Abstract
The small Maf proteins, MafF, MafG, and MafK, possess a leucine zipper (Zip) domain that is required for homodimer or heterodimer complex formation with other bZip transcription factors. In this study we sought to determine the identity of the specific constituent that collaboratively interacts with Nrf2 to bind to the Maf recognition element in vivo. Studies in vitro suggested that Nrf2 forms heterodimers with small Maf proteins and then bind to Maf recognition elements, but the bona fide partner molecules supporting Nrf2 activity in vivo have not been definitively identified. Nrf2 activity is usually suppressed by a cytoplasmic repressor, Keap1, so disruption of the keap1 gene causes constitutive activation of Nrf2. Nrf2 hyperactivity results in hyperproliferation of keratinocytes in the esophagus and forestomach leading to perinatal lethality. However, simultaneous disruption of nrf2 rescued keap1-null mice from the lethality. We exploited this system to investigate whether small Mafs are required for Nrf2 function. We generated keap1 and small maf compound mutant mice and examined whether keratinocyte abnormalities persisted in these animals. The data show that loss of mafG and mafF in the keap1-null mice reversed the lethal keratinocyte dysfunction and rescued the keap1-null mutant mice from perinatal lethality. This rescue phenotype of mafG::mafF::keap1 triple compound mutant mice phenocopies that of the nrf2::keap1 compound mutant mice, indicating that the small Maf proteins MafG and MafF must functionally cooperate with Nrf2 in vivo.
Acentral issue in deciphering the regulatory mechanisms mediated by the activity of transcription factors is how to best evaluate the in vivo contribution of each protein–protein and transcription factor–DNA interaction that is defined in vitro. Transcription factors that interact with Maf recognition elements (MARE) possess a basic region-leucine zipper (bZip) domain and form dimers that can be characterized by a potentially enormous combinatorial array (1). In vitro analysis has shown that MARE binding complexes consist mainly of: (i) four large Maf or three small Maf protein homodimers; (ii) heterodimer complexes containing a small Maf with any of six different Cap-N-Collar (CNC) family proteins; and (iii) homo- or heterodimers composed of Jun and Fos family members (2–4). To elucidate MARE-dependent gene regulatory mechanisms, we need to identify the major participants among all these possible interacting molecules in an in vivo context.
The small Maf proteins, MARE-binding components that were originally identified as cellular homologs of the v-maf oncogene (4–6), dimerize among themselves and with other bZip factors, usually CNC or Bach family proteins (7–11). The small Maf family consists of only three members, MafF, MafG, and MafK, but to date, other than their differential tissue distribution (12), no functional differences among the three have been revealed. The CNC family includes NF-E2 p45, Nrf1, Nrf2, and Nrf3 (7, 9, 13, 14), and Bach family proteins are closely related to CNC members (10). While small Maf proteins lack any recognizable transcriptional effector domains, CNC and Bach families possess transactivation or -repression domains unique to each molecule. Through heterodimerization, the small Maf protein confers DNA-binding specificity to its CNC or Bach partner molecule on the MARE sequence, and enables these heterodimers to execute differential activating or repressing activities as dictated by their encoded functional domains.
The Maf proteins recognize either a T-MARE, containing a TPA responsive element (TRE), or a C-MARE, containing a cAMP responsive element (CRE) as a core sequence. In these MAREs, the core consensus motifs are flanked on each side by three conserved residues “TGC” and “GCA” at the 5′ and 3′ ends, respectively. The DNA binding specificity of Maf proteins is achieved through their inherent recognition of these flanking sequences, whereas the other bZip factors, such as Nrf2 and Fos, recognize primarily the TRE or CRE core sequences. A previous NMR study revealed that the structural basis for the unique “GC” requirement of Maf proteins for DNA binding is caused by the presence of an extended homology region, which is conserved only within the Maf family (15).
Germ-line mutagenesis of the nrf2 gene revealed that Nrf2 is an essential component for antioxidant and detoxification enzyme gene expression (16). Nrf2 transcriptional activity is controlled by an interaction between Nrf2 and the cytoplasmic regulatory protein Keap1 (17). When cells are exposed to electrophiles or reactive oxygen species (ROS), Nrf2 is released from Keap1 cytoplasmic capture, leading to its translocation to the nucleus, where Nrf2 activates transcription of target genes. The marked susceptibility of nrf2-null mutant mice to the toxicity of electrophiles and ROS demonstrates the importance of Nrf2 for protection against oxidative stress (18–20). We therefore generated keap1-null mutant mice, anticipating that Nrf2 might be constitutively activated in the absence of Keap1, thereby conferring resistance to electrophilic stress. To our surprise, the keap1-null mutant mice died before weaning due to a hyperkeratotic proliferative disorder (21). The cornified layers of esophageal and forestomach stratified squamous epithelia were abnormally thickened, thereby obstructing the lumen. We found that this epithelial phenotype was completely rescued by the additional disruption of nrf2, indicating that the keap1-null phenotype reflects a gain of Nrf2 function.
There has been some uncertainty regarding the identity of the heterodimeric partner molecule of Nrf2 in vivo. In vitro studies have shown strong DNA binding activity of Nrf2-small Maf heterodimers (16, 22, 23), which supported our contention that this complex actually functions as a major transcriptional activator in vivo. However, although strong transactivation activity was observed when Nrf2 was overexpressed in culture cells, addition of small Maf to the transfection reaction led to reporter gene repression in most cases (22, 24–26). Hence, the question we sought to answer was whether or not the Nrf2/small Maf heterodimer is the functionally active species that acts at MAREs in vivo.
