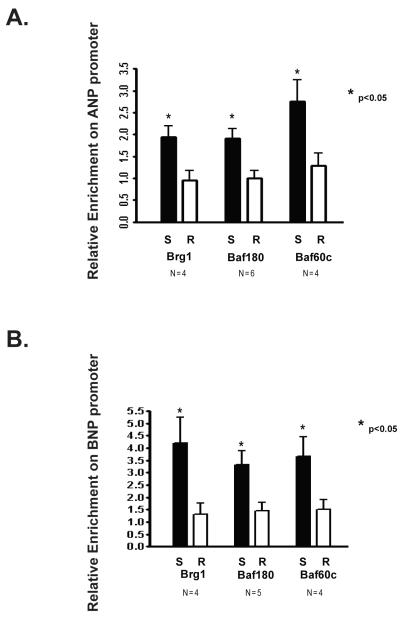
In order to determine if there is an association between salt induced cardiac hypertrophy and SWI/SNF recruitment to cardiac fetal gene promoters, we performed chromatin immunoprecipitations (ChIPs) to detect Brg1, Baf180 and Baf60c occupancy on the ANP and BNP promoters from left ventricles obtained from S and R rats fed a high salt diet. We found that enrichment of each of these SWI/SNF subunits was significantly greater at both promoters in ventricles from S rats compared to those from R rats (Fig. 3 A and B). Thus, in addition to high levels of SWI/SNF subunit expression, there is significantly enhanced recruitment of these proteins to fetal cardiac gene promoters in hypertrophic hearts from S rats compared with R rats.
Figure 3. Differential enrichment of SWI/SNF components on the ANP and BNP promoter in left ventricles from Dahl salt-sensitive and Dahl salt-resistant rats.
Male Dahl salt-sensitive (S) and salt-resistant (R) rats were given a 2% NaCl diet at 6 weeks of age for another 6 weeks. Chromatin immunoprecipitations (ChIPs) were performed on the left ventricles isolated from S and R rats. ChIP enrichment was quantified by qPCR, is expressed relative to IgG, and further normalized to a control region (the myelin protein zero intron). A. Relative enrichment of Brg1, Baf180, and Baf60c on the ANP promoter. B. Relative enrichment of Brg1, Baf180, and Baf60c on the BNP promoter. Bars represent the average obtained from independent experiments performed with the indicated number of rat hearts (N). Error bars represent SEM.
Fetal cardiac gene promoters are associated with accessible chromatin structure in hearts from genetically hypertensive S rats
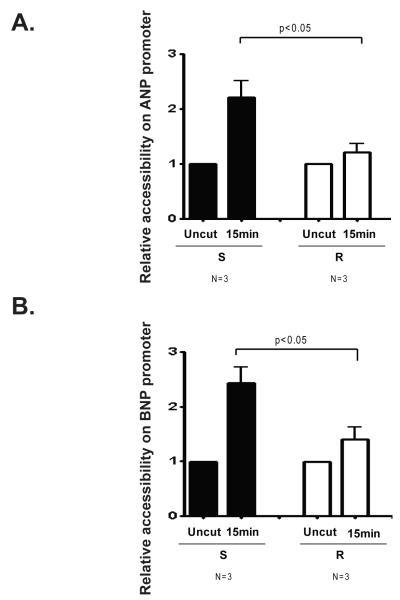
The correlation between high levels of ANP and BNP expression and high levels of SWI/SNF promoter occupancy of the ANP and BNP promoters in hypertrophic hearts from S rats indicated that SWI/SNF enzymes may activate the expression of these genes by remodeling chromatin structure. Therefore one would expect that the ANP and BNP promoters would exhibit highly accessible chromatin structure in hearts from S rats. To test this prediction, we digested nuclei from S and R rat left ventricles with micrococcal nuclease (MNase). MNase digests chromatin in regions that are nucleosome-free and can be used to determine the accessibility of chromatin structure. We found that the regions of the ANP and BNP promoters that were associated with SWI/SNF subunit occupancy were also more accessible to MNase I in hearts from S rats compared with those from R rats (Figure 4A and B). Thus, high levels of ANP and BNP expression in hypertrophic hearts from genetically hypertensive S rats is highly correlated with SWI/SNF recruitment and open chromatin structure.