Alternative candidates for heterodimeric partner molecule of Nrf2 have been suggested. For example, c-Jun and ATF-4 were reported to cooperate with Nrf2 for gene activation in transfecto (27, 28). However, because disruption of c-jun or atf-4 does not cause a defect similar to that observed in nrf2-null mutant animals (29–31), it remains to be clarified whether these factors can heterodimerize with Nrf2 to transduce transcriptional responses from MAREs in vivo. Similarly, a functional contribution of small Maf proteins to Nrf2 activity has not been well documented in small maf mutant mice.
Because disruption of the keap1 gene causes severe dysfunction of keratinocytes that leads to perinatal lethality, but simultaneous disruption of nrf2 rescued keap1-null mice from the lethality, we exploited this compound knockout–rescue approach to investigate whether small Mafs actually function cooperatively with Nrf2 and activate transcription in vivo. To this end, we generated keap1::small maf compound knockout mutant mice and examined whether a reduction in small Maf activity, as does the loss of Nrf2, mitigates the keap1-null phenotype. We show here that simultaneous disruption of the mafG and mafF genes rescued the keap1-null pups from perinatal lethality, allowing them to survive to adulthood. Thus, the small maf::keap1 mutant mice phenocopy the rescue phenotype of nrf2::keap1 compound mutant mice, demonstrating that the small Maf proteins cooperatively function with Nrf2 in vivo.
Materials and Methods
Generation of the Small maf::keap1 Compound Mutant Mice. Germline mutagenesis of the murine mafF, mafG, mafK, nrf2, and keap1 genes has been described (12, 16, 21, 32). All of the mice examined in this study were of mixed genetic background with contributions from 129Sv/J, C57BL/6J, and ICR. Genotypes were determined by PCR. The body weight of each mouse was measured weekly. More than three independent animals of each genotype were first weighed on postnatal day 7, and then followed to the 6th week.
Histological Analysis. Two-day-old pups, 10- to 12-day-old pups, and 4-month-old mice were killed, and the forestomach was dissected. Samples for staining with hematoxylin and eosin were fixed in 3.7% formaldehyde overnight and then embedded in paraffin. LacZ staining was performed as described (33). Samples for immunostaining with antibodies against Nrf2 or keratin 6 were fixed in PBS containing 1% formaldehyde, 0.2% glutaraldehyde, and 0.02% Nonidet P-40 for 30 min, embedded in OCT compound (Tissue-tek, Sakura Finetechnical, Chuo-ku, Tokyo), followed by frozen sectioning with a cryostat. The antibody against Nrf2 (C-20, Santa Cruz Biotechnology) was used at a 1:400 dilution; immunoreactivity was visualized with an avidin-biotin-peroxidase kit (Vector Laboratories). The antibody recognizing keratin 6 (PRB-169P, Covance, Princeton) was used at a 1:500 dilution.
Quantitative Real-Time PCR. Total RNA was extracted from the forestomach of 10- to 12-day-old pups using ISOGEN (Nippon Gene, Toyama, Japan). Random cDNA was synthesized from the isolated RNAs, and real-time PCR (ABI PRISM 7700) was performed as described (12) with minor modifications. To measure the copy number of each mRNA, plasmids harboring each cDNA were used as standards. Oligonucleotide sequences used for detecting MafF mRNA are available upon request.
RNA Blot Analysis. Total RNA was prepared as described above. Total RNA (2 μg per lane) was used for electrophoresis, followed by hybridization to radiolabeled probes. The keratin 6 probe was generated by PCR (primer sequences are available upon request). The PCR product was cloned, sequenced, and used as a probe for detecting the transcripts of both keratin 6 genes (i.e., keratin 6a and keratin 6b).
Results
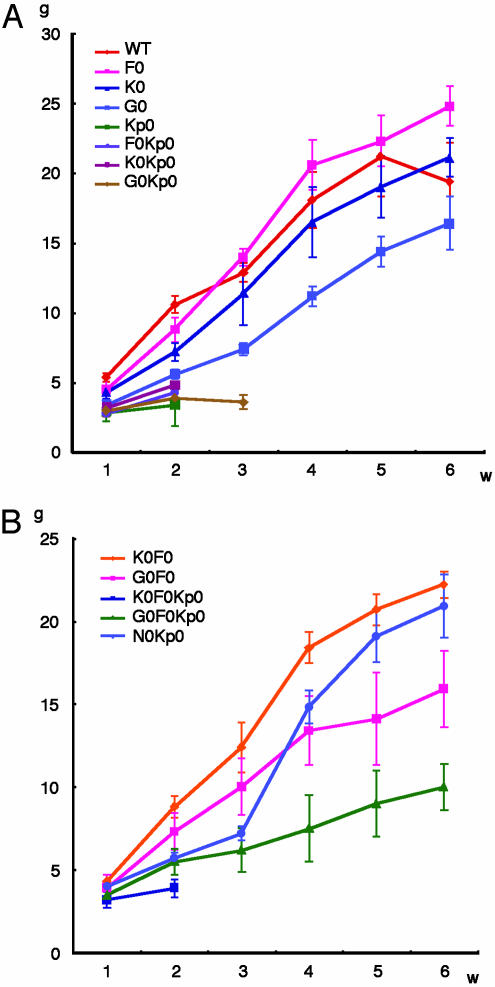
Simultaneous Loss of mafG and mafF Rescues keap1-Null Mutant Postnatal Lethality. To ask whether a reduction in small Maf activity mitigates the keap1-null phenotype, we crossed small maf mutant mice with keap1 mutants to generate compound mutant animals. When mafK or mafF was deleted in addition to keap1, no pups survived beyond weaning (Fig. 1A; precisely the same phenotype as in the keap1 mutants), but the life span of mafG::keap1 compound mutant mice was approximately one week longer than mice bearing only the keap1 mutant alleles (Fig. 1A, G0Kp0 mice), suggesting that the mafG contribution to keratinocytic homeostasis is greater than that of either MafK or MafF.
Fig. 1.
Small maf::keap1 compound mutant mice survive beyond weaning. (A) Body weight change for mice bearing single small maf gene disruptions in the keap1-null background. Deletion of either mafK or mafF did not extend the lifespan of keap1-null mutant pups. When mafG alone was disrupted in addition to keap1, pups survived one week longer. (B) Body weight change for mice with compound small maf gene disruptions in the keap1-null mutant background. Simultaneous deletion of mafG and mafF rescued the lethality of keap1-null mutant pups. The body weight of nrf2::keap1 mice was examined as a control. WT indicates wild-type; F0, mafF-/-; K0, mafK-/-; G0, mafG-/-; N0, nrf2-/-; Kp0, keap1-/-.
Because the results of small maf compound mutant analyses indicate that three small Mafs are capable of compensating for one another (34), we next further reduced the number of active small maf alleles in the keap1-null background. Whereas mice bearing disruptions in all three small maf genes might have been preferred for this analysis, mice lacking all small Mafs die by midgestation (unpublished observations). Similarly, mafG::mafK compound mutant mice expire before weaning (34). We therefore examined instead two genotypes that were healthy and fertile (i.e., mafG::mafF and mafF::mafK compound mutants) in combination with the mutant keap1-null alleles (Fig. 1B). Whereas disruption of both mafK and mafF did not rescue keap1 mutant from lethality (K0F0Kp0 mice), when the mafG and mafF genes were simultaneously deleted in the keap1-null background the mice survived for more than four months (G0F0Kp0 mice in Fig. 1B).
Because we previously found that the simultaneous deletion of nrf2 with keap1 genes almost completely reversed the keap1-null mutant phenotype (21), we compared the body weight gain of G0F0Kp0 mice with that of nrf2::keap1 compound mutant mice (N0Kp0 mice) to evaluate the efficiency of the rescue by simultaneous mafG and mafF deletion. Whereas N0Kp0 mice showed a similar body weight gain to that of wild-type mice, G0F0Kp0 mice were much smaller than both of these at 6 weeks after birth (Fig. 1). G0F0Kp0 mice were also smaller than G0F0 mice (G0F0Kp0 vs. G0F0 in Fig. 1B), indicating that the effect of the Keap1 deficiency is only partially overcome in G0F0Kp0 mice. These results demonstrate that Keap1 deficiency provokes constitutive activation of Nrf2 and the growth retardation of mice, but this deleterious effect of Keap1 loss can be partially circumvented by the simultaneous loss of MafG and MafF.
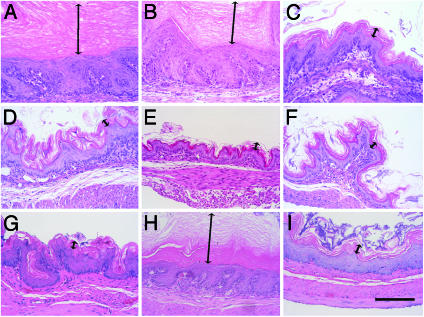
Improvement of Hyperkeratosis in Forestomach and Esophagus of mafG::mafF::keap1 Triple Mutant Mice. We next performed histological analysis of the forestomach and esophagus of the rescued triple compound mutant (i.e., G0F0Kp0) animals at 10 days after birth (Fig. 2 and data not shown). Heavily thickened cornified layers, convoluted basal layers, and thickened spinous and granular layers were observed in the forestomach of keap1-null mutant and F0Kp0 mice (Fig. 2 A and B), whereas G0F0Kp0 mice displayed a normal stratified squamous epithelium (Fig. 2C), similar to the control samples (Fig. 2 D–F).
Fig. 2.
Histological examination of forestomach. Hematoxylin/eosin staining of 10-day-old forestomach thin sections from Kp0 (A), F0Kp0 (B), G0F0Kp0 (C), WT (D), F0 (E), and G0F0 (F) pups are shown. Sections of the forestomach from 4-month-old mice of WT (G), G0F0Kp0 (H), and N0Kp0 (I) genotypes are also presented. Double-ended arrows indicate the cornified layer. (Scale bar, 150 μm.)
Because the body weight difference was apparent between G0F0Kp0 and N0Kp0 mice at 6 weeks of age, we hypothesized that the constitutive elevation of Nrf2 activity in the keap1-null background was only partially overcome in G0F0Kp0 mice and that the rescue of the keap1-null phenotype might be incomplete. In support of this contention, we found that the forestomach epidermal layers of G0F0Kp0 mice were hyperkeratotic, whereas those of the N0Kp0 forestomach were almost indistinguishable from wild type (Fig. 2 G–I). We envisage that in contrast to keap1-null mutant mice, thickening of the cornified epithelial layers develop more gradually in the G0F0Kp0 forestomach as a consequence of residual elevated Nrf2 activity.
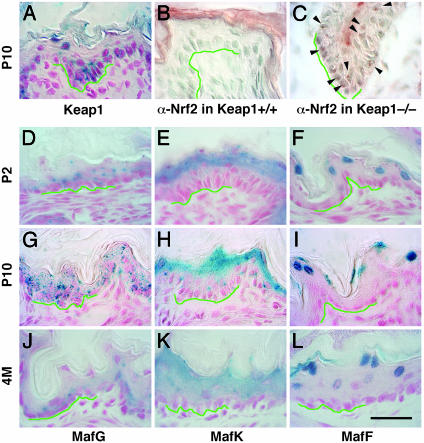
Keratinocyte Abnormality Is Involved in the Hyperkeratosis Observed in keap1-Null Mice. To determine whether diminished Keap1 expression in keratinocytes might lead to the observed hyperkeratosis, we examined keap1 gene expression in the stratified squamous epithelium by monitoring the expression of the LacZ gene that was inserted into the keap1 locus when the gene was disrupted (21). β-Galactosidase activity was observed primarily in keratinocytes of whole layers of stratified squamous epithelium, from the immature cells in basal layers to the granular layers composed of more differentiated cells (Fig. 3A). The green lines in Fig. 3 define the position of the basement membrane. These data indicate that Keap1 is predominantly expressed in keratinocytes, regardless of differentiation stage.