Figure 4. Differential chromatin accessibility on the ANP and BNP promoters in left ventricles from Dahl salt-sensitive and Dahl salt-resistant rats.
Male Dahl salt-sensitive (S) and salt-resistant (R) rats were given a 2% NaCl diet at 6 weeks of age for another 6 weeks. Nuclei were isolated from the left ventricles of S and R rats and digested with MNase I. Accessibility was quantified by qPCR by calculating (CT undigested-CT digested), normalizing to undigested samples and also by further normalizing to the values obtained with primers to a control region (the myelin protein zero intron) after 5 and 15 minutes of MNase treatment. A. Chromatin accessibility on the ANP promoter. B. Chromatin accessibility on the BNP promoter. Bars represent the average obtained from independent experiments performed with the indicated number of rat hearts (N). Error bars represent SEM.
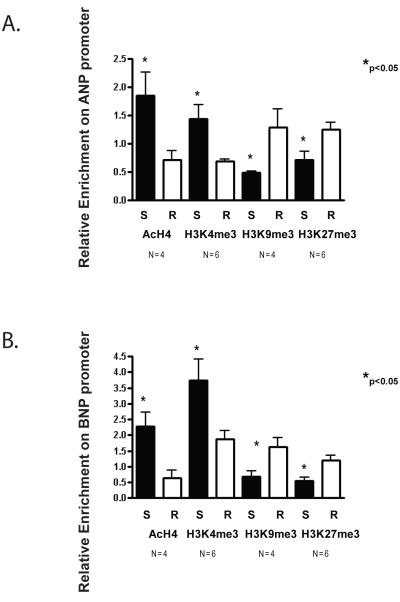
Covalent histone modifications on fetal cardiac gene promoters in hearts from genetically hypertensive S rats are consistent with transcriptionally active chromatin structure
Activation of ANP and BNP expression in immortalized cardiomyocytes is associated with increased histone H4 acetylation (AcH4) and histone H3 at lysine 4 dimethylation (H3K4me2) at these loci (Bingham et al., 2007). Furthermore, genome wide changes in histone H3 tri-methylation at lysine 4 (H3K3me3) and lysine 9 (H3K9me3) have been reported to occur in failing hearts from Dahl salt-sensitive rats switched from a low salt to a high salt diet (Kaneda et al., 2009). To determine if SWI/SNF occupancy on the ANP and BNP promoters correlates with covalent histone modifications that are indicative of active gene expression, we assayed AcH4, H3K4me3, and H3K9me3 as well as histone H3 tri-methylation at lysine 27 (H3K27me3) levels on the ANP and BNP promoters by ChIP analysis. Levels of AcH4 and H3K4me3 on these promoters were significantly higher in S rat hearts than in R rat hearts (Fig. 5 A and B) whereas H3K27me3 and H3K9me3 levels were significantly lower in S rat hearts than in R rat hearts (Fig. 5C and D). Thus, consistent with the observed higher mRNA levels of ANP and BNP, greater enrichment of SWI/SNF subunits, and significantly more accessible chromatin structure, histone modifications indicative of active transcription were also highly enriched on the ANP and BNP promoters in S rats compared to R rats, whereas inhibitory histone modifications were reduced.
Figure 5. Differential covalent histone modifications on the ANP and BNP promoters in left ventricles from Dahl salt-sensitive and Dahl salt-resistant rats.