Fig. 3.
Expression profiles of Keap1, Nrf2, and small Mafs in the forestomach. (A) LacZ staining of squamous cell epithelia in the forestomach of keap1 heterozygous mutant mice. Blue staining is observed in LacZ-expressing cells. (B and C) Immunohistochemistry with anti-Nrf2 antibody. Brown punctate staining, indicated by arrowheads, is observed within nuclei of keap1-null mutant cells (C), whereas no staining develops in wild-type keratinocytes (B). Nonspecific staining is observed in the cornified epithelial layers. (D–L) LacZ staining of squamous cell epithelia of the forestomach in sections prepared from mafG (D, G, and J), mafK (E, H, and K), and mafF (F, I, and L) heterozygous mutant mice. Samples were prepared from 2-day-old (D–F), 10-day-old (G–I), or 4-month-old (J–L) mice. The green lines indicate the position of basement membranes. (Scale bar, 30 μm throughout.)
We next examined Nrf2 activation in the unstimulated, but pathologically expanded, forestomach epithelium of keap1-null mutant mice by examining the epithelial cells for nuclear accumulation of Nrf2 protein. Keap1 sequesters Nrf2 in the cytoplasm and prevents it from traversing into the nucleus (17). Recent reports showed that Nrf2 is constantly degraded by the proteasome when cells are not stimulated (35–38), thus keeping Nrf2 concentration low, whereas Nrf2 is translocated into the nucleus and is stabilized upon exposure to electrophilic reagents or oxidative stress. We performed immunohistochemical examination of the keratinocytes of keap1-null animals using an antibody that specifically recognizes Nrf2. As expected, intranuclear foci were observed in the forestomach epithelium of the keap1-null mutant mice (Fig. 3C, arrowheads); also as anticipated, no Nrf2 was detected in comparable cells of a wild-type littermate (Fig. 3B), consistent with the notion that proteasome-mediated turnover leads to rapid degradation of Nrf2 in the unstimulated cytoplasm. These results thus demonstrate that the control processes mediated by Nrf2 and Keap1 are actually extant in forestomach keratinocytes. The results also suggest that gene dysregulation within keratinocytes is involved in the hyperkeratosis observed in keap1-null mutant mice.
Small Maf Proteins Are Expressed in Forestomach Keratinocytes. The rescue experiments described above suggest that diminished small Maf activity is required for correction of the keap1-induced keratinocytic abnormality, and that the small Mafs must therefore act in the same genetic pathway as Nrf2 to execute the transcriptional program in keratinocyte differentiation (Fig. 1). However, none of the small maf mutant phenotypes described to date include keratinocytic abnormalities; no report has emerged demonstrating any functional contribution of small Mafs to keratinocyte biology.
Therefore, we examined the expression of the three small maf genes in the forestomach epithelium by monitoring LacZ activity in the small maf mutants, because each of the small maf knockouts was generated by simultaneous insertion of a LacZ gene. While we inserted the normal LacZ gene into the mafK and mafG loci, the mafF insertion contained lacZ linked to a nuclear localization signal (NLS). At 10 days after birth, β-galactosidase activity was observed throughout the layers of stratified squamous epithelium in the mafG mutant mice (Fig. 3G). Expression of mafK is weaker in the basal cells and stronger in more differentiated cells of the spinous and granular layers (Fig. 3H). Interestingly, mafF is expressed almost exclusively in the most differentiated granular layer cells (Fig. 3I). β-Galactosidase activity was localized in the cytoplasm of keratinocytes in mafK and mafG mutant mice, which gave rise to both diffuse and punctate staining (Fig. 3 G for mafG and H for mafK), but the activity was exclusively nuclear in keratinocytes of mafF mutant mice (Fig. 3I). These expression patterns were reproducible when 2-day-old pups and 4-month-old adult mice were analyzed for each small maf gene (Fig. 3 D–F and J–L). β-Galactosidase staining was especially intense in the granular layer of mafK mutant mice, regardless of developmental stage (Fig. 3 E, H, and K). The abundance of all three small maf gene mRNAs was almost constant from neonatal stages to adulthood, when monitored by quantitative real-time PCR (data not shown). Thus, all three small Maf proteins are expressed in the forestomach squamous epithelium, and the expression patterns are indeed overlapping with those of Nrf2 and Keap1.
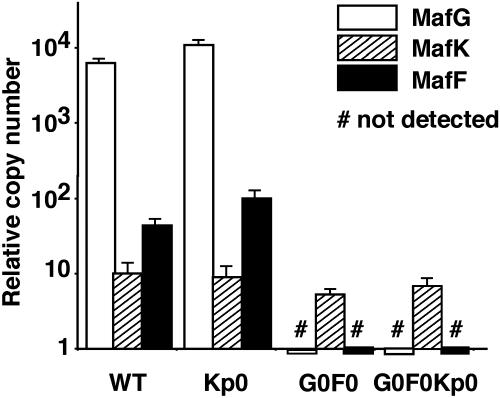
MafG Is a Major Small Maf Protein Species in Keratinocytes. To determine whether there were any correlations between the mRNA abundance and the functional contribution of each small maf gene, the copy numbers of each small Maf mRNA expressed in the forestomach at 10 days after birth were quantified by quantitative real-time PCR, and MafG mRNA was found to be the most abundant small Maf (Fig. 4). The observed abundant MafG expression in the forestomach shows very good agreement with the data in the small maf gene knockout–rescue experiments (Fig. 1), which suggest that MafG makes the largest contribution to normal keratinocyte function among the three small Maf proteins. We conclude that mafG and mafF disruption attained a significant enough reduction in the total amount of small Maf protein that there was no longer sufficient Maf activity present in cells to effectively execute Nrf2 function.
Fig. 4.
Quantification of small Maf mRNA abundance in forestomach. cDNA was synthesized from total RNA prepared from WT, Kp0, G0F0, and G0F0Kp0 forestomach at 10 days after birth. MafG, MafK, and MafF mRNA levels were quantified by quantitative real-time PCR using a plasmid containing each cDNA as the abundance standard.
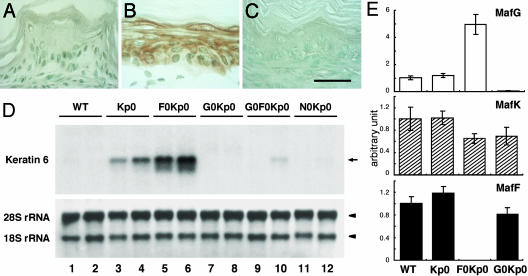
Increased Keratin 6 Expression Returns to Normal Levels in the Forestomach of mafG::mafF::keap1 Triple Mutant (Rescued) Mice. Keratin 6 is strongly induced in the keap1-null mutant esophagus, and it has been suggested to be a direct Nrf2 target gene in keratinocytes. The expression of keratin 6 was dramatically induced in the forestomach epithelium of the keap1-null mice, judging from the strong immunohistochemical anti-keratin 6 staining (Fig. 5 A and B). Consistent with the distribution of constitutively activated Nrf2 in the keap1 mutant mice (see Fig. 3C), keratin 6 was induced in all keratinocyte layers (Fig. 5B). To investigate whether increased keratin 6 expression in the keap1-null mutant forestomach is caused by constitutive activation of Nrf2, we analyzed keratin 6 expression in N0Kp0 compound mutant mice. Keratin 6 mRNA was abundant in the keap1 mutant (Fig. 5D, lanes 3 and 4), but was scarcely detectable in either the wild-type or N0Kp0 animals (Fig. 5D, lanes 1, 2, 11, and 12), thus confirming the Nrf2-dependency of keratin 6 gene activation.
Fig. 5.
Expression of keratin 6 as a marker of Nrf2-mediated transcriptional activity. (A–C) Immunohistochemistry with anti-keratin 6 antibody. Keratin 6 is highly expressed in keap1-null keratinocytes in the forestomach (B), whereas no signals were observed in the wild-type (A) or rescued G0F0Kp0 (C) mice. (Scale bar, 30 μmin A–C.) (D) Keratin 6 expression levels were examined by RNA blot analysis. Total RNA prepared from wild-type (lanes 1 and 2), Kp0 (lanes 3 and 4), F0Kp0 (lanes 5 and 6), G0Kp0 (lanes 7 and 8), G0F0Kp0 (lanes 9 and 10), and N0Kp0 (lanes 11 and 12) mice forestomachs are shown. The arrow and arrowheads indicate keratin 6 mRNA and ribosomal RNAs (18S and 28S), respectively. (E) Relative expression levels of the three small maf genes in the forestomach of wild-type, Kp0, F0Kp0, and G0Kp0 at 10 days after birth (compared with the levels in wild-type mice).
We then examined the expression of keratin 6 in the rescued G0F0Kp0 mice. Application of anti–keratin 6 antibody did not generate a signal in the forestomach epithelial layers of G0F0Kp0 mice (Fig. 5C), and, as in the N0Kp0 mutant, the G0F0Kp0 mutant forestomach expressed the same low levels of keratin 6 mRNA as wild-type mice (Fig. 5D, lanes 9 and 10). These results thus demonstrate that the small Maf proteins are required for Nrf2-dependent transcriptional activation of the keratin 6 gene.
Although the double deletion of mafF and mafG rescued the lethality of keap1 null mutants, mafF deletion alone did not affect the lifespan of keap1-null pups (F0Kp0 in Fig. 1), and ablation of mafG extended their lifespan by only one week (G0Kp0 in Fig. 1). These results suggested that the rescue efficiency of each compound mutant mouse inversely correlates with the remaining small Maf activity; Kp0 mice possess full activity, and small Maf abundance in keratinocytes appears to be reduced in the order: F0Kp0 > G0Kp0 > G0F0Kp0. Hence we hypothesized that the expression of keratin 6 mRNA would be higher in the order: Kp0 > F0Kp0 > G0Kp0 > G0F0Kp0. In the hope of detecting graded expression of keratin 6 mRNA, we further examined G0Kp0 and F0Kp0 mice.
Contrary to our expectation, keratin 6 mRNA was hardly detectable except in the Kp0 and F0Kp0 mutants (Fig. 5D, lanes 1, 2, and 7–12). Considering the similarity in low-level expression of keratin 6 in G0Kp0 and G0F0Kp0 mice, some additional factors may be involved in the large difference in weight gain and viability between the mice of two genotypes. A second unexpected result was that keratin 6 is more abundant in mafF::keap1 double mutant than keap1 single mutant forestomach (Fig. 5D, lanes 3–6). Interestingly, when small maf expression levels were examined, mafG was found to be highly induced in mafF::keap1 double mutant forestomach (Fig. 5E).