Male Dahl salt-sensitive (S) and salt-resistant (R) rats were given a 2% NaCl diet at 6 weeks of age for another 6 weeks. Chromatin immunoprecipitations (ChIPs) were performed on the left ventricles isolated from S and R rats. ChIP enrichment was quantified by qPCR, is expressed relative to IgG, and further normalized to a control region (the myelin protein zero intron). A. ChIP analysis of tetra acetylated H4 (AcH4), H3 lysine 4 tri-methylation (H3K4me3), H3 lysine 9 tri-methylation (H3K9me3) and H3 lysine 27 tri-methylation (H3K27me3) on the ANP promoter B. ChIP analysis of tetra-acetylated H4(AcH4), H3 lysine 4 tri-methylation (H3K4me3), H3 lysine 9 trimethylation (H3K9me3) and H3 lysine 27 tri-methylation (H3K27me3) on the BNP promoter. Bars represent the average obtained from independent experiments performed with the indicated number of rat hearts (N). Error bars represent SEM.
DISCUSSION
In this study we demonstrated that expression of Brg1, Baf180, and Baf60c, components of the SWI/SNF complex required for heart development, are significantly higher in hypertrophic hearts obtained from genetically hypertensive S rats that were fed a high salt diet compared with hearts from genetically normotensive R rats fed the same diet. We also observed that enrichment of these SWI/SNF subunits on the promoters of two genes expressed in the fetal cardiac gene program, ANP and BNP, is significantly higher in hypertrophic hearts from S rats compared to R rats. Furthermore, the ANP and BNP promoters from S rats are in a chromatin configuration that is highly accessible and associated with covalent histone modifications that are indicative of highly active transcription. These findings are novel in linking SWI/SNF enzymes and open chromatin structure with the critical changes in gene expression that promote cardiac hypertrophy in the Dahl genetic model of hypertension.
Our findings are consistent with prior work in demonstrating a role for Brg1, an ATPase subunit of the SWI/SNF chromatin remodeling enzymes, in heart muscle development and disease (Hang et al., 2010). In this previous study, Brg1 expression in the myocardium of both mouse and human hypertrophic hearts was found to be increased compared to normal adult hearts (Hang et al., 2010). Importantly, in a surgical model of hypertrophy, conditional deletion of Brg1 in the adult myocardium significantly attenuated left ventricular hypertrophy (Hang et al., 2010). Brg1 was found to activate fetal genes and repress adult genes in the hypertrophic heart, thus providing strong evidence that Brg1 plays a causal role in cardiac hypertrophy. A second study implicated the Brm ATPase as well as Brg1, and a different SWI/SNF subunit, Baf57, in cardiac hypertrophy. Interestingly, this previous study also demonstrated that administration of a histone deacetylase inhibitor prevented SWI/SNF subunit recruitment to fetal cardiac promoters and prevented left ventricular hypertrophy, suggesting that SWI/SNF enzymes may be effective therapeutic targets.
Cardiac hypertrophy in humans is often associated with chronic hypertension, a clinical condition that is associated with dietary sodium consumption (Joe and Shapiro, 2012). Understanding salt-sensitive hypertension in humans has been facilitated by the use of inbred rat models, including the Dahl salt-sensitive and salt-resistant rats. Thus, we complement the existing genetic and physiological information that has been gathered from this extremely important model with epigenetic data and provide insight into the mechanisms by which salt induced hypertension leads to cardiac hypertrophy. Further investigation will be required to assess any causal role of SWI/SNF components in promoting cardiac hypertrophy in this rat model of hypertension and to investigate the therapeutic implications.
Acknowledgments
GRANT SUPPORT: ILD was supported by National Institute of Health ARO59379. BJ was supported by National Institute of Health HL020176 and HL112641.