Discussion
It has been postulated that the small Maf proteins function as major heterodimeric partner molecules of Nrf2 based principally on in vitro DNA binding evidence. Heterodimer formation between Nrf2 and the small Mafs was detected by using bacterially expressed proteins or in vitro translated proteins (14, 16, 22, 24, 28, 39, 40), and substantial efforts have been made to document endogenous Nrf2-small Maf complexes capable of interacting with MAREs (23, 25, 41, 42). Although these studies have shown Nrf2-small Maf heterodimer formation, the question remained whether Nrf2-small Maf heterodimer actually functions as a transcription activator or whether Nrf2 might require an alternative partner to activate MARE-dependent target genes, because in many transfection assays cooperative activation of Nrf2 and small Mafs was not observed (22, 24–26). Hence the aim of this study was to test the contention that small Mafs cooperate with Nrf2 to activate transcription in an in vivo experimental system.
To address this issue, we generated keap1 and small maf compound gene knockout mice and asked whether the loss of one or two small maf genes in concert with the keap1 gene affected the lethal phenotype of Keap1-null mutant mice caused by the hyperactivity of Nrf2. We found that loss of mafG and mafF indeed rescued the keap1-null mutant mice from perinatal lethality. Because the rescue phenotype of mafG::mafF::keap1 triple compound mutant mice is similar to that of the nrf2::keap1 compound mutant mice, we conclude that small Maf proteins are indispensable components for Nrf2-dependent transcription in vivo. These results further support the contention that Nrf2-small Maf heterodimers serve as indispensable transcriptional regulators of keratinocytic gene expression.
We suggested previously that loricrin and keratin 6 are potential Nrf2 target genes in keratinocytes (21). There is one potential MARE in loricrin “CCATGGTGACATAGCTTGAA”, and two MAREs in keratin 6, “TGATGGTGAGCTTGCAGAGT,” and “GTGTGGTGAGGGGGCACACA,” ≈40 bp and 300 bp 5′ to the TATA boxes, respectively (nucleotides corresponding to a typical T-MARE, “TGCTGAG/CTCAGCA,” are italicized). These sequences correspond closely to the consensus sequence of antioxidant responsive elements (ARE), well characterized Nrf2 target sites that exist in the regulatory regions of phase 2 detoxifying enzyme genes and oxidative stress-inducible genes (43). In this study, we monitored keratin 6 as a parameter for measuring Nrf2 transcriptional activity, because the expression profile of keratin 6 corresponds closely to the distribution of Keap1-Nrf2 system in forestomach (see Fig. 5 A and B). Importantly, the increase in expression of keratin 6 observed in the keap1 mutant background returned to normal levels by simultaneous deletion of either nrf2 or deletion of both mafG and mafF.
When we examined mafF::keap1 compound mutant forestomach in the hope of detecting an intermediate expression level of keratin 6, an unexpected result emerged, in that the level of keratin 6 mRNA in F0Kp0 pups was higher than in the Kp0 pups (Fig. 5D). One intriguing explanation for this result can be drawn from the unanticipated induction of mafG expression in the F0Kp0 forestomach (Fig. 5E). The regulatory mechanisms controlling mafG levels in the forestomach seem to be sensitive to the total amount of small Mafs, whereas those controlling mafK and mafF do not. MafG mRNA expression might be elevated because of compensatory mechanisms whose nature is unknown at present, but may involve autoregulation of a subset of the small Mafs. Consequently, more small Maf proteins are produced in the F0Kp0 forestomach, ending in higher expression of keratin 6 mRNA than in Kp0 mice. This result, although surprising, nonetheless further supports the contention that small Maf cooperatively activates Nrf2-dependent transcription in vivo.
Given the complexity of MARE-dependent gene regulation, germ-line mutagenesis and loss of function studies of each gene have proven to be a powerful approach to glean insight into the function of the molecules that interact with this site in vivo. Because small Maf proteins are capable of heterodimerizing with many bZip factors, including the CNC and Bach proteins, diminished small Maf abundance should be reflected by a functional defect in the activity of these multiple partner molecules. Megakaryocytic defects result as a consequence of defective p45 function or to a small Maf deficiency (32, 44–46). Similarly, aberrant constitutive induction of heme oxygenase-1 can be attributed to a functional defect of Bach1 with insufficient small Maf partner molecules (47, 48). The other phenotypes observed in compound small maf mutant mice are red cell membrane abnormalities and a profound neurological disorder (34, 48), neither of which have been encountered in the analysis of Nrf2-deficient mice. This observation likely indicates that the small Mafs also regulate batteries of genes that are not under Nrf2 regulatory influence. However, it does not exclude the possible involvement of Nrf2, because double or triple mutations including nrf2 and other CNC transcription factor genes may recapitulate these phenotypes.
We have shown in this study that both nrf2-null mutant mice and small maf compound null mutant mice confer a similar rescue phenotype to keap1-null mutant mice, demonstrating that Nrf2-small Maf heterodimers play indispensable roles in keratinocytic gene expression. Considering the fact that excess small Mafs have been shown to repress transcription by forming inactive homodimers, keratinocytic overexpression of small Mafs would also be predicted to rescue keap1-null lethality. A recent study showed, using chromatin immunoprecipitation, that Nrf2 and small Maf are recruited to a MARE element in the mouse quinone reductase gene promoter when the gene is activated (43). This result is in very good agreement with the cooperative gene activation model executed by Nrf2 and small Maf that we propose here. In addition to oxidative stress-inducible genes, Nrf2 has been recognized as a critical regulator of other biological processes, including wound healing (49), endoplasmic reticulum stress response (40), inflammation resolution (50), and apoptosis (51). We suggest that the contribution of small Mafs to each of these Nrf2-dependent processes should be individually evaluated; some of the processes may depend on both Nrf2 and small Mafs, whereas the others may be independent of any small Maf contribution. Finally, the compound knockout–rescue approach exploited in the present study is an effective system for evaluating the in vivo contribution of test regulatory factors to Nrf2 activity, as long as mice deficient in these test factors are available. It would be interesting to apply this system to other candidate molecules that have been touted to be required for Nrf2 transcription activity, including other factors in the transcriptional machinery itself as well as specific signal transduction pathways.
Acknowledgments
We thank Ms. N. Kaneko and R. Kawai for technical assistance and Ms. K. Tong for critical reading. This work was supported in part by grants from the National Institutes of Health (CA80088 to J.D.E.), Exploratory Research for Advanced Technology (to M.Y.), the Ministry of Education, Science, Sports, and Technology (to H.M. and M.Y.), the Ministry of Health, Labor, and Welfare (to H.M. and M.Y.), the Japan Society for the Promotion of Science (to F.K.), Core Research for Evolutional Science and Technology (to H.M.), the Naito Foundation (to M.Y.), and the Special Coordination Fund for Promoting Science and Technology (to H.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MARE, Maf recognition element; CNC, Cap-N-Collar.
References
- 1.Motohashi, H., Shavit, J. A., Igarashi, K., Yamamoto, M. & Engel, J. D. (1997) Nucleic Acids Res. 25, 2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerppola, T. K. & Curran, T. (1994) Oncogene 9, 3149-3158. [PubMed] [Google Scholar]
- 3.Kataoka, K., Noda, M. & Nishizawa, M. (1994) Mol. Cell. Biol. 14, 700-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kataoka, K., Igarashi, K., Itoh, K., Fujiwara, K. T., Noda, M., Yamamoto, M. & Nishizawa, M. (1995) Mol. Cell. Biol. 15, 2180-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai, S., Goto, N., Kataoka, K., Saegusa, T., Shinno-Kohno, H. & Nishizawa, M. (1992) Virology 188, 778-784. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara, K. T., Kataoka, K. & Nishizawa, M. (1993) Oncogene 8, 2371-2380. [PubMed] [Google Scholar]
- 7.Andrews, N. C., Erdjument-Bromage, H., Davidson, M. B., Tempst, P. & Orkin, S. H. (1993) Nature 362, 722-728. [DOI] [PubMed] [Google Scholar]
- 8.Igarashi, K., Kataoka, K., Itoh, K., Hayashi, N., Nishizawa, M. & Yamamoto, M. (1994) Nature 367, 568-572. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, K., Igarashi, K., Hayashi, N., Nishizawa, M. & Yamamoto, M. (1995) Mol. Cell. Biol. 15, 4184-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oyake, T., Itoh, K., Motohashi, H., Hayashi, N., Hoshino, H., Nishizawa, M., Yamamoto, M. & Igarashi, K. (1996) Mol. Cell. Biol. 16, 6083-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marini, M. G., Chan, K., Casula, L., Kan, Y. W., Cao, A. & Moi, P. (1997) J. Biol. Chem. 272, 16490-16497. [DOI] [PubMed] [Google Scholar]
- 12.Onodera, K., Shavit, J. A., Motohashi, H., Katsuoka, F., Akasaka, J. E., Engel, J. D. & Yamamoto, M. (1999) J. Biol. Chem. 274, 21162-21169. [DOI] [PubMed] [Google Scholar]
- 13.Chan, J. Y., Han, X. L. & Kan, Y W. (1993) Proc. Natl. Acad. Sci. USA 90, 11371-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi, A., Ito, E., Toki, T., Kogame, K., Takahashi, S., Igarashi, K., Hayashi, N. & Yamamoto, M. (1999) J. Biol. Chem. 274, 6443-6452. [DOI] [PubMed] [Google Scholar]
- 15.Kusunoki, H., Motohashi, H., Katsuoka, F., Morohashi, A., Yamamoto, M. & Tanaka, T. (2002) Nat. Struct. Biol. 9, 252-256. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, K., Chiba, T., Takahashi, S., Ishii, T., Igarashi, K., Katoh, Y., Oyake, T., Hayashi, N., Satoh, K., Hatayama, I., et al. (1997) Biochem. Biophys. Res. Commun. 236, 313-322. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., Igarashi, K., Engel, J. D. & Yamamoto, M. (1999) Genes Dev. 13, 76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enomoto, A., Itoh, K., Nagayoshi, E., Haruta, J., Kimura, T., O'Connor, T., Harada, T. & Yamamoto, M. (2001) Toxicol. Sci. 59, 169-177. [DOI] [PubMed] [Google Scholar]
- 19.Aoki, Y., Sato, H., Nishimura, N., Takahashi, S., Itoh, K. & Yamamoto, M. (2001) Toxicol. Appl. Pharmacol. 173, 154-160. [DOI] [PubMed] [Google Scholar]
- 20.Ramos-Gomez, M., Kwak, M. K., Dolan, P. M., Itoh, K., Yamamoto, M., Talalay, P. & Kensler, T. W. (2001) Proc. Natl. Acad. Sci. USA 98, 3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakabayashi, N., Itoh, K., Noda, S., Wakabayashi, J., Motohashi, H., Imakado, S., Kotsuji, T., Otsuka, F., Roop, D. R., Harada, T., et al. (2003) Nat. Genet. 35, 238-245. [DOI] [PubMed] [Google Scholar]
- 22.Dhakshinamoorthy, S. & Jaiswal, A. K. (2000) J. Biol. Chem. 275, 40134-40141. [DOI] [PubMed] [Google Scholar]