LITERATURE CITED
- Barry SP, Davidson SM, Townsend PA. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol. 2008;40(10):2023–2039. doi: 10.1016/j.biocel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Weeks KL, Pretorius L, McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128(1):191–227. doi: 10.1016/j.pharmthera.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Bingham AJ, Ooi L, Kozera L, White E, Wood IC. The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes. Mol Cell Biol. 2007;27(11):4082–4092. doi: 10.1128/MCB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Kiriazis H, Gao XM, Du XJ, El-Osta A. Cardiac genes show contextual SWI/SNF interactions with distinguishable gene activities. Epigenetics. 2011;6(6):760–768. doi: 10.4161/epi.6.6.16007. [DOI] [PubMed] [Google Scholar]
- Cruickshank M, Fenwick E, Abraham LJ, Ulgiati D. Quantitative differences in chromatin accessibility across regulatory regions can be directly compared in distinct cell-types. Biochem Biophys Res Commun. 2008;367(2):349–355. doi: 10.1016/j.bbrc.2007.12.121. [DOI] [PubMed] [Google Scholar]
- Dahl LK, Heine M, Tassinari L. Effects of chronia excess salt ingestion. Evidence that genetic factors play an important role in susceptibility to experimental hypertension. J Exp Med. 1962a;115:1173–1190. doi: 10.1084/jem.115.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl LK, Heine M, Tassinari L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature. 1962b;194:480–482. doi: 10.1038/194480b0. [DOI] [PubMed] [Google Scholar]
- de La Serna IL, Carlson KA, Hill DA, Guidi CJ, Stephenson RO, Sif S, Kingston RE, Imbalzano AN. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20(8):2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- El-Osta A. Remodeling is at the heart of chromatin: the heartaches of chromatin. Epigenetics. 2011;6(7):884–887. doi: 10.4161/epi.6.7.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Cooper RS, Bilal U, Fuster V. Challenges and opportunities for cardiovascular disease prevention. Am J Med. 2011;124(2):95–102. doi: 10.1016/j.amjmed.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- Gaspar-Pereira S, Fullard N, Townsend PA, Banks PS, Ellis EL, Fox C, Maxwell AG, Murphy LB, Kirk A, Bauer R, Caamano JH, Figg N, Foo RS, Mann J, Mann DA, Oakley F. The NF-kappaB subunit c-Rel stimulates cardiac hypertrophy and fibrosis. Am J Pathol. 2012;180(3):929–939. doi: 10.1016/j.ajpath.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008;319(2):258–266. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- Joe B, Shapiro JI. Molecular mechanisms of experimental salt-sensitive hypertension. J Am Heart Assoc. 2012;1(3):e002121. doi: 10.1161/JAHA.112.002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda R, Takada S, Yamashita Y, Choi YL, Nonaka-Sarukawa M, Soda M, Misawa Y, Isomura T, Shimada K, Mano H. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14(1):69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2010;29(1):81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei I, Gao X, Sham MH, Wang Z. SWI/SNF component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J Biol Chem. 2012 doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Maillet M, van Berlo JH, Molkentin JD. Molecular basis of physiological heart growth: fundamental concepts and new players. Nat Rev Mol Cell Biol. 2013;14(1):38–48. doi: 10.1038/nrm3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Olson EN. Cardiac histone acetylation--therapeutic opportunities abound. Trends Genet. 2004;20(4):206–213. doi: 10.1016/j.tig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Mu S, Shimosawa T, Ogura S, Wang H, Uetake Y, Kawakami-Mori F, Marumo T, Yatomi Y, Geller DS, Tanaka H, Fujita T. Epigenetic modulation of the renal beta-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat Med. 2011;17(5):573–580. doi: 10.1038/nm.2337. [DOI] [PubMed] [Google Scholar]
- Patten RD, Hall-Porter MR. Small animal models of heart failure: development of novel therapies, past and present. Circ Heart Fail. 2009;2(2):138–144. doi: 10.1161/CIRCHEARTFAILURE.108.839761. [DOI] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167(8):4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev. 2000;80(1):135–172. doi: 10.1152/physrev.2000.80.1.135. [DOI] [PubMed] [Google Scholar]
- Saladi SV, Wong PG, Trivedi AR, Marathe HG, Keenen B, Aras S, Liew ZQ, Setaluri V, de la Serna IL. BRG1 promotes survival of UV-irradiated melanoma cells by cooperating with MITF to activate the melanoma inhibitor of apoptosis gene. Pigment Cell Melanoma Res. 2013;26(3):377–391. doi: 10.1111/pcmr.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, Zhou YQ, Yeh RF, Henkelman RM, Harvey RP, Metzger D, Chambon P, Stainier DY, Pollard KS, Scott IC, Bruneau BG. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang C, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18(24):3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]