- 23.Gong, P., Hu, B., Stewart, D., Ellerbe, M., Figueroa, Y. G., Blank, V., Beckman, B. S. & Alam, J. (2001) J. Biol. Chem. 276, 27018-27025. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, T., Huang, H. C. & Pickett, C. B. (2000) J. Biol. Chem. 275, 15466-15473. [DOI] [PubMed] [Google Scholar]
- 25.Wild, A. C., Moinova, H. R. & Mulcahy, R. T. (1999) J. Biol. Chem. 274, 33627-33636. [DOI] [PubMed] [Google Scholar]
- 26.Alam, J., Stewart, D., Touchard, C, Boinapally, S., Choi, A. M. & Cook, J. L. (1999) J. Biol. Chem. 274, 26071-26078. [DOI] [PubMed] [Google Scholar]
- 27.Venugopal, R. & Jaiswal, A. K. (1998) Oncogene 17, 3145-3156. [DOI] [PubMed] [Google Scholar]
- 28.He, C. H., Gong, P., Hu, B., Stewart, D., Choi, M. E., Choi, A. M. & Alam, J. (2001) J. Biol. Chem. 276, 20858-20865. [DOI] [PubMed] [Google Scholar]
- 29.Hilberg, F., Aguzzi, A., Howells, N. & Wagner, E. F. (1993) Nature 365, 179-181. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, T., Tsujimura, T., Takeda, K., Sugihara, A., Maekawa, A., Terada, N., Yoshida, N. & Akira, S. (1998) Genes Cells 3, 801-810. [DOI] [PubMed] [Google Scholar]
- 31.Masuoka, H. C. & Townes, T. M. (2002) Blood 99, 736-745. [DOI] [PubMed] [Google Scholar]
- 32.Shavit, J. A., Motohashi, H., Onodera, K., Akasaka, J., Yamamoto, M. & Engel, J. D. (1998) Genes Dev. 12, 2164-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsuoka, F., Motohashi, H., Onodera, K., Suwabe, N., Engel, J. D. & Yamamoto, M. (2000) EMBO J. 19, 2980-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onodera, K., Shavit, J. A., Motohashi, H., Yamamoto, M. & Engel, J. D. (2000) EMBO J. 19, 1335-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart, D., Killeen, E., Naquin, R., Alam, S. & Alam, J. (2003) J. Biol. Chem. 278, 2396-2402. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, T., Sherratt, P. J., Huang, H. C., Yang, C. S. & Pickett, C. B. (2003) J. Biol. Chem. 278, 4536-4541. [DOI] [PubMed] [Google Scholar]
- 37.Itoh, K., Wakabayashi, N., Katoh, Y., Ishii, T., O'Connor, T. & Yamamoto, M. (2003) Genes Cells 8, 379-391. [DOI] [PubMed] [Google Scholar]
- 38.McMahon, M., Itoh, K., Yamamoto, M. & Hayes, J. D. (2003) J. Biol. Chem. 278, 21592-21600. [DOI] [PubMed] [Google Scholar]
- 39.Huang, H.-C., Nguyen, T. & Pickett, C. B. (2002) J. Biol. Chem. 277, 42769-42774. [DOI] [PubMed] [Google Scholar]
- 40.Cullinan, S. B., Zhang, D., Hannink, M., Arvisais, E., Kaufman, R. J. & Diehl, J. A. (2003) Mol. Cell. Biol. 23, 7198-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishii, T., Itoh, K., Takahashi, S., Sato, H., Yanagawa, T., Katoh, Y., Bannai, S. & Yamamoto, M. (2000) J. Biol. Chem. 275, 16023-16029. [DOI] [PubMed] [Google Scholar]
- 42.Kataoka, K., Handa, H. & Nishizawa, M. (2001) J. Biol. Chem. 276, 34074-34081. [DOI] [PubMed] [Google Scholar]
- 43.Nioi, P., McMahon, M., Itoh, K., Yamamoto, M. & Hayes, J. D. (2003) Biochem. J. 374, 337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shivdasani, R. A., Rosenblatt, M. F., Zucker-Franklin, D., Jackson, C. W., Hunt, P., Saris, C. J. & Orkin, S. H. (1995) Cell 81, 695-704. [DOI] [PubMed] [Google Scholar]
- 45.Lecine, P., Blank, V. & Shivdasani, R. (1998) J. Biol. Chem. 273, 7572-7578. [DOI] [PubMed] [Google Scholar]
- 46.Motohashi, H., Katsuoka, F., Shavit, J. A., Engel, J. D. & Yamamoto, M. (2000) Cell 103, 865-875. [DOI] [PubMed] [Google Scholar]
- 47.Sun, J., Hoshino, H., Takaku, K., Nakajima, O., Muto, A., Suzuki, H., Tashiro, S., Takahashi, S., Shibahara, S., Alam, J., Taketo, M. M., Yamamoto, M. & Igarashi, K. (2002) EMBO J. 21, 5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsuoka, F., Motohashi, H., Tamagawa, Y., Kure, S., Igarashi, K., Engel, J. D. & Yamamoto, M. (2003) Mol. Cell. Biol. 23, 1163-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braun, S., Hanselmann, C., Gassmann, M. G., auf dem Keller, U., Born-Berclaz, C., Chan, K., Kan, Y. W. & Werner, S. (2002) Mol. Cell. Biol. 22, 5492-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itoh, K., Mochizuki, M., Ishii, Y., Ishii, T., Shibata, T., Kawamoto, Y., Kelly, V., Sekizawa, K., Uchida, K. & Yamamoto, M. (2004) Mol. Cell. Biol. 24, 36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morito, N., Yoh, K., Itoh, K., Hirayama, A., Koyama, A., Yamamoto, M. & Takahashi, S. (2003) Oncogene 22, 9275-9281. [DOI] [PubMed] [Google Scholar